Abstract
• Background and Aims China is one of the countries most severely affected by desertification. Haloxylon ammodendron (Amaranthaceae) is an ecologically important component of the desert ecosystem and is one of the main tree species used for restoration, yet we know little about its genetic structure.
• Methods Genetic variation within and between nine populations of H. ammodendron from two regions of China was investigated using ISSR (inter-simple sequence repeat) markers.
• Key Results Eight primers used in this study amplified 219 reproducible bands of which 184 (84 %) were polymorphic. Analysis of molecular variance (AMOVA) revealed high genetic variation within populations (97·63 %) and low genetic differentiation between regions (0·62 %) and among populations (1·75 %).
• Conclusions It is suggested that the present genetic structure could have arisen by high levels of gene flow. The gene flow among populations observed here is probably mainly attributable to pollen movement. The genetic structure also has important implications in ecological restoration practice.
Keywords: Haloxylon ammodendron, ISSR, genetic structure, gene flow, ecological restoration
INTRODUCTION
China is one of the countries most severely affected by desertification. The total area of desertified land in China is approx. 2 622 300 km2, or 27·32 % of the total land territory. Desertification is still occurring at a rapid pace (CCICCD, 2000), and rehabilitation and ecological restoration are major tasks. A primary goal of restoration is the rapid establishment of long-term viable populations that will help restore ecosystem functions and processes, prevent erosion and protect biological diversity (Bradshaw, 1987; Lesica and Allendorf, 1999). The success of a vegetation restoration project might depend on choosing the appropriate genetic sources, and the use of native plants in ecological restoration has received much attention recently (Knapp and Rice, 1994; Linhart, 1995; Lesica and Allendorf, 1999), especially for use in stressful environments.
It is often stated that only by possessing genetic variation will a given species be able to respond to environmental pressure, evolve, and survive in the long term (e.g. Ayala and Kiger, 1984). The amount and partitioning of genetic variation among and within populations is correlated with the life history characteristics and population history of a species, and environmental factors (Hamrick and Godt, 1989). Hoffmann and Parsons (1991) stated that severe environmental stress could be regarded as an environmental probe, leading to useful evolutionary insights that are difficult to reveal under less extreme conditions. Despite extensive ecological research on plants from arid regions, there is a lack of information concerning levels of genetic variation in these systems (Hu and Wang, 1998). Knowledge of the genetic structure of desert plants not only permits us to estimate the importance of some evolutionary forces, such as selection, mutation, migration and genetic, drift under stressful environmental conditions, but also provides fundamental information for plans for restoration and rational exploitation.
Haloxylon ammodendron is a xerophytic desert tree. Due to its great drought resistance and saline tolerance, H. ammodendron occurs naturally in various habitats, including gravel desert, clay desert, fixed and semi-fixed sand, and saline land in the Asian and African deserts (Chen et al., 1983; Tobe et al., 2000). In China, about 56 % of H. ammodendron is found in Xinjiang, 40 % in Inner Mongolia, and the remaining 4 % in Qinghai and Gansu Provinces (Ma et al., 2000). As a dominant desert plant, H. ammodendron plays an important role in the maintenance of the structure and function of the whole ecosystem, reducing wind speed and ameliorating the forest microclimate, thus facilitating the settlement and growth of other desert plants (Shamsutdinov and Ubaidullaev, 1988). During the last five decades, land reclamation and cultivation, over-grazing, over-cutting and digging have resulted in the destruction of H. ammodendron vegetation and, as a result, have increased mobility of sand dunes that are no longer stabilized by root systems. Compared with a field survey in 1958, over 50 % of the natural populations of the species have become extinct or degraded (Huang, 2002). Meanwhile, the genetic diversity and population genetic structure of H. ammodendron are largely unknown, making the conservation and utilization in ecological restoration of this important desert species difficult.
In recent years, the technique of inter-simple sequence repeats (ISSR) has been used in the detection of genetic diversity in many species (Robinson et al., 1997; Wolfe et al., 1998a, b; Esselmann et al., 1999; Qian et al., 2001). The ISSR technique has several advantages including a relatively unbiased sampling of the genome, simplicity, relatively low cost, and the requirement for only a small amount of plant material. In the present study, ISSR markers were used to examine the genetic structure of natural populations of H. ammodendron in China. Of particularly interest were the level of genetic diversity within populations and the level of genetic differentiation among populations, information that might provide important insights into the population genetics of the species and facilitate its use in vegetation restoration.
MATERIALS AND METHODS
Plant material
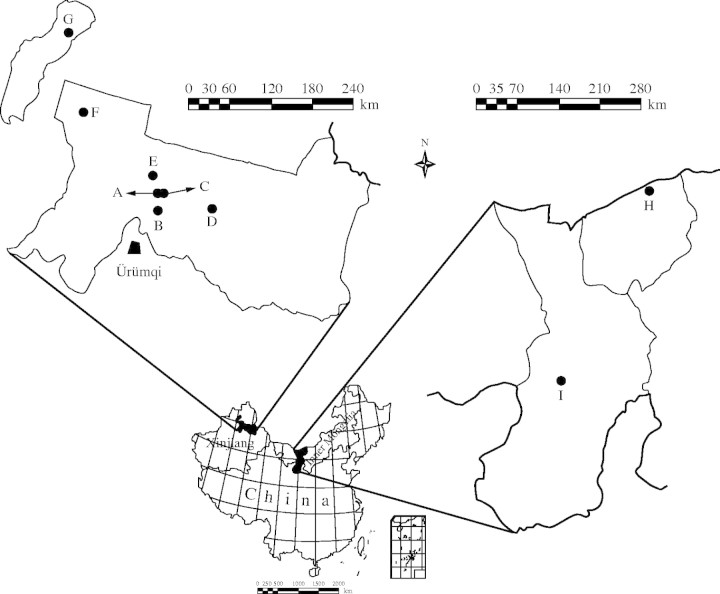
Nine populations of Haloxylon ammodendron (C. A. Mey.) Bunge (Saxaul; Amaranthaceae), which could be grouped into two regions, Xinjiang (Fig. 1A–G) and Inner Mongolia (Fig. 1H and I), were sampled. Because the leaves of H. ammodendron are reduced to scales (Pyankov et al., 1999), the young annual cylindrical shoots were used as material for DNA extraction. In each of the nine populations, shoot tissue from 20 mature individuals was collected (Table 1). All tissue was dried at room temperature (Thomson and Henry, 1993) and transported back to the Institute of Botany, Beijing.
Fig. 1.
Map showing Haloxylon ammodendron populations analysed in this study. See Table 1 for reference to the population locations.
Table 1.
Populations of Haloxylon ammodendron for ISSR analysis
| Population no. |
Location |
Latitude (°N) |
Longitude (°E) |
Samples size |
Voucher* |
|---|---|---|---|---|---|
| A | Beishawo, Fukang City, Xinjiang | 44·24 | 87·52 | 20 | Qin RC 623 |
| B | Xingonglu, Fukang City, Xinjiang | 44·13 | 87·50 | 20 | Qin RC 623 |
| C | Beishawo, Fukang City, Xinjiang | 44·25 | 87·55 | 20 | Qin RC 623 |
| D | Wucaiwuan, Jimusaer County, Xinjiang | 44·50 | 88·55 | 20 | Qin RC 623 |
| E | Dongdaohaizi, Miquan City, Xinjiang | 44·41 | 87·33 | 20 | Qin RC 623 |
| F | Baikouquan, Kelamayi City, Xinjiang | 45·53 | 85·24 | 20 | Li AR, Zhu AN 5877 |
| G | Mosuowan, Manasi County, Xinjiang | 45·03 | 86·09 | 20 | XZ 0054 |
| H | Wulatehou, Bayannaoer, Inner Mongolia | 42·10 | 108·35 | 20 | 0027 |
| I | Alashanzuo, Alashan, Inner Mongolia | 39·30 | 104·59 | 20 | 0066 |
Vouchers are deposited in the Chinese National Herbarium, Institute of Botany, Beijing.
Genomic DNA extraction
Genomic DNA was extracted from approx. 1 g of dried shoot material using a modification of the 2× CTAB method of Doyle and Doyle (1987). The tissue was ground to fine powder in liquid nitrogen and incubated at 65 °C for 60 min in 2 mL of 2× CTAB isolation buffer [100 mM Tris–HCl, pH 8·0, 1·4 m NaCl, 20 mm EDTA, 2 % hexadecyltrimethyl-ammonium bromide (CTAB), 0·2 % β-mercaptoethanol]. The sample was mixed with an equal volume of Tris–phenol, centrifuged at 10 000 g for 10 min and then the supernatant was collected in a clean tube. An equal volume of chloroform–isoamyl alcohol (24:1) was added and mixed by inversion for 10 min. Then the mixture was centrifuged at 10 000 g for 10 min. The supernatant was mixed with 0·5 mL ice-cold isopropanol and 0·1 mL 2 m potassium acetate to precipitate the DNA. The DNA was pelleted by centrifugation at 10 000 g for 5 min, washed twice with 0·6 mL of 70 % ethanol, air-dried at room temperature and resuspended in 0·2 mL of 0·1× TE buffer (10 mM Tris–HCl, 1 mm EDTA; pH 8·0). All DNA samples were purified using the Wizard DNA Clean-Up System (Promega, Madison, WI, USA). After visual quantification by comparison with standard DNA concentrations, DNA samples of approx. 50 ng µl−1 were prepared.
ISSR amplification
Twenty-one primers from Shanghai Sangon Biologic and Engineering Technology and Service Co. Ltd (Shanghai, China) were screened initially against four plants from four populations. Primer design was based on SSR motifs reported for flowering plants (reviewed in Wolfe and Liston, 1998; Wolfe et al., 1998a, b) and recommended on at http://www.biosci.ohio-state.edu/~awolfe/ISSR/ISSR.html. To avoid biasing estimates of polymorphism, the selection of primers for band scoring was dependent only on the clearness and reproducibility of ISSR fragments, not on the level of polymorphism. Reactions were carried out in a volume of 25 µl consisting of 1·5 mm MgCl2, 0·2 mM dNTPs, 1·25 μm of primer, 1·25 units of Taq DNA polymerase (Promega), 1× Taq DNA polymerase buffer and approx. 20 ng of template DNA. PCR amplifications were performed in a PTC-200 thermocycler (MJ Research, Watertown, MA, USA) under the following conditions: 94 °C for 1·5 min; 35 cycles of 94 °C for 40 s, 49 °C for 45 s, 72 °C for 1·5 min; linked to 94 °C for 45 s, 44 °C for 45 s, 72 °C for 5 min. PCR products were then stored at 4 °C. The amplification products were separated by electrophoresis on 2 % agarose gels in 1× TBE (Tris–EDTA–borate) buffer, stained with ethidium bromide. After running for approx. 6 h at 50 V (2·5 V cm−1) the gel was photographed by an Alpha Ease FC Imaging System (Alpha Innotech Corporation, San Leandro, CA, USA). Scoring of the bands was performed by visual analysis of the gel photographs. Molecular size of the fragments was estimated using a 200-bp DNA ladder.
Data analysis
Amplified fragments were scored for presence (1) or absence (0) of putatively homologous bands and a matrix of ISSR phenotypes was assembled. Homology was assessed based on co-migration of bands on the gels. Only data from intensely stained, unambiguous, clear bands were used for statistical analysis. An ISSR marker was determined to be polymorphic when it was found in <95 % of the individuals sampled (i.e. absent in eight or more individuals). Genetic diversity was measured as the percentage of polymorphic loci. Analysis of molecular variance (AMOVA) was conducted using DCFA 1.1 (Zhang and Ge, 2002) and WINAMOVA 1.55 (Excoffier et al., 1992; Excoffier, 1993) to calculate variance components and their statistical significance levels for variation among and within the nine populations. Euclidean squared distances (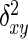 ) instead of Jaccard similarity coefficients were used in AMOVA as suggested by Zhang and Ge (2002) and Excoffier (1993). The
) instead of Jaccard similarity coefficients were used in AMOVA as suggested by Zhang and Ge (2002) and Excoffier (1993). The 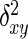 was obtained by the following equation:
was obtained by the following equation:
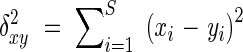 |
where for two individuals x and y, xi and yi represent presence or absence of a particular molecular marker, and s is the total number of markers in the AMOVA. Pairwise genetic distances (ΦST) among the nine populations were obtained from the AMOVA and were used to construct a UPGMA tree (routine UPGMA in the NTSYSpc package; Rohlf, 1994). A nonparametric test for homogeneity of molecular variance (HOMOVA), based on the Bartlett statistic, was also applied to test whether the populations have different amounts of ISSR variation (Stewart and Excoffier, 1996). Both HOMOVA and AMOVA were performed using the WINAMOVA 1.5 program (obtained at ftp: 129.194.113.1). The Mantel test (Mantel, 1967) was used to test whether matrices of genetic distances between populations were significantly correlated with matrices of geographic distances (1000 permutations; routine MXCOMP in NTSYSpc).
RESULTS
ISSR diversity
Of the 21 ISSR primers screened, eight produced clear and repeatable fragments and were selected for further analysis (Table 2). These primers consistently amplified a total of 219 scorable markers of 165–2300 base pairs (bp) in size. Individual primers generated 18–36 bands, with a mean of 27·4. Of the 219 markers, 184 (84 %) were polymorphic across the 180 plants, and each individual had a unique multilocus ISSR phenotype. Some primers were more efficient in differentiating between individuals than others. For example, ISSR-7 and ISSR-12 identified 179 unique phenotype profiles, whereas ISSR-6 only revealed 124 profiles (Table 2).
Table 2.
Primers for ISSR analysis
| Primer |
Primer sequence* |
Band size range (bp) |
Tm (°C) |
Total no. of loci |
Total no. of polymorphic loci (%) |
Total no. of distinct phenotypes |
|---|---|---|---|---|---|---|
| ISSR-4 | (AC)8YT | 190–1000 | 44 | 22 | 19 (86·4) | 160 |
| ISSR-6 | (CA)6RY | 180–1200 | 44 | 18 | 15 (83·3) | 124 |
| ISSR-7 | (CA)6RG | 180–2300 | 43 | 36 | 30 (83·3) | 179 |
| ISSR-9 | (CTC)4RC | 210–2150 | 48 | 32 | 24 (75·0) | 169 |
| ISSR-11 | BDB(ACA)5 | 185–1050 | 49 | 24 | 21 (87·5) | 146 |
| ISSR-12 | BBB(GAAA)3GAA | 165–1800 | 43 | 33 | 27 (81·8) | 179 |
| ISSR-13 | (AC)8YG | 190–1700 | 53 | 30 | 27 (90·0) | 178 |
| ISSR-14 | (CAC)4RC | 180–1000 | 60 | 24 | 21 (87·5) | 171 |
Y = G/C; R = A/T; B = C/G/T; D = A/G/T.
Percentages of polymorphic loci per population and per region are shown in Table 3. The percentage of polymorphic loci within populations ranged from 48·9 % (A) to 74·9 % (E), with the mean percentage of 57·7 %. Within regions, the percentage of polymorphic loci of the populations from Xinjiang was 78·5 %, a little higher than that from Inner Mongolia (70·8 %).
Table 3.
Percentage of polymorphic ISSR loci in each Haloxylon ammodendron population and region (see Fig. 1)
| A |
B |
C |
D |
E |
F |
G |
Xinjiang Province |
H |
I |
Inner Mongolia |
Total |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of PL | 107 | 110 | 110 | 109 | 164 | 146 | 122 | 172 | 127 | 142 | 155 | 184 |
| Percentage of PL | 48·9 | 50·2 | 50·2 | 49·8 | 74·9 | 66·7 | 55·7 | 78·5 | 58·0 | 64·8 | 70·8 | 84·0 |
PL, polymorphic loci.
Genetic structure
The AMOVA of the distance matrix for the 180 individuals permitted a partitioning of the overall variation into three levels (Table 4). The proportion of variation attributable to within-population differences was high (97·63 %), whereas only 0·62 % and 1·75 % occurred between regions and among populations, respectively. Genetic differences between Xinjiang and Inner Mongolia were not significant (P = 0·202), whereas those between populations were significant (P < 0·05). Bartlett's test for the homogeneity of the ISSR variance indicated significant differences in the amount of genetic variability present in the nine populations (B = 0·194, P < 0·001).
Table 4.
Analysis of molecular variance (AMOVA) for 180 individuals in nine populations of Haloxylon ammodendron
| Source of variance |
d.f. |
SSD |
MSD |
Variance component |
%Total |
P |
|---|---|---|---|---|---|---|
| Among regions | 1 | 0·6079 | 0·608 | 0·002 | 0·62 | 0·202 |
| Among populations | 7 | 3·2983 | 0·471 | 0·006 | 1·75 | <0·05 |
| Within populations | 171 | 59·2825 | 0·347 | 0·347 | 97·63 | <0·001 |
SSD, sum of squared deviation; MSD, mean squared deviation.
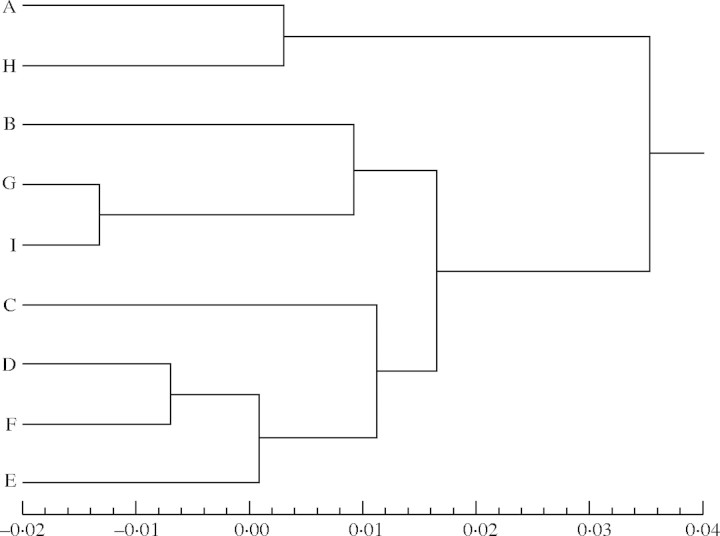
Pairwise ΦST values between populations and their significance are presented in Table 5. Values of ΦST ranged from −0·0069 (D–F) to 0·07 (C–H), indicating that populations D and F were the most similar and populations C and H were the most different. An UPGMA dendrogram of the nine populations based on the pairwise ΦST values is shown in Fig. 2.
Table 5.
Pairwise genetic distances (ΦST) among nine populations of Haloxylon ammodendron
| Population |
A |
B |
C |
D |
E |
F |
G |
H |
I |
|---|---|---|---|---|---|---|---|---|---|
| A | – | ||||||||
| B | 0·0374 | – | |||||||
| C | 0·0453*** | 0·0154 | – | ||||||
| D | 0·0311* | 0·0061 | 0·0030 | – | |||||
| E | 0·0365* | 0·0310*** | 0·0196 | 0·0051 | – | ||||
| F | 0·0143 | 0·0106 | 0·0111* | −0·0069 | −0·0035 | – | |||
| G | 0·0152 | 0·0092 | 0·0280 | 0·0150 | 0·0206* | −0·0052 | – | ||
| H | 0·0030 | 0·0556*** | 0·0700*** | 0·0610*** | 0·0474*** | 0·0224 | 0·0128* | – | |
| I | 0·0224* | 0·0094 | 0·0293*** | 0·0159*** | 0·0278*** | 0·0050 | –0·0134 | 0·0229 | – |
P values indicate the probability that a random genetic distance is larger than the observed distance and are based on 1000 permutations.
P < 0·05;
P < 0·001.
Fig. 2.
Dendrogram of the nine Haloxylon ammodendron populations obtained by UPGMA cluster analysis of a matrix of pairwise ΦST values.
Correlation of geographical and genetic (ISSR) distance
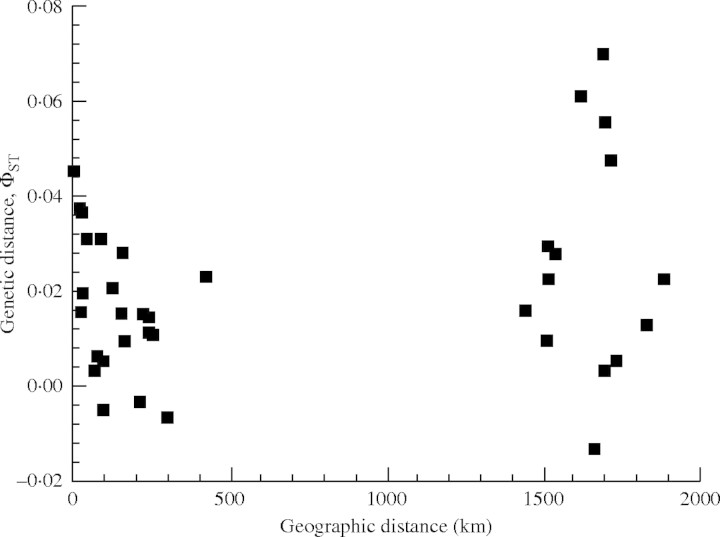
The matrix of pairwise genetic distances was not significantly correlated with the corresponding matrix of geographic distances (Mantel tests; r = 0·2230, P = 0·8448; Fig. 3). Thus, differentiation observed among populations (Table 4) did not directly correspond to the geographic distance.
Fig. 3.
The relationship between pairwise geographic and pairwise genetic distances (ΦST) among nine populations of H. ammodendron. Distances >1000 km denote pairs with one population in Xinjiang and the other at Inner Mongolia.
DISCUSSION
Genetic diversity and population structure
Using ISSR markers, 84 % of loci were found to be polymorphic in H. ammodendron. Levels of genetic variation within populations established for H. ammodendron can be compared with other woody plants with similar life histories. Ge and Sun (1999) revealed a surprisingly low genetic variation with ISSR (P = 16·18 %) in Aegiceras corniculatum (Myrsinaceae), a widespread, long-lived, woody, perennial species with mixed-mating to outcrossing systems. Lacerda et al. (2001) using RAPD markers, found considerable levels of intrapopulation genetic variation (P = 70·8 %) for Plathymenia reticulata (Fabaceae), a tree from the Brazilian Cerrado. Shrestha et al. (2002) used 18 RAPD primers to detect the genetic diversity of 12 populations of Acacia raddiana (Mimosaceae), a keystone tree species in the Negev desert of Israel. Of the 290 markers revealed, 90·69 % were polymorphic. By comparison, it can be estimated that H. ammodendron presents high levels of intrapopulation genetic variation.
In H. ammodendron, ISSR markers revealed that the majority of the variation was found within populations rather than among populations. The genetic differentiation among regions was not significant. Long-lived, woody plants typically harbour a greater percentage of their variation within populations (Hamrick and Loveless, 1989; Hamrick et al., 1992). According to Hamrick and Godt (1989), reproductive biology is the most important factor in determining the genetic structure of plant populations. They showed that outcrossing plant species tend to have 10–20 % of the genetic variation among populations, whereas selfing species have, on average, 50 % of the variation among populations. Studies of the biology of flowering and pollination in H. ammodendron indicate that it is an outcrosser (Tursunov et al., 1989). Despite several possible pollination mechanisms, such as xenogamy, geitonogamy and autogamy, the presence of three groups of bisexual flowers that differ in time of maturation of the anthers and stigma promotes cross-pollination by anemophily (Tursunov et al., 1989). However, Hamrick and Godt (1996) have pointed out that life history traits alone explain a relatively low amount of the variation in genetic structure that is seen among species. The high intrapopulation variability and genetic homogeneity across populations could have arisen by high levels of gene flow. Alternatively, it may be that these populations have simply not been separated long enough to accumulate detectable genetic differences.
Haloxylon ammodendron generally reproduces sexually (Song and Jia, 2000), although in the young forest, vegetative propagation from buds on the trunk or branches has been reported when the plant is damaged (Huang, 2002). In the mature natural populations studied, however, vegetative propagation is infrequent. Seeds are small (2·5mm in diameter, 1000 seeds weighing 2·99 ± 0·04 g; Huang, 2002), and bear wing-like membranes. Occasionally seeds can fall >5 miles away from the canopies of their maternal trees (Mamedov and Osipov, 1985; Y. Sheng et al., unpubl. res.). Most of the seeds are consumed by ants and small mammals, which is a major cause of seed mortality in the desert (Brown and Davidson, 1977). Seed longevity of H. ammodendron is short under natural conditions, and few seeds survive in the soil for >10months (Huang et al., 2002). In deserts, the water necessary for seedling establishment is only available transiently following precipitation (Ellner and Shmida, 1981), and the dispersed seeds must geminate quickly after precipitation during April and May, or they will die the following summer (Yang et al., 1995). Dispersal and germination investigations, as presented above, suggest that long-distance seed dispersal is rare in H. ammodendron. Absence of adaptation for long-distance dispersal in desert plants has long been recognized. Ellner and Shmida (1981) argue that atelechory (the absence of dispersal-enhancing characters) in desert species is an adaptive response to the extremely low benefit of long-range dispersal mechanisms in deserts. Pollen is small (19·1µm in diameter; Pan, 1993), round or nearly round, and wind-dispersed (Wu et al., 1992). Due to the frequently strong winds and the low coverage of vegetation in the desert, long-distance dispersal of pollen by wind is possible. After pollination and fertilization in May, the ovaries do not begin to develop until September, when the temperature and moisture levels are suitable (Song and Jia, 2000). This special habit, summer dormancy, ensures effective pollen flow. Therefore, the high levels of gene flow among populations observed here might be mainly attributed to pollen movement. Further studies of pollination biology and parentage analysis using co-dominant markers are needed to clarify this point. For most biological systems, the most powerful genetic tools for parentage analysis will be microsatellite markers, and most of the recent advances in techniques of data analysis have been aimed at studies employing microsatellites (Jones and Ardren, 2003). Using microsatellite analysis, Dow and Ashley (1998) characterized pollen dispersal in a stand of 62 adult bur oaks (Quercus macrocarpa), providing direct evidence for high levels of long-distance pollination in this wind-pollinated species. Konuma et al. (2000) using microsatellite polymorphism suggested that long-distance gene flow and seed migration were responsible for the poorly developed genetic structure of the tropical rainforest tree Neobalanocarpus heimii (Dipterocarpaceae). Recent review papers (Estoup and Angers, 1998; Squirrell et al., 2003; Zhang and Hewitt, 2003) have addressed the advantages and disadvantages of the approach and have suggested methods for isolation of microsatellite markers at relatively low cost.
Restoration implications
The genetic structure of H. ammodendron revealed in this study is important in ecological conservation and restoration practice. The fact that an overwhelming proportion of the variability is present within populations suggests that smaller numbers of populations will be required for effective conservation compared with island endemics with many, strongly isolated populations. Likewise, the high genetic homogeneity among populations of H. ammodendron suggests that the exact population used as a source of material for restoration may be less crucial in this species, compared with other species with strong genetic structure and a high potential for local adaptation. Population size is also an important restoration consideration in H. ammodendron. It is thought that small populations can lose large amounts of genetic variation due to of genetic drift and thus have reduced probabilities of long-term viability (Lande and Barrowclough, 1987). Moreover, small populations are more prone to extinction from random environmental fluctuations (Allendorf, 1986). Therefore, maintaining a proper population size should be emphasized in planning restoration and rational exploitation of H. ammodendron.
Acknowledgments
We thank the Key Project of the Chinese Academy of Sciences (KZCX1-10-05) for financial support, Dr J. Jiang for help with sampling and Drs X. Q. Wang and W. Qian for valuable comments on an earlier draft of this manuscript.
LITERATURE CITED
- Allendorf FW. 1986. Genetic drift and the loss of alleles versus heterozygosity. Zoo Biology 5: 181–190. [Google Scholar]
- Ayala FJ, Kiger JA. 1984.Modern genetics, 2nd edn. Menlo Park: Benjamin/Cumings. [Google Scholar]
- Bradshaw AD. 1987. The reclamation of derelict land and the ecology of ecosystems. In: Jordan WR, Gilpin ME, Aber JD, eds. Restoration ecology: a synthetic approach to ecological research. Cambridge: Cambridge University Press, 53–83. [Google Scholar]
- Brown JH, Davidson D. 1977. Granivory in desert ecosystems. Annual Review of Ecology and Systematics 10: 201–227. [Google Scholar]
- CCICCD. 2000. China national report on the implementation of United Nations convention to combat desertification and national action programme to combat desertification. 2000. Secretariat of China National Committee for the Implementation of the United Nations Convention to Combat Desertification (CCICCD). http://www.unccd.int/cop/reports/asia/national/2000/china-eng.pdf. 11 Oct. 2003. [Google Scholar]
- Chen CD, Zhang LY, Hu WK. 1983. The basic characteristics of plant communities, flora and their distribution in the sandy district of Gurbantungut. Acta Phytoecologica et Geobotanica Sinica 7: 89–99. [Google Scholar]
- Dow BD, Ashley MV. 1998. High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. Journal of Heredity 89: 62–70. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America 19: 11–15. [Google Scholar]
- Ellner S, Shimida A. 1981. Why are adaptations for long-range seed dispersal rare in desert plants? Oecologia 51: 133–144. [DOI] [PubMed] [Google Scholar]
- Esselman EJ, Jiangiang L, Crawford DJ, Windus JL, Wolfe AD. 1999. Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results of allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Molecular Ecology 8: 443–451. [Google Scholar]
- Estoup A, Angers B. 1998. Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In: Carvalho GR, ed. Advances in molecular ecology. Amsterdam: IOS Press, 55–86. [Google Scholar]
- Excoffier L. 1993.Analysis of molecular vaiance (AMOVA) ver 1·55. Genetics and Biometry Laboratory, University of Geneva, Switzerland. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XJ, Sun M. 1999. Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and intersimple sequence repeat (ISSR) analysis. Molecular Ecology 8: 2061–2069. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. 1989. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer, 43–63. [Google Scholar]
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London: Series B 351: 1291–1298. [Google Scholar]
- Hamrick JL, Loveless MD. 1989. The genetic structure of tropical tree populations: associations with reproductive biology. In: Bock JH, Linhart YB, eds. The evolutionary ecology of plants. Boulder: Westview Press, 139–146. [Google Scholar]
- Hamrick JL, Godt MJW, Sherman-Broyles SL. 1992. Factors influencing levels of genetic diversity in woody plant species. New Forests 6: 95–124. [Google Scholar]
- Hoffmann AA, Parsons PA. 1991.Evolutionary genetics and environmental stress. Oxford: Oxford University Press. [Google Scholar]
- Hu ZA, Wang HX. 1998. Advances in molecular ecology. Acta Ecologica Sinica 18: 565–574. [Google Scholar]
- Huang PX. 2002.No-irrigation vegetation and its restoration in arid area. Beijing: Science Press. [Google Scholar]
- Huang ZY, Zhang XS, Zheng GH, Jing XM, Lin J. 2002. Increased storability of Haloxylon ammodendron seeds in ultra-drying storage. Acta Botanica Sinica 44: 239–241. [Google Scholar]
- Jones AG, Ardren WR. 2003. Methods of parentage analysis in natural populations. Molecular Ecology 12: 2511–2523. [DOI] [PubMed] [Google Scholar]
- Knapp EE, Rice KJ. 1994. Starting from seed: genetic issues in using native grasses for restoration. Restoration Management Notes 12: 40–45. [Google Scholar]
- Konuma A, Tsumura Y, Lee CT, Lee SL, Okuda T. 2000. Estimation of gene flow in the tropical-rainforest tree Neobalanocarpus heimii (Dipterocarpaceae), inferred from paternity analysis. Molecular Ecology 9: 1843–1852. [DOI] [PubMed] [Google Scholar]
- Lacerda DR, Acedo MDP, Lemos Filho JP, Lovato MB. 2001. Genetic diversity and structure of natural populations of Plathymenia reticulata (Mimosoideae), a tropical tree from the Brazilian Cerrado. Molecular Ecology 10: 1143–1152. [DOI] [PubMed] [Google Scholar]
- Lande R, Barrowclough GF. 1987. Effective population size, genetic variation and their use in population management. In: Soulé ME, ed. Viable populations for conservation. Cambridge: Cambridge University Press, 87–123. [Google Scholar]
- Lesica P, Allendorf FW. 1999. Ecological genetics and the restoration of plant communities: mix or match? Restoration Ecology 7: 42–50. [Google Scholar]
- Linhart YB. 1995. Restoration, revegetation, and the importance of genetic and evolutionary perspectives. In: Roundy BA, McArthur ED, Haley JS, Mann DK, eds. Proceedings of the Wildland Shrub and Arid Land Restoration Symposium, 19–21 Oct. 1993, Las Vegas, Nevada, 271–287. General Technical Report INT-GTR-315. US Forest Service, Ogden, UT, USA. [Google Scholar]
- Ma HB, Bao GX, Ma WD, Rong ZJ, WanG XY, Li B. 2000. The resource, protection and utilization of Haloxylon ammodendron deserted grassland in Inner Mongolia. Pratacultural Science 17: 1–5. [Google Scholar]
- Mamedov P, Osipov YS. 1985. Results of tests of pneumatic collection of saxaul and salt-tree seeds. Problems of Desert Development, 2: 87–89. [Google Scholar]
- Mantel NA. 1967. The detection of disease clustering and a generalized regression approach. Cancer Research 27: 209–220. [PubMed] [Google Scholar]
- Pan AD. 1993. Studies on the pollen morphology of Chenopodiaceae from Xinjiang. Arid Land Geography 16: 22–27. [Google Scholar]
- Pyankov VI, Black JCC, Artyusheva EG, Voznesenskaya EV, Ku MSB, Edwards GE. 1999. Features of photosynthesis in Haloxylon species of Chenopodiaceae that are dominant plants in central Asian deserts. Plant Cell Physiology 40: 125–134. [Google Scholar]
- Qian W, Ge S, Hong DY. 2001. Genetic variation within and among populations of a wild rice Oryza granulata from China detected by RAPD and ISSR markers. Theoretical and Applied Genetics 102: 440–449. [Google Scholar]
- Robinson WA, Liston A, Doescher PS, Svejcar T. 1997. Using ISSR markers to quantify clonal vs. sexual reproduction in Festuca idahoensis (Poaceae). American Journal of Botany 84: 89 (abstract). [Google Scholar]
- Rohlf FJ. 1994.NTSYS. Numerical taxonomy and multivariate analysis system. New York: Exeter Ltd, Setauket. [Google Scholar]
- Shamsutdinov ZS, Ubaidullaev SR. 1988. Distribution of Poa bulbosa L. and Carex pachystylis within the phytogenous field of black saxaul. Problems of Desert Development 1: 38–43. [Google Scholar]
- Shrestha MK, Golan-Goldhirsh A, Ward D. 2002. Population genetic structure and the conservation of isolated populations of Acacia raddiana in the Negev Desert. Biological Conservation 108: 119–127. [Google Scholar]
- Song CS, Jia KF. 2000.Scientific survey of the Wulate Haloxylon ammodendron Nature Reserve. Beijing: China Forestry Publishing House. [Google Scholar]
- Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W. 2003. How much effort is required to isolate nuclear microsatellites from plants? Molecular Ecology 12: 1339–1687. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Excoffier L. 1996. Assessing population genetic structure and variability with RAPD data: application to Vaccinium macrocarpon (American cranberry). Journal of Evolutionary Biology 9: 153–171. [Google Scholar]
- Thomson D, Henry R. 1993. Use of DNA from dry leaves for PCR and RAPD analysis. Plant Molecular Biological Report 11: 202–206. [Google Scholar]
- Tobe K, Li XM, Omasa K. 2000. Effects of sodium chloride on seed germination and growth of two Chinese desert shrubs, Haloxylon ammodendron and H. persicum (Chenopodiaceae). Australian Journal of Botany 48: 455–460. [Google Scholar]
- Tursunov Z, Matyunina TE, Kiseleva GK, Abdullaena AT. 1989. Seed reproduction of the main forest-forming species of the central Asian deserts. Problems of Desert Development 2: 53–57. [Google Scholar]
- Wolfe AD, Liston A. 1998. Contributions of the polymerase chain reaction to plant systematics. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants. Vol. I. DNA sequencing. New York: Kluwer, 43–86. [Google Scholar]
- Wolfe AD, Xiang QY, Kephart SR. 1998. Diploid hybrid speciation in Penstemon (Scrophulariaceae). Proceedings of the National Academy of Sciences of the USA 95: 5112–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Xiang QY, Kephart SR. 1998. Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable inter-simple sequence repeat markers. Molecular Ecology 7: 1107–1125. [DOI] [PubMed] [Google Scholar]
- Wu GF, Feng ZJ, Ma WL, Zhou XJ, Lang QC, Hu RL, Wang CJ, Li RG. 1992.Botany. Beijing: Higher Education Press. [Google Scholar]
- Yang MX, Zou SY, Zhao XY. 1995. Natural regeneration of cakcaryr forest in Ji LanTai. Journal of Inner Mongolia Forestry College 17: 74–86. [Google Scholar]
- Zhang DX, Hewitt GM. 2003. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Molecular Ecology 12: 563–584. [DOI] [PubMed] [Google Scholar]
- Zhang FM, Ge S. 2002. Data analysis in population genetics. I. Analysis of RAPD data with AMOVA. Biodiversity Science 10: 438–444. [Google Scholar]