Abstract
• Background and Aims The phylogenetic relationships of Petunia sensu Jussieu (Petunia sensu Wijsman plus Calibrachoa) are unclear. This study aimed to resolve this uncertainty using molecular evidence.
• Methods Phylogenetic trees of 52 taxa of Petunia sensu Jussieu were constructed using restriction fragment length polymorphism (RFLP) of chloroplast DNA digested with 19 restriction enzymes and hybridized with 12 cloned Nicotiana chloroplast DNA fragments as probes.
• Key Results In all, 89 phylogenetically informative RFLPs were detected and one 50 % majority consensus tree was obtained, using the maximum parsimony method, and one distance matrix tree, using the neighbour joining method. Petunia sensu Wijsman and Calibrachoa were monophyletic sister clades in both trees. Calibrachoa parviflora and C. pygmaea, previously thought to differ from the other species in terms of their cross-compatibility, seed morphology, and nuclear DNA content, formed a basal clade that was sister to the remainder of Calibrachoa. Several clades found in the phylogenetic trees corresponded to their distribution ranges, suggesting that recent speciation in the genus Petunia sensu Jussieu occurred independently in several different regions.
• Conclusions The separation of Petunia sensu Wijsman and Calibrachoa was supported by chloroplast DNA analysis. Two groups in the Calibrachoa were also recognized with a high degree of confidence.
Keywords: Calibrachoa, cpDNA, distribution, Petunia, phylogeny, RFLP, Solanaceae
INTRODUCTION
In 1803, Jussieu established the genus Petunia (Solanaceae), with two species as types: P. parviflora and P. nyctaginiflora [=P. axillaris (Lam.) Britton, Sterns & Poggenb.]. Fries (1911) wrote the most recent formal generic treatment on Petunia. More recently, Sink (1984) provided a comprehensive list of taxonomic works by that time. Hereafter, the genus Petunia circumscribed by Jussieu (1803) is referred to as Petunia sensu Jussieu. Wijsman and de Jong (1985) and Wijsman (1990) divided Petunia sensu Jussieu into two genera, Petunia and Calibrachoa La Llave & Lex., and transferred 15 species of Petunia sensu Jussieu to Calibrachoa. Stehmann and Semir (1997) transferred nine additional species to Calibrachoa in a work that described a species of Calibrachoa using the circumscription of Wijsman (1990). In this paper, Petunia as circumscribed by Wijsman (1990) is referred to solely as Petunia.
Several attempts have been made to understand parts of the phylogeny of Petunia sensu Jussieu using molecular data, such as restriction fragment length polymorphism (RFLP) analysis of ribosomal DNA (Kabbaj et al., 1995), DNA amplification fingerprinting (Cerny et al., 1996) and polymorphism in the chalcone synthase intron sequence (Griesbach et al., 2000); however, no attempts have been made to analyse the phylogeny of the entire genus. The objective of the present study was to explore the phylogeny of Petunia sensu Jussieu using all available taxa of wild origin to elucidate the existence of clades equivalent to Petunia, Calibrachoa and their subgroups, and to understand their evolutionary history. Recent studies indicate that Calibrachoa can be divided into two subgroups based on cross-compatibility (Watanabe et al., 1997), seed surface morphology (Watanabe et al., 1999) and nuclear DNA content (Mishiba et al., 2000). In all the analyses, C. parviflora (Juss.) Wijsman and C. pygmaea (R. E. Fr.) Wijsman formed one group, and the remaining Calibrachoa formed the other. Since C. parviflora is the type species of the genus Calibrachoa, it was attempted to resolve the relationships of these species to the other members of Petunia sensu Jussieu for nomenclatural reasons.
In this study, chloroplast DNA (cpDNA) RFLP was chosen to construct the phylogenetic trees. Contiguous clones are readily available for the entire cpDNA of Nicotiana tabacum L. (Sugiura et al., 1986), which is in the same family as Petunia sensu Jussieu.
MATERIALS AND METHODS
Plant materials
Table 1 shows the taxa used in this study. These 52 taxa cover almost the entire genus Petunia sensu Jussieu. Ando and Hashimoto (e.g. 1993, 1998) described most of the new taxa of Petunia, except unnamed taxon P1, while many Calibrachoa remain undescribed (unnamed taxa C1 to C9). Nicotiana langsdorffii Weinm. was used as the outgroup. The seeds were collected randomly from respective native populations and were raised in a greenhouse using standard cultivation techniques for garden petunias.
Table 1.
Names and voucher specimens of Petunia sensu Jussieu taxa used in the cpDNA RFLP analysis
| Taxa |
Botanical name, collection locality, voucher specimen number and herbaria code* |
|
|---|---|---|
| Petunia sensuWijsman (1990), 2n = 2x = 14 | ||
| 1 | P. altiplana T.Ando & Hashim. BRAZIL: Rio Grande do Sul, Ando et al. B319 (S, US) | |
| 2 | P. axillaris (Lam.) Britton, Sterns & Poggenb. subsp. axillaris. URUGUAY: Montevideo, Ando & Kokubun U1 (MVFA, Ando) | |
| 3 | P. axillaris subsp. parodii (Steere) Cabrera. URUGUAY: Artigas, Ando & Kokubun U27 (MVFA, Ando) | |
| 4 | P. axillaris subsp. subandina T.Ando. ARGENTINA: Salta, Ando & Iida A100 (S, SI, Ando) | |
| 5 | P. bajeensis T.Ando & Hashim. BRAZIL: Rio Grande do Sul, Hashimoto et al. B662 (BM, MBM, MVFA, R, S, SI, SP, U, US, GHSP, Ando) | |
| 6 | P. bonjardinensis T.Ando & Hashim. BRAZIL: Rio Grande do Sul, Ando et al. B174 (S, GHSP, Ando) | |
| 7 | P. exserta Stehmann BRAZIL: Rio Grande do Sul, Hashimoto et al. B931 (GHSP) | |
| 8 | P. guarapuavensis T.Ando & Hashim. BRAZIL: Paraná, Hashimoto et al. B65 (MBM, BM, HBR, S, US, GHSP, Ando) | |
| 9 | P. inflata R.E.Fr. ARGENTINA: Misiones, Ando & Buto A7 (SI, GHSP, Ando) | |
| 10 | P. integrifolia (Hook.) Schinz & Thell. subsp. integrifolia. URUGUAY: Río Negro, Ando & Watanabe U106 (MVFA, S, SI, GHSP, Ando) | |
| 11 | P. integrifolia subsp. depauperata R.E.Fr. BRAZIL: Rio Grande do Sul, Hashimoto et al. B59 (MVFA, SI, GHSP, Ando) | |
| 12 | P. interior T.Ando & Hashim. BRAZIL: Santa Catarina, Hashimoto et al. B569 (MBM, BM, R, S, US, GHSP, Ando) | |
| 13 | P. littoralis L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B29 (MVFA, GHSP, Ando) | |
| 14 | P. mantiqueirensis T.Ando & Hashim. BRAZIL: Minas Gerais, Hashimoto et al. B357 (S, BM, K, SP, U, US, GHSP, Ando) | |
| 15 | P. occidentalis R.E.Fr. ARGENTINA: Jujuy, Ando & Iida A109 (MVFA, S, SI, GHSP, Ando) | |
| 16 | P. reitzii L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B28 (GHSP, Ando) | |
| 17 | P. riograndensis T.Ando & Hashim. BRAZIL: Rio Grande do Sul, Hashimoto et al. B860 (MBM, S, GHSP, Ando) | |
| 18 | P. saxicola L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B113 (GHSP, Ando) | |
| 19 | P. scheideana L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B109 (BM, MBM, MVFA, S, SI, US, GHSP, Ando) | |
| 20 | Unnamed taxon P1. BRAZIL: Rio Grande do Sul, Hashimoto et al. B990 (GHSP, Ando) | |
| Petunia sensuJussieu (1803) with 2n = 2x = 18 chromosomes sharing similar morphological characters with Calibrachoa | ||
| 21 | P. alpicola L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B130 (BM, MBM, MVFA, S, SI, US, GHSP, Ando) | |
| 22 | P. helianthemoides Sendtn. ARGENTINA: Misiones, Ando & Buto A4 (SI, Ando) | |
| 23 | P. kleinii L.B.Sm. & Downs. BRAZIL: Santa Catarina, Hashimoto et al. B105 (MBM, GHSP, Ando) | |
| 24 | P. pubescens (Spreng.) R.E.Fr. URUGUAY: Durazno, Ando & Kokubun U23 (S, MVFA, Ando) | |
| 25 | P. variabilis R.E.Fr. URUGUAY: Tacuarembó, Ando & Kokubun U6 (MVFA, Ando) | |
| Calibrachoa La Llave & Lexarza (1825), 2n = 2x = 18 | ||
| 26 | C. calycina (Sendtn.) Wijsman. ARGENTINA: Corrientes, Ando & Buto A15 (MBM, GHSP, Ando) | |
| 27 | C. dusenii (R.E.Fr.) Stehmann & Semir. BRAZIL: Paraná, Hashimoto et al. B484 (MBM, S, GHSP, Ando) | |
| 28 | C. eglandulata Stehmann & Semir. BRAZIL: Santa Catarina, Hashimoto et al. B917 (GHSP, Ando) | |
| 29 | C. elegans (Miers) Stehmann & Semir. BRAZIL: Minas Gerais, Hashimoto et al. B489 (GHSP, Ando) | |
| 30 | C. ericaefolia (R.E.Fr.) Wijsman. BRAZIL: Paraná, Hashimoto et al. B63 (MBM, GHSP, Ando) | |
| 31 | C. heterophylla (Sendtn.) Wijsman. BRAZIL: Rio Grande do Sul, Hashimoto et al. B60 (BM, MBM, GHSP, Ando) | |
| 32 | C. linearis (Hook.) Wijsman. URUGUAY: Río Negro, Ando & Buto U165 (MVFA, Ando) | |
| 33 | C. linoides (Sendtn.) Wijsman. BRAZIL: Rio Grande do Sul, Hashimoto et al. B282 (GHSP, Ando) | |
| 34 | C. macrodactylon (L.B.Sm. & Downs) Wijsman. BRAZIL: Santa Catarina, Hashimoto et al. B138 (Ando) | |
| 35 | C. micrantha (R.E.Fr.) Stehmann & Semir. BRAZIL: Paraná, Hashimoto et al. B486 (BM, MBM, S, US, GHSP, Ando) | |
| 36 | C. parviflora (Juss.) Wijsman. URUGUAY: Tacuarembó, Ando & Kokubun U42 (MVFA, Ando) | |
| 37 | C. pygmaea (R.E.Fr.) Wijsman. URUGUAY: Artigas, Ando & Kokubun U39 (BM, MVFA, S, US, Ando) | |
| 38 | C. rupestris (Dusén) Wijsman. BRAZIL: Paraná, Hashimoto et al. B479 (BM, MBM, GHSP, Ando) | |
| 39 | C. selloviana (Sendtn.) Wijsman. BRAZIL: Rio Grande do Sul, Hashimoto et al. B892 (MBM, S, US, GHSP, Ando) | |
| 40 | C. sendtneriana (R.E.Fr.) Stehmann & Semir. BRAZIL: Santa Catarina, Hashimoto et al. B447 (BM, US, GHSP, Ando) | |
| 41 | C. serrulata (L.B.Sm. & Downs) Stehmann & Semir. BRAZIL: Santa Catarina, Hashimoto et al. B1031 (BM, S, US, U, GHSP, Ando) | |
| 42 | C. spathulata (L.B.Sm. & Downs) Stehmann & Semir. BRAZIL: Santa Catarina, Hashimoto et al. B90 (GHSP, Ando) | |
| 43 | C. thymifolia (A.St.-Hil.) Stehmann & Semir. BRAZIL: Santa Catarina, Hashimoto et al. B36 (BM, MBM, MVFA, S, SI, US, GHSP, Ando) | |
| 44 | unnamed taxon C1. BRAZIL: Rio Grande do Sul, Hashimoto et al. B7 (GHSP, Ando) | |
| 45 | unnamed taxon C2. BRAZIL: Rio Grande do Sul, Hashimoto et al. B201 (GHSP, Ando) | |
| 46 | unnamed taxon C3. BRAZIL: Santa Catarina, Hashimoto et al. B215 (GHSP, Ando) | |
| 47 | unnamed taxon C4. BRAZIL: Rio Grande do Sul, Hashimoto et al. B755 (GHSP, Ando) | |
| 48 | unnamed taxon C5. BRAZIL: Rio Grande do Sul, Hashimoto et al. B789 (GHSP, Ando) | |
| 49 | unnamed taxon C6. BRAZIL: Santa Catarina, Hashimoto et al. B1126 (GHSP, Ando) | |
| 50 | unnamed taxon C7. BRAZIL: Rio Grande do Sul, Hashimoto et al. B1176 (GHSP, Ando) | |
| 51 | unnamed taxon C8. BRAZIL: Santa Catarina, Hashimoto et al. B1248 (GHSP, Ando) | |
| 52 | unnamed taxon C9. URUGUAY: Salto, Ando & Iida U229 (GHSP, Ando) | |
| Nicotiana L. (1735) 2n = 18, 20, 24, 32, 36, 38, 40, 42, 44, 46, 48 | ||
| 55 | Nicotiana langsdorffii Weinm. (2n = 2x = 18). BRAZIL: Rio Grande do Sul, Hashimoto et al. HB622 (Ando) | |
Abbreviations of herbaria are after Holmgren et al. (1990) except GHSP (Centro de Pesquisas História Natural, São Paulo, Brazil) and Ando (temporary collection of Toshio Ando).
Petunia alpicola L. B. Sm. & Downs, P. helianthemoides Sendtn., P. kleinii L. B. Sm. & Downs, P. pubescens (Spreng.) R. E. Fr. and P. variabilis R. E. Fr. are five species of Petunia sensu Jussieu that share the same morphology and chromosome count (2n = 2x = 18) with Calibrachoa (Watanabe et al., 1996, 1997). Here, they are tentatively treated as Calibrachoa (C. alpicola, C. helianthemoides, C. kleinii, C. pubescens and C. variabilis), except in Table 1.
Southern hybridization
Total Petunia DNA was extracted from approx. 5 g of fresh leaves using a modified CTAB method (Doyle and Doyle, 1987). However, an unidentified substance, possibly a polysaccharide, prevented the use of this method with many of the Calibrachoa species. For these samples, the modified CTAB method described by Lassner et al. (1989) was used.
Approximately 1 µg each of extracted DNA was digested using 19 endonucleases that recognize four to six base pairs (AluI, AvaI, AvaII, BamHI, BcnI, BglII, ClaI, DraI, EcoO109I, EcoRI, EcoRV, EcoT22I, HaeIII, HhaI, HincII, HindIII, StyI, VspI, XbaI) according to the manufacturers' instructions. The recognition sequence of four base pair recognition enzymes was not included in those of six base pair recognition enzymes. DNA fragments were separated by electrophoresis in 1.5 % agarose gels and transferred onto Hybond N+ nylon membrane (Amersham, Little Chalfont, Buckinghamshire, UK) by capillary blotting with 20× SSC.
Twelve contiguous clones of Nicotiana tabacum cpDNA (pTBa1, pTBa2, pTB7, pTB8, pTB10, pTB15, pTB19, pTB20, pTB22, pTB25, pTB28, pTB29; Sugiura et al., 1986), which cover 99.4 % of the total sequence, were used as probes to detect RFLP. These probes were labelled with the ECL™ direct nucleic acid labelling and detection system (Amersham), according to the manufacturer's instructions, and hybridized with given cpDNA fragments for 12 h at 42°C. The hybridized blot was washed in 0.5× SSC with 0.4 % SDS for 2 × 20 min at 42°C, then in 5× SSC for 2 × 5 min at room temperature before detection.
Phylogenetic analysis
The data set was constructed by scoring the presence and absence of a restriction site as 1 and 0, respectively. When the site mutation was not unambiguously determined due to the detection limit of approx. 300 base pairs (bp), length mutation of a particular fragment was scored 1 and 0, according to the length. Other fragments that cannot be reasonably explained were recorded as presence/absence of the fragments. The most-parsimonious trees were sought using PAUP* version 4.0b10 (Swofford, 1998), using a heuristic search with the MULPARS and TBR branch-swapping options, following the strategy of Maddison et al. (1992), as adopted by Cantino et al. (1998), to search as many islands as practical. In this analysis, the 50 % majority consensus tree was calculated from the set of equally most-parsimonious trees obtained. To test the support for each clade in the parsimony tree, bootstrap analysis (Felsenstein, 1985) was performed using 1000 replicates with PAUP*. A decay analysis (Bremer, 1988) was also performed using TreeRot version 2 (Sorenson, 1999) in conjunction with PAUP*.
Another phylogenetic tree was also reconstructed for comparison using a neighbour joining (NJ) distance matrix method, which will be referred to as the distance tree. The distance matrix consisting of the number of nucleotide substitutions per site was calculated from only the unambiguously identified site mutations. Calculation of the matrix was done with the RESTDATA program (Ota, 1994) that employs the estimation method of Nei and Tajima (1983). The matrix was reformatted as NEXUS file, then the distance tree was calculated using the NJ option of PAUP*. The purpose of this analysis was to assess overall sequence similarity while avoiding the effect of homoplasy.
RESULTS
RFLPs
A total of 212 RFLPs was detected with all 19 restriction enzymes distributed in all 12 Nicotiana cpDNA clone regions for all 20 taxa of Petunia and 32 taxa of Calibrachoa. Nicotiana langsdorffii produced sufficiently similar RFLPs to Petunia sensu Jussieu for site mutations to be inferred without exact restriction site maps. Of the 212 mutations, 89 were shared by more than one taxon and were phylogenetically informative. Tree search for the parsimony tree was carried out using these 89 site mutations.
Of the 212 RFLPs, 85 (41 %) were unambiguously identified as gain or loss of a particular restriction site with this method. These site mutation data were used to calculate a distance matrix. Due to the length of the fragments produced and the detection limit of approx. 300 bp, it was not possible to unambiguously identify such restriction sites for some enzymes. There were 83 such cases (39 %), and the mutation was recorded as length mutation of a fragment when applicable. When this failed, a presence of unexplained fragment was recorded as 1.
Most of the taxa in Petunia sensu Jussieu that were studied had at least one unique site mutation that distinguished that taxon from all others. However, two pairs of taxa shared exactly the same site mutations. The first pair was Calibrachoa macrodactylon (L. B. Sm. & Downs) Wijsman and C. selloviana (Sendtn.) Wijsman, and the second pair was C. thymifolia (A. St.-Hil.) Stehmann & Semir and unnamed taxon C3. Both pairs of species had overlapping or similar distribution ranges [high altitude plateau of Santa Catarina (SC) and Rio Grande do Sul (RS) for the former pair and a localized area on the Atlantic coast in SC for the latter].
Deletion, insertion and reversion of the cpDNA
Two cases of deletion of a DNA fragment were detected in two taxa, Petunia altiplana T. Ando & Hashim. and unnamed taxon C1, both in the cpDNA overlap region of pTB7 and pTB20. The estimated length of the deletion was approx. 0.2 kilobase pairs (kbp) for P. altiplana and approx. 0.4 kbp for taxon C1. No insertion or reversion of a DNA fragment was unambiguously identified in any taxon studied.
Phylogenetic trees
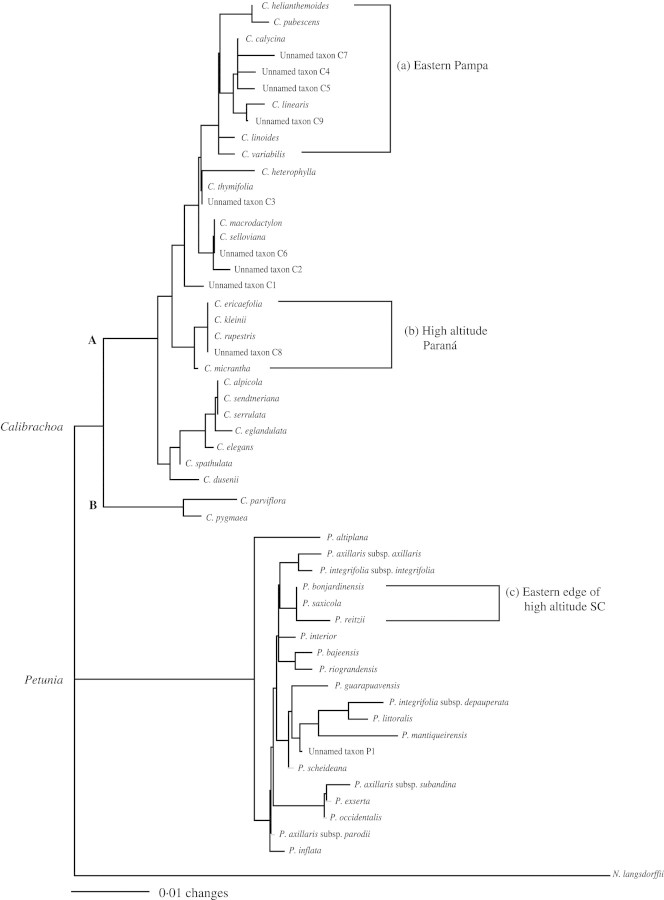
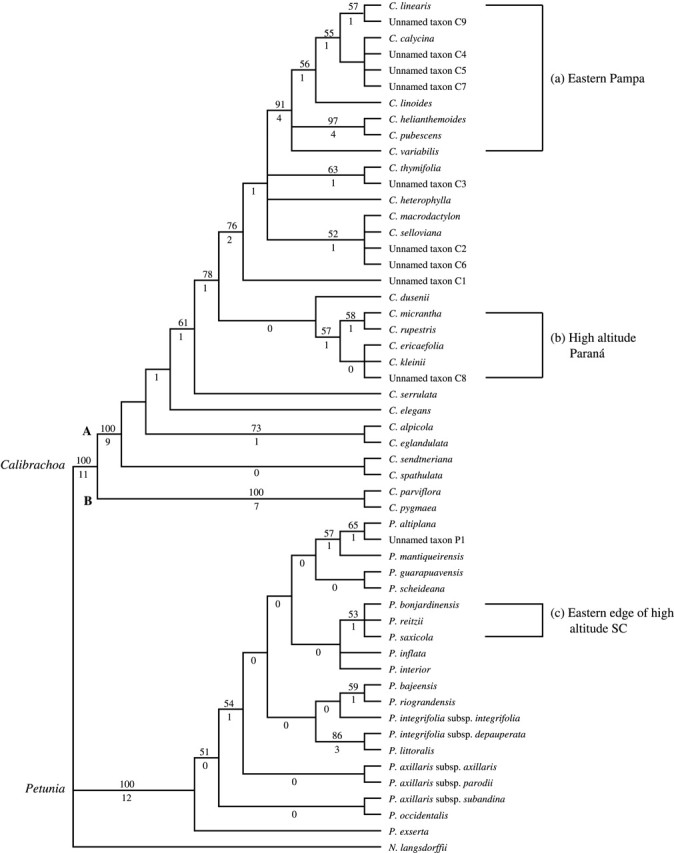
The heuristic search with PAUP* found 3420 equally parsimonious trees. From these trees, the 50 % majority consensus tree was calculated (Fig. 1). Due to the paucity of synapomorphic site mutations, many clades contained polytomies, so that the relationships of the terminal taxa in some clades were not resolved completely. However, both Petunia and Calibrachoa were monophyletic, and Calibrachoa was further subdivided into two clades: one consisted of C. parviflora and C. pygmaea (clade B in Fig. 1) and the other comprised the remaining Calibrachoa (clade A in Fig. 1). All of these clades had 100 % bootstrap support (Fig. 1). The five species of Petunia sensu Jussieu that have 2n = 2x = 18 chromosomes and morphological characters of Calibrachoa (Table 1) were included in the clade of Calibrachoa. In the distance tree (Fig. 2), the two genera, Petunia and Calibrachoa, and the two subgroups within Calibrachoa were again clearly identified.
Fig. 1.
Fifty per cent majority consensus tree calculated from 3420 equally most-parsimonious trees resulting from RFLP data obtained in this study with a heuristic search using the island strategy. This tree is identical to one of the most-parsimonious trees with tree length = 271, CI = 0.78, RI = 0.95. Numbers above the clades are the bootstrap confidence values (%, shown if >50 %), and numbers below the clades are decay values.
Fig. 2.
Distance matrix tree calculated using the NJ method. The distance matrix was calculated from 85 unambiguous site mutation data with the method of Nei and Tajima (1983) as implemented in RESTDATA program (Ota, 1994).
The most significant difference between the 50 % majority consensus tree and the distance tree was the position of P. altiplana. This was the taxon in which a deletion of part of the cpDNA was detected. The other taxon with the deletion (unnamed taxon C1) was found at the same place in both trees.
DISCUSSION
Phylogeny of Petunia sensu Jussieu
The phylogenetic trees obtained in this study (Figs 1 and 2) clearly identify two major clades in Petunia sensu Jussieu: the clades Petunia and Calibrachoa. Bootstrap support for both the Petunia and Calibrachoa clades in the 50 % majority consensus tree was 100 %, with 12 and 11 decay values, respectively (Fig. 1). This further validates the treatment dividing Petunia and Calibrachoa (Wijsman, 1990). Watanabe et al. (1996, 1997, 2001) and Ando et al. (2001) have confirmed that species of Petunia and Calibrachoa are genetically isolated from each other.
The Calibrachoa clade was further divided into two clades: the first with two species (C. parviflora and C. pygmaea, Figs 1 and 2; clade B), and the second with the remaining Calibrachoa (clade A). Each clade had a bootstrap probability of 100 % with decay values of seven or nine, respectively (Fig. 1).
Calibrachoa parviflora and C. pygmaea are unique in Petunia sensu Jussieu in terms of seed morphology (Watanabe et al., 1999) and nuclear DNA content (Mishiba et al., 2000). Moreover, these two species cross only with each other, and are cross-incompatible with all other Calibrachoa. In contrast, all Calibrachoa species except these two are cross-compatible with C. pubescens (Watanabe et al., 1997). In other words, the Calibrachoa species in clades A and B (Figs 1 and 2) are genetically isolated from each other. The great difference in the site mutations among the three subgroups of Petunia sensu Jussieu described above may reflect prolonged genetic isolation from one another.
When Petunia and Calibrachoa were compared in the distance tree (Fig. 2), larger genetic distances were found within the genus Calibrachoa than within Petunia. The average genetic distances within Calibrachoa and Petunia calculated from the distance matrix were 0·0144 and 0·0112, respectively. This suggests that divergence is further advanced in the genus Calibrachoa than in the genus Petunia. If the genetic distance is proportional to time, this may indicate that the genus Calibrachoa is older than the genus Petunia. However, the differences between genetic distances are only very small, hence more molecular phylogenetic data would be needed to be able to either accept or reject this hypothesis.
Clades associated with geographic distribution
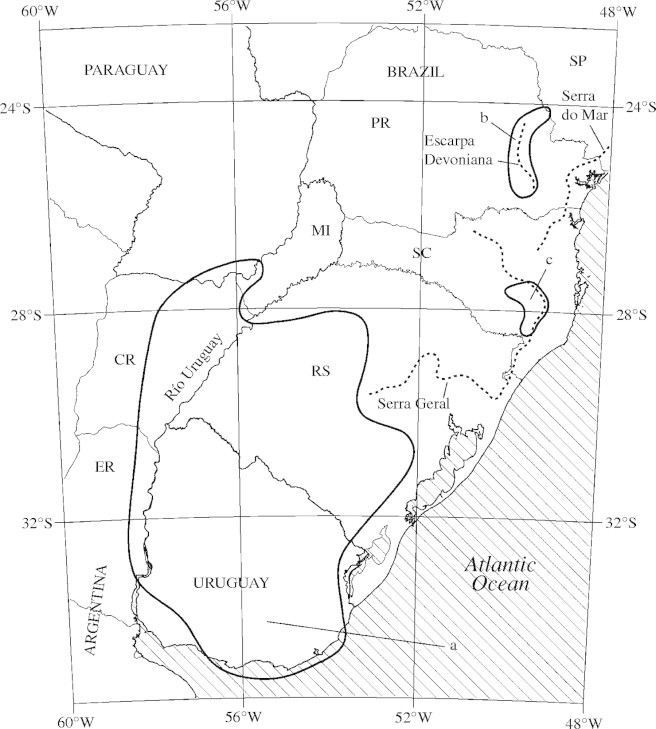
The native habitats of Petunia sensu Jussieu in South America have been studied for 17 seasons since 1988. A start is being made to understand the distributions and geographical proximities of the members of Petunia sensu Jussieu, and overall distribution maps of Petunia (Tsukamoto et al., 1998) and Calibrachoa (Tsukamoto et al., 2002) have been published. In the 50 % majority consensus tree (Fig. 1) and the distance tree (Fig. 2), the three clades marked a, b and c are considered to be associated with geographic distribution enclosing species endemic to their respective regions. Based on the present field observation data, a distribution map illustrating the distributions of these clades was made (Fig. 3).
Fig. 3.
Distribution of selected clades (a, b and c, corresponding to the previous figures) in Petunia sensu Jussieu found in this study. Abbreviation of states and provinces: Argentina, CR = Corrientes, ER = Entre Ríos, MI = Misiones; Brazil, PR = Paraná, RS = Rio Grande do Sul, SC = Santa Catarina, SP = São Paulo.
Clade a (Figs 1 and 2) includes Calibrachoa calycina (Sendtn.) Wijsman, C. helianthemoides, C. linealis (Hook.) Wijsman, C. linoides (Sendtn.) Wijsman, C. pubescens, C. variabilis and unnamed taxa C4, C5, C7 and C9. This clade was monophyletic in both trees (Figs 1 and 2) with 91 % bootstrap support in the 50 % majority consensus tree (Fig. 1) and two unique site mutations (Table 2). Except for Calibrachoa linoides, which has a wide distribution in southern Brazil and adjacent regions, these Calibrachoa taxa exclusively occupy the most south-western territory of the Calibrachoa, from southern Uruguay (C. pubescens) to southern Paraguay (C. calycina; region a in Fig. 3). Because these hilly lowlands are a continuation of the Pampa, the vast grassland extending widely in temperate Argentina, this region can be summarized as the Eastern Pampa.
Table 2.
Synapomorphic site mutations for selected clades in the 50 % majority consensus tree
| Clade |
Number of taxa |
Synapomorphic site mutations* |
||
|---|---|---|---|---|
| Petunia (sensu Wijsman) | 20 | AluI+pTB19, AvaI+pTB19, AvaII+pTBa1, AvaII+pTBa2, AvaII+pTB25, BamHI+pTBa1, BamHI+pTB19, BcnI+pTB25, ClaI+pTB28, EcoT22I+pTB25, HhaI+pTB8, Hind III+pTBa1 | ||
| Calibrachoa | 32 | AluI+pTB19, AvaII+pTBa2, AvaII+pTB8, AvaII+pTB19, BamHI+pTB29, BglII+pTB25, ClaI+pTBa1, DraI+pTBa2, EcoO109I+pTBa1, EcoO109I+pTB8, EcoO109I+pTB22, EcoRV+pTB25, HincII+pTB15, HincII+pTB25, HindIII+pTB8 | ||
| Clade A | ||||
| Calibrachoa other than C. parviflora and C. pygmaea | 30 | AluI+pTBa2, AluI+pTB7, AluI+pTB25, BamHI+pTB20, BcnI+pTB10, BcnI+pTB29, EcoRI+pTBa2, HhaI+pTB7, HhaI+pTB19 | ||
| Clade B | ||||
| C. parviflora and C. pygmaea | 2 | AluI+pTB20, AvaII+pTB20, BglII+pTBa2, EcoRV+pTB7-20, EcoT22I+pTBa1, EcoT22I+pTB20, XbaI+pTB22 | ||
| Clades with geographic association | ||||
| Clade a | ||||
| Eastern Pampa—Calibrachoa | 10 | AvaII+pTB19, AvaII+pTB25, EcoO109I+pTB15-28, HhaI+pTB19 | ||
| Clade b | ||||
| High altitude Paraná—Calibrachoa | 5 | HhaI+pTBa1 | ||
| Clade c | ||||
| Eastern edge of high altitude Santa Catarina—Petunia | 3 | ClaI+pTB15-28 | ||
The underline indicates that the mutation occurred exclusively in that clade.
Clade b in both trees (Figs 1 and 2) included C. ericaefolia (R. E. Fr.) Wijsman, C. kleinii, C. micrantha (R. E. Fr.) Stehmann & Semir, C. rupestris (Dusén) Wijsman and unnamed taxon C8. Although the bootstrap value for the clade was relatively low (57 %), and characterized with only one unambiguous, unique site mutation (Fig. 1 and Table 2), these taxa are exclusively found in eastern Paraná (PR) in Brazil (region b in Fig. 3), except C. kleinii and unnamed taxon C8. Calibrachoa kleinii is distributed more widely along the border of PR and SC in Brazil, the province of Misiones in Argentina, and Paraguay, and taxon C8 was from SC. Region b is highland near an escarpment called Escarpa Devoniana (the broken line in region b in Fig. 3), which is approx. 1000 m a.s.l., and is the eastern edge of a second high plateau (Segundo Planalto) that declines gradually to the west (Maack, 1968). This region, one may call it high-altitude Paraná, represents the most northern territory of Calibrachoa, except C. elegans (Miers) Stehmann & Semir, which is isolated far north in Minas Gerais in Brazil (Fries, 1911; Tsukamoto et al., 2002). Although Calibrachoa kleinii has a wider range, it is apparently a northern element. Calibrachoa species are abundant in region b, while no species of Petunia is found there.
Clades c in both trees (Figs 1 and 2) with weak bootstrap support of 53 % included three Petunia species: P. bonjardinensis T. Ando & Hashim., P. reitzii L. B. Sm. & Downs and P. saxicola L. B. Sm. & Downs. These are rather rare species endemic to region c (Fig. 3) along the Serra Geral in south-eastern SC, where the altitude exceeds 1000 m. Serra Geral (dotted line in eastern SC and RS in Fig. 3) is the edge of a lava plateau that covers much of eastern SC and north-eastern RS. The highest regions on the plateau are located near Serra Geral, where the altitude drops abruptly toward the eastern lowlands. The western side of the plateau declines gradually toward the River Uruguay (Fig. 3). This region can suitably be called the eastern edge of high-altitude SC. This region is also the endemic distribution range of some Calibrachoa species [C. alpicola, C. eglandulata Stehmann & Semir, C. sendtneriana (R.E. Fr.) Stehmann & Semir and C. serrulata (L.B.Sm. & Downs) Stehmann & Semir].
Compared with the total ranges of Petunia (Tsukamoto et al., 1998) and Calibrachoa (Tsukamoto et al., 2002), the species enclosed in clades b and c seem to be concentrated at the eastern margin of the territories (Fig. 3). It is assumed that the large amount of precipitation delimiting the highland regions (Maack, 1968) is one of the reasons why many species of both Petunia and Calibrachoa are found in such small areas. In contrast, clade a is characterized by the members with wider distributions (Fig. 3). The hilly lowland of this region may have allowed rapid dispersal or active genetic exchange among the members.
Infrageneric relationships of Petunia
In the 50 % majority consensus tree, Petunia axillaris together with the infraspecific taxa, P. exserta Stehmann and P. occidentalis R. E. Fr. formed the most basal branch in the Petunia clade (Fig. 1). They have a thick, consistently inflexed pedicel in contrast to the remaining members of Petunia, which have a thin, deflexed pedicel in the fruiting state. An exception is P. inflata R. E. Fr., whose thin pedicel is inflexed at the upper stem, but is often deflexed at the lower stem in younger plants.
The RFLP data did not appear particularly useful for resolving infraspecific relationships in Petunia, since there was some incongruity between the present results and the traditional infraspecific taxonomy. Petunia axillaris occurs widely in southern Bolivia, Paraguay, RS (Brazil), Uruguay, and the northern half of Argentina. Three allopatric subspecies are known today: subsp. axillaris, subsp. parodii (Steere) Cabrera and subsp. subandina T. Ando (Ando, 1996). In the phylogenetic trees produced in the present study, these three subspecies never formed a clade (Figs 1 and 2). The two most likely causes are convergence (three subspecies appear similar, but represent three distinctive lineages) and geographic isolation (they share a common ancestor, but have been separated in each region for enough time not to appear monophyletic when analysed).
Petunia inflata has been treated in various ways taxonomically. Fries (1911) described it as an independent species, while Smith and Downs (1966) treated it as synonymous to P. integrifolia, and Wijsman (1982) regarded it an extreme form of P. integrifolia. In the present study, P. inflata was distinguished from P. integrifolia by eight site mutations (AvaII+pTB7, BamHI+pTB7-20, BcnI+pTB15-28, BglII+pTBa1-15, DraI+pTB19, EcoRI+pTB25, StyI+pTB7, XbaI+pTB19), and the two taxa were situated in different locations in the phylogenetic trees (Figs 1 and 2). The present results seem to support the treatment of Fries (1911).
As shown here, the phylogenetic trees constructed from cpDNA RFLP did not necessarily correspond to the current infrageneric taxonomy of Petunia. Additional molecular data from nuclear DNA or a more rapidly changing region of cpDNA should be analysed to explain this discrepancy.
CONCLUSIONS
It is concluded that Petunia sensu Jussieu is a monophyletic clade when rooted using Nicotiana langsdorffii, and that it consists of two major monophyletic sister clades: Petunia and Calibrachoa. Two unusual Calibrachoa species, C. parviflora (the type species) and C. pygmaea, form a basal clade that is sister to the remaining Calibrachoa. Therefore, it is not necessary to consider a different nomenclature for the remaining Calibrachoa for now. Some of the infrageneric clades seemed to be tied with geographic distribution, although the support from the RFLP data was weak.
Supplementary Material
Acknowledgments
We thank Mr Tsuguyoshi Aoki (Buenos Aires, Argentina), Mr Sebastião T. Nagase, Mr Nobuyuki Hiranaka, Mr Tomio Koshizawa, Mr Hideo Ohkubo and Mr Roberto H. Ohkubo (São Paulo, Brazil) and Mr Masao Udagawa (Montevideo, Uruguay) for help in surveying the natural habitat. We also thank the DNA Bank of the Center for Gene Research of Nagoya University for kindly providing the tobacco cpDNA clones. This work was partly supported by a Grant-in-Aid for Scientific Research (B; project number 08456016) from the Ministry of Education, Science, Sports and Culture of Japan.
LITERATURE CITED
- Ando T. 1996. Distribution of Petunia axillaris (Solanaceae) and its new subspecies in Argentina and Bolivia. Acta Phytotaxonomica et Geobotanica 44: 19–30. [Google Scholar]
- Ando T, Hashimoto G. 1993. Two new species of Petunia (Solanaceae) from southern Brazil. Botanical Journal of the Linnean Society 111: 265–280. [Google Scholar]
- Ando T, Hashimoto G. 1998. Two new species of Petunia (Solanaceae) from southern Rio Grande do Sul, Brazil. Brittonia 50: 483–492. [Google Scholar]
- Ando T, Nomura M, Tsukahara J, Watanabe H, Kokubun H, Tsukamoto T, Hashimoto G, Marchesi E, Kitching IJ. 2001. Reproductive isolation in a native population of Petunia sensu Jussieu (Solanaceae). Annals of Botany 88: 403–413. [Google Scholar]
- Bremer K. 1988. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795–803. [DOI] [PubMed] [Google Scholar]
- Cantino PD, Wagstaff SJ, Olmstead RG. 1998.Caryopteris (Lamiaceae) and the conflict between phylogenetic and pragmatic considerations in botanical nomenclature. Systematic Botany 23: 369–386. [Google Scholar]
- Cerny TA, Caetano-Anollés G, Trigiano RN, Starman TW. 1996. Molecular phylogeny and DNA amplification fingerprinting of Petunia taxa. Theoretical and Applied Genetics 92: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Fries RE. 1911. Die Arten der Gattung Petunia Kungliga Svenska Vetenskapsakademiens Handlingar 46: 1–72. [Google Scholar]
- Griesbach RJ, Beck RM, Stehmann JR. 2000. Molecular heterogeneity of the chalcone synthase intron in Petunia HortScience 35: 1347–1349. [Google Scholar]
- Jussieu AL. 1803. Sur le Pétunia, genre nouveau de la famille des plantes solanées. Annales du Muséum National d'Histoire Naturelle 2: 214–216. [Google Scholar]
- Holmgren PK, Holmgren NH, Barnett LC. 1990.Index Herbariorum Part I: the herbaria of the world. Bronx, NY: New York Botanical Garden. [Google Scholar]
- Kabbaj A, Zeboudj F, Peltier D, Tagmount A, Tersac M, Dulieu H, Berbillé A. 1995. Variation and phylogeny of the ribosomal DNA unit types and 5 S DNA in Petunia Jussieu. Genetic Resources and Crop Evolution 42: 311–325. [Google Scholar]
- Lassner MW, Peterson P, Yoder JI. 1989. Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Molecular Biology Reporter 7: 116–128. [Google Scholar]
- Maack R. 1968. As zonas das paisagens naturais. In: Maack R. Geografia fisica do estado do Paraná. Curitiba: Banco de Desenvolvimento do Paraná, 85–88. [Google Scholar]
- Maddison DR, Ruvolo M, Swofford DL. 1992. Geographic origins of human mitochondrial DNA: phylogenetic evidence from control region sequences. Systematic Biology 41: 111–124. [Google Scholar]
- Mishiba K, Ando T, Mii M, Watanabe H, Kokubun H, Hashimoto G, Marchesi E. 2000. Nuclear DNA content as an index character discriminating taxa in the genus Petunia sensu Jussieu (Solanaceae). Annals of Botany 85: 665–673. [Google Scholar]
- Nei M, Tajima F. 1983. Maximum likelihood estimation of the number of nucleotide substitutions for restriction sites data. Genetics 105: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T. 1994. RESTDATA: restriction data and phylogenetic analysis (http://mep.bio.psu.edu). [Google Scholar]
- Sink KC. 1984. Taxonomy. In: Sink KC. ed. Petunia. Berlin: Springer, 3–9. [Google Scholar]
- Smith LB, Downs RJ. 1966. Petunia. In: Reitz PR. ed. Flora Ilustrada Catarinense. Solanáceas. Itajai: Herbario Barbosa Rodrigues, 261–291. [Google Scholar]
- Sorenson MD. 1999. TreeRot, version 2. Boston: Boston University. [Google Scholar]
- Stehmann JR, Semir J. 1997. A new species and new combinations in Calibrachoa (Solanaceae). Novon 7: 417–419. [Google Scholar]
- Sugiura M, Shinozaki K, Zaita N, Kusuda M, Kumano M. 1986. Clone bank of the tobacco (Nicotiana tabacum) chloroplast genome as a set of overlapping restriction endonuclease fragments: mapping of eleven ribosomal protein genes. Plant Science 44: 211–216. [Google Scholar]
- Swofford DL. 1998.PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tsukamoto T, Ando T, Kokubun H, Watanabe H, Tanaka R, Hashimoto G, Marchesi E, Kao T. 1998. Differentiation in the status of self-incompatibility among all natural taxa of Petunia (Solanaceae). Acta Phytotaxonomica et Geobotanica 49: 115–133. [Google Scholar]
- Tsukamoto T, Ando T, Watanabe H, Kokubun H, Hashimoto G, Sakazaki U, Suárez E, Marchesi E, Oyama K, Kao T-H. 2002. Differentiation in the status of self-incompatibility among Calibrachoa species (Solanaceae). Journal of Plant Research 115: 185–193. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Ando T, Iida S, Suzuki A, Buto K, Tsukamoto T. 1996. Cross-compatibility of Petunia cultivars and P. axillaris with native taxa of Petunia in relation to their chromosome number. Journal of the Japanese Society for Horticultural Science 65: 625–634. [Google Scholar]
- Watanabe H, Ando T, Iida S, Buto K, Tsukamoto T, Kokubun H, Hashimoto G, Marchesi E. 1997. Cross-compatibility of Petunia pubescens and P. pygmaea with native taxa of Petunia Journal of the Japanese Society for Horticultural Science 66: 607–612. [Google Scholar]
- Watanabe H, Ando T, Nishino E, Kokubun H, Tsukamoto T, Hashimoto G, Marchesi E. 1999. Three groups of species in Petunia sensu Jussieu (Solanaceae) inferred from the seed morphology. American Journal of Botany 86: 302–305. [PubMed] [Google Scholar]
- Watanabe H, Ando T, Tsukamoto T, Hashimoto G, Marchesi E. 2001. Cross-compatibility of Petunia exserta with other Petunia taxa. Journal of the Japanese Society for Horticultural Science 70: 33–40. [Google Scholar]
- Wijsman HJW. 1982. On the interrelationships of certain species of Petunia I. Taxonomic notes on the parental species of Petunia hybrida. Acta Botanica Neerlandica 31: 477–490. [Google Scholar]
- Wijsman HJW. 1990. On the inter-relationships of certain species of Petunia VI. New names for the species of Calibrachoa formerly included into Petunia (Solanaceae). Acta Botanica Neerlandica 39: 101–102. [Google Scholar]
- Wijsman HJW, de Jong JH. 1985. On the interrelationships of certain species of Petunia IV. Hybridization between P. linearis and P. calycina and nomenclatural consequences in the Petunia group. Acta Botanica Neerlandica 34: 337–349. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.