Significance
Human respiratory syncytial virus (RSV) is the most important viral agent of serious pediatric respiratory-tract disease. We designed new live attenuated RSV vaccine candidates by codon-pair deoptimization (CPD). Specifically, viral ORFs were recoded to increase the usage of underrepresented codon pairs, leaving amino acid coding unchanged. CPD viruses were temperature-sensitive and grew less efficiently in vitro than wild-type RSV. In addition, the CPD viruses exhibited a range of restriction in mice and African green monkeys that compared favorably with existing attenuated strains presently in clinical studies. This study produced examples of a new type of vaccine candidate for RSV and showed that CPD of a nonsegmented negative-strand RNA virus can rapidly generate vaccine candidates with a range of attenuation.
Keywords: negative strand RNA virus, pneumovirus, live attenuated vaccine
Abstract
Human respiratory syncytial virus (RSV) is the most important viral agent of serious pediatric respiratory-tract disease worldwide. A vaccine or generally effective antiviral drug is not yet available. We designed new live attenuated RSV vaccine candidates by codon-pair deoptimization (CPD). Specifically, viral ORFs were recoded by rearranging existing synonymous codons to increase the content of underrepresented codon pairs. Amino acid coding was completely unchanged. Four CPD RSV genomes were designed in which the indicated ORFs were recoded: Min A (NS1, NS2, N, P, M, and SH), Min B (G and F), Min L (L), and Min FLC (all ORFs except M2-1 and M2-2). Surprisingly, the recombinant CPD viruses were temperature-sensitive for replication in vitro (level of sensitivity: Min FLC > Min L > Min B > Min A). All of the CPD mutants grew less efficiently in vitro than recombinant wild-type (WT) RSV, even at the typically permissive temperature of 32 °C (growth efficiency: WT > Min L > Min A > Min FLC > Min B). CPD of the ORFs for the G and F surface glycoproteins provided the greatest restrictive effect. The CPD viruses exhibited a range of restriction in mice and African green monkeys comparable with that of two attenuated RSV strains presently in clinical trials. This study provided a new type of attenuated RSV and showed that CPD can rapidly generate vaccine candidates against nonsegmented negative-strand RNA viruses, a large and expanding group that includes numerous pathogens of humans and animals.
Human respiratory syncytial virus (RSV) is a negative-strand RNA virus of genus Pneumovirus, family Paramyxoviridae. RSV is the most important viral agent of serious respiratory tract illness in infants and children worldwide (1–3). Worldwide, nearly all children are infected by RSV at least once by the age of 2 y. RSV disease ranges from rhinitis to bronchiolitis or pneumonia. The RSV genome consists of a single-stranded negative-sense 15.2-kb RNA and has 10 genes in the order 3′ NS1-NS2-N-P-M-SH-F-G-M2-L 5′ (for a review, see ref. 4). The M2 gene encodes two separate proteins, M2-1 and M2-2, from overlapping ORFs.
RSV vaccines and new antiviral drugs are in preclinical and clinical development; however, no RSV vaccines or antiviral drugs suitable for routine use are commercially available. The goal of the present study was to design and generate new vaccine candidates for RSV using the recently described strategy of codon-pair deoptimization (CPD) (5). By this strategy, one or more ORFs in a virus or other pathogen are recoded by rearranging existing synonymous codons so as to increase the presence of underrepresented codon pairs within the ORF. CPD can be done without changing codon use although, in the present study, codon use was occasionally changed slightly when we manually edited the sequence to remove features such as long homo-oligomers. Amino acid coding was completely unaffected, and nontranslated genome regions were unchanged. A major effect of CPD is thought to be a reduction in the efficiency of translation although this effect is not well-defined (6–11). Because CPD involves hundreds or thousands of nucleotide substitutions, deattenuation is thought to be unlikely. Because amino acid coding is unaffected, CPD strains provide the same repertoire of epitopes for inducing cellular and humoral immunity as the WT pathogen. Recently, the CPD approach has been used successfully to attenuate poliovirus, influenza A virus, Streptococcus pneumonia, and HIV type 1 (5, 10–13).
In the present work, four CPD RSV genomes were designed, synthesized, and recovered by reverse genetics. The CPD recombinant (r) RSVs were attenuated and temperature-sensitive in vitro. Furthermore, we demonstrated that the CPD rRSVs were attenuated and immunogenic in mice and African green monkeys (AGMs). In the AGM model, CPD rRSVs replicated at a level comparable with two pediatric RSV vaccine candidates, which are currently being tested in clinical phase I studies in infants and young children. Thus, CPD represents a new strategy for the generation of novel live-attenuated RSV vaccine candidates.
Results
Replication of CPD rRSVs Is Restricted in Cell Culture.
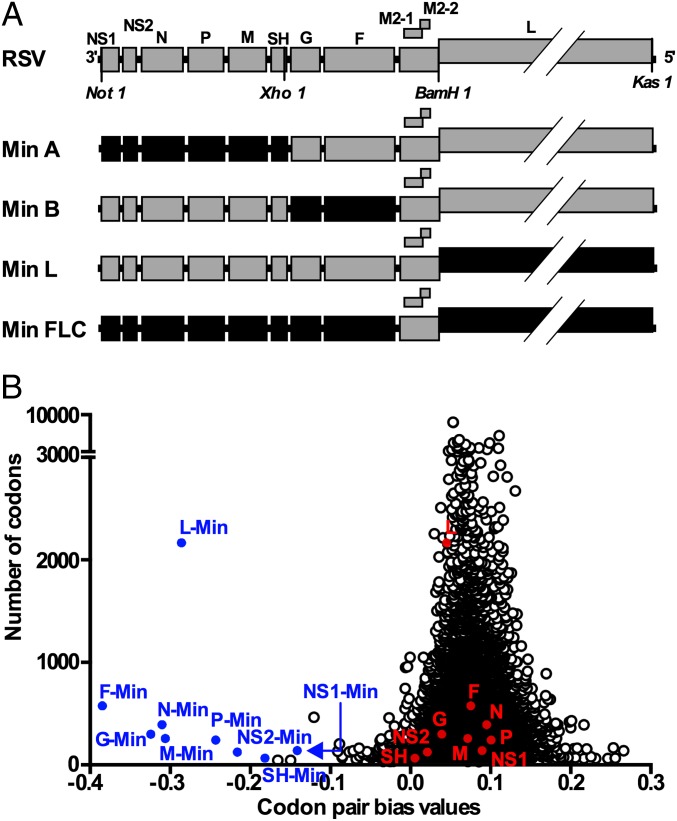
We designed and generated, by reverse genetics, four synthetic rRSVs, named Min A, Min B, Min L, and Min FLC, in which various genome regions were subjected to CPD (Fig. 1A). The designation “Min” (for “minimized”) signifies that the ORFs were designed with the most negative achievable codon pair bias score. The M2-1 and M2-2 ORFs were not subjected to CPD because these overlapping ORFs engage in coupled stop–start translation that depends on sequence—and possibly secondary structure—located in the M2-1 ORF ∼150 nt upstream of the M2-2 ORF (14). Given the incompletely defined nature of the cis-acting element(s) in the M2 gene, we left it undisturbed.
Fig. 1.
Generation of codon pair-deoptimized (CPD) rRSVs. (A) Gene maps of the CPD rRSVs Min A, Min B, Min L, and Min FLC. CPD ORFs are shown as black boxes; WT ORFs are shown as gray boxes. Restriction sites used for the constructions are indicated. (B) Codon-pair bias (CPB) values plotted against ORF lengths in codons, shown for WT RSV ORFs (red circles), CPD RSV ORFs (blue circles), and a set of 14,795 National Center for Biotechnology Information (NCBI)-annotated human protein-coding sequences [Consensus Coding Sequence (CCDS) database published March 2005, open circles]. As described previously (5), the CPB score for each ORF is the mean of the codon-pair scores (CPSs) for all of its codon pairs. In turn, the CPS for each codon pair is the natural log of the ratio of the observed versus expected frequency of that codon pair in the human ORFeome.
The process of CPD mainly involved rearranging existing synonymous codons, and codon use was only occasionally changed. As noted, there were no changes in amino acid coding. On the other hand, the percent nucleotide sequence identity between the WT and CPD RSV ORFs ranged from 78% (F) to 92% (SH), and the number of nucleotide differences increased with increasing ORF length and ranged from 23 (SH) to 1,378 (L) (Table S1). The effect on codon-pair bias (CPB) is shown in Fig. 1B (5); although the ORFs of WT RSV have CPB values comparable with those of the human ORFeome, the CPB values of the CPD RSV ORFs were much lower.
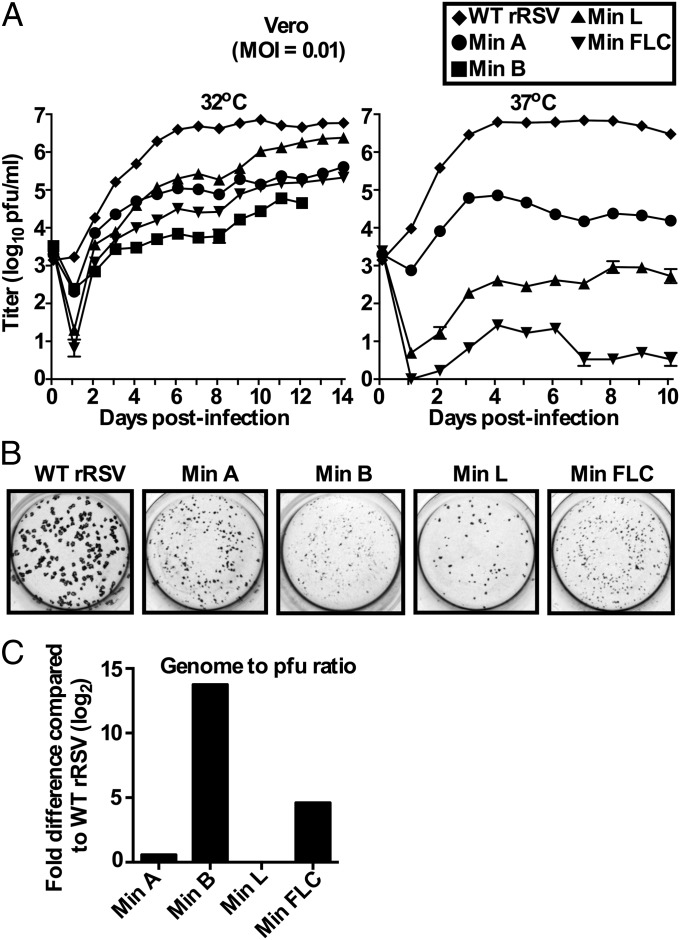
Each of the CPD rRSVs was readily recovered from cDNA. The genome sequences of the recovered viruses were free of adventitious mutations. At 32 °C, the growth of all of the CPD viruses was delayed and reduced compared with WT rRSV (Fig. 2A): WT rRSV reached its maximum titer of ∼107 pfu/mL on day 7 whereas Min A, Min L, and Min FLC reached their maximum titers on day 14. The maximum titer of Min L at 32 °C was only threefold lower than that of WT rRSV. Min A and Min FLC were slightly more restricted in growth, with maximum titers that were ∼15- and 30-fold lower than WT rRSV, respectively. Min B was the most restricted for replication (100-fold lower than that of WT rRSV).
Fig. 2.
Replication of WT and CPD rRSVs on Vero cells. (A) Multicycle growth kinetics of WT and CPD rRSVs on Vero cells. Replicate cell monolayers were infected with the indicated WT or CPD rRSVs at a multiplicity of infection (MOI) of 0.01 and incubated at 32 °C (Left) or 37 °C (Right). Triplicate monolayers per virus were harvested daily from day 1–14 by collecting the monolayers with the media and preparing clarified supernatants. Note that Min B replication was assayed only for days 1–12 at 32 °C because of limited virus inoculum due to its poor replication. The triplicate samples for each time point were titered by plaque assay in duplicate at 32 °C, and the results are expressed as mean values with SD. (B) Plaque-size phenotype on Vero cells of WT and CPD rRSVs, visualized by immunostaining following incubation for 12 d under methylcellulose at 32 °C. (C) Specific infectivity of CPD viruses compared with WT rRSV. RNA was isolated from virus stocks grown at 32 °C, and the amount of genomic RNA in samples corresponding to 2.0 × 102 pfu of WT or CPD rRSV was quantified by qRT-PCR specific for M2 sequence as a target in negative-sense RSV RNA. The results are expressed as log2 fold difference in the amount of genomic RNA compared with WT rRSV.
WT rRSV replicated faster at 37 °C than at 32 °C, but it reached similar peak titers at either temperature. Min A replicated slightly less efficiently at 37 °C than at 32 °C (∼105 pfu/mL after 4 d at 37 °C). In contrast, replication of Min L and Min FLC was more restricted at 37 °C than at 32 °C. Compared with WT rRSV, Min L and Min FLC titers were 10,000- and 1,000,000-fold reduced, respectively, at 37 °C. Min B was not evaluated at this temperature due to its poor replication.
We also compared plaque phenotypes on Vero cells at 32 °C. Compared with WT rRSV, Min A and Min L induced small plaques, and Min B and Min FLC induced microplaques (Fig. 2B).
Evaluation of the Temperature Sensitivity of the CPD rRSVs.
We next evaluated the ability of these viruses to form plaques on Vero and HEp-2 cells at temperatures ranging from 32 °C to 40 °C (Table 1). As expected, WT rRSV was not temperature-sensitive (ts) at physiological temperatures. The temperature shut-off (TSH) (see Table 1 for a definition) of Min A was determined to be 40 °C on both Vero and HEp-2 cells, indicating that this virus was minimally ts. The other CPD viruses exhibited a range of increasing temperature sensitivity, with the following TSH values on Vero cells and HEp-2 cells, respectively: Min B, 38 °C and 39 °C; Min L, 37 °C and 38 °C; and Min FLC, 35 °C and 36 °C.
Table 1.
Temperature sensitivity of the CPD RSVs on Vero and HEp-2 cells
| Virus | Cell line | Temperature, °C* | TSH | ||||||
| 32 | 35 | 36 | 37 | 38 | 39 | 40 | |||
| Min A | Vero | 7.2 | 6.6 | 6.2 | 6.1 | 5.7 | 5.4 | 4.4 | 40 |
| Min B | Vero | 4.2 | 3.1 | 3.2 | 2.8 | 2.0 | 1.7 | <1 | 38 |
| Min L | Vero | 7.5 | 7.1 | 6.5 | 5.2 | 4.2 | <1 | <1 | 37 |
| Min FLC | Vero | 6.7 | 4.5 | 3.4 | <1 | <1 | <1 | <1 | 35 |
| WT rRSV | Vero | 8.4 | 8.3 | 8.3 | 8.0 | 8.1 | 8.1 | 8.1 | >40 |
| Min A | HEp-2 | 7.1 | 6.7 | 6.6 | 6.4 | 6.3 | 5.6 | 4.8 | 40 |
| Min B | HEp-2 | 4.0 | 4.0 | 4.0 | 4.0 | 3.7 | 2.0 | <1 | 39 |
| Min L | HEp-2 | 7.6 | 7.1 | 6.7 | 6.1 | 4.5 | 3.2 | 2.3 | 38 |
| Min FLC | HEp-2 | 6.5 | 5.0 | 4.0 | 2.0 | <1 | <1 | <1 | 36 |
| WT rRSV | HEp-2 | 8.3 | 8.1 | 8.3 | 8.3 | 8.1 | 8.3 | 8.1 | >40 |
Virus titer (log10 pfu/mL) at indicated temperature (°C).
The ts phenotype for each virus was evaluated by assessing virus growth on Vero and HEp-2 cells at the indicated temperatures using temperature-controlled water baths. For viruses with a ts phenotype, the shut-off temperatures (TSH) are listed in boldface at the right. TSH is defined as the lowest restrictive temperature resulting in ≥100-fold reduced titer (underlined) compared with that virus’s growth at the permissive temperature of 32 °C. The ts phenotype is defined as having a TSH of 40 °C or less.
Effects on Specific Viral Infectivity.
We evaluated the specific infectivity of WT or CPD rRSVs by measuring the ratio of genomic RNA to pfu in virus preparations using quantitative (q)RT-PCR for the M2 gene, which was unchanged and identical in all of the viruses (Fig. 2C). Min A and Min L exhibited the same abundance of genomic RNA per 2.0 × 102 pfu as WT rRSV. In contrast, 2.0 × 102 pfu of Min FLC and Min B, respectively, contained 24- and 13,900-fold more genomic RNA than WT rRSV, suggesting that, despite their reduced plaque titers, CDP viruses ultimately produce similar amounts of viral particles compared with wild type.
Effects of CPD on the Production of Viral RNA, Protein, and Infectious Virus in Vitro.
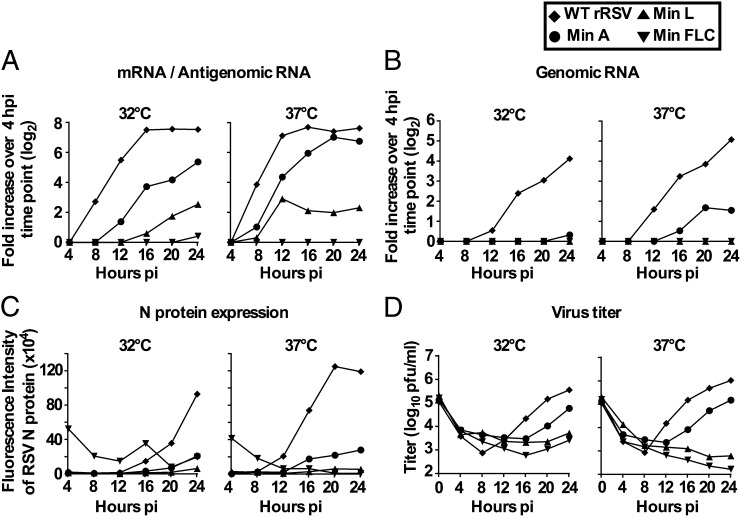
We monitored the production of cell-associated viral RNA, protein, and infectious virus in Vero cells infected with WT and CPD rRSVs in a single-step growth experiment (Fig. 3).
Fig. 3.
Single-cycle replication of Min A, Min L, Min FLC, and WT rRSV in Vero cells. Replicate wells of Vero cell monolayers were mock-infected or infected at an MOI of 1 pfu per cell with the indicated virus and incubated at 32 °C or 37 °C, as indicated. At 4-h intervals up to 24 hpi, one well was harvested and processed for analysis of cell-associated RNA and protein, and a second well was harvested and processed for virus titration. (A and B) Analysis of cell-associated RNA by strand-specific qRT-PCR, with primers and probe to the M2 gene, to detect (A) positive-sense RSV RNA (antigenome/mRNA) or (B) negative-sense RSV RNA (genome), with results normalized to 18S rRNA and expressed as fold increase compared with the 4-h time point. (C) Analysis of cell-associated proteins by Western blotting. Ten micrograms of cell lysates were separated on 4–12% SDS/PAGE gels, and proteins were transferred to PVDF membranes. The blots were analyzed with rabbit anti-RSV–specific polyclonal Ab and mouse monoclonal anti-tubulin Ab (loading control), followed by secondary antibodies labeled with an infrared fluorophore. Membranes were scanned on the Odyssey Infrared Imaging System. Data were analyzed using Odyssey software. For quantification of RSV N protein, background fluorescence was corrected, and values indicate the median fluorescence intensity per protein band. (D) Virus titers from 4 hpi to 24 hpi are expressed in log10 pfu/mL. At each time point, the cell monolayer was collected with the media, and clarified supernatants were prepared and analyzed by plaque assay. The mean of the duplicate values is shown.
WT rRSV exhibited extensive accumulation of cell-associated positive-sense RNA that was detected by 8 h postinfection (hpi) and was maximal by 16–24 hpi (Fig. 3A). Typically, for WT RSV, ∼95% of positive-sense cell-associated RNA is mRNA, and the remainder is antigenome RNA (15). The CPD viruses exhibited reduced accumulation of mRNA/antigenome. In the case of Min A, mRNA/antigenome was first detected at 12 hpi, and the maximum accumulation was lower than that of WT rRSV at both 32 °C and 37 °C. The accumulation of Min L mRNA/antigenome also was reduced and was first detected only at 16 hpi at 32 °C at very low levels. The accumulation of Min L mRNA/antigenome was not more efficient at 37 °C. The accumulation of Min FLC mRNA/antigenome was the least efficient, with only a small increase of positive-sense RNA detected at 24 hpi at 32 °C and no detectable positive-sense RNA at 37 °C.
The production of cell-associated genomic RNA by WT rRSV (Fig. 3B) was delayed compared with mRNA/antigenome, as is typically observed. WT rRSV genomic RNA was detected as early as 12 hpi and increased until 24 hpi. Min A produced a substantial but reduced amount of genomic RNA at 37 °C and a greatly reduced amount at 32 °C. Min L and Min FLC produced very low levels of genomic RNA at either temperature.
The observed reduction in viral gene transcription in CPD virus-infected cells was associated with reduced viral protein synthesis (Fig. 3C and Fig. S1). In WT rRSV-infected cells, an increase in RSV N protein was detectable at 12 hpi and 16 hpi at 37 °C and 32 °C, respectively. In the case of Min FLC, a substantial background of viral protein was associated with the infected cells at the beginning of the infection. This background likely was a consequence of the lower specific infectivity of this virus, necessitating an inoculum with a much higher content of viral protein. At 20 hpi, N protein expressed by Min FLC was evident at 32 °C but not at 37 °C. The synthesis of N protein by Min A virus was evident at both temperatures whereas little N protein synthesis was evident for Min L at either temperature.
The production of infectious WT rRSV was first observed at 12 hpi at 37 °C and 32 °C (Fig. 3D) and thus was approximately concurrent with the accumulation of genomic RNA. All of the tested CPD viruses exhibited delayed and reduced production of virus particles. An increase in Min A virus particles was detected at 16 hpi and 20 hpi at 37 °C and 32 °C, respectively, and thus lagged behind WT rRSV. The production of Min L virus was more strongly reduced and delayed: a small increase in virus particles was detected at 24 hpi at 32 °C and 37 °C. Finally, a small increase in FLC virus particles was observed at 24 hpi at 32 °C whereas no increase in particles was detected at 37 °C.
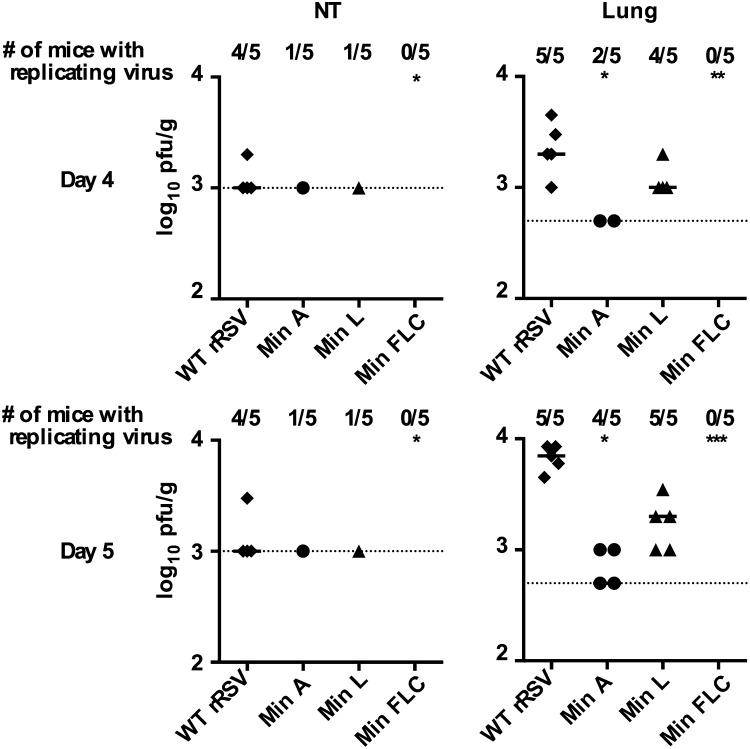
Reduced Replication of CPD rRSVs in BALB/c Mice.
We next evaluated the replication of the CPD viruses compared with WT rRSV in the respiratory tract of mice following intranasal inoculation (Fig. 4). WT rRSV was detected in the nasal turbinates (NTs) in 8 out of 10 mice on days 4 and 5 (median titer of 103 pfu/g). Min A and Min L were each detected in the NT in 2 out of 10 mice whereas Min FLC was not detected in the NT of any mouse on any day (P < 0.05 compared with WT rRSV). In the lungs, the titers of WT rRSV reached 103 pfu/g and 104 pfu/g on days 4 and 5, respectively. In comparison, Min A replication was detected in 2 out of 5 mice on day 4 and 4 out of 5 mice on day 5, and the titers were about 10-fold lower than those of WT rRSV (P < 0.05). Min L replication was detected in 4 of 5 mice on day 4 and in all 5 mice on day 5, and its titer was only twofold lower than that of WT rRSV on day 4, and fivefold lower on day 5. Finally, Min FLC was not detected in any mouse on day 4 or 5 (P < 0.01 and P < 0.001 at day 4 and day 5, respectively), showing that this virus is strongly attenuated in mice.
Fig. 4.
Replication of Min A, Min L, Min FLC, and WT rRSV in the respiratory tract of BALB/c mice. Virus replication was evaluated in the upper and lower respiratory tract (URT and LRT) of mice as described previously (24). Six-week-old BALB/c mice in groups of 10 were inoculated intranasally under methoxyflurane anesthesia on day 0 with 4.5 × 105 pfu of the indicated virus. On days 4 and 5, 5 mice per group were killed per day. Nasal turbinates (NTs) and lung tissue were harvested and homogenized separately. Virus titers were determined in duplicate by plaque assay on Vero cells at 32 °C and expressed as log10 pfu/g tissue. The number of mice per group in which virus was detected is indicated at the top, the symbols indicate virus titers for individual animals, and the bars indicate median titers. The limit of detection is indicated by a dotted line (1 × 103 pfu/g and 5 × 102 pfu/g in the NT and lungs, respectively). Datasets were assessed for significance using nonparametric Kruskal–Wallis with Dunn’s post hoc test. Data were considered significant at P < 0.05 (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 compared with WT rRSV).
Attenuation and Immunogenicity of CPD rRSVs in AGMs.
We also evaluated the replication of CPD viruses versus WT rRSV in nonhuman primates. For comparison, we also evaluated, in two separate experiments, two previously described (16, 17) attenuated strains of RSV called RSV cps2 and RSV ΔNS2 Δ1313 I1314L (Table 2). These two viruses are currently being evaluated in clinical phase I trials in children and infants (ClinicalTrials.gov identifiers NCT01893554 and NCT01852266, respectively). These vaccine candidates also were previously shown to be attenuated in chimpanzees and are moderately (RSV ΔNS2 Δ1313 I1314L) or highly (RSV cps2) ts, with TSH values of 38–39 °C and 36 °C, respectively (16, 17).
Table 2.
Viral titers of nasopharyngeal swab specimens and tracheal lavage samples from AGMs inoculated with CPD viruses RSV cps2, RSV ΔNS2 Δ1313 I1314L, or WT rRSV
| Virus | Nasopharyngeal swab (URT) | Tracheal lavage (LRT) | ||||
| Duration of sheddinga | Peak virus titer | Sum of daily titersb | Duration of sheddinga | Peak virus titer | Sum of daily titersb | |
| Exp. 1 | ||||||
| Min A | 2.3 (4) | 2.2 | 7.7 | 6.0 (4) | 1.8 | 6.6 |
| Min L | 5.0 (4) | 2.4 | 10.4 | 4.0 (4) | 1.4 | 5.8 |
| Min FLC | 1.3* (1) | 0.7** | 4.8** | 0.3** (1) | 1.1** | 4.6** |
| WT rRSV | 6.3 (4) | 3.1 | 15.0 | 10.0 (4) | 3.6 | 13.0 |
| Exp. 2 | ||||||
| RSV cps2 | 1.8* (1) | 0.7* | 5.3* | —* (0) | —* | 4.2* |
| WT rRSV | 9.0 (4) | 4.3 | 26.2 | 9.0 (4) | 4.6 | 16.5 |
| Exp. 3 | ||||||
| RSV ΔNS2 Δ1313 I1314L | 7.3* (4) | 3.3 | 13.6 | 1.3* (3) | 1.3* | 4.9* |
| WT rRSV | 9.3 (3)c | 3.5 | 21.9 | 8.0 (4) | 3.6 | 13.9 |
Four AGMs were inoculated by the combined intranasal and intratracheal routes with 106 pfu of the indicated virus (total dose 2 x 106 pfu per animal). Three independent experiments (Exp. 1, Exp. 2, and Exp. 3) were performed. Nasopharyngeal (NP) swabs were collected every day from day 0–10 and on day 12 (Exp. 1) or day 14 (Exp. 2 and Exp. 3) postinoculation. The tracheal lavage (TL) was done every other day from day 2–12 (Exp. 1) or day 14 (Exp. 2 and Exp. 3) postinoculation (see SI Materials and Methods for details). Virus titrations were performed in duplicate on Vero cells at 32 °C and are expressed in log10 pfu/mL. The lower limits of detection were 0.7 log10 pfu/mL and 1 log10 pfu/mL for NP and TL samples, respectively. Samples with no detectable virus are represented as “—“. Mean values from four AGMs per group are shown (with the exception of WT rRSV in Exp. 3, for which only three AGMs have been considered; see footnote “c” for explanation). Data were considered significant only at P ≤ 0.05 (*P ≤ 0.05, **P ≤ 0.01 compared with WT rRSV).
The period of days from the first to the last day on which virus was detected, including negative days (if any) in between. The number of animals shedding virus is indicated in parentheses.
Sum of daily titers is used as an estimate for the magnitude of shedding (area under the curve). A value of 0.4 and 0.7 for NP and TL, respectively, was used for samples with no detectable virus.
One AGM reacted to anesthesia by emesis, which likely interfered with the efficacy of the intranasal inoculation. Based on this assumption, NP shedding results from this AGM were excluded from calculation of means and from statistical analysis.
Shedding of WT rRSV from the upper respiratory tract (URT) was detectable over an average of 6.3–9.3 d from the three experiments, with mean peak titers of ∼103 to 2 × 104 pfu/mL. Shedding of the Min A and Min L viruses from the URT was somewhat delayed and of shorter duration (2.3 d and 5.0 d, respectively), and the peak titers were ∼10- to 100-fold lower than those detected for WT rRSV. Shedding of Min FLC from the URT was detected at very low titer in a single animal. Thus, all three of the CPD viruses appeared to be somewhat more attenuated in the URT than the RSV ∆NS2 ∆1313 I1314L virus, and the Min FLC virus appeared to be similar in attenuation to the more highly attenuated RSV cps2 virus.
Shedding of WT rRSV from the lower respiratory tract (LRT) was detected over an average of 8–10 d, with mean peak titers of 4 × 103 to 4 × 104 pfu/mL of tracheal lavage (TL) specimen. Shedding of Min A and Min L was of shorter duration (6.0 d and 4.0 d, respectively), and the titers were reduced ∼100- to 1,000-fold. Shedding of Min FLC from the LRT was detected only at low titer from a single animal. The somewhat greater restriction of Min L compared with Min A in the LRT is the converse of what was observed in the URT. This result might reflect the greater degree of temperature sensitivity of Min L because the LRT is warmer than the URT (the AGM LRT was reported to be 37.2–39.2 °C in the Association of Primate Veterinarians formulary, compared with 35.6–36.7 °C in the URT as measured from three AGM at the National Institutes of Health animal center). Based on the peak titer of shedding from the LRT, the CPD viruses were very similar in attenuation to RSV ∆NS2 ∆1313 I1314 whereas RSV cps2 was somewhat more attenuated.
Despite their high level of restriction, the Min A and Min L viruses induced titers of RSV-neutralizing serum antibodies that were not statistically different from those induced by WT rRSV (Table S2). Similarly, RSV ΔNS2 Δ1313 I1314L was statistically indistinguishable from WT rRSV in the ability to induce RSV-neutralizing serum antibodies. On the other hand, the titers of RSV-neutralizing serum antibodies induced by Min FLC or RSV cps2 were significantly lower than WT rRSV (P < 0.05).
Discussion
We investigated the recently described strategy of CPD (5, 10) as a means to develop new attenuated versions of RSV as potential live vaccine candidates. Four synthetic live RSV mutants were designed and readily recovered. It is noteworthy that Min FLC was readily recovered even though 9 of the 11 RSV ORFs (representing almost 94% of the protein-coding sequence) in this virus had been subjected to CPD; for comparison, to date, CPD has been reported for substantially smaller regions of the genomes of poliovirus or HIV-1 (∼23% and 4% of the protein-coding sequence, respectively) or influenza virus (three segments out of eight) (5, 10, 11, 13).
Each of the CPD RSVs replicated less efficiently in vitro than WT rRSV. Min L was the least restricted of the CPD mutants. This result suggested that suboptimal expression of L could be tolerated to some extent. If so, this result would imply that expression of L is not limiting in a normal RSV infection. Evaluation of virus replication and specific infectivity showed that Min B was the most restricted CPD virus, which indicates that recoding the G and/or F ORFs had the greatest effect on restricting RSV replication in cell culture. This result implies that optimal expression of these two glycoproteins is limiting for efficient RSV replication. Surprisingly, Min B was even more restricted than Min FLC because this latter virus contained the same versions of CPD G and F ORFs, but in combination with seven other CPD ORFs. CPD of G and F in an otherwise WT genome may have resulted in inappropriate molar ratios relative to one or more other viral proteins that heightened the restrictive effect.
The factors involved in CPD are incompletely understood. A primary effect is thought to be on the efficiency and fidelity of translation (6–9). This effect was illustrated recently with influenza virus. However, mRNA synthesis also was affected for influenza virus, consistent with the expectation that CPD can have multiple effects (10, 11). The effects of CPD on RSV protein synthesis in the present study were somewhat more complicated to characterize. For example, expression of the N protein (and other RSV proteins) (Fig. S1) by Min L was strongly reduced even though only the L ORF had been subjected to CPD in Min L. A likely explanation is that CPD of the L ORF resulted in less efficient translation of the CPD L mRNA, resulting in reduced synthesis of L polymerase protein. The reduction in L polymerase protein in turn would reduce viral RNA synthesis, reducing transcription of all of the viral genes. This reduced transcription would result in reduced production of all of the viral mRNAs, leading to reduced production of all of the viral proteins. It also would reduce viral RNA replication, due to the reduced synthesis of L polymerase protein and other viral proteins. Thus, CPD of the L ORF is thought to have a direct effect, namely reduced efficiency of translation of the CPD L mRNA, and also would have indirect effects, including global reductions in viral RNA synthesis and viral protein synthesis. These direct and indirect effects cannot be easily dissociated in the context of an RSV infection due to the intertwined nature of viral transcription, protein synthesis, and RNA replication for this type of virus. CPD of ORFs encoding other proteins involved in RNA synthesis, such as N, P, and M2-1 (the last of which was not subjected to CPD in the present study), similarly might have direct and indirect effects.
Surprisingly, we found that each of the CPD RSV mutants was ts, with the order of increasing sensitivity being Min A < Min B < Min L < Min FLC. Temperature sensitivity was unexpected because the amino acid sequences of each CPD virus and of WT rRSV were identical. The observation that Min L and Min FLC were more ts than Min A and Min B suggests that CPD of the L ORF was a major contributor to temperature sensitivity. However, the basis for the temperature sensitivity of any of these CPD viruses remains to be directly identified.
Attenuation of viruses due to CPD is thought to be the aggregate effect of hundreds or thousands of nucleotide changes so that the possibility of deattenuation should be extremely low. In previous work, serial passage of CPD poliovirus in cell culture showed that these viruses indeed were genetically stable (5). Preliminary experiments showed that Min FLC did not lose any temperature sensitivity after serial passage at increasing temperatures and thus appeared phenotypically stable.
We found that the CPD rRSVs were restricted for replication in mice and nonhuman primates. We observed increased attenuation of Min FLC compared with Min A and Min L. The level of restriction of the Min viruses in AGMs was similar to that of two live-attenuated pediatric RSV vaccine candidates presently in clinical trials in infants and young children. It should be noted that the body temperature of the AGM is higher than that of the human, and thus viruses that are highly ts likely would be disproportionately restricted in AGMs versus humans. Thus, the level of attenuation of the highly ts Min FLC and RSV cps2 viruses likely was exaggerated in the present study. This exaggerated restriction likely accounts for the poor immunogenicity of these viruses in the present study because a virus that is nearly identical phenotypically to RSV cps2 was previously shown to be immunogenic in infants and young children (18).
The nonsegmented negative-strand virus group (order Mononegavirales) represented by RSV includes a number of pathogens of humans and animals, including highly pathogenic agents such as Ebola, Nipah, and Hendra viruses. In addition, new members of this virus type continue to be identified, such as in recent field studies in bats (19), and these viruses may include species pathogenic to animals and humans. The results of the present study suggest that the strategy of CPD could readily be applied to any pathogen of this virus type by developing a reverse genetics system (which is facilitated by the present ease of commercial DNA synthesis) and generating an array of attenuated vaccine candidates with differing extents of CPD.
Materials and Methods
See SI Materials and Methods for detailed protocols.
Design of the Codon Pair-Deoptimized RSV Sequences.
The theoretical basis of codon-pair deoptimization leading to viral phenotypes is described in SI Materials and Methods. Briefly, codon pair-deoptimized (CPD) ORFs were designed using previously described computational algorithms (5, 10) and were based on the RSV strain A2 (Fig. 1A). The first 10 codons of each ORF were left unchanged to avoid effects on possible cis-acting sequences. Runs of more than six identical nucleotides and sequences resembling RSV gene-start or gene-end signals were removed from the computer-generated CPD sequences by manual editing.
Construction of cDNAs Encoding CPD Recombinant RSVs.
Recombinant RSVs were constructed by reverse genetics (20) using the antigenome cDNA D46/6120, a derivative of the rA2 cDNA plasmid (21). Four full-length cDNA plasmids were generated (Fig. 1A), and viruses were rescued as described previously (20, 22).
Virus Growth and Titration.
Following rescue, WT and CPD rRSVs were propagated once on Vero cells at 32 °C. Viral RNA was isolated from all virus stocks, and sequence analysis of the viral genomes was performed from overlapping RT-PCR fragments, confirming that the genomic sequences of the recombinant viruses were correct and free of adventitious mutations. Virus titers were determined by plaque assay on Vero cells as described previously (23). Titers were expressed as plaque forming units (pfu) per mL.
Evaluation of Replication and Immunogenicity of CPD rRSVs in Mice and AGMs.
All animal experiments were approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (C.L.N., L.G.B., C.L., T.M., L.Y., M.M., P.L.C., U.J.B., and J.M.D.) and in part by NIH Grant R01 AI07521901 A1 (to E.W.).
Footnotes
Conflict of interest statement: C.L.N., L.G.B., E.W., S.M., P.L.C., U.J.B., and J.M.D. are coinventors on a patent application for the development of respiratory syncytial virus (RSV) vaccines by codon-pair deoptimization.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ817798, KJ817799, KJ817800, and KJ817801).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411290111/-/DCSupplemental.
References
- 1.Collins PL, Karron RA. 2013. Respiratory syncytial virus and metapneumovirus. Fields Virology, eds Knipe DM, Howley PM (Lippincott, Williams and Wilkins, Philadelphia), 6th Ed, Vol 1, pp 1086–1123.
- 2.Collins PL, Graham BS. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol. 2008;82(5):2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall CB, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360(6):588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res. 2011;162(1-2):80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320(5884):1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folley LS, Yarus M. Codon contexts from weakly expressed genes reduce expression in vivo. J Mol Biol. 1989;209(3):359–378. doi: 10.1016/0022-2836(89)90003-x. [DOI] [PubMed] [Google Scholar]
- 7.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci USA. 1989;86(10):3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan JR, Aucott LS, Stansfield I. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 2006;34(3):1015–1027. doi: 10.1093/nar/gkj488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moura G, et al. Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS ONE. 2007;2(9):e847. doi: 10.1371/journal.pone.0000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28(7):723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Skiena S, Futcher B, Mueller S, Wimmer E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc Natl Acad Sci USA. 2013;110(23):9481–9486. doi: 10.1073/pnas.1307473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman JR, Papamichail D, Yano M, García-Suárez MdelM, Pirofski LA. Designed reduction of Streptococcus pneumoniae pathogenicity via synthetic changes in virulence factor codon-pair bias. J Infect Dis. 2011;203(9):1264–1273. doi: 10.1093/infdis/jir010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martrus G, Nevot M, Andres C, Clotet B, Martinez MA. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology. 2013;10:78. doi: 10.1186/1742-4690-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould PS, Easton AJ. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J Biol Chem. 2005;280(23):21972–21980. doi: 10.1074/jbc.M502276200. [DOI] [PubMed] [Google Scholar]
- 15.Fearns R, Peeples ME, Collins PL. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236(1):188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 16.Luongo C, Winter CC, Collins PL, Buchholz UJ. Increased genetic and phenotypic stability of a promising live-attenuated respiratory syncytial virus vaccine candidate by reverse genetics. J Virol. 2012;86(19):10792–10804. doi: 10.1128/JVI.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luongo C, Winter CC, Collins PL, Buchholz UJ. Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol. 2013;87(4):1985–1996. doi: 10.1128/JVI.02769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karron RA, et al. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis. 2005;191(7):1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 19.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins PL, et al. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92(25):11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukreyev A, Belyakov IM, Berzofsky JA, Murphy BR, Collins PL. Granulocyte-macrophage colony-stimulating factor expressed by recombinant respiratory syncytial virus attenuates viral replication and increases the level of pulmonary antigen-presenting cells. J Virol. 2001;75(24):12128–12140. doi: 10.1128/JVI.75.24.12128-12140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8(5):497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 24.Whitehead SS, Firestone CY, Collins PL, Murphy BR. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotypes. Virology. 1998;247(2):232–239. doi: 10.1006/viro.1998.9248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.