Significance
Voltage-gated sodium (Nav) channels control neuronal signaling and are key targets for local anesthetics, antiepileptics, and therapeutics for a range of disorders. Multimicrosecond Anton simulations have provided completely unbiased molecular-level views of the interactions of lipophilic drugs with the recently solved bacterial channel, NavAb from Arcobacter butzleri. Newly parameterized benzocaine and phenytoin molecules exhibit different membrane partition coefficients, crossing rates and distributions around the channel, leading to the identification of distinct high- and low-affinity sites. We observe a minimum free energy pathway through membrane-bound fenestrations to a pore-blocking location, or from aqueous solution directly through the (closed) intracellular gate. These observations help explain experimental data and provide insight into Nav inhibition processes that will assist future drug development.
Keywords: bacterial sodium channel, drug binding
Abstract
Voltage-gated sodium (Nav) channels are important targets in the treatment of a range of pathologies. Bacterial channels, for which crystal structures have been solved, exhibit modulation by local anesthetic and anti-epileptic agents, allowing molecular-level investigations into sodium channel-drug interactions. These structures reveal no basis for the “hinged lid”-based fast inactivation, seen in eukaryotic Nav channels. Thus, they enable examination of potential mechanisms of use- or state-dependent drug action based on activation gating, or slower pore-based inactivation processes. Multimicrosecond simulations of NavAb reveal high-affinity binding of benzocaine to F203 that is a surrogate for FS6, conserved in helix S6 of Domain IV of mammalian sodium channels, as well as low-affinity sites suggested to stabilize different states of the channel. Phenytoin exhibits a different binding distribution owing to preferential interactions at the membrane and water–protein interfaces. Two drug-access pathways into the pore are observed: via lateral fenestrations connecting to the membrane lipid phase, as well as via an aqueous pathway through the intracellular activation gate, despite being closed. These observations provide insight into drug modulation that will guide further developments of Nav inhibitors.
Voltage-gated sodium (Nav) channel inhibitors can modulate sensory or motor activity without threatening vital bodily functions, enabling a wide range of therapeutic applications. In particular, the ability to control Nav channel current allows for effective use as local anesthetic (LA) and antiepileptic (AE) drugs, normalizing function in conditions of hyperexcitability, such as epilepsy, cardiac arrhythmias, hyperalgesia, and myotonia (1, 2). Recent breakthroughs in the solution of atomic structures of the bacterial NavBac family (3–6) provide an excellent opportunity to describe drug binding at the molecular level.
Pharmacological Nav channel modulation is a complex phenomenon involving voltage- and state-dependent block of current, as well as prolongation of recovery from inactivated states, with interrelated observations that have implicated different active sites, pathways and inactivated channel states (7). Despite historical focus on fast inactivation (8), it has now been discovered that slow inactivation is affected by LA/AE drugs (9) and that mutation of key residues involved in inhibition impairs slow inactivation (10). The NavBac family lacks the Domain (D) III–DIV linker responsible for fast inactivation (3), enabling molecular-level investigation into this common modulation process. Moreover, key binding regions are well conserved in bacterial and mammalian channels (3, 11), and it has been shown that bacterial channels interact with common inhibitory drugs (11–14). However, questions remain as to how structurally disparate inhibitor molecules can cause qualitatively similar modulation of different mammalian and bacterial Nav channels. Here, we explore the binding of LA benzocaine (BZC) and AE phenytoin (PHT) to the bacterial NavAb channel (3) (Fig. 1), providing detailed insight into Nav modulation mechanisms.
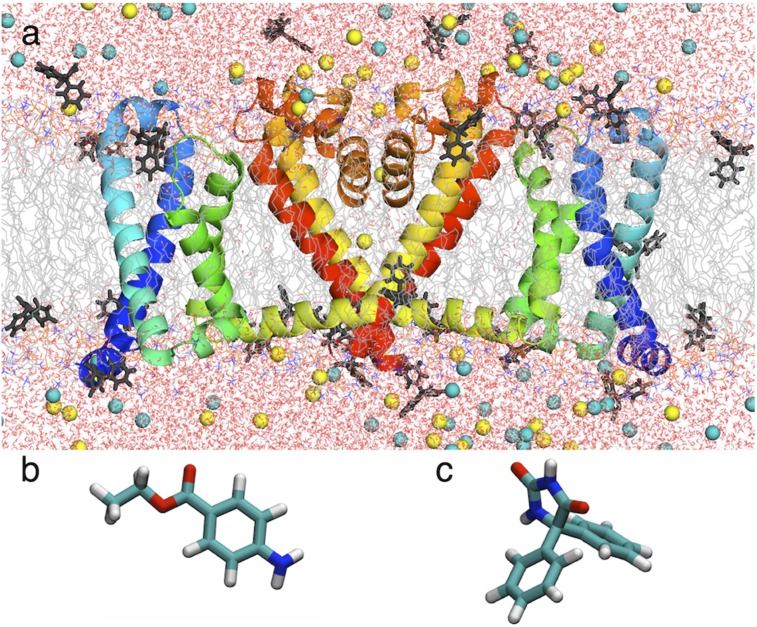
Fig. 1.
(A) Simulation system showing NavAb (two of four subunits as ribbons, with voltage sensor domain (VSD) S1 and S2 in blue and S3 and S4 in green, pore domain (PD) S5 in yellow and S6 in red, P-loop P1, P2, and selectivity filter (SF; orange) in a hydrated lipid bilayer (chains as gray lines; water as red/white sticks; NaCl as yellow/cyan balls) and PHT (dark gray sticks). (B and C) Stick representations of BZC (B) and PHT (C).
Functional studies have suggested at least two major LA/AE binding sites: a high-affinity, state- and voltage-dependent site; and a low-affinity, less state-dependent site (15). The high-affinity site has been the focus of considerable investigation (16) and appears to be shared by many drugs (2). Studies of mammalian Nav have suggested this site is on helix S6 of DIV, involving residue FS6 [F1764 in Nav1.2 (16), F1579 in Nav1.4, F1759 in Nav1.5 (17)], as well as a Tyr (Y1771 in Nav1.2, Y1586 in Nav1.4, Y1766 in Nav1.5). Binding at this site is use-dependent, such that under brief repetitive stimuli that open the channel, the channel enters a stable nonconducting state, with a cumulative reduction of current (18), signifying preferential binding to an inactivated state (10). Binding to FS6 appears to involve cation–π interaction for charged lidocaine (19), but π-stacking for neutral agents such as BZC and PHT. In contrast, little information exists to pinpoint the low-affinity site(s), whose binding may result from a “combination of hydrophobic interactions” occurring when the channel is maintained at hyperpolarized voltages (15). Complete sampling of the drug-channel binding distribution is needed to define these binding modes.
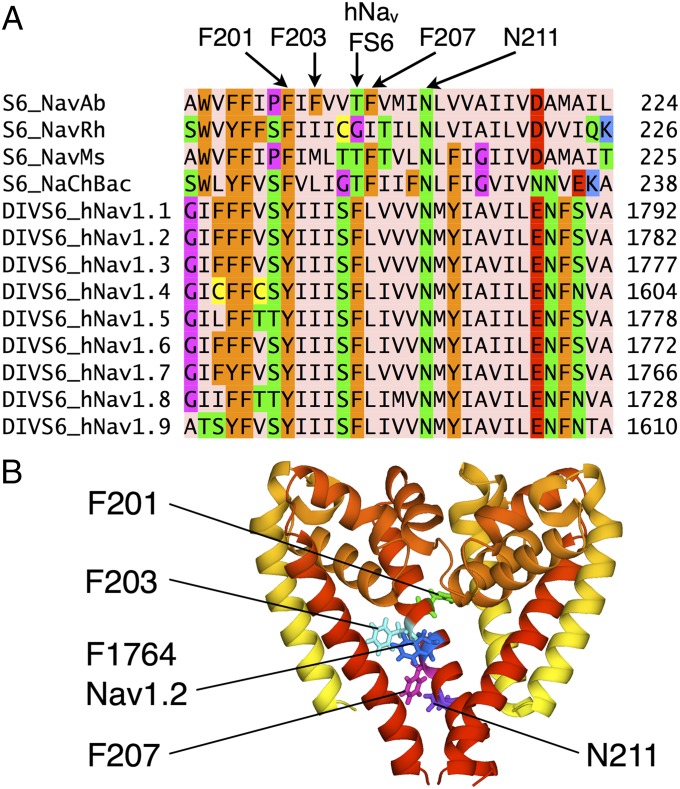
Residue FS6 on domain IV of the mammalian channels is a critical feature in drug modulation, but does not exist at that position in the bacterial channels (Fig. 2A; with full comparison in SI Appendix, Fig. S1). The absence of this important residue may contribute to low LA affinities observed for NaChBac (12). However, the recently solved NavAb (3) possesses a nonconserved F203, which appears to mimic FS6. Fig. 2B shows a simple overlay of the hNav1.2 sequence on the NavAb structural template, with FS6 location and orientation similar to F203 (shifted only slightly along the S6 helix), and thus could play a similar role in drug binding. Furthermore, mammalian Nav channels possess a second Phe in domain I that is equivalent to F203 (SI Appendix, Fig. S1). In contrast, the other bacterial Nav channels only possess two conserved Phe (F201 and F207 in NavAb), with orientations inconsistent with an FS6 site (participating in interactions away from the pore; Fig. 2B). NavAb may therefore offer unique similarity to mammalian Nav for the investigation of LA/AE binding (we note, however, that binding may still occur in the absence of an FS6-like residue, as seen for brominated drug-like compounds to more distant F214 on S6 in NavMs; ref. 11).
Fig. 2.
(A) Sequence alignment of segment S6 of NavAb to other Nav channels (showing only FS6-containing domain IV; see SI Appendix, Fig. S1, for full comparison). F201, F203, F207, and N211 in NavAb and F1764 in Nav1.2 are indicated. Amino acids were colored with Jalview (46, 47) using the Zappo scheme. (B) Aligned structures (showing three subunits) of NavAb and model Nav1.2 (based on NavAb) comparing NavAb F201, F203, and F207 to Nav1.2 F1764. Conserved N211 (NavAb numbering) is also indicated.
In the 1970s, Hille proposed two separate pathways for inhibitor binding, with charged drugs such as titratable amines and quaternary ammonium derivatives requiring an open channel, and neutral drugs such as BZC and PHT entering via a lipophilic pathway somewhere in the membrane (allowing for binding and dissociation, even when the pore is nonconducting) (20). Our previous simulations revealed dynamic interplay between the protein and lipids, with fenestrations allowing lipid tails to enter the pore (21), representing a potential drug pathway (20, 22). However, access to these openings, and subsequent binding, have yet to be described. Here we use the Anton supercomputer (23) to carry out extensive unbiased fully atomistic simulations of BZC and PHT binding to the NavAb channel to reveal the distribution of binding sites and observe their pathways to shed light on inhibition mechanisms and aid future Nav-drug development.
Results and Discussion
Membrane Partitioning of BZC and PHT.
To examine the abilities of these amphiphilic drug molecules to move within the membrane, we first carried out Umbrella Sampling (US) simulations for a pure lipid bilayer, using newly parameterized standard CHARMM models (SI Appendix). SI Appendix, Fig. S2A shows that BZC binds at the membrane interface by −3.3 ± 0.4 kcal/mol, with center of mass (COM) at |z|∼8.6 ± 0.1 Å from the bilayer center, interacting primarily with lipid carbonyls and phosphates via its amine H (SI Appendix, Fig. S3A). The corresponding membrane partition coefficient for BZC, Pb, is 38.7 ± 6.3, being less than the experimental values [ranging from 186 to 202 (24, 25) at 310–313 K]. This comparison with a range of results indicates ∼1 kcal/mol error, and suggests somewhat too weak binding to the membrane interface, perhaps due to the slightly overestimated dipole moment (SI Appendix, Table SA4), chosen to ensure correct condensed phase properties (26, 27) and accurate interaction energies (SI Appendix, Table SA6). However, SI Appendix, Table S1 shows that the model reproduces water–hydrocarbon partitioning free energies to within 0.2–0.3 kcal/mol. We consider these levels of discrepancy to be acceptable, being comparable to typical kcal/mol-order statistical errors, and expected to be largely cancelled for relative occupation of binding sites (see Benzocaine Binding to the Channel). BZC faces a smooth barrier of +2.7 ± 0.4 kcal/mol (relative to interfacial minima; −0.6 ± 0.1 kcal/mol relative to water). Applying Kramer’s transition rate theory (SI Appendix), we obtain a crossing rate of 1.8 ± 0.5 μs−1, indicating rapid submicrosecond membrane transfer.
PHT has a similar dipole moment to BZC (SI Appendix, Tables SA4 and SA5), yet experiences stronger binding at the membrane interface (SI Appendix, Fig. S2A). The minimum of −4.1 ± 0.1 kcal/mol (at |z| ∼ 9.59 ± 0.03 Å), allows both aromatic rings to reside in hydrocarbon while satisfying H-bonds at the interface, to a greater extent than BZC (SI Appendix, Fig. S3B). There is a similar discrepancy between calculated (139 ± 25) and experimental (657 ± 33) (28) partition coefficients for PHT, although those experiments were done using a different membrane (egg phosphatidylcholine) and temperature (298 K). The partition coefficient is 3.6 times higher than that for BZC with our models, consistent with experiments (24, 25, 28). PHT experiences a high and sharp barrier of +4.8 ± 0.1 kcal/mol relative to the interfacial minima (+0.66 ± 0.07 kcal/mol with respect to water). We compute a crossing rate of 35 ± 3 ms−1 (SI Appendix); being ∼2 orders of magnitude smaller than that for BZC, and approaching the millisecond time scale. This crossing rate suggests that PHT binding deep in the membrane may be restricted, even with multimicrosecond simulations.
Benzocaine Binding to the Channel.
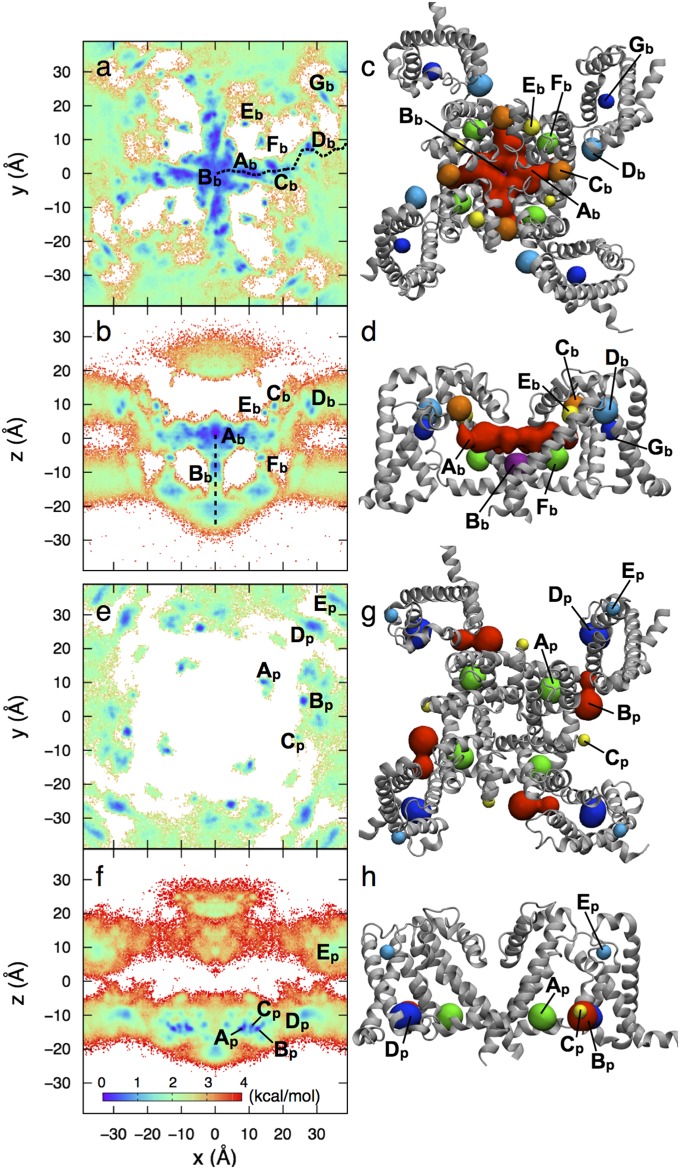
Unbiased ∼2-μs fully atomistic simulation in equilibrium with a bulk solution containing ∼10 mM BZC (SI Appendix, Fig. S4) has yielded a complete mapping of BZC locations throughout the membrane (consistent with the above calculations) and around the NavAb channel. Fig. 3 A and B show top and side projections of the free energy surface, respectively, allowing identification of seven binding sites (Ab–Gb; Figs. 3 C and D and 4). The highest affinity site, Ab, at the level of F203 and fenestrations, is seen to directly block the pore, as recently shown for volatile general anesthetics (14) and for brominated inhibitor analogs (11). This finding is consistent with our observation that F203 mimics mammalian FS6 (Fig. 2), which is crucial to inhibition (10, 16, 19, 29). Site Ab has a surprising expanse throughout the pore, leading to a KD of 141 ± 28 μM (SI Appendix, Table S2; potentially reduced by BZC accumulation, discussed below). This value is ∼4 times lower (stronger) than the experimental estimate for NaChBac (12), which lacks an FS6 mimic, possibly suggesting that NavAb may provide a better model for mammalian channel inhibition.
Fig. 3.
Free energy surfaces (Left) and binding sites (Right) for BZC (A–D) and PHT (E–H), with color scale as inset. For clarity, B for BZC includes only −10 < y < 10 Å (full data in SI Appendix, Fig. S5). Binding sites for BZC (C and D) and PHT (G and H), identified from free energy minima, are shown as spheres (colored for clarity, not by free energy).
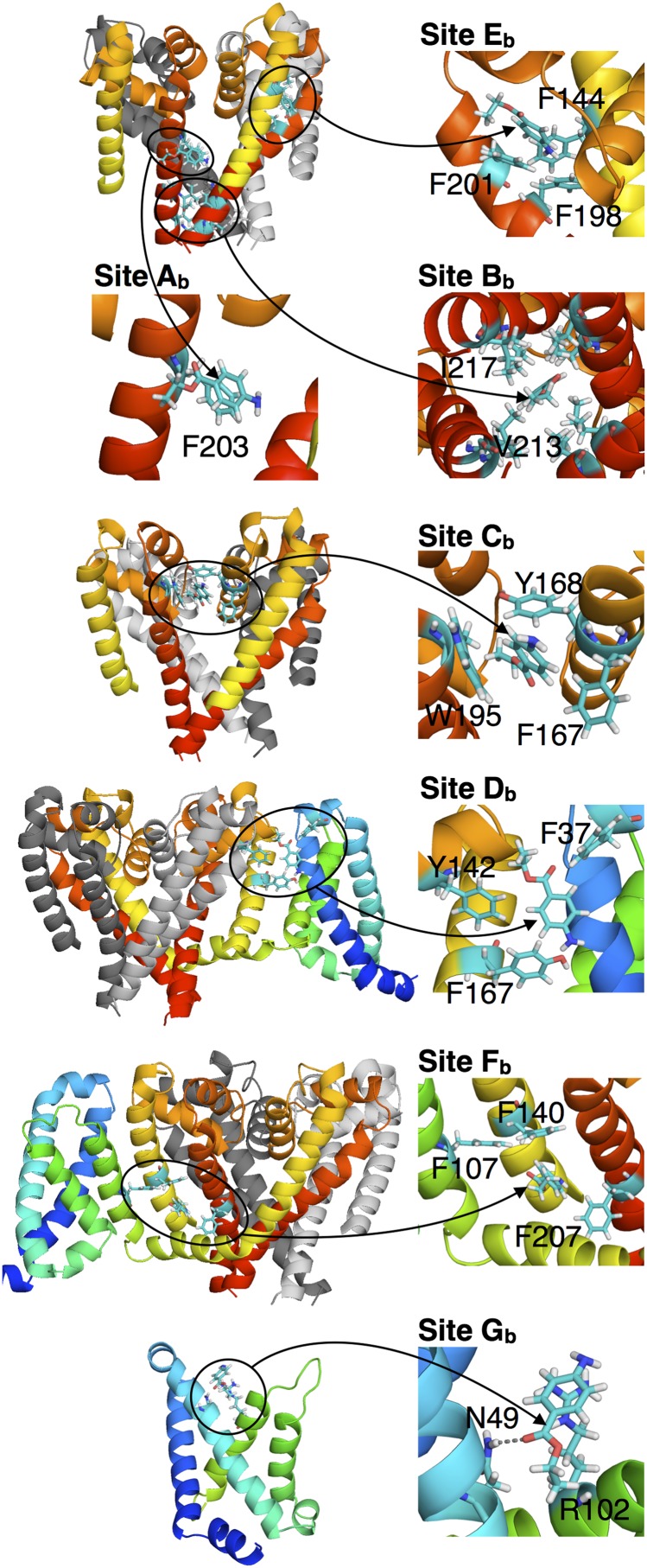
Fig. 4.
Interactions involved in binding sites Ab to Gb of BZC, with segments colored as in Fig. 1. (Insets) Arrows point directly at the BZC molecule.
SI Appendix, Fig. S6 shows that the fenestration radius is affected noticeably by the presence of BZC, owing to its binding to F203 at this location (SI Appendix, Fig. S6A). It has previously been proposed that mammalian FS6 and LA aromatic rings interact via π-stacking (19). In NavAb, we observe that F203 is more often oriented upward, apparently closing the fenestration pathway between membrane and central cavity of the ion conduction pathway. SI Appendix, Fig. S6B shows the free energy as a function of F203 side chain χ1 and χ2 dihedral angles in the presence of BZC, compared with the drug-free (and PHT) simulations. These results reveal that the biggest shift occurs in the upper right rotamer basin that corresponds to an open fenestration (SI Appendix, Fig. S6C) in the presence of BZC. BZC binding thus tends to favor the rotamer that opens the pathway (or that its binding requires a more open fenestration). BZC binding is either from the inside or the outside of the pore at F203 (SI Appendix, Fig. S6C). The distribution of the distance d between the center of the aromatic rings of F203 and BZC (SI Appendix, Fig. S6E) displays a plateau for d ∼ 5–7 Å, typical of π-stacking in protein (30) (with a small peak at ∼6.5 Å arising from BZC confinement in the central cavity and not due to direct stacking). Analysis of π-stacking conformations for outer and inner binding (SI Appendix, Fig. S6 F and G) shows that both favor more stable parallel displaced and T-shaped conformations (31). However, shorter distances are achieved from the outside (SI Appendix, Fig. S6 E and F), suggesting stronger interactions for initial binding. The ability of BZC to π-stack with F203 supports our assertion that F203 in NavAb plays a similar role to mammalian FS6, leading to a pore-blocking site.
Below this high-affinity site is Bb, deep in the intracellular gate, created by hydrophobic interactions with I217 and V213 (Figs. 3C and 4). Its KD of 407 ± 54 µM is higher (weaker) than Ab, and may be specific to the closed gate. Although both Ab and Bb evidence the ability for BZC to directly block the channel, the nonspecific binding in Bb may dominate closed-state low-affinity block, involving stabilization of the S6 bundle crossing via a hydrophobic seal (15).
Stacks of aromatics formed by pore helices create localized sites higher in the P-loop; this includes: Cb [intersubunit, F167(P1)–Y168(P1) and W195(S6), KD = 460 ± 62 µM], potentially linked to low-affinity binding (32); Db at the PD–voltage sensor domain (VSD) interface [F167(P1), Y142(S5) and F37(S1–S2), KD = 811 ± 77 µM]; and Eb [intrasubunit; F144(S5)–F198(S6)–F201(S6), KD = 4790 ± 830 µM]. Sites Cb and Db, connected via F167, appear to be important in the access pathway (see Drug Access Pathways).
At a level just below fenestrations and between S5/S6 and the VSD, BZC binds to aromatic pocket site Fb, created by F107(S4), F140(S5), and F207(S6). This site may help explain block by voltage-sensor inhibition, suggested to occur close to FS6, restricting the bottom of the PD and S4–S5 linker, reducing the effect of voltage on the activation gate (15, 33, 34). Even more directly influencing activation, low-affinity site Gb involves H-bonding to N49 and interaction via cation–π to the voltage-sensing R102 (S4; see Fig. 4). This finding is consistent with evidence for stabilization of the depolarized up state by LAs (33, 35).
Phenytoin Binding to the Channel.
Unbiased ∼3-μs simulation has been used to sample PHT binding to NavAb, leading to a distribution (Fig. 3 E and F) that is very different to BZC. PHT entered the bilayer and protein within ∼3 μs, but was unable to penetrate into the pore (although approached fenestrations on two occasions). This result may be explained by the greater binding to the membrane interface and slower time scales to enter the membrane core, calculated above. It is also consistent with experiments that demonstrate mutation of mammalian FS6 reduces PHT efficacy only marginally (36), suggesting other binding sites may play a greater role for this drug. Interestingly, the KD for PHT sites Ap–Ep, (Figs. 3 G and H and 5) range from as little (as strong) as 19 µM, to as high as 294 µM (SI Appendix, Table S3). Thus, the dominant PHT sites exhibit lower KD values than seen for BZC, consistent with the higher affinity for PHT compared with BZC seen experimentally for mammalian channels (37). However, binding of PHT is of higher affinity than expected, given the closed state of the channel’s activation gate [e.g., KD(resting) ∼ 600 µM for mammalian Nav (8)], perhaps indicating increased binding to non-FS6-like sites in NavAb due to sequence differences and fourfold symmetry, or due to increased interfacial preference for the PHT model used.
Fig. 5.
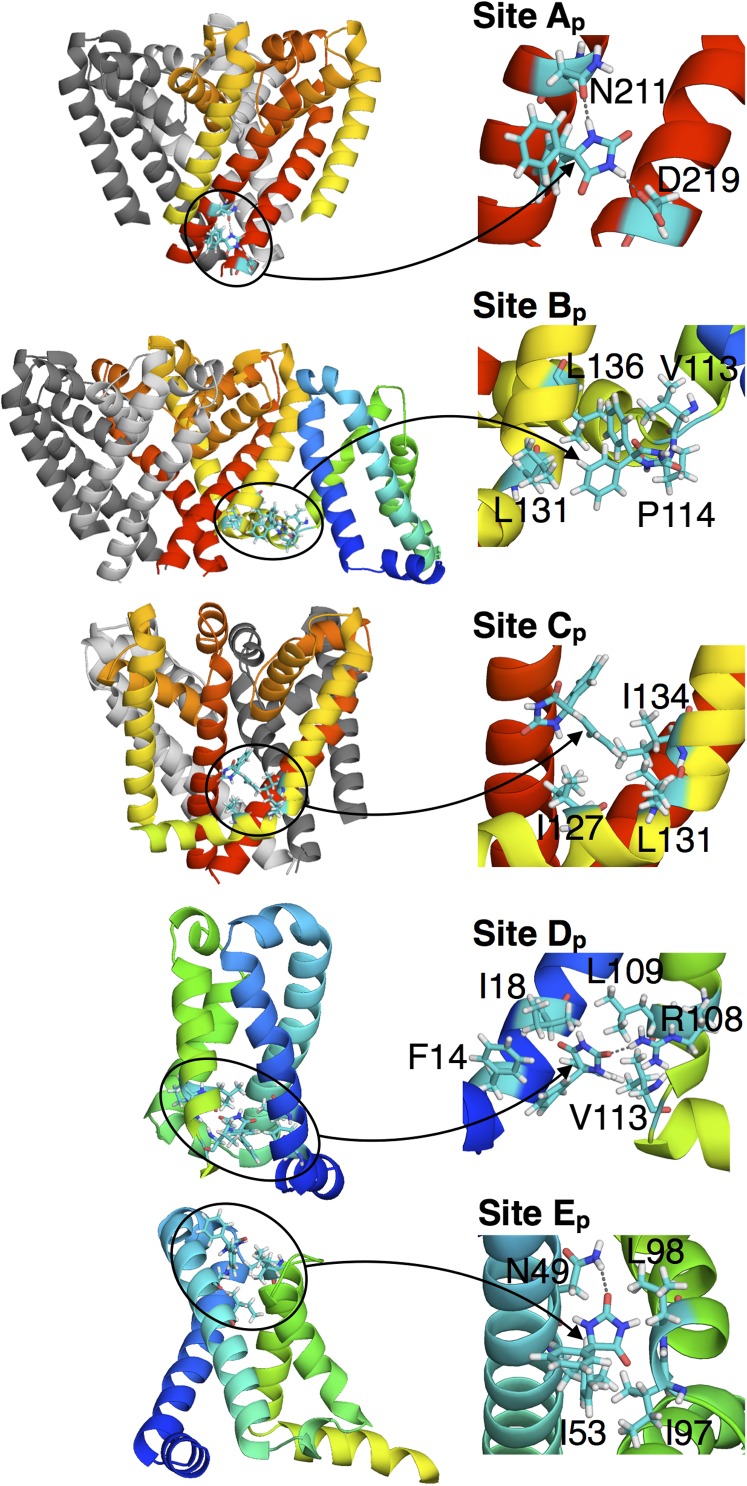
Interactions involved in the binding sites Ap to Fp of PHT. (Insets) Arrows point directly at the PHT molecule.
PHT site Ap is formed by residues in S6, involving H-bonding between imide H and conserved N211 side chain amide O, and D219 carboxyl O (conserved as D or E in mammalian channels) of adjacent subunits. A similar binding site has also been identified for a volatile general anesthetic in a NaChBac homology model, involving H-bonding to the equivalent residue N225 (14). Interestingly, mutation of N418 in DI, N1466 (DIII), and N1769 (DIV) in Nav1.2, which aligns with N211 (Fig. 2 A and B), leads to reduced affinity for LAs (38). This site has previously been suggested to be pore-facing and to involve hydrophobic residues at the bottom of S6 (38). However, we observe that N211 forms Ap that is not pore facing in NavAb, and instead associate the nearby hydrophobic residues forming Bb with such a pore-facing site.
The binding of PHT at site Ap relies on the gate being in its closed, nonconducting state, locking the helices in place and effectively blocking the activation gate from both collapse, associated with slow inactivation (6, 21), and opening. SI Appendix, Fig. S7 shows that the presence of PHT in the simulation drastically modifies the behavior of the S6 helices at the level of the activation gate by limiting the natural fluctuations observed in the absence of drug (21) or in the presence of BZC. In particular, it can be seen that PHT reduces asymmetry of the gate (measured as the difference between the distances separating the COM of the Cα of residues 215–218 on pairs of opposite monomers), due to binding at Ap involving H-bonding to N211 and D219. The KD for this site (73 µM) is consistent with the experimental value for binding to the NaChBac resting state (120 µM) (39).
Site Bp involves hydrophobic interactions with residues connecting the PD and VSD [L131(S5), L136(S5), V113(S4), and P114(S4–S5) of adjacent subunits, as well as Y20(S1)] and may contribute to low-affinity bundle crossing stabilization (37). Site Cp is the closest to the fenestrations (and F203), but is made up of nonspecific interactions with I127(S4–S5), L131(S5) and I134(S5). Site Dp is within the VSD and involves H-bonding of hydantoin carbonyl O to R108, with PHT phenyls interacting with F14(S1), I18(S1), L109(S4), and V113(S4). Similar site Ep involves imine H-bonding with N49(S2) and phenyl interactions with I53(S2), I97(S3), and L98(S4) (Fig. 5). SI Appendix, Table S3 summarizes the KD values for PHT, revealing surprisingly high-affinity binding around the PD, VSD, and PD–VSD interface, without binding inside the pore. Again, this may explain the modest effect of FS6 mutation on PHT efficacy in mammalian channels (36).
Drug Access Pathways.
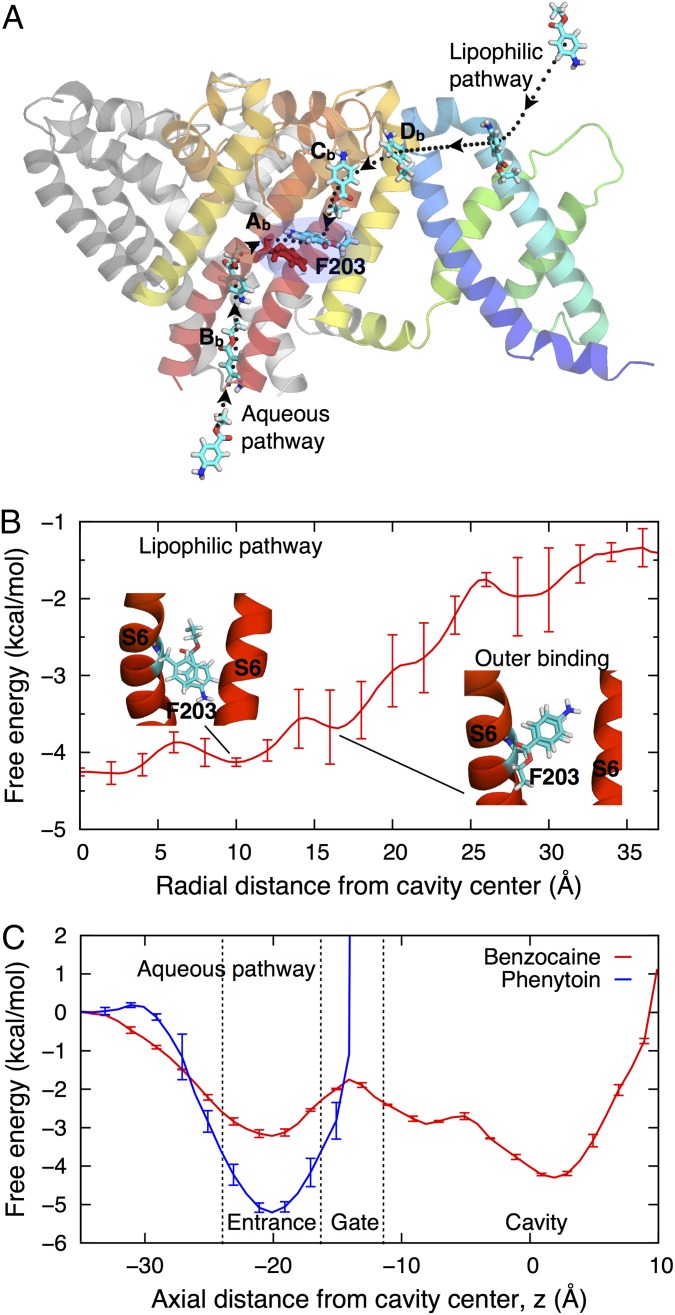
Given the complete sampling of BZC throughout the membrane and NavAb protein, we have extensive trajectories to examine the possible drug-binding processes. On seven occasions, BZC moved from the extracellular side through the intersubunit space between helices P1 and S6, via sites Db and Cb, and passed the fenestration into the pore. Fig. 6A visualizes the minimum free energy pathway by showing configurations from successful binding event trajectories from bulk water into the dominant site Ab in the central cavity (see also sample trajectory in SI Appendix, Fig. S8 and Movie S1). The minimum free energy path (Fig. 3A) involves low barriers (<1.5 kcal/mol), due to P-loop and fenestration flexibility (21), and stabilization by −2.5 kcal/mol, as can also be seen in the 1D free energy profile shown in Fig. 6B. This finding demonstrates that the hypothesized lipophilic drug access via fenestrations, initially proposed by Hille (20) and identified in the crystal structures (3), is a low free energy pathway.
Fig. 6.
Drug-binding pathways. (A) Side view of the channel showing movement of BZC from water to the central cavity along lipophilic and aqueous pathways (dotted curves). The full PD and only one VSD subunit are shown. (B and C) The free energy profile for BZC movement through a fenestration (B) and those for BZC (red) and PHT (blue) movements through the activation gate (C). The free energy is set to zero in water, using SI Appendix, Fig. S2A to set the membrane reference for B.
Surprisingly, BZC was also seen to enter up through the closed intracellular gate on 17 occasions, as indicated in Fig. 3B; this is also illustrated in the time series of SI Appendix, Fig. S9, with SI Appendix, Fig. S9B showing that the initially water-filled central cavity can accommodate up to ∼9 BZC molecules. Similar accumulation due to unbiased drug-binding simulation was also observed in recent simulations of isofluorane (14), although no permeation through the gate was observed in that study. After binding at the entrance (z ∼ −21 Å), BZC experiences a ∼1 kcal/mol barrier to cross the closed gate, before being stabilized in the central cavity by ∼−4 kcal/mol (Fig. 6C; although it may have been lowered due to BZC accumulation in the closed pore). This alternative aqueous route for BZC is therefore a viable alternative to the lipophilic pathway.
We showed above that PHT faces a greater barrier to enter the membrane, slowing down membrane entry by almost 2 orders of magnitude. As a result, sampling of PHT approaching the fenestrations was limited. These observations are consistent with slower rates of binding seen experimentally for PHT (8). PHT also experienced a greater barrier than BZC to pass the closed intracellular gate (Fig. 6C), being unable to completely pass into the pore within ∼3 μs, which may be consistent with experiments that suggest lower PHT affinity for the NaChBac resting state (39). Collectively, these observations reveal a greater energetic cost and time scales for PHT to leave the membrane or protein interfacial regions to bind inside a Nav channel.
Conclusions
We have carried out large-scale unbiased exploration of drug binding to a bacterial Nav channel that exhibits activation and pore-based inactivation processes in common with mammalian Nav channels. These shared functional characteristics, combined with crucial sequence similarity for LA and AE modulation, allow for molecular-level insight into Nav inhibition mechanisms from a structurally defined and functionally simpler bacterial model.
These simulations have led to the determination of a high-affinity pore-blocking site at the level of fenestrations in NavAb, stabilized by π-stacking to the FS6 mimic, F203 (unique to NavAb within the bacterial family), that is consistent with the accepted channel block mechanism (12). We have also identified sites that either participate in access pathways, or may be associated with low-affinity Nav block (15), previously uncharacterized at the molecular level. These sites involve interactions between the PD and VSD, potentially affecting transmission of voltage-dependent movements to the activation gate (33).
PHT experiences stronger binding than BZC at the membrane interface, with sites across the PD and VSD, but little binding at the level of F203 in unbiased ∼3-μs simulation, either because its onset rate is too low for binding to occur in this timeframe, or because the resting state of the channel impairs its binding (39). We have suggested that one site, formed by PHT imide–N211 H-bonding, may prevent gate collapse, consistent with mutagenesis of conserved residues (N418-DI; N1466-DIII and N1769-DIV in Nav1.2) that reduce LA affinity (38).
We have observed a minimum free energy pathway from the extracellular side involving clusters of aromatic and hydrophobic residues in the P-loops and PD–VSD interface, before passing through fenestrations and blocking the pore. Both BZC and PHT were seen to associate with the external pore vestibule (free energy ∼2 kcal/mol above the identified binding sites), but entry into the pore via the SF is forbidden. Surprisingly, however, we observed a second pathway from the intracellular side through the gate, despite being closed; opposing the prevailing view of binding to the resting-state via a lipophilic channel within the membrane (12, 20). In contrast, PHT was unable to pass through the closed gate, consistent with its lower binding rates (8) and reduced resting-state binding (39).
Knowledge of the LA and AE binding sites has provided the necessary first step in understanding the molecular origins of Nav channel inhibition. The next stage is to explore the state dependence of these interactions. Such studies are promising, given that F203 orientation and accessibility have been linked to structural changes associated with slow-inactivation (21), suggesting conformation-dependent drug binding within NavAb (10, 36, 40). Furthermore, by having uncovered a range of lower-affinity sites and the pathways of drug access, and not just the more conserved high-affinity blocking site, these results suggest new targets for therapeutically selective Nav inhibitor compounds.
Methods
The NavAb protein (Protein Data Bank ID code 3RVY; ref. 3) was embedded in a bilayer of dipalmitoylphosphatidylcholine (DPPC) with explicit TIP3P water molecules and 150 mM of NaCl. High initial concentrations of 500 mM BZC (Fig. 1B) or 75 mM PHT (Fig. 1C; lower due to the tendency for PHT to aggregate in solution in trial µs-long trajectories) were added, with both systems experiencing rapid partitioning of drugs to the membrane, such that the membrane-channel system was in equilibrium with 5–10 mM bulk drug concentrations after ∼0.2 µs (SI Appendix, Fig. S4C). Systems were built and preequilibrated with CHARMM (41) and then further equilibrated using NAMD (42). Anton software from D. E. Shaw Research was then used for production runs, using the purpose-built Anton supercomputer (23) for additional 1.8 and 2.9 µs in the presence of BZC and PHT, respectively. BZC and PHT topology and parameters were developed de novo, consistent with the CHARMM force field, with initial guesses for partial atomic charges and other force field parameters obtained using the CGENFF program (43, 44), optimized and validated using QM calculations and experimental partitioning free energies, as described in SI Appendix.
Free energy maps for drug binding to NavAb were calculated from unbiased Anton simulations as , where ρ is the unbiased probability distribution as a function of reaction coordinates Ri (position vectors of the COM of the drug molecule relative to the COM of the PD, translated along z to align with the COM of the membrane), with C a constant. Binding sites were identified when lower than −1 kcal/mol in free energy relative to the membrane reference, with calculation of KD values described in SI Appendix. Separate simulations of membrane partitioning were performed in dimyristoylphosphatidylcholine (DMPC) in 150 mM of NaCl. US (45) simulations for BZC and PHT were performed along the z axis with the drug COM held with respect to membrane COM near each US window center by a 2.5 kcal/mol/Å2 force constant for 13–25 ns each. Full details of US simulations, partition coefficient, and rate calculations are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Andrew Hung for helpful interactions. This work was supported by Australian Research Council Grant DP120103548, National Science Foundation Grant MCB1052477, DE Shaw Anton (PSCA00061P; National Resource for Biomedical Supercomputing, through National Institutes of Health Grant RC2GM093307), the Victorian Life Sciences Computation Initiative (VR0200), and the National Computational Infrastructure (dd7).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408710111/-/DCSupplemental.
References
- 1.England S, de Groot MJ. Subtype-selective targeting of voltage-gated sodium channels. Br J Pharmacol. 2009;158(6):1413–1425. doi: 10.1111/j.1476-5381.2009.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mike A, Lukacs P. The enigmatic drug binding site for sodium channel inhibitors. Curr Mol Pharmacol. 2010;3(3):129–144. doi: 10.2174/1874467211003030129. [DOI] [PubMed] [Google Scholar]
- 3.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille B. Ionic Channels of Excitable Membranes. 3rd Ed. Sunderland: Sinauer; 2001. [Google Scholar]
- 8.Kuo CC, Bean BP. Slow binding of phenytoin to inactivated sodium channels in rat hippocampal neurons. Mol Pharmacol. 1994;46(4):716–725. [PubMed] [Google Scholar]
- 9.Fozzard HA, Lee PJ, Lipkind GM. Mechanism of local anesthetic drug action on voltage-gated sodium channels. Curr Pharm Des. 2005;11(21):2671–2686. doi: 10.2174/1381612054546833. [DOI] [PubMed] [Google Scholar]
- 10.Carboni M, Zhang Z-S, Neplioueva V, Starmer CF, Grant AO. Slow sodium channel inactivation and use-dependent block modulated by the same domain IV S6 residue. J Membr Biol. 2005;207(2):107–117. doi: 10.1007/s00232-005-0805-0. [DOI] [PubMed] [Google Scholar]
- 11.Bagnéris C, et al. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci USA. 2014;111(23):8428–8433. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Goodchild SJ, Ahern CA. Local anesthetic inhibition of a bacterial sodium channel. J Gen Physiol. 2012;139(6):507–516. doi: 10.1085/jgp.201210779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber AF, Carnevale V, Klein ML, Eckenhoff RG, Covarrubias M. Modulation of a voltage-gated Na+ channel by sevoflurane involves multiple sites and distinct mechanisms. Proc Natl Acad Sci USA. 2014;111(18):6726–6731. doi: 10.1073/pnas.1405768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raju SG, Barber AF, LeBard DN, Klein ML, Carnevale V. Exploring volatile general anesthetic binding to a closed membrane-bound bacterial voltage-gated sodium channel via computation. PLOS Comput Biol. 2013;9(6):e1003090. doi: 10.1371/journal.pcbi.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheets MF, Fozzard HA, Lipkind GM, Hanck DA. Sodium channel molecular conformations and antiarrhythmic drug affinity. Trends Cardiovasc Med. 2010;20(1):16–21. doi: 10.1016/j.tcm.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265(5179):1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 17.McNulty MM, et al. Charge at the lidocaine binding site residue Phe-1759 affects permeation in human cardiac voltage-gated sodium channels. J Physiol. 2007;581(Pt 2):741–755. doi: 10.1113/jphysiol.2007.130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courtney KR. Mechanism of frequency-dependent inhibition of sodium currents in frog myelinated nerve by the lidocaine derivative GEA. J Pharmacol Exp Ther. 1975;195(2):225–236. [PubMed] [Google Scholar]
- 19.Ahern CA, Eastwood AL, Dougherty DA, Horn R. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ Res. 2008;102(1):86–94. doi: 10.1161/CIRCRESAHA.107.160663. [DOI] [PubMed] [Google Scholar]
- 20.Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69(4):497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111(9):3454–3459. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morello RS, Begenisich T, Yeh JZ. Determination of the active form of phenytoin. J Pharmacol Exp Ther. 1984;230(1):156–161. [PubMed] [Google Scholar]
- 23.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 24.Ávila CM, Martínez F. Thermodynamics of partitioning of benzocaine in some organic solvent/buffer and liposome systems. Chem Pharm Bull (Tokyo) 2003;51(3):237–240. doi: 10.1248/cpb.51.237. [DOI] [PubMed] [Google Scholar]
- 25.Lukacova V, et al. Drug-membrane interactions studied in phospholipid monolayers adsorbed on nonporous alkylated microspheres. J Biomol Screen. 2007;12(2):186–202. doi: 10.1177/1087057106297063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang KY, Braswell LM, Chang L, Sommer TJ, Miller KW. The perturbation of lipid bilayers by general anesthetics: A quantitative test of the disordered lipid hypothesis. Mol Pharmacol. 1980;18(1):84–90. [PubMed] [Google Scholar]
- 29.Li HL, Galue A, Meadows L, Ragsdale DS. A molecular basis for the different local anesthetic affinities of resting versus open and inactivated states of the sodium channel. Mol Pharmacol. 1999;55(1):134–141. doi: 10.1124/mol.55.1.134. [DOI] [PubMed] [Google Scholar]
- 30.McGaughey GB, Gagné M, Rappé AK. π-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273(25):15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 31.Sinnokrot MO, Valeev EF, Sherrill CD. Estimates of the ab initio limit for π-π interactions: The benzene dimer. J Am Chem Soc. 2002;124(36):10887–10893. doi: 10.1021/ja025896h. [DOI] [PubMed] [Google Scholar]
- 32.Yamagishi T, et al. Novel molecular determinants in the pore region of sodium channels regulate local anesthetic binding. Mol Pharmacol. 2009;76(4):861–871. doi: 10.1124/mol.109.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muroi Y, Chanda B. Local anesthetics disrupt energetic coupling between the voltage-sensing segments of a sodium channel. J Gen Physiol. 2009;133(1):1–15. doi: 10.1085/jgp.200810103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arcisio-Miranda M, Muroi Y, Chowdhury S, Chanda B. Molecular mechanism of allosteric modification of voltage-dependent sodium channels by local anesthetics. J Gen Physiol. 2010;136(5):541–554. doi: 10.1085/jgp.201010438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheets MF, Hanck DA. Molecular action of lidocaine on the voltage sensors of sodium channels. J Gen Physiol. 2003;121(2):163–175. doi: 10.1085/jgp.20028651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, et al. Lidocaine induces a slow inactivated state in rat skeletal muscle sodium channels. J Physiol. 2000;524(Pt 1):37–49. doi: 10.1111/j.1469-7793.2000.t01-1-00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo C-C. A common anticonvulsant binding site for phenytoin, carbamazepine, and lamotrigine in neuronal Na+ channels. Mol Pharmacol. 1998;54(4):712–721. [PubMed] [Google Scholar]
- 38.Yarov-Yarovoy V, et al. Role of amino acid residues in transmembrane segments IS6 and IIS6 of the Na+ channel alpha subunit in voltage-dependent gating and drug block. J Biol Chem. 2002;277(38):35393–35401. doi: 10.1074/jbc.M206126200. [DOI] [PubMed] [Google Scholar]
- 39.Pavlov E, Bladen C, Dhaliwal PPS, French RJ. Block of a prokaryotic voltage-gated sodium channel (NaChBac) by phenytoin and lidocaine. Biophys J. 2003;84:66a. [Google Scholar]
- 40.Sheets PL, Jarecki BW, Cummins TR. Lidocaine reduces the transition to slow inactivation in Na(v)1.7 voltage-gated sodium channels. Br J Pharmacol. 2011;164(2b):719–730. doi: 10.1111/j.1476-5381.2011.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanommeslaeghe K, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J Chem Inf Model. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J Chem Inf Model. 2012;52(12):3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comput Phys. 1977;23(2):187–199. [Google Scholar]
- 46.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20(3):426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 47.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.