Abstract
• Background and Aims Genetic diversity in Castilleja grisea, an endangered, perennial herb endemic to San Clemente Island, California was investigated. Subsequent to the elimination of goats from the island in 1992, many populations of C. grisea have reappeared and have been increasing in size.
• Methods Nineteen populations were surveyed for their genotype at 19 allozyme loci.
• Key Results At the taxon level, 57·9 % of loci are polymorphic with AP = 3·09 and HE = 0·137. Populations averaged 33·0 % polymorphic loci with AP = 2·43 and HE = 0·099. Most variation is found within rather than among populations (GST = 0·128), although differentiation among populations is significant. Genetic identities range from I = 0·960 to I = 1·000 with mean I = 0·990. There is no significant relationship between genetic and geographic distance. Gene flow among populations is Nm = 2·50 based on private alleles and Nm = 1·70 based on FST. Outcrossing rates based on fixation indices average t = 1·01, indicating a primarily outcrossed mating system.
• Conclusions The observed genetic variation is moderately high, unusually so for an insular endemic species, suggesting that C. grisea may not have lost substantial genetic variation during 150 years of overgrazing, and indicating that it is unlikely to be endangered by genetic factors.
Keywords: Allozymes, conservation, Castilleja grisea, endangered species, endemic, genetic diversity, Scrophulariaceae, San Clemente Island
INTRODUCTION
We live in an age of unprecedented threats to biodiversity. Rates of extinction and endangerment are currently higher than at any time in the recent past, and they continue to increase (IUCN, 2003). Most endangerment in the United States occurs through habitat loss (Wilcove et al., 1998), and this is a factor that can be difficult to reverse. Sometimes, however, it is possible to minimize or eliminate current threats to endangered species. In particular, when endangerment is caused by introduced predators, their removal may allow endangered species to recover.
San Clemente Island, one of California's Channel Islands, has 272 native vascular plant species, 47 of which (13 %) are endemic (Junak et al., 1995). Starting in the mid-1800s, sheep, cattle, pigs and goats were introduced to the island by ranchers. Overgrazing by this combination of species continued until the early 1930s, when the US Government bought out the ranchers and the US Navy began to use the island for military training. Livestock were removed to the mainland, but feral goats remained and their populations increased dramatically to the detriment of native plant populations. An extirpation programme was initiated in 1962, but progress was uneven due to inconsistent funding. Goats were finally eliminated in 1992 (Kellogg and Kellogg, 1994). Some of the rare species have recovered substantially, in spite of the existence of other threats such as introduced plant and pathogen species, and increased incidence of fire, soil erosion, trampling, and bombardment due to Navy activities. Currently, six plant taxa are federally listed as endangered, and another 20 are considered Species of Concern by the US Fish and Wildlife Service (Junak and Wilken, 1998).
In spite of apparent population recovery, it is possible for populations of formerly rare species to remain highly susceptible to extirpation. Long periods of small population size are predicted to cause loss of genetic variation through genetic drift. On San Clemente Island, in particular, more than 150 years of overgrazing would be expected to have reduced levels of genetic variation substantially. Loss of genetic variation contributes to endangerment through reduction of both long-term evolutionary potential (adaptation to a changing environment is only possible with genetic variation; Frankel et al., 1995) and short-term fitness (in spatially or temporally heterogeneous environments, by increased inbreeding, and by other mechanisms; Huenneke, 1991).
Although it is not possible to compare current levels of genetic variation with those that existed before grazers were introduced to San Clemente Island, current levels of allozyme variation can be evaluated by comparison to related taxa or to taxa sharing characteristics that affect genetic variation. For example, species with a narrow geographic range have fewer polymorphic loci and alleles per locus, and lower expected heterozygosity than species with larger ranges (Hamrick and Godt, 1989). Similarly, island populations have less genetic variation (Barrett and Husband, 1989; Frankham, 1997) and generally experience more inbreeding than mainland populations (Frankham, 1998), and taxa endemic to islands appear to be especially genetically depauperate (DeJoode and Wendel, 1992; Frankham, 1997; Stuessy et al., 1998, but see Francisco-Ortega et al., 2000).
Castilleja grisea Dunkle (Orobanchaceae), the San Clemente Island Indian Paintbrush, is endemic to San Clemente Island (Fig. 1). Castilleja consists of approx. 200 species, primarily in the New World and especially in western North America (Junak and Wilken, 1998). Of the 36 species in California, five are endemic to the Channel Islands (including Guadalupe Island; Wallace, 1985). Castilleja grisea is a perennial herb or subshrub (Chuang and Heck, 1993), and it is the only representative of this genus found on San Clemente Island. Most populations occur in the shrubland/woodland communities found in steep canyons, but some are found in maritime desert scrub communities dominated by Lycium californicum, Opuntia littoralis and O. prolifera (Kellogg and Kellogg, 1994). Although, presumably, hemiparasitic like other members of the genus (Junak and Wilken, 1998), little is known about host species.
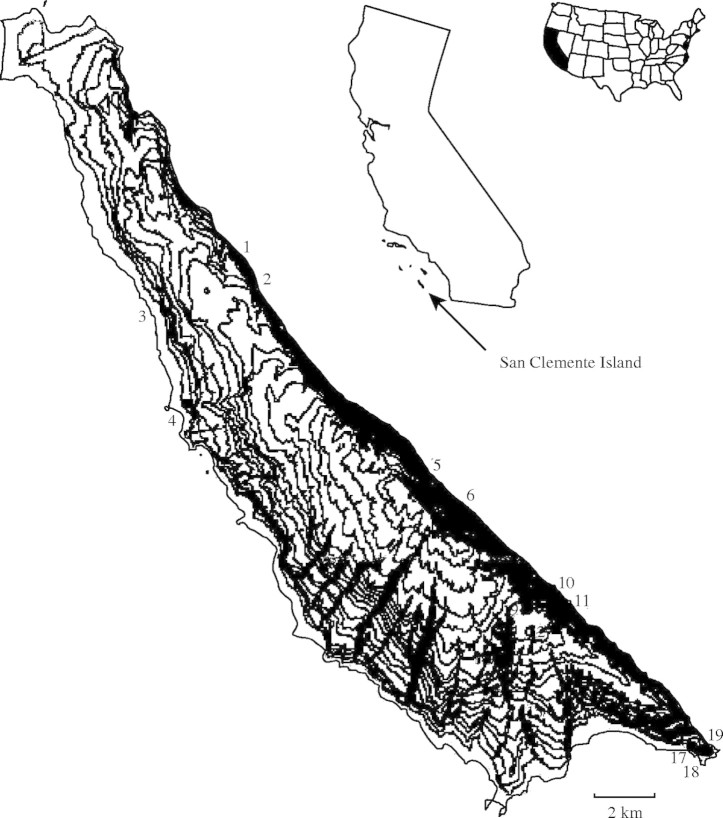
Fig. 1.
Locations of 19 sampled populations of Castilleja grisea on San Clemente Island, California.
Reported to be relatively common in the 1930s, C. grisea underwent a steady decline over the next half century. By 1963 it was reported as rare or occasional, and by 1978 very few individuals were observed (Kellogg and Kellogg, 1994). It was federally listed as endangered in 1977 by the US Fish & Wildlife Service, and was state-listed as endangered in 1982 by the California Department of Fish and Game; it is also on the California Native Plant Society List 1B (plants rare, threatened or endangered in California or elsewhere; Skinner and Pavlik, 1994). Subsequent to the elimination of goats from San Clemente Island in 1992 (Kellogg and Kellogg, 1984) the species has recovered to more than 3500 individuals (Junak and Wilken, 1998). Many populations consist of more than 500 individuals, and most consist of individuals of a range of sizes, suggesting that consistent recruitment has been occurring.
This article reports on the moderately high levels of genetic polymorphism in C. grisea on San Clemente Island. The goals of the study were (a) to determine the overall level of genetic variation, (b) to assess genetic structure and (c) to provide management guidelines based on population genetic data. This study was initiated as part of an investigation of the conservation genetics of rare and endangered plant species of San Clemente Island.
MATERIALS AND METHODS
Plant material
Leaf tissue was sampled from 19 populations spanning the range of C. grisea on San Clemente Island (Fig. 1 and Table 1). Seventeen to 55 individuals were haphazardly sampled from each population. Leaves were stored in plastic bags, and kept moist and cool until they were transported to the laboratory, where they were stored at 5 °C.
Table 1.
Genetic variability at 19 loci in 19 populations of Castilleja grisea on San Clemente Island
| Population |
N |
P |
A |
AP |
AE |
HO |
HE |
F |
t |
|---|---|---|---|---|---|---|---|---|---|
| 1. Ashtree | 27 | 26·3 | 1·32 | 2·20 | 1·11 | 0·090 | 0·099 | 0·083 | 0·85 |
| 2. Triangulation Point Jack | 36 | 31·6 | 1·42 | 2·33 | 1·12 | 0·114 | 0·109 | −0·057 | 1·12 |
| 3. Exclosure Cage | 45 | 42·1 | 1·68 | 2·62 | 1·16 | 0·114 | 0·140 | 0·182 | 0·69 |
| 4. Bunker | 37 | 33·3 | 1·50 | 2·50 | 1·14 | 0·131 | 0·123 | −0·029 | 1·06 |
| 5. North Boulders | 38 | 52·6 | 1·63 | 2·20 | 1·15 | 0·115 | 0·131 | 0·060 | 0·89 |
| 6. South Boulders | 35 | 38·9 | 1·50 | 2·29 | 1·08 | 0·075 | 0·072 | −0·027 | 1·06 |
| 7. Norton Canyon | 28 | 31·6 | 1·37 | 2·17 | 1·12 | 0·132 | 0·107 | −0·176 | 1·43 |
| 8. Box Canyon | 37 | 33·3 | 1·39 | 2·17 | 1·09 | 0·068 | 0·080 | 0·070 | 0·87 |
| 9. Upper Bryce Canyon | 30 | 36·8 | 1·42 | 2·14 | 1·09 | 0·077 | 0·081 | 0·036 | 0·93 |
| 10. Triangulation Point Malo | 17 | 27·8 | 1·33 | 2·20 | 1·10 | 0·092 | 0·092 | −0·035 | 1·07 |
| 11. Grassy Knoll | 33 | 42·1 | 1·68 | 2·62 | 1·14 | 0·112 | 0·124 | 0·045 | 0·91 |
| 12. Dead Man's Curve | 48 | 22·2 | 1·33 | 2·50 | 1·08 | 0·090 | 0·076 | −0·141 | 1·33 |
| 13. Upper China Canyon | 55 | 31·6 | 1·42 | 2·33 | 1·07 | 0·066 | 0·069 | 0·017 | 0·97 |
| 14. South China Chalk | 22 | 36·8 | 1·42 | 2·14 | 1·15 | 0·122 | 0·127 | 0·012 | 0·98 |
| 15. Horse Beach Canyon | 38 | 18·8 | 1·44 | 3·33 | 1·08 | 0·058 | 0·071 | 0·147 | 0·74 |
| 16. Sibara Ridge | 44 | 33·3 | 1·56 | 2·67 | 1·10 | 0·087 | 0·094 | 0·055 | 0·90 |
| 17. North Pyramid Head | 36 | 16·7 | 1·22 | 2·33 | 1·04 | 0·054 | 0·041 | −0·177 | 1·43 |
| 18. Pyramid Head 2 | 44 | 42·1 | 1·74 | 2·75 | 1·14 | 0·112 | 0·124 | 0·070 | 0·87 |
| 19. Pyramid Head 1 | 24 | 29·4 | 1·47 | 2·60 | 1·13 | 0·113 | 0·115 | −0·025 | 1·05 |
| Mean | 33·0 | 1·47 | 2·43 | 1·11 | 0·096 | 0·099 | 0·006 | 1·01 | |
| s.e. | 2·0 | 0·03 | 0·07 | 0·01 | 0·006 | 0·006 | 0·022 | 0·05 | |
| Species | 674 | 57·9 | 2·21 | 3·09 | 1·16 | 0·100 | 0·137 | − | – |
N, sample size; P, proportion of polymorphic loci; A, alleles per locus; AP, alleles per polymorphic locus; AE, effective number of alleles; HO, observed heterzygosity; HE, expected heterozygosity; F, fixation index; t, outcrossing rate.
Electrophoresis
Electrophoretic methods followed Soltis et al. (1983). Leaf tissue was crushed in phosphate extraction buffer (Conkle et al., 1982) and stored at −80 °C until electrophoresis was conducted. Two buffer systems were used to resolve loci coding for 16 enzymes. Aspartate aminotransferase (AAT; EC 2.6.1.1), diaphorase (DIA; EC 1.6.99.–), menadione reductase (MR; EC 1.6.99.3), phosphoglucose isomerase (PGI; EC 5.3.1.9), and phosphoglucomutase (PGM; EC 5.4.2.2) were resolved using a TRIS-citrate, pH 6·3/6·7 buffer system (Selander et al., 1971) with 13·0 % starch gels. Aconitase (ACO; EC 4.2.1.3), alcohol dehydrogenase (ADH; EC 1.1.1.1), aldolase (ALD; EC 4.1.2.13), fructose-1,6-diphosphatase (FDP; EC 3.1.3.11), glyceraldehyde-3-phosphate dehydrogenase (G3P; EC 1.2.1.12), isocitrate dehydrogenase (IDH; EC 1.1.1.42), malate dehydrodenase (MDH; EC 1.1.1.37), malic enzyme (ME; EC 1.1.1.40), mannose phosphate isomerase (MPI; EC 5.3.1.8), 6-phosphogluconate dehydrogenase (PGD; EC 1.1.1.44) and shikimate dehydrogenase (SKD; EC 1.1.1.25) were resolved using a morpholine citrate, pH 6·1 buffer system (Clayton and Tretiak, 1972) with 13 % starch gels. Staining recipes for all enzymes followed Soltis et al. (1983), except for diaphorase (Murphy et al., 1990). Loci were numbered sequentially with the most anodally migrating enzyme designated ‘1’.
Data analysis
Data were analysed using the computer program Genestrut (Constantine et al., 1994). Percentage of polymorphic loci (P), mean number of alleles per locus (A) and per polymorphic locus (AP), effective number of alleles (AE), observed heterozygosity (HO), and expected heterozygosity (HE) were calculated. Loci were considered polymorphic if more than one allele was detected. Levels of genetic variation were calculated for individual populations and for all individuals pooled across all populations. Fixation indices (F), reflecting deviations from Hardy–Weinberg equilibrium, were calculated and outcrossing rates (t) were estimated using t = (1 − F)/(1 + F) (Weir, 1990). The partitioning of genetic diversity within and among all populations was analysed using F-statistics (Nei, 1973). Nei's unbiased genetic identity (I) (Nei, 1978) was calculated for pairwise comparisons of populations. The relationship between genetic and geographic distance on San Clemente Island was tested using Mantel (1967) analysis. A cluster analysis was performed using UPGMA and Rogers' genetic distance (Rogers, 1972). Gene flow was estimated using Wright's formula (Wright, 1951), Nm = (1 − FST)/4FST, with FST considered equivalent to GST (Nei, 1977). A second estimate was based on the frequency of private alleles (alleles found in a single population; Slatkin, 1985).
RESULTS
Loci and alleles scored
Enzyme electrophoresis resulted in clear and consistent staining for 16 enzymes encoded by 19 putative loci: Aat, Aco, Adh, Ald, Dia-1, Dia-2, Fdp, G3p, Idh, Mdh-1, Mdh-2, Mdh-3, Me, Mpi, Mr, Pgd, Pgi, Pgm and Skd. All enzymes migrated anodally.
Eleven loci are polymorphic in at least one population (57·9 %; Table 1). Totalled across all populations, 42 alleles were detected on San Clemente Island. Seven loci have one common allele in all populations and one or two alternative alleles. Three loci (Adh, Mpi and Pgm) have two common alleles and one to three alternative alleles. One locus (Skd) has three common alleles and one alternative allele. The average number of alleles per polymorphic locus is 3·09, and overall expected heterozygosity for San Clemente Island is 0·137.
Measures of genetic variability
Populations contain from three to ten polymorphic loci with an average number of alleles per polymorphic locus of 2·43, and average expected heterozygosity of 0·099 (Table 1). Populations contain 14–25 alleles ranging in frequency from 0·01 to 0·30 in populations (mean q = 0·086). Only five alleles are unique to single populations (five different ones).
Fixation indices and outcrossing rates
Mean fixation indices (F) range from F = −0·177 to F = 0·147 with mean F = 0·006 (s.e. = 0·022; n = 19), indicating an overall conformance to Hardy–Weinberg expectations. Nineteen of 116 fixation indices for individual loci (ranging from F = −0·461 to F = 0·512) are significant; 14 of these (73·7 %) are positive. Outcrossing rates (t) based on fixation indices range from 0·69 to 1·43 (mean t = 1·01; Table 1), indicating a predominantly outcrossed mating system.
Genetic identity measures
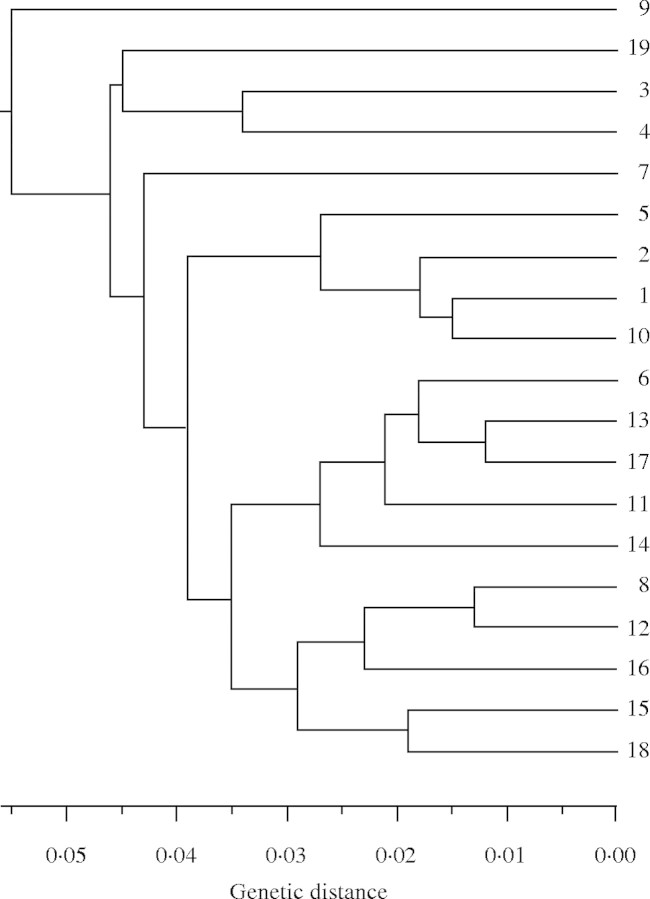
Genetic identities for pairs of populations are generally high, ranging from I = 0·960 to I = 1·000 with a mean of I = 0·990 (s.e. = 0·001). The phenogram produced by UPGMA cluster analysis depicts a relatively close genetic relationship among all populations (Fig. 2). No significant relationship exists between genetic and geographic distance (P = 0·6640, n = 19; Mantel, 1967).
Fig. 2.
Cluster analysis of 19 populations of C. grisea on San Clemente Island, California, using the UPGMA method and Rogers' genetic distance values (Rogers, 1972).
F-statistics
Mean FIS, representing average deviation from Hardy–Weinberg expectations within populations, is not significant (mean FIS = −0·056; Table 2). Differentiation of populations is relatively low but significantly different from 0 (mean FST = 0·128, P < 0·01). Of the total gene diversity found on San Clemente Island, 87·3 % is found within populations and 12·7 % is found among populations (mean HS = 0·131, mean DST = 0·019, mean HT = 0·150, GST = 0·127).
Table 2.
Summary of F-statistics at seven polymorphic loci of Castilleja grisea on San Clemente Island
| Locus |
FIS |
FST |
FIT |
|---|---|---|---|
| Idh | −0·139* | 0·070* | −0·059 |
| Mdh-1 | −0·166 | 0·130* | −0·015 |
| Mdh-2 | −0·193 | 0·131* | −0·037 |
| Pgd | −0·012 | 0·070* | 0·059 |
| Skd | −0·005 | 0·143* | 0·139 |
| Ald | 0·002 | −0·002 | 0·000 |
| Mr | −0·069 | 0·057* | −0·008 |
| Mean | −0·056 | 0·128* | 0·080 |
FST values significantly different from 0 (P < 0·01).
Gene flow
Gene flow among San Clemente Island populations is Nm = 2·50 using Slatkin's method (Slatkin, 1985) based on five private alleles with an average frequency of 0·046. Wright's method (Wright, 1951) yielded an estimate of Nm = 1·70.
DISCUSSION
Genetic variation
Castilleja grisea exhibits moderately high levels of genetic variation, especially at the taxon level. Standard measures of genetic variation are considerably higher in C. grisea (P = 57·9 %, A = 2·21, HE = 0·137) than in endemic species in general (P = 40·0 %, A = 1·80, HE = 0·096; Hamrick and Godt, 1989) and rival values for widespread species (P = 58·9 %, A = 2·29, HE = 0·202). At the population level, C. grisea values (P = 33·0 %, A = 1·47, HE = 0·099) are comparable to those in the average plant species (P = 34·2 %, A = 1·53, HE = 0·113), considerably higher than those for endemic plant species (P = 26·3 %, A = 1·39, HE = 0·063), and lower than those for widespread species (P = 43·0 %, A = 1·72, HE = 0·159).
Several characteristics of C. grisea are associated with higher levels of genetic variation in plants: sexual reproduction, a long-lived perennial herbaceous habit, late successional status, and an outcrossing, animal-pollinated, mating system (Hamrick and Godt, 1989). Although pollinators have not been observed on this species, the yellow flowers suggest that insect pollination is likely to occur. Both insect and hummingbird pollination have been reported for other species in the genus (Grant, 1994). The present estimates of outcrossing rate, based on three to ten polymorphic loci per population, clearly indicate that C. grisea produces seeds primarily through outcrossing.
The moderately high level of genetic variation observed in C. grisea is surprising for an insular endemic species with a recent history of extreme rarity and endangerment. The rapid recovery of populations subsequent to the removal of goats suggests that considerable recruitment has occurred from the seed bank, and our genetic data suggest that the seed bank maintains substantial genetic diversity. Although innate dormancy does not appear to occur in C. grisea (Junak and Wilken, 1998), seeds may remain viable in the soil for many years through induced or enforced dormancy.
Genetic differentiation
Most variation is found within rather than among populations of C. grisea on San Clemente Island (GST = 0·127), although differentiation among populations is significant. This level of differentiation among populations is low for plant species (mean GST = 0·224; Hamrick and Godt, 1989), but perhaps not surprising because all 19 populations are found within 23 km of each other. Low genetic differentiation is also consistent with characteristics such as an outcrossing breeding system and late successional status. Moreover, the indirect estimate of gene flow based on private alleles (Nm = 2·50) indicates sufficient historical gene flow to reduce population differentiation.
The high genetic identity values of C. grisea populations also indicate close similarity among all populations. Strikingly, adjacent populations such as Triangulation Point Malo and Grassy Knoll (populations 10 and 11), or Sibara Ridge, North Pyramid Head, Pyramid Head 2 and Pyramid Head 1 (populations 16–19), usually are widely distributed in the UPGMA phenogram, consistent with the absence of a correlation between genetic and geographic distance among pairs of populations. Gene flow may have occurred at a broad enough scale to reduce correlation with distance.
The occurrence of C. grisea populations in two distinct communities (canyon shrubland/woodland and maritime desert scrub) may be of genetic and ecological significance. Different host species of hemiparasites have been shown to affect rate of attachment of haustoria and subsequent nutrient uptake, as well as interactions with pollinators and herbivores, and, ultimately, fitness (Helton et al., 2000; Adler, 2002, 2003). Populations of a hemiparasitic species in different habitats might therefore be expected to be genetically differentiated for different host species. However, the present data do not suggest any differentiation at alloyzme loci correlated with habitat. The maritime desert scrub populations (3, 4 and 16–19) are scattered throughout the UPGMA phenogram, and virtually no genetic diversity is attributable to habitat in a genetic diversity analysis (87·2 % of the genetic variation occurs within populations, 12·3 % among populations within habitat types, and only 0·50 % among habitat types).
Comparison with other plants
Island plants generally have been found to have reduced levels of genetic variation. DeJoode and Wendel (1992) reviewed data from 69 insular endemic plants in 16 genera, mainly from Pacific archipelagos, and found lower than average species-level values of genetic diversity (P = 0·25, A = 1·32, HT = 0·064). Frankham (1997) reviewed comparisons of closely related insular endemic and mainland plant taxa, and found that the insular endemic species is nearly always less heterozygous than its mainland congener. Endemic species of the Juan Fernandez archipelago, Chile, generally exhibit low genetic variation (Stuessy et al., 1998). Curiously, endemic plants of the Canary Islands are more genetically variable (HT = 0·186 for 69 species in 18 genera) than species of other island archipelagos (HT = 0·064) possibly due to the greater age of these islands compared with their Pacific counterparts and to proximity to a continental source of migrants (Francisco-Ortega et al., 2000).
Other endemic plant species of San Clemente Island exhibit a wide range of genetic diversity. Jepsonia malvifolia maintains extraordinarily high levels of genetic variation, both at the species (P = 95·2 %, A = 2·81, HE = 0·179) and population levels (P = 60·2 %, A = 1·85, HE = 0·158; Helenurm, 2001). In contrast, the rare taxa Camissonia guadalupensis ssp. clementina (P = 37·5 %, A = 1·69, HE = 0·088; K. Helenurm and S. Hall, unpubl. res.), Cryptantha traskiae (P = 18·8 %, A = 1·31, HE = 0·003; K. Helenurm and S. Hall, unpubl. res.) and Sibara filifolia (P = 6·9 %, A = 1·10, HE = 0·010; Helenurm, 2003) exhibit low levels of genetic diversity that are consistent with their recent and continued rarity, and with their narrow range and insular endemic status. The taxon that perhaps most closely resembles C. grisea on San Clemente Island is Delphinium variegatum ssp. variegatum. This taxon is also federally endangered, but many populations have reappeared and population sizes have increased subsequent to the removal of goats in 1992. Delphinium v. variegatum has levels of genetic variation comparable to C. grisea (P = 55·6 %, A = 2·61, HE = 0·127 for the species; P = 33·6 %, A = 1·52, HE = 0·064 for populations; Dodd and Helenurm, 2002). Thus, the moderately high level of genetic diversity reported here for C. grisea may be characteristic of recovering species on San Clemente Island.
Levels of genetic diversity in plant species are also associated with family. A review of genetic diversity in 507 plant species (Hamrick and Godt, 1996) indicates that families with predominantly herbaceous species, such as the Scrophulariaceae (including genera now transferred to the Orobanchaceae), have less genetic diversity and higher genetic differentiation than families with predominantly long-lived, woody perennials. The 16 species of Scrophulariaceae included in this analysis averaged P = 37·2, HE = 0·123 and GST = 0·372; C. grisea thus exhibits relatively high levels of genetic variation but relatively little genetic differentiation in comparison to related species.
Management guidelines
San Clemente Island has a long history of sheep, cattle and pig ranching and feral goat grazing, resulting in overgrazing, subsequent erosion and loss of shrub and tree cover. It has been owned by the United States Government and operated by the United States Navy since the early 1930s, providing active naval training and support activities. The elimination of goats in 1992 has permitted vegetative recovery (Kellogg and Kellogg, 1994), but other threats remain. Introduced plant species threaten the native vegetation (including grasses such as Avena spp., Bromus spp., Ehrharta calycina and Piptantherum miliaceum, and other species such as Asphodelus fistulosus, Foeniculum vulgare and Carpobrotus spp.), and military activities have increased the frequency of fires and trampling of vegetation. Because the island will continue to experience heavy use for military purposes, management guidelines are required to safeguard rare and threatened plants. Understanding the pattern of genetic variation within and among populations of rare species is critical for prioritizing populations for protection, for guiding collection of ex situ material and for choosing seed sources for reintroduction and restoration.
Overall, C. grisea on San Clemente Island does not appear to be threatened by genetic factors. Populations maintain moderately high levels of genetic diversity in comparison to most plants, indicating greater than average evolutionary potential for an endemic insular species with a narrow geographic range. Moreover, the observed levels of variation in combination with an outcrossing mating system suggest that potential problems with inbreeding are unlikely. If population sizes (currently ranging from about 30 to more than 500 individuals) can be maintained or increased, there would appear to be little cause for concern on genetic grounds regarding the long-term persistence of this species.
Differences in levels of genetic diversity among populations of C. grisea provide a criterion for prioritization for conservation or management. Populations vary more than two-fold in most measures of genetic variation. Exclosure Cage, North Boulders, Grassy Knoll, and Pyramid Head 2 are high in proportion of polymorphic loci and expected heterozygosity, also contain most variant alleles, and together contain all but two of the alleles (both private alleles) identified in this study. These populations are all relatively large (100 to more than 500 individuals), and are distributed throughout the range of C. grisea. Although the conservation of these populations seems especially important, continued presence of several intervening populations may also be essential. Loss of intervening populations may reduce overall levels of gene flow, potentially causing loss of variation through genetic drift occurring in newly isolated populations.
The distribution of genetic variation among populations provides few guidelines for prioritizing populations. If alleles are categorized as widespread versus local (found in only one or a few adjacent populations) and as common versus rare (having frequencies always less than 0·05) following Marshall and Brown (1975), then all four categories of alleles are found in C. grisea, although far from equally. Thirty-six of the total 42 alleles (86 %) are widespread on San Clemente Island; 35 of these are common and only one is always rare. Only six alleles (14 %) are locally distributed; two of these are found at frequencies >0·05 in some populations while only four are always rare. These locally distributed alleles are found throughout the range of C. grisea, indicating that populations selected for protection (or for collection of material for ex situ conservation) should not be restricted to particular areas of San Clemente Island. However, the loss of any one population is unlikely to represent a significant loss of genetic diversity because the majority of alleles are found in more than one population and locally distributed alleles tend to occur at low frequencies.
Conclusions
Levels of genetic variation are moderately high in C. grisea, especially considering its endangered status and its narrow geographic range as an endemic species of a small island. It appears to be in no special danger of extinction due to genetic factors. Although populations vary about two-fold in most measures of genetic diversity, differentiation among them is low. The present genetic data, in combination with the ecological recovery of C. grisea since the removal of goats from San Clemente Island, suggest that this taxon is in no urgent danger of extinction.
Acknowledgments
We thank Jonathan Dunn, Steve Junak, Jan Larson, Kim O'Connor and David Pivorunas for help with various aspects of this study. This study was funded by the Natural Resources Office, Staff Civil Engineer, Naval Air Station, North Island, San Diego, California.
LITERATURE CITED
- Adler LS. 2002. Host effects on herbivory and pollination in a hemiparasitic plant. Ecology 83: 2700–2710. [Google Scholar]
- Adler LS. 2003. Host species affects herbivory, pollination and reproduction in experiments with parasitic Castilleja Ecology 84: 2083–2091. [Google Scholar]
- Barrett SCH, Husband BC. 1989. The genetics of plant migration and colonization. In: Brown AHD, Clegg MT, Kahler AT, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer Associates, 254–277. [Google Scholar]
- Chuang TI, Heck LR. 1993.Castilleja In: Hickman, JC, ed. The Jepson manual: higher plants of California. Berkeley, California: University of California Press, 916–922. [Google Scholar]
- Clayton JW, Tretiak DN. 1972. Amine-citrate buffers for pH control in starch gel electrophoresis. Journal of the Fisheries Research Board of Canada 29: 1169–1172. [Google Scholar]
- Conkle MT, Hodgskiss PD, Nunnally LB, Hunter SC. 1982.Starch gel electrophoresis of conifer seeds: a laboratory manual. USDA Forest Service General Technical Report PSW-64. Berkeley, CA: Pacific Southwest Forest and Range Experiment Station. [Google Scholar]
- Constantine CC, Hobbs RP, Lymbery AJ. 1994. FORTRAN programs for analyzing population structure from multilocus genotypic data. Journal of Heredity 85: 336–337. [Google Scholar]
- DeJoode DR, Wendel JF. 1992. Genetic diversity and origin of the Hawaiian Island cotton, Gossypium tomentosum American Journal of Botany 79: 1311–1319. [Google Scholar]
- Dodd SC, Helenurm K. 2002. Genetic diversity in Delphinium variegatum (Ranunculaceae): a comparison of two insular endemic subspecies and their widespread mainland relative. American Journal of Botany 89: 613–622. [DOI] [PubMed] [Google Scholar]
- Francisco-Ortega J, Santos-Guerra A, S-C. K, Crawford DJ. 2000. Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany 87: 909–919. [PubMed] [Google Scholar]
- Frankel OH, Brown AHD, Burdon JJ. 1995.The conservation of plant biodiversity. Cambridge: Cambridge University Press. [Google Scholar]
- Frankham R. 1997. Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327. [DOI] [PubMed] [Google Scholar]
- Frankham R. 1998. Inbreeding and extinction: island populations. Conservation Biology 12: 665–675. [Google Scholar]
- Grant V. 1994. Historical development of ornithophily in the western North American flora. Proceedings of the National Academy of Sciences of the USA 91: 10407–10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. 1989. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AT, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer Associates, 43–63. [Google Scholar]
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London, Series B 351: 1291–1298. [Google Scholar]
- Helenurm K. 2001. High levels of genetic polymorphism in the insular endemic herb Jepsonia malvifolia Journal of Heredity 92: 427–432. [DOI] [PubMed] [Google Scholar]
- Helenurm K. 2003. Genetic diversity in the rare insular endemic Sibara filifolia (Brassicaceae). Madrono 50: 181–186. [Google Scholar]
- Helton RC, Kirkman LK, Musselman LJ. 2000. Host preference of the federally endangered hemiparasite Schwalbea americana L. (Scrophulariaceae). Journal of the Torrey Botanical Society 127: 300–306. [Google Scholar]
- Huenneke LF. 1991. Ecological implications of genetic variation in plant populations. In: Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants. New York: Oxford University Press, 31–44. [Google Scholar]
- IUCN. 2003. IUCN red list of threatened species. http://www.redlist.org/info/tables/table1.html. 2 Feb. 2004. [Google Scholar]
- Junak SA, Wilken DH. 1998.Sensitive plant status survey, Naval Auxiliary Landing Field, San Clemente Island, California. Santa Barbara, CA: Santa Barbara Botanic Garden. [Google Scholar]
- Junak S, Ayers T, Scott R, Wilken D, Young D. 1995.A flora of Santa Cruz Island. Santa Barbara, CA: Santa Barbara Botanic Garden. [Google Scholar]
- Kellogg EM, Kellogg JL. 1994.San Clemente Island vegetation condition and trend and the elements of ecological restoration. Reedley, California: Tierra Data Systems. [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Research 27: 209–220. [PubMed] [Google Scholar]
- Marshall DR, Brown AHD. 1975. Optimum sampling strategies in genetic conservation. In: Frankel OH, Hawkes JG, eds. Crop genetic resources for today and tomorrow. Cambridge: Cambridge University Press, 53–80. [Google Scholar]
- Murphy RW, Sites JW, Buth DB, Haufler CH. 1990. Proteins I: isozyme electrophoresis. In: Hillis DM, Moritz C, eds. Molecular systematics. Sunderland, MA: Sinauer Associates, 45–127. [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1977. F-statistics and analysis of gene diversity in subdivided populations. Annals of Human Genetics 41: 225–233. [DOI] [PubMed] [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JS. 1972. Measures of genetic similarity and genetic distance. Studies in Genetics VII, University of Texas Publication No. 7213, 145–153. [Google Scholar]
- Selander RK, Smith MH, Yang SY, Johnson WE, Gentry JB. 1971. Biochemical polymorphism and systematics in the genus Peromyscus 1. Variation in the old-field mouse (Peromyscus polionotus). Studies in Genetics VI. University of Texas Publication 7103: 49–90. [Google Scholar]
- Skinner MW, Pavlik BM. 1994.California Native Plant Society inventory of rare and endangered vascular plants of California. Sacramento, CA: California Native Plant Society. [Google Scholar]
- Slatkin M. 1985. Rare alleles as indicators of gene flow. Evolution 39: 53–65. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Haufler CH, Darrow DC, Gastony GH. 1983. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal 73: 9–27. [Google Scholar]
- Stuessy TF, Swenson U, Crawford DJ, Anderson G, Silva OM. 1998. Plant conservation in the Juan Fernandez archipelago, Chile. Aliso 16: 89–101. [Google Scholar]
- Wallace GD. 1985.Vascular plants of the Channel Islands of southern California and Guadalupe Island, Baja California, Mexico. Natural History Museum of Los Angeles County, Contributions in Science No. 365, 1–136. [Google Scholar]
- Weir BS. 1990.Genetic data analysis. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Wilcove DS, Rothstein D, Dubow J, Phillips A, Losos E. 1998. Quantifying threats to imperiled species in the United States. Bioscience 48: 607–615. [Google Scholar]
- Wright S. 1951. The genetic structure of populations. Annals of Eugenics 15: 323–354. [DOI] [PubMed] [Google Scholar]