Significance
Epithelial–mesenchymal transition (EMT) is a transdifferentiation program implicated in tumor cell dissemination, controlled by networks of transcription complexes responsive to paracrine factors, such as TGF-β. Pre–B-cell leukemia homeobox (Pbx)-regulating protein-1 (PREP1) is a ubiquitous homeodomain transcription factor involved in early development, genomic stability, insulin sensitivity, and hematopoiesis. PREP1 is a haploinsufficient oncosuppressor in mouse tumorigenesis. By characterizing PREP1 as a novel regulator of EMT in human lung adenocarcinoma, we show that PREP1 also harbors prometastatic properties. While autosustaining its activity by stabilizing its transcriptional partner PBX1, PREP1 modulates the responsiveness of lung cancer cells to TGF-β by controlling the expression of two proinvasive transcription factors (SMAD3 and Fos-related antigen 1) implicated in metastasis mechanisms. Thus, PREP1 represents a novel, promising therapeutic target in non-small cell lung cancer.
Keywords: TALE proteins, PTGFβ
Abstract
Pre–B-cell leukemia homeobox (Pbx)-regulating protein-1 (Prep1) is a ubiquitous homeoprotein involved in early development, genomic stability, insulin sensitivity, and hematopoiesis. Previously we have shown that Prep1 is a haploinsufficient tumor suppressor that inhibits neoplastic transformation by competing with myeloid ecotropic integration site 1 for binding to the common heterodimeric partner Pbx1. Epithelial–mesenchymal transition (EMT) is controlled by complex networks of proinvasive transcription factors responsive to paracrine factors such as TGF-β. Here we show that, in addition to inhibiting primary tumor growth, PREP1 is a novel EMT inducer and prometastatic transcription factor. In human non-small cell lung cancer (NSCLC) cells, PREP1 overexpression is sufficient to trigger EMT, whereas PREP1 down-regulation inhibits the induction of EMT in response to TGF-β. PREP1 modulates the cellular sensitivity to TGF-β by inducing the small mothers against decapentaplegic homolog 3 (SMAD3) nuclear translocation through mechanisms dependent, at least in part, on PREP1-mediated transactivation of a regulatory element in the SMAD3 first intron. Along with the stabilization and accumulation of PBX1, PREP1 induces the expression of multiple activator protein 1 components including the proinvasive Fos-related antigen 1 (FRA-1) oncoprotein. Both FRA-1 and PBX1 are required for the mesenchymal changes triggered by PREP1 in lung tumor cells. Finally, we show that the PREP1-induced mesenchymal transformation correlates with significantly increased lung colonization by cells overexpressing PREP1. Accordingly, we have detected PREP1 accumulation in a large number of human brain metastases of various solid tumors, including NSCLC. These findings point to a novel role of the PREP1 homeoprotein in the control of the TGF-β pathway, EMT, and metastasis in NSCLC.
PREP1 [pre–B-cell leukemia homeobox (Pbx)-regulating protein-1], also known as “PKNOX1” (PBX/knotted homeobox 1), belongs to the TALE (three-amino acid-loop-extension) family of homeodomain transcription factors, along with myeloid ecotropic integration site (MEIS) and PBX proteins. As indicated by its name, PREP1 retains PBX1 in the nucleus and induces its binding to DNA (1).
PREP1 is essential in embryonic development. Although Prep1-null embryos die before gastrulation from apoptosis of the epiblast (2), hypomorphic Prep1-mutant mice (Prepi/i), expressing severely decreased Prep1 levels (3–10% of wild-type) exhibit incompletely penetrant embryonic phenotypes, mainly characterized by alterations in hematopoiesis and angiogenesis (3).
Prep1 deficiency affects cell proliferation and survival in various biological contexts. Decreased Prep1 expression results in increased apoptosis in Prep1i/i embryos and mouse embryonic fibroblasts (MEFs) (4). In Prep1−/− embryos, early preimplantation lethality is consequent to increased apoptosis in mouse pluripotent epiblast cells. Genetic analysis suggests that the lack of Prep1 confers increased sensitivity to DNA damage by Atm- and p53-mediated mechanisms (2).
TALE homeoproteins are strongly implicated in leukemia- and lymphomagenesis. The t (1, 19) translocation forming the PBX1-E2A chimeric transcription factor is a cause of pediatric acute pre–B-lymphoblastic leukemia (5, 6), and in myeloid leukemias MEIS1 is a relevant oncogene, acting in tight cooperation with HOXA9 (7).
Unlike PBX1 and MEIS1, PREP1 is able to inhibit neoplastic transformation. Indeed, Eμ-myc–induced lymphomagenesis is accelerated by Prep1 haploinsufficiency. In addition, surviving Prepi/i mice develop spontaneous pretumoral lesions and eventually lymphomas and carcinomas. Accordingly, we have detected very low PREP1 expression in a majority of specimens representing a wide range of human tumors (8).
In both Prepi/i MEFs and human PREP1 down-regulated fibroblasts, decreased Prep1 expression causes increased susceptibility to RAS transformation by mechanisms mediated by the Prep1-dependent inhibition of oncogene-induced senescence (OIS). The increased resistance to OIS is associated with the accumulation of DNA damage and chromosomal instability (9).
TGF-β and the tumor microenvironment exert both positive and negative influences on cancer development. In premalignant stages, TGF-β represents a major tumor suppressor cytokine. During tumor progression, loss of the TGF-β–dependent inhibition of cell growth is associated with the gain of TGF-β–dependent control of the invasive phenotype (10).
TGF-β is a master regulator of epithelial–mesenchymal transition (EMT), which, in addition to its roles in embryonic morphogenetic programs, is responsible for the transformation of polarized epithelial cancer cells into highly motile mesenchymal derivatives responsible for tumor invasion, intravasation, extravasation, and metastatic dissemination (11). EMT recently has been implicated in the acquisition of cancer stem cell properties (12) and resistance to conventional therapies (13). TGF-β inhibits the expression of epithelial genes, such as E-cadherin, and induces the expression of mesenchymal genes such as vimentin and extracellular matrix-degrading proteases (14), including the urokinase-type plasminogen activator (uPA) (15) and various matrix metallopeptidases (MMPs) (16).
Transcription factors of the SMAD (small mothers against decapentaplegic) family, SMAD2 and SMAD3, forming heteromeric complexes with SMAD4, represent key regulators of EMT in response to TGF-β. Increased expression of SMAD2 or SMAD3, along with SMAD4, enhances the EMT induction by the activated form of type-1 TGF-β receptor (TGFBR1). SMADs, in turn, control their transcriptional targets by cooperating with a variety of nuclear factors, including E-cadherin repressors [snail family zinc finger 1 and 2 (SNAI1/2), zinc finger E-box binding homeobox 1 and 2 (ZEB-1/2), and others], activator protein 1 (AP-1) components, c-MYC, and HMGA2 (17). In contrast with the broad knowledge of posttranscriptional mechanisms (heterodimerization, nucleo–cytoplasmic shuttling, phosphorylation, ubiquitylation) controlling SMAD’s activity, very little is known about the transcriptional regulation of SMAD genes.
Lung cancer is the greatest cause of cancer-related death in Western countries, and metastasis is the most common cause of death in lung cancer patients. Eighty-five percent of lung cancers are non-small cell lung cancers (NSCLC). Only 20–30% of treated NSCLC patients exhibit clinically significant therapeutic responses. No certain criteria are available to distinguish a good prognosis from a bad prognosis in patients with stage I NSCLC.
Here, we characterize the role of PREP1 as a novel regulator of EMT. Despite its inhibitory action on primary tumor growth, PREP1 triggers EMT, invasion, and metastasis in lung adenocarcinoma cells. Accordingly, we show that PREP1 represents a strong negative prognostic factor in patients with stage 1 NSCLC and is overexpressed in brain metastasis of lung adenocarcinoma.
Results
PREP1 Triggers the Inhibition of Tumor Growth Associated with EMT Induction in Lung Adenocarcinoma Cells.
We recently have shown that PREP1 is a haploinsufficient tumor suppressor and that PREP1 expression is absent or down-regulated in a large variety of solid tumors (8). Having observed that PREP1 was very low or absent in a large fraction (313/445) of lung cancer specimens (8), we decided to investigate the consequence of PREP1 re-expression in a non-small cell lung adenocarcinoma line expressing PREP1 at very low level. As compared with control cells (A549pBABE), A549PREP1 cells displayed slightly decreased proliferation, correlating with signs of increased apoptosis (i.e., increased Caspase-3/7 activity) (SI Appendix, Fig. S1 A–C). However, after injection into immunocompromised mice, the A549PREP1 cells formed fewer tumors, and these tumors were significantly smaller than the xenografts formed by the control A549pBABE cell line (SI Appendix, Fig. S1 D and E). These data suggested that PREP1 is able to suppress neoplastic growth in lung cancer cells.
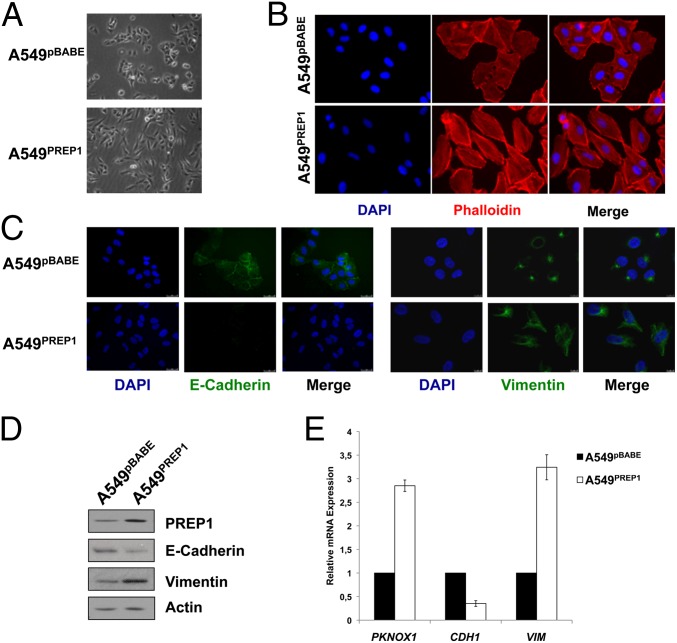
However, the A549PREP1 cells exhibited striking morphological modifications. The ectopic PREP1 triggered the transition from a cobblestone-like morphology to an elongated shape associated with increased cell scattering. Fibroblast-like stress fibers were increased, and cortical actin was decreased (Fig. 1 A and B). Loss of intercellular adhesion and cytoskeletal remodeling suggested that PREP1 could induce EMT. Accordingly, we observed nearly complete loss of E-cadherin immunostaining in A549PREP1 cells as compared with control cells. Vimentin staining revealed changes in protein distribution from a localized perinuclear signal to a diffuse filamentous distribution (Fig. 1C). In addition, the E-cadherin protein level was down-regulated, and vimentin expression was increased significantly in response to PREP1. These modifications reflected consistent changes in transcript levels in A549PREP1 as compared with A549pBABE cells (Fig. 1 D and E).
Fig. 1.
PREP 1 induces EMT in A549 cells. (A–C) Cell morphology under bright-field (A) and immunofluorescence (B and C) microscopy. Actin microfilaments were stained by rhodamine-conjugated phalloidin. E-cadherin and vimentin were visualized by FITC immunostaining. Cell nuclei were visualized by DAPI. (D) Western blotting analysis of E-cadherin and vimentin in total extracts from A549PREP1 and A549pBABE cells. Anti-actin was used to normalize for protein loading. (E) Quantitative RT-PCR (qRT-PCR) analysis of the PREP1 (PKNOX1), E-cadherin (CDH1), and vimentin (VIM) transcripts. GAPDH was used as an internal control. Expression levels in A549PREP1 cells are shown relative to the levels in A549pBABE cells. Error bars represent the mean ± SD of three independent experiments.
PREP1-Induced Cell Motility and Invasion Are Associated with the Accumulation of Proinvasive AP-1 Components.
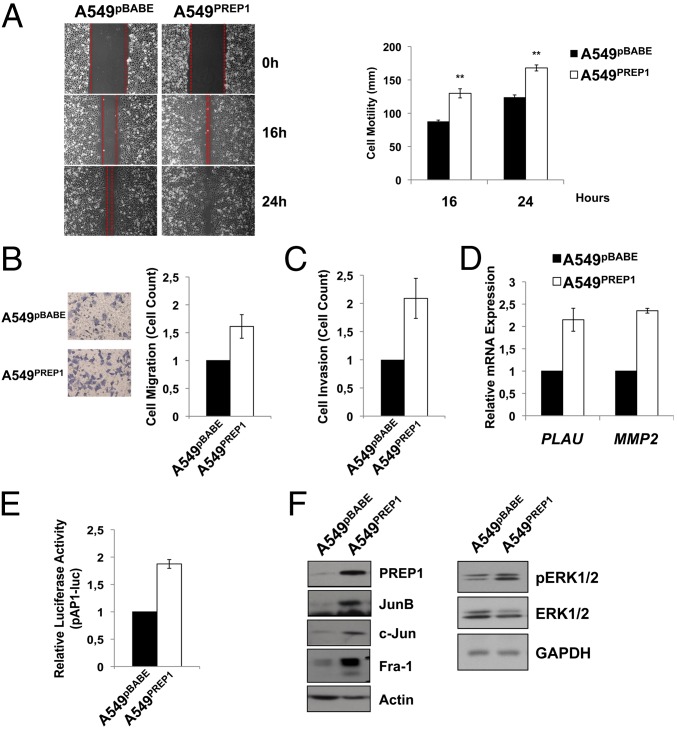
According to the EMT, A549PREP1 cells exhibited increased motility in wound-healing (Fig. 2A) and Transwell (Fig. 2B) assays. In addition, PREP1 was able to stimulate the invasion of an artificial basement membrane, as shown by the increased migration of A549PREP1 cells through Matrigel-coated Transwell inserts (Fig. 2C). Because Matrigel invasion requires proteolytic degradation of extracellular matrix components, we analyzed the expression of two secreted proteases strongly implicated in proinvasive extracellular proteolysis. Both uPA and matrix metallopeptidase 2 (MMP-2; 72-kDa type IV collagenase) mRNA levels were increased significantly (more than twofold) in A549PREP1 cells as compared with control cells (Fig. 2D). Both uPA- and MMP-2–coding genes (PLAU and MMP2, respectively) represent prototype targets of the AP-1 complex, a master transcriptional regulator of tumor cell migration and invasion (18, 19). Therefore we analyzed the AP-1 activity and composition in the A549 cells overexpressing PREP1. The activity of an AP-1–luciferase reporter was increased significantly in A549PREP1 cells as compared with control cells (Fig. 2E), and the expression of multiple AP-1 components was up-regulated. Specifically, jun proto-oncogene (c-JUN), jun B proto-oncogene (JUNB), and Fos-related antigen 1 (FRA-1) were strongly up-regulated (Fig. 2F). The MAPK pathway is one of the major positive regulators of AP-1 activity and composition. Accordingly, we observed that the accumulation of FRA-1 and JUN family members correlated with increased phospho-ERK1/2 levels in the A549PREP1 cells as compared with A549pBABE cells. These results (Fig. 2F) suggest that PREP1 can induce the AP-1 activity and target genes through the activation of the mitogen-activated protein kinase kinase (MEK)/ERK pathway.
Fig. 2.
PREP1 induces cell migration, invasion, and accumulation of AP-1 components. (A) Wound-healing assay. (Left) Three different fields were taken. (Right) The graph shows the width (in millimeters) of the wound 16 and 24 h after the scratch. Error bars represent the mean ± SD of three independent experiments. *P < 0.01. (B and C) Equal numbers of viable cells were seeded on the top of Transwell inserts and incubated for 5 h in serum-containing medium. Cells that migrated through filters were stained with crystal violet and then were examined under a microscope. The graph shows the cell count in five different fields. Error bars represent the mean ± SD of three independent experiments. (C) Invasion was measured by coating the Transwell inserts with a layer of Matrigel (50 μg/mL). (D) PLAU and MMP2 transcript levels were quantitated by real-time RT-PCR analysis using GAPDH as internal control. Error bars represent the mean ± SD of three independent experiments. (E) Luciferase activity of an AP-1 reporter construct (pAP-1-luc) transfected in A549pBABE and A549PREP1 cells. Firefly luciferase activity was normalized by cotransfecting a Renilla luciferase reporter. Error bars represent the mean ± SD of three independent experiments. (F) Western blot analysis of AP-1 components and phosph-ERK1/2 in A549PREP1 cells and A549pBABE cells. β-Actin or GAPDH was used to normalize for equal loading.
PREP1 Accumulation Is Required for the TGF-β–Induced EMT.
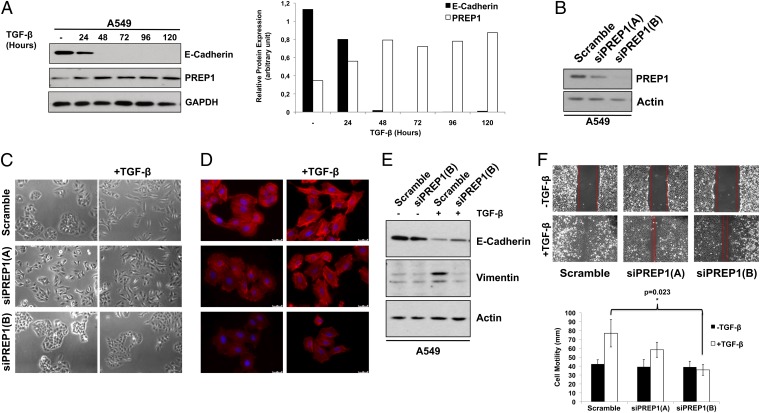
TGF-β induces EMT in various systems, including A549 cells (20). Therefore we decided to investigate the role of PREP1 in response to TGF-β. Indeed, in unmodified A549 cells PREP1 expression was induced almost threefold by TGF-β, with a peak between 24 and 48 h, intriguingly coinciding with the loss of E-cadherin (Fig. 3A).
Fig. 3.
PREP1 accumulation is required for the induction of EMT and cell motility in response to TGF-β. (A) Western blot analysis of E-cadherin and PREP1 expression in response to TGF-β (5 ng/mL) induction in A549 cells at different time points, as indicated. GAPDH was used to normalize for equal loading. The histograms represent the PREP1 expression level quantified by densitometry and normalized for GAPDH. (B) Two distinct PREP1-targeting siRNAs (si-PREP1-A and si-PREP1-B) were evaluated by immunoblot analysis of PREP1 72 h after transfection in A549 cells. β-Actin was used to normalize for equal loading. (C and D) Twenty-four hours after transfection of the two PREP1-targeting siRNAs, A549 cells were treated with TGF-β for 48 h and analyzed by bright-field microscopy (C) and immunofluorescent staining of actin cytoskeleton by rhodamine-conjugated phalloidin and nuclei by DAPI (D). (E) Immunoblotting analysis of E-cadherin and vimentin in A549 cells transfected with the PREP1-specific siRNA (5 d ± TGF-β treatment). β-Actin was used to normalize for equal loading. (F) Wound-healing assays were performed 24 h after transfecting the PREP1-specific siRNAs. (Upper) Wound closure was determined 24 h after the scratch. (Lower) Graphs show the results of three independent experiments. Error bars indicate mean ± SD; *P < 0.05.
We therefore tested the reverse correlation between PREP1 and E-cadherin in the TGF-β–induced EMT by downregulating PREP1 with two distinct siRNAs, siRNA-A and siRNA-B. The TGF-β–induced A549 cell scattering, elongation, and cytoskeletal remodeling were inhibited partially by siRNA-A knockdown of PREP1 and were inhibited completely by siRNA-B knockdown of PREP1. The extent of the effects reflected the degree of PREP1 down-regulation by the two distinct siRNAs (siPREP1-B > siPREP1-A) (Fig. 3 B–D).
Then we tested the effect of PREP1 down-regulation on the TGF-β–induced changes of E-cadherin and vimentin. TGF-β–induced down-regulation of E-cadherin was partially antagonized, whereas the accumulation of vimentin was abrogated completely by the knockdown of PREP1 (Fig. 3E).
Finally, cell-wounding and Transwell assays showed that the TGF-β–induced motility was inhibited significantly by PREP1 knockdown, with a fold-decrease reflecting the relative efficacy of the two distinct siRNAs (Fig. 3 F).
When propagated in presence of low serum (0.1%), A549PREP1 cells lost the mesenchymal appearance, reverting to the aspect of A549pBABE control cells (SI Appendix, Fig. S2 A and B). We postulated that low concentrations of TGF-β in serum-supplemented medium might be sufficient to trigger the EMT phenotype in A549PREP1 cells but not in control cells. To test if PREP1 actually is able to enhance the TGF-β responsiveness of A549 cells, we treated the A549PREP1 and A549pBABE cells with increasing concentrations of TGF-β in serum-free medium. A TGF-β concentration as low as 0.1 ng/mL was sufficient to induce the scattering of A549PREP1 cells, whereas a fivefold-higher cytokine concentration was required to induce similar changes in A549pBABE cells (SI Appendix, Fig. S2A). Correspondingly, only the maximal cytokine concentration caused a partial down-regulation of E-cadherin in A549pBABE cells, whereas a 10× lower concentration almost completely inhibited E-cadherin expression in A549PREP1 cells (SI Appendix, Fig. S2C). In summary, these results show that PREP1 is required for the responses to TGF-β and that it acts by modulating the threshold of sensitivity to the cytokine.
PREP1 Transcriptionally Induces SMAD3 by Interacting with an Intronic Enhancer Element.
Next, we tested the effects of PREP1 on the key components of the TGF-β pathway. By analyzing multiple SMADs, we found that the SMAD3 mRNA was increased significantly in A549PREP1 cells as compared with A549pBABE cells, but smaller changes in SMAD1, SMAD2, SMAD4, SMAD5, and SMAD7 transcripts did not reach statistical significance (Fig. 4A). The increased SMAD3 mRNA level actually resulted in the increased accumulation of the protein, whereas SMAD2 expression was essentially unchanged in A549PREP1 cells as compared with A549pBABE cells (Fig. 4B).
Fig. 4.
PREP1 induces the expression of SMAD3. (A) SMAD family transcripts were quantitated by real-time RT-PCR analysis of RNA from A549pBABE and A549PREP1 stable transfectants. GAPDH was used as internal control. Error bars represent the mean ± SD of three independent experiments; *P < 0.05. (B and C) Western blotting analysis of Smad2 and Smad3 in total extracts from A549PREP1 and A549pBABE cells (B) and in A549 cells transfected with two PREP1-specific siRNAs, siPREP1(A) and siPREP1(B) (C). β-Actin was used to normalize for equal loading. (D) qRT-PCR analysis of SMAD3 and SMAD2 in A549 cells transfected with the two PREP1-specific siRNAs. GAPDH was used as an internal control. Error bars represent the mean ± SD of three independent experiments. (E) Diagram of amplicons used for ChIP analysis. TSS, the major SMAD3 transcription start site on human chromosome 15. The Inset shows the PREP1 site in the first intron of the gene. The endpoints of amplicons (a, b, c, d, and e) are relative to the position of TSS, according to the coordinates on University of California, Santa Cruz Genome Browser (hg19, February 2009) (F) Representative semiquantitative PCR analysis of PREP1 binding to fragments a, c, and e, in A549PREP1 cells. (G) Real-time qPCR analysis in A549PREP1 cells of PREP1 binding to the amplicons shown in SI Appendix, Fig. S2C. Fold enrichment represents the percent of the signal detected as compared with the input chromatin. Error bars represent the mean ± SD of three independent experiments.
These findings suggested that SMAD3 might be controlled by PREP1 in A549 cells. Indeed, SMAD3 was decreased significantly in response to PREP1 down-regulation by the two distinct siRNAs, whereas SMAD2 was unaffected. Accordingly, the SMAD3, but not the SMAD2, transcript was down-regulated by PREP1 knockdown (Fig. 4 C and D).
Is SMAD3 a transcriptional target of PREP1 in lung adenocarcinoma cells? SMAD3 contains a conserved decameric PREP1 target sequence in the first intron (SI Appendix, Fig. S3 A and B). Semiquantitative PCR of anti-PREP1–immunoprecipitated chromatin in A549PREP1 cells enriched the +1002/+1150 amplicon encompassing the putative PREP1 element, but no amplification product was detected by using primers complementary to adjacent regions (Fig. 4 E and F). These results were confirmed by real-time PCR, which also showed that both an upstream (−1074/−916) and a far downstream (+28456/+28565) amplicon exhibited near-background levels, comparable to the signals generated by control antibodies (Fig. 4G). These findings were confirmed in HeLa cells that express high PREP1 levels. The SMAD3 transcript was affected by the knockdown of PREP1 (SI Appendix, Fig. S4A), and the highest fold-enrichment observed in ChIP with the +1002/+1150 amplicon was reconfirmed with a distinct overlapping (+996/+1133) amplicon (SI Appendix, Fig. S4B).
To test if the binding site identified by ChIP could be transactivated by PREP1, we subcloned a 247-bp fragment encompassing the putative regulatory element upstream of an enhancerless luciferase reporter. When coexpressed with a PREP1 expression vector, the SMAD3 reporter exhibited a twofold increase of luciferase activity, which was abrogated when the PREP1 sequence was mutated by double nucleotide substitution (SI Appendix, Fig. S5). These data indicate that PREP1 binding to a regulatory element in the first SMAD3 intron results in the transactivation of the SMAD3 promoter.
PREP1 Is Required for the Nuclear Accumulation of SMAD3 in Response to TGF-β.
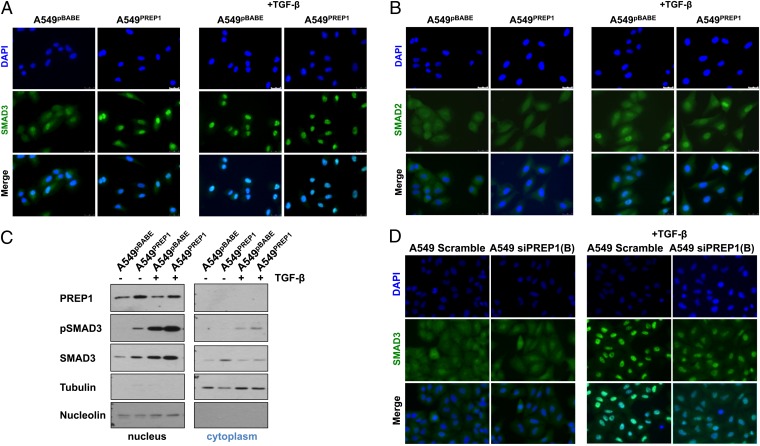
Nuclear localization of regulatory SMADs (R-SMADs) requires TGFBR1-mediated C-terminal phosphorylation and formation of heteromeric complexes with SMAD4. We analyzed the subcellular localization of SMAD2 and SMAD3 in untreated and TGF-β–induced PREP1 cells overexpressing A549. Unlike the homogeneous staining in the control cell line, A549PREP1 cells exhibited predominantly nuclear SMAD3 localization. Strikingly, the intensity of the nuclear SMAD3 signal in uninduced A549PREP1 cells was comparable to that of nuclear SMAD3 after induction with TGF-β in control cells (Fig. 5A). The effect was specific for SMAD3, because the SMAD2 signal remained uniformly distributed in the cytoplasm of both A549pBABE and A549PREP1 cells. As expected, TGF-β treatment induced the nuclear localization of SMAD2 in both control and PREP1-overexpressing cell lines (Fig. 5B).
Fig. 5.
PREP1 induces the nuclear accumulation of SMAD3. (A and B) Immunofluorescence analysis of SMAD3 (A) and SMAD2 (B) in A549PREP1 and A549pBABE cells. SMAD3 was stained with FITC-conjugated antibodies. Cell nuclei were visualized by DAPI staining. TGF-β (5 ng/mL) induction was for 1 h. (C) Western blotting analysis of PREP1, phospho-Smad3 (pSMAD3; Ser423/425), and total Smad3 in nuclear (Left) and cytoplasmic (Right) extracts of A549PREP1 and A549pBABE cells treated with TGF-β (5 ng/mL for 1 h) or not treated. Equal loading and nucleo–cytoplasmic fractionation were monitored by anti-nucleolin and anti-tubulin antibodies. (D) Immunofluorescence analysis of Smad3 in A549 cells transfected with the negative control (Scramble) or PREP1-specific siRNA [si-PREP1(B)] and treated for 1 h with TGF-β (5 ng/mL).
Immunoblotting analysis confirmed that the nuclear fraction of unstimulated A549PREP1 cells contained phospho-SMAD3, which was proportionally increased in the nuclei of TGF-β–induced A549PREP1 cells as compared with control cells. The antibody against total SMAD3 gave similar results, showing that SMAD3 was more abundant in nuclei from cells overexpressing PREP1 (Fig. 5C), in agreement with the increase observed in unfractionated cell extracts (Fig. 4B).
Next, we tested whether inhibition of PREP1 could affect the nuclear accumulation of SMAD3 in response to the cytokine. Immunostaining showed a strong decrease in the TGF-β–induced SMAD3 nuclear signal in cells treated with the PREP1-targeting siRNA as compared with the nontargeting control siRNA (Fig. 5D). Finally, we investigated whether the PREP1-dependent control of SMAD3 could affect the induction of known SMAD-regulated transcripts, such as plasminogen activator inhibitor 1 (PAI-1) (encoded by SERPINE1), representing a well-characterized target of the TGF-β–SMAD pathway (21). Both basal and TGF-β–induced PAI-1 mRNA levels were increased about 2.5-fold in A549PREP1 cells as compared with control A549pBABE cells. On the other hand, the PREP1-specific siRNA-B, but not the control siRNA, decreased (from eightfold to 2.5-fold) the induction of PAI-1 mRNA in A549 cells treated with TGF-β (SI Appendix, Fig. S6). Therefore the level of PREP1 expression affects the TGF-β–dependent transactivation of SMAD-responsive genes in A549 cells. The increase in PAI-1 agrees with the induction of uPA and increased motility, because the uPA–PAI-1 complex is required for cell migration (22).
Both SMAD3 and FRA-1 Are Required for PREP1-Dependent EMT, Cell Motility, and Induction of ECM-Degrading Proteases.
After nuclear translocation, complexes formed by R-SMADs (SMAD2 and SMAD3) and the common mediator SMAD (SMAD4) interact with their target promoters in cooperation with a variety of transcription factors, including members of the AP-1 complex. The results shown in Fig. 2F suggest that FRA-1 might represent a major heterodimeric partner of c-JUN and JUNB in cells overexpressing PREP1. In addition, SMAD2/3–FRA-1 complexes have been strongly implicated in the TGF-β–induced invasion program (23). Therefore, we investigated the cooperation between SMAD3 and FRA-1 in A549PREP1 cells.
Down-regulation of either SMAD3 or FRA-1 resulted in a similar restoration of intercellular adhesions and actin cytoskeleton in A549PREP1 cells (SI Appendix, Fig. S7 A and B). However, SMAD3 and FRA-1 did not affect each other's expression at either the RNA or protein level (SI Appendix, Fig. S7 C and D). PLAU and MMP2 transcripts were decreased significantly in response to both SMAD3 and FRA-1 knockdown, whereas E-cadherin mRNA and protein expression was up-regulated in response to SMAD3 down-regulation but was slightly down-regulated by FRA-1 inhibition in A549PREP1 cells (SI Appendix, Fig. S7 C–E).
Motility of A549PREP1 cells was decreased in response to SMAD3 or FRA-1 inhibition, whereas the SMAD3 and FRA-1 double knockdown resulted in a significantly stronger inhibition of cell migration (SI Appendix, Fig. S7F). Interestingly, we also observed that the SMAD3/FRA-1 double knockdown resulted in a further decrease in the expression of MMP2 and PLAU as compared with the individual down-regulation of the two transcription factors (SI Appendix, Fig. S7G).
Therefore, both SMAD3 and FRA-1 are implicated in the changes in cell morphology and motility triggered by PREP1 in A549 cells. Although SMAD3 and FRA-1 cooperate for transcriptional induction of PLAU and MMP2 in response to PREP1, SMAD3, but not FRA-1, is involved in the PREP1-dependent down-regulation of E-cadherin in A549 cells.
PBX1 Cooperates with PREP1 for the Induction of EMT in A549 Cells.
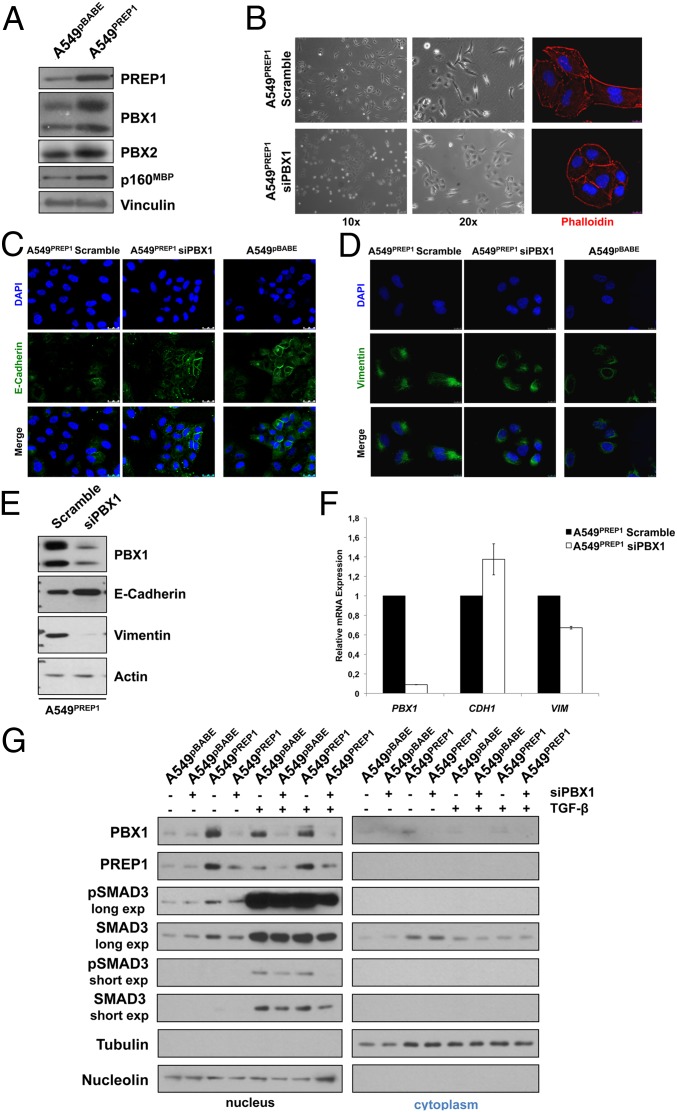
Because Prep1 interaction stabilizes both Pbx2 (24) and Pbx1 (25), we asked if the ectopic PREP1 affected the PBX proteins in A549 cells. Both PBX family members, along with another previously reported PREP1 interactor, Myb-binding-protein 1a (MBP1a) (26), were accumulated in A549PREP1 cells as compared with A549pBABE control cells (Fig. 6A). Because of the strong accumulation of PBX1 in response to the ectopic PREP1, we decided to analyze the effect of PBX1 inhibition in A549PREP1 cells. We observed that, after PBX1 expression was knocked down, A549PREP1 cells reverted toward the epithelioid morphology (Fig. 6B). Restoration of E-cadherin pericellular staining and intercellular contacts, along with modifications of vimentin microfilaments, rendered the PBX1-knockdown A549PREP1 cells more similar to the parental A549 cell line (Fig. 6 C and D). Accordingly, the increased E-cadherin and strongly decreased vimentin at both the protein and mRNA levels (Fig. 6 E and F) indicated that the decrease in PBX1 induced mesenchymal-to-epithelial transition in A549PREP1 cells.
Fig. 6.
PBX1 expression is required for the PREP1-mediated induction of EMT. (A) Western blotting analysis of PREP1, PBX1, PBX2, p160MBP, and vinculin in A549PREP1 and A549pBABE cells. (B) Bright-field (Left and Center) and immunofluorescence (Right) microscopy of A549PREP1 transfected with the PBX1 or control (Scramble) siRNAs. The actin cytoskeleton was visualized by rhodamine-conjugated phalloidin. Cell nuclei were visualized by DAPI staining. (C and D) Immunofluorescence analysis of E-cadherin (C) and vimentin (D) in A549PREP1 cells transfected with PBX1-specific or control (Scramble) siRNAs and in A549pBABE cells. Both proteins were detected by indirect FITC staining. Cell nuclei were visualized by DAPI. (E) Immunoblotting analysis of PBX1, E-cadherin, and vimentin in A549PREP1 cells transfected with PBX1-specific or control (Scramble) siRNA. β-Actin was used to normalize for equal loading. (F) qRT-PCR analysis of PBX1, E-cadherin (CDH1), and vimentin (VIM) mRNAs, in the same samples of shown in SI Appendix, Fig. S6E. Gapdh was used as internal control. Error bars represent the mean ± SD of three independent experiments. (G) Western blotting analysis of PBX1, PREP1, phospho-Smad3 (pSMAD3; Ser423/425), and total Smad3 in nuclear (Left) and. cytoplasmic (Right) extracts of A549PREP1 and A549pBABE cells treated with TGF-β (5 ng/mL for 1 h) after the PBX1-specific or control-siRNA transfection. Equal loading and nucleo–cytoplasmic fractionation were monitored by anti-nucleolin and anti-tubulin antibodies.
To understand whether, like PREP1, PBX1 affects the activity of SMAD3, we then analyzed the effect of PBX1 knockdown on SMAD3 intracellular distribution. Western blot analysis of SMAD3 in fractionated cell extracts showed that nuclear SMAD3 accumulation in response to PREP1 overexpression and/or TGF-β treatment was decreased when PBX1 expression was silenced (compare lanes 3 vs. 4, 5 vs. 6, and 7 vs. 8 in Fig. 6G). As expected, the levels of phospho-SMAD3 paralleled the levels of SMAD3 accumulation in the nuclear fraction. Although PREP1 expression was undetectable in the cytoplasm in both A549pBABE and A549PREP1 cells, nuclear PREP1 accumulation was inhibited by PBX1 knockdown, thus suggesting that the effect on SMAD3 resulted from the down-regulation of both PREP1 and PBX1. As suggested by the effect also detected on the ectopically expressed PREP1, PBX1 controlled PREP1 at the posttranslational level. Immunoblotting results (Fig. 6G) were confirmed by immunofluorescence microscopy, showing that the nuclear signal of both basal and TGF-β–induced SMAD3 was reduced when PBX1 was silenced in A549PREP1 cells (SI Appendix, Fig. S8).
In summary, the data presented here show that PBX1 cooperates with PREP1 in inducing EMT by supporting the nuclear accumulation of SMAD3 in response to TGF-β.
Aberrant expression of embryonic EMT-inducing transcription factors frequently is associated with EMT and metastasis. Therefore, we investigated the expression of multiple EMT inducers (SNAI1, SNAI2, ZEB1, and ZEB2) in response to TGF-β induction or ectopic PREP1. Although SNAIL (encoded by SNAI1) was strongly up-regulated in parental A549 cells in response to the cytokine treatment (SI Appendix, Fig. S9A), we did not detect significant up-regulation for any of the EMT inducers in A549PREP1 cells as compared with A549pBABE cells (SI Appendix, Fig. S9B). These results indicated that the PREP1-mediated EMT does not require the transcriptional induction of E-cadherin repressors.
PREP1 Overexpression Promotes Lung Metastases in Mice.
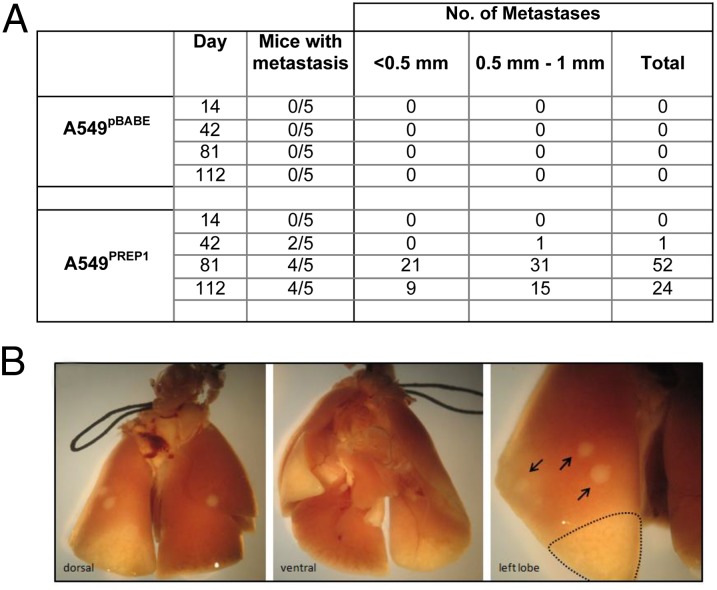
The mesenchymal transformation, along with the increased motility and invasiveness of A549PREP1 cells, prompted us to analyze the effects of PREP1 overexpression in an in vivo metastasis assay. Starting at 6 wk after tail-vein injection, metastatic nodules could be detected in the lungs of 10 of the 15 mice injected with A549PREP1 cells, whereas no nodules were found in any of the 15 control mice injected with the A549pBABE cells (Fig. 7 A and B). Therefore, we conclude that PREP1 is able to increase the metastatic potential of lung adenocarcinoma cells.
Fig. 7.
PREP1 overexpression promotes the formation of lung metastases in immunocompromised mice. Lungs were harvested at various time points after the injection of A549PREP1 or A549pBABE in the dorsal caudal vein of two groups of immunocompromised mice. (A) Counts of visible lesions present on the external surface of the lungs from mice (n = 20 mice per group) harvested at the four indicated time points after tail-vein injection. (B) Representative image of lung nodules formed by A549PREP1 cells on day 112 after tail-vein injection. Several white circles (arrows) with an elevated appearance typical of early metastases were found on all lobes of the lungs. Two necrotic areas are visible on the lower part of left lobe and on the right middle lobe.
PREP1 Negatively Correlates with Survival in NSCLC Patients and Is Overexpressed in Brain Metastases.
The discrepancy between the PREP1-dependent inhibition of primary tumor growth (SI Appendix, Fig. S1) and the PREP1-dependent increase of metastases (Fig. 7) in immunocompromised mice called our attention to the possible prognostic correlations of PREP1.
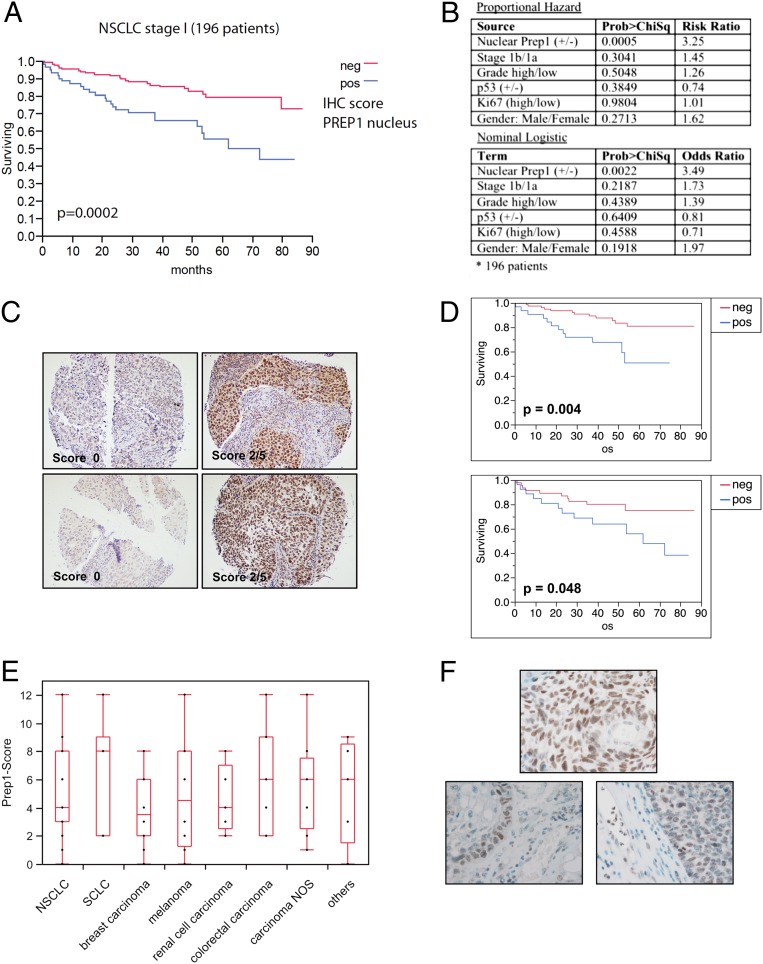
In the previously analyzed cohort of 445 patients representing all four stages of NSCLC, the presence or absence of PREP1 did not correlate with increased/decreased survival (8). However, when the analysis was focused on the patients with stage 1 NSCLC (n = 196), Kaplan–Meier curves showed that nuclear PREP1 expression clearly correlated with survival rates. Patients were divided in two groups on the basis of the PREP1 immunohistochemistry (IHC) score (Fig. 8A). The high-PREP1 group was associated with a statistically significant (P = 0.0002) decrease in overall survival time, with a 3.5-fold higher risk of death. Other parameters (such as sex, tumor stage and grade, p53 expression, and Ki67-positivity) did not change the risk or odds ratios appreciably or were of minimal statistical significance (Fig. 8B). When patients were grouped on the basis of tumor histology, the negative prognostic correlation was retained, with higher statistical significance in lung adenocarcinomas than in squamous cell carcinomas (Fig. 8 C and D). Next, we asked if the negative prognostic correlation corresponded with the expression of Prep1 in metastatic lesions. We analyzed 104 brain metastases from different kinds of human cancers, including 33 NSCLC. Among the analyzed metastases, only three of the 104 specimens were PREP1−. The box plot in Fig. 8E shows that essentially all metastases expressed PREP1; overall expression was strongest in small cell lung carcinoma, lowest in breast cancer, and intermediate in NSCLC and others. Interestingly, several NSCLC brain metastases stained strongly positive at the tumor–stroma interface, whereas the fibrovascular stroma stained negative, except for the strongly PREP+ leukocytes (Fig. 8F). In the cohort of 196 patients with stage I NSCLC (Fig. 8A), about 50% of the specimens had no or very low PREP1 expression; in the overall cohort of patients with lung cancer (n = 454) this frequency reached 70% (8). On this basis, the presence of PREP1 in all metastatic lesions indicates a very significant enrichment despite the relatively small number of available specimens.
Fig. 8.
PREP1 correlates negatively with survival in patients who have NSCLC and is expressed in brain metastases of lung cancer. (A) Kaplan–Meier survival curve for 196 patients with stage 1 NSCLC who were categorized into two groups on the basis of the nuclear PREP1 IHC score. Blue trace, positive (≥1); red trace, negative (<1). (B) Multivariate analysis of PREP1 expression in patients with stage I NSCLC. The risk ratio (proportional hazard) or odds ratio (nominal logistic) were calculated with respect to the following parameters: ± nuclear PREP1 expression; stage (1b/1a); grade (high/low); ± p53 wild-type or mutated status; Ki67 (high/low); sex (male/female). (C) Representative IHC images of PREP1− (Left) and PREP1+ (Right) NSCLC tumor specimens. (Upper Row) Adenocarcinoma. (Lower Row) Squamous cell carcinoma. (D) Kaplan–Meier survival curve for the subsets of NSCLC patients affected by adenocarcinoma (Upper) or squamous cell carcinoma (Lower). (E) Summary of PREP1 expression in 104 brain metastasis from different kinds of tumors, as indicated. NOS, not otherwise specified. Scores were calculated by multiplying the PREP1-staining intensity by the frequency of positive cells (SI Appendix, SI Experimental Procedures). (F) Representative IHC images showing PREP1 expression in brain metastasis. (Upper) Strongly PREP1+. (Lower Left) Squamous cell carcinoma metastasis exhibiting strong reactivity at the tumor-stroma border. (Lower Right) Note the PREP1-negative fibrovascular stroma, whereas leukocytes show focally strong reactivity. Of 104 metastases, 101 exhibited various degrees of PREP1 immunoreactivity, from low-level staining in a few tumor cells to high uniform expression across the entire metastasis or a salt-and-pepper pattern of high- and low-expressing tumor cells.
Discussion
In this study we show that the tumor suppressor PREP1 also has properties that favor tumor malignancy.
Although the effect of PREP1 on the growth of primary tumors (SI Appendix, Fig. S1) agrees with our previous findings (8, 27, 28), the proinvasive and prometastatic functions of PREP1 are novel. PREP1 induces the EMT hallmarks—down-regulation of E-cadherin and up-regulation of vimentin—along with cytoskeleton remodeling, increased motility, and invasiveness in lung adenocarcinoma cells. Mesenchymal changes are associated with the increased lung colonization by cells overexpressing PREP1. The high frequency of PREP1+ brain metastases of NSCLC, as opposed to the high level of PREP1− primary NSCLC, agrees with this overall interpretation. Indeed, PREP1 represents a negative prognostic factor in patients who have stage 1 NSCLC.
Mechanistically, our data indicate that PREP1 represents a novel modulator of cell responsiveness to TGF-β: PREP1 is necessary for the induction of the EMT in response to TGF-β, which, in turn, positively controls PREP1 accumulation.
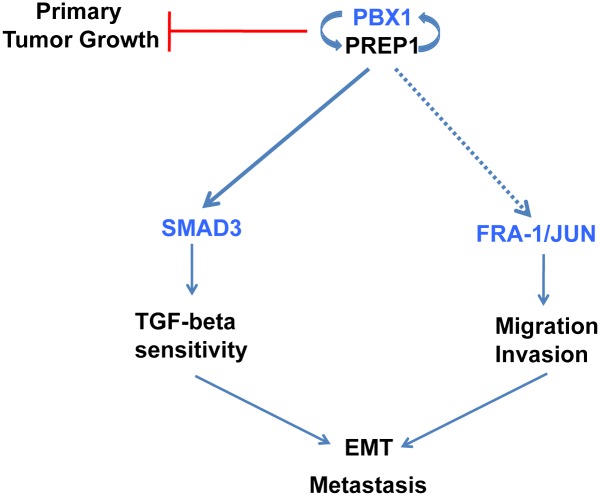
The effects of PREP1 on EMT and motility require the PREP1-mediated induction of three transcription factors: PBX1, SMAD3, and FRA-1 (diagrammed in Fig. 9). The PREP1 and PBX1 proteins are mutually required for their nuclear localization. Because of the lack of a nuclear localization signal, PREP1 is unable to enter the nucleus and bind DNA in absence of its PBX-family heteromeric partners. Dimerization with PREP1, in turn, protects PBX from nuclear export (29) and increases the half-life of both PBX1 and PBX2 (24, 25). Recent genome-wide analysis of mouse Prep1- and Pbx1-binding sites shows that most Prep1-binding sites are also occupied by Pbx1 or Pbx2 (30). Therefore PREP1 up-regulation might be functionally irrelevant in the absence of a concomitant increase of PBX.
Fig. 9.
Scheme of the regulatory interactions connecting PREP1 and PBX1 expression with the TGF-β–SMAD3 pathway, cellular invasion, and EMT.
PBX1 is likely to assist PREP1 in modulating the responses to TGF-β in lung adenocarcinoma cells. Remarkably, analysis of Pbx1−/− long-term hematopoietic stem cells showed that a significant fraction of Pbx1-regulated genes was associated with the TGF-β pathway (31).
Although preventing PBX1 accumulation in A549 cells overexpressing PREP1inhibits EMT induction (Fig. 6), we cannot rule out the possibility that other PBX proteins also collaborate with PREP1 in lung tumor progression. Interestingly, based on the IHC analysis of a large number of patients with NSCLC, a negative prognostic role has been proposed for PBX2 (32).
SMAD3, an important metastasis-related gene and a key transducer of TGF-β signaling, is a direct target of PREP1 (Fig. 4 and SI Appendix, Figs. S3–S5). PREP1-PBX dimers can bind the promoter-proximal (+1 kb) (30) decanucleotide (TGATTGACAG) in a region exhibiting histone marks of transcriptional enhancers and identified by ChIP-seq in A549 cells as a PBX3 DNA-binding site (33) within the highly conserved, large (∼100 kb) SMAD3 first intron (SI Appendix, Fig. S3). Although the down-regulation of PBX1 in A549PREP1 cells has a mild effect on the SMAD3 protein level, the amount of phosphorylated SMAD3 is strongly decreased under those conditions. Therefore, rather than having a direct transcriptional effect, a signaling mechanism seems operative in the PBX1–down-regulated cells. Indeed, the activity of SMAD proteins is modulated by multiple mechanisms, including nucleo–cytoplasmic shuttling, phosphorylation by nonreceptor kinases, ubiquitylation, and feedback control by inhibitory SMADs (reviewed in ref. (17), in addition to the transcriptional regulation.
Our results show that PREP1-PBX dimers modulate the cellular responses to TGF-β. The effect of PREP1 appears specific for SMAD3, because SMAD2 expression was not affected. Interestingly, SMAD3, but not SMAD2, expression is required for the induction of EMT in A549 cells (34).
Another possible link between PREP1 and SMADs is based on the DNA-independent interactions between PREP1 and PBX1 with both SMAD2 and SMAD3 (35). However, because we observe the PREP1-dependent nuclear accumulation of SMAD3, but not SMAD2, these interactions do not appear to be implicated in the PREP1-dependent control of SMADs in A549 cells.
In addition to PBX1/2 (Fig. 6) and SMAD3 (Fig. 4), PREP1 controls the expression of several AP-1 family transcription factors, including FRA-1, c-JUN, and JUNB, which are strongly increased in response to ectopic PREP1, possibly as a consequence of the PREP1-induced ERK1/2 activity (Fig. 2). Although the exact link between PREP1 and the induction of AP-1 components needs to be elucidated, a relationship between PREP/PBX transcription factors and AP-1–dependent transactivation was established previously. Indeed, the PREP1-binding element was shown to be required for the FRA-1/AP-1–dependent induction of uPA transcription (36). The cooperation between PREP1 and AP-1 might involve their functional interaction at regulatory elements of other proinvasive genes, in addition to uPA (37).
FRA-1 represents a key regulator of tumor cell proliferation, motility, and invasion (38). In A549 cells, FRA-1 overexpression induces in vitro invasion and anchorage-independent growth, along with in vivo metastasis (39). More recently, FRA-1 has been implicated in the control of EMT in response to the RAS–MEK–ERK pathway in multiple tumor cell systems (40–43). Interactions between SMAD3/4 and AP-1 components are well-characterized examples of cooperation between transcription factors (44). Indeed, interactions between SMAD3, FRA-1, c-JUN, and JUNB are essential for the TGF-β–induced binding of SMAD3 to several AP-1–regulated genes critically involved in breast cancer cell invasion (23). Our data show that the FRA-1–SMAD3 cooperation triggers mesenchymal changes and activation of proinvasive genes in response to PREP1, whereas FRA-1 seems to be dispensable for the PREP1-dependent inhibition of E-cadherin in A549 cells.
In contrast with the transcriptional induction of SMAD3, PREP1 did not affect the expression of any of the EMT-inducing transcription factors (SI Appendix, Fig. S9), thus suggesting possible posttranscriptional mechanisms. Given the previously demonstrated role of the SNAIL1–SMAD3/4 corepressor complex in downregulating multiple epithelial promoters (E-cadherin, occludin, and claudin-3) (45), we envisage that the accumulation and nuclear translocation of SMAD3/4 results in biochemical and functional interaction with promoter-bound SNAIL1 and consequent inhibition of epithelial hallmarks in response to the ectopic PREP1.
Cancer-associated EMT results from the hijacking of embryonic programs involved in gastrulation and critical steps of morphogenesis, including cardiac formation and secondary palate fusion in response to TGF-β signaling (46, 47). Although Prep1-dependent roles in mouse embryonic craniofacial development have not been characterized, Pbx1 has been shown to regulate fusion of the frontonasal processes in the mouse embryo (48). In addition, given the role of TGF-β–induced EMT in inflammatory diseases resulting in organ (such as lung and kidney) fibrosis (49), it will be important to understand the possible roles of Prep1 and Pbx1 in mouse models of fibrosis.
The invasive and metastatic phenotype triggered by PREP1 overexpression in lung adenocarcinoma cells is at odds with the tumor-suppressive role of PREP1 (8, 28). However, the novel function of PREP1 as regulator of the TGF-β pathway can explain the apparent discrepancy. In line with its tumor-suppressor activity, PREP1 overexpression induces a decrease in A549 cell viability associated with increased apoptosis. The stronger growth inhibition observed in tumor xenografts as compared with cultured cells suggests that paracrine factors might contribute to the effect of PREP1 on A549 cell viability (SI Appendix, Fig. S1). These data indicate that the oncosuppressor activity of PREP1 is exerted also in lung tumor cells, in agreement with our previous data in a system of MYC-induced lymphomagenesis (8, 28).
Because the EMT at the invasive front of carcinomas depends on tumor–stroma paracrine interactions involving TGF-β secretion by tumor-infiltrating inflammatory cells, PREP1 overexpression might be induced by TGF-β at the tumor–stroma interface (Fig. 8F).
The significant enrichment of PREP1 expression in brain metastases (which are all PREP1+, whereas 50% or more of all primary lung tumors are PREP1−) supports a causal role for PREP1 in the metastatic spread of human lung cancer. These findings harbor important prognostic relevance in a cancer in which prognostic markers are badly needed. The positive staining of PREP1 in brain metastasis derived from several kinds of human cancer suggests that PREP1 might be an EMT inducer in different cellular contexts.
In this respect, a recently reported 10-gene prognostic signature resulting from microarray-based expression profiling of noninvolved tissue in 282 patients with NSCLC includes PREP1/PKNOX1 as a negative prognosticator (with a hazard ratio of 6.5; P value 1.5E-04) (50). This finding might be evidence of a PREP1-dependent (perhaps TGF-β–mediated) stroma–tumor interaction, which would be important to investigate in further detail.
We speculate that the worse prognosis associated with PREP1 expression in patients with stage 1 NSCLC (Fig. 8) might result from the increased metastatic potential of PREP1+ lung tumors, despite the inhibitory effect of PREP1 on primary tumor growth suggested by our present (SI Appendix, Fig. S1) and previous (8, 27, 28) findings.
Materials and Methods
Detailed information on in vitro experimental procedures and reagents is provided in SI Appendix, Experimental Procedures.
Human Samples and IHC.
General tissue microarrays were specifically designed with normal and malignant tumor tissues and prepared as described with minor modifications (8). IHC analysis was performed on NSCLC cases having undergone surgical resection at the European Institute of Oncology (IEO) from June, 1998 to December, 2002. Written informed consent was obtained from all patients, and ethical approval was given by the IEO Institutional Review Board.
Statistical Analysis.
Kaplan–Meier survival curves and univariate–multivariate analyses were done using JMP-IN 5.1 software (SAS). For Kaplan–Meier curves, P values were computed using a log-rank test. For univariate and multivariate analyses, nominal logistic regression and Wald χ2 tests were used.
Supplementary Material
Acknowledgments
We thank L. Selleri (Weill Cornell Medical College) for personal communication on Pbx1 mouse models. We also thank the Institute of Genetics and Biophysics (IGB) Microscopy Facility and I. Iaccarino (IGB) for critical discussions. This work was supported Associazione Italiana per la Ricerca sul Cancro Grant IG 10489, Association for International Cancer Research (AICR) Grant 08-0182, and Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) (MERIT Grants RBNE08NKH7 and RBNE08YFN3) (to P.V.); Associazione Italiana per la Ricerca sul Cancro (AIRC) Grant IG12829 and Ministero della Sanità Grant 2008-1228056 (to F. Blasi); and Cariplo.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407074111/-/DCSupplemental.
References
- 1.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17(5):1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Diaz LC, et al. The absence of Prep1 causes p53-dependent apoptosis of mouse pluripotent epiblast cells. Development. 2010;137(20):3393–3403. doi: 10.1242/dev.050567. [DOI] [PubMed] [Google Scholar]
- 3.Ferretti E, et al. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol. 2006;26(15):5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Micali N, et al. Down syndrome fibroblasts and mouse Prep1-overexpressing cells display increased sensitivity to genotoxic stress. Nucleic Acids Res. 2010;38(11):3595–3604. doi: 10.1093/nar/gkq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60(4):547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 6.Nourse J, et al. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60(4):535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 7.Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: Potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98(23):13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longobardi E, et al. Prep1 (pKnox1)-deficiency leads to spontaneous tumor development in mice and accelerates EmuMyc lymphomagenesis: A tumor suppressor role for Prep1. Mol Oncol. 2010;4(2):126–134. doi: 10.1016/j.molonc.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iotti G, et al. Homeodomain transcription factor and tumor suppressor Prep1 is required to maintain genomic stability. Proc Natl Acad Sci USA. 2011;108(29):E314–E322. doi: 10.1073/pnas.1105216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massagué J. TGFbeta in Cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heldin C-H, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollier BG, Evans K, Mani SA. The epithelial-to-mesenchymal transition and cancer stem cells: A coalition against cancer therapies. J Mammary Gland Biol Neoplasia. 2009;14(1):29–43. doi: 10.1007/s10911-009-9110-3. [DOI] [PubMed] [Google Scholar]
- 14.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 15.Keski-Oja J, Blasi F, Leof EB, Moses HL. Regulation of the synthesis and activity of urokinase plasminogen activator in A549 human lung carcinoma cells by transforming growth factor-beta. J Cell Biol. 1988;106(2):451–459. doi: 10.1083/jcb.106.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiercinska E, et al. The TGF-β/Smad pathway induces breast cancer cell invasion through the up-regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Res Treat. 2011;128(3):657–666. doi: 10.1007/s10549-010-1147-x. [DOI] [PubMed] [Google Scholar]
- 17.Heldin CH, Moustakas A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012;347(1):21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 18.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 19.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26(1):1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 20.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua X, Miller ZA, Wu G, Shi Y, Lodish HF. Specificity in transforming growth factor beta-induced transcription of the plasminogen activator inhibitor-1 gene: Interactions of promoter DNA, transcription factor muE3, and Smad proteins. Proc Natl Acad Sci USA. 1999;96(23):13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conese M, Blasi F. Urokinase/urokinase receptor system: Internalization/degradation of urokinase-serpin complexes: Mechanism and regulation. Biol Chem Hoppe Seyler. 1995;376(3):143–155. [PubMed] [Google Scholar]
- 23.Sundqvist A, et al. Specific interactions between Smad proteins and AP-1 components determine TGFβ-induced breast cancer cell invasion. Oncogene. 2013;32(31):3606–3615. doi: 10.1038/onc.2012.370. [DOI] [PubMed] [Google Scholar]
- 24.Longobardi E, Blasi F. Overexpression of PREP-1 in F9 teratocarcinoma cells leads to a functionally relevant increase of PBX-2 by preventing its degradation. J Biol Chem. 2003;278(40):39235–39241. doi: 10.1074/jbc.M304704200. [DOI] [PubMed] [Google Scholar]
- 25.Oriente F, et al. Prep1 controls insulin glucoregulatory function in liver by transcriptional targeting of SHP1 tyrosine phosphatase. Diabetes. 2011;60(1):138–147. doi: 10.2337/db10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Oriente F, et al. Prep1 deficiency induces protection from diabetes and increased insulin sensitivity through a p160-mediated mechanism. Mol Cell Biol. 2008;28(18):5634–5645. doi: 10.1128/MCB.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dardaei L, Longobardi E, Blasi F. Prep1 and Meis1 competition for Pbx1 binding regulates protein stability and tumorigenesis. Proc Natl Acad Sci USA. 2014;111(10):E896–E905. doi: 10.1073/pnas.1321200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iotti G, et al. Reduction of Prep1 levels affects differentiation of normal and malignant B cells and accelerates Myc driven lymphomagenesis. PLoS ONE. 2012;7(10):e48353. doi: 10.1371/journal.pone.0048353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13(8):946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penkov D, et al. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Reports. 2013;3(4):1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2(5):484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu Y, et al. Prognostic significance of pre B cell leukemia transcription factor 2 (PBX2) expression in non-small cell lung carcinoma. Cancer Sci. 2009;100(7):1198–1209. doi: 10.1111/j.1349-7006.2009.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium EP, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reka AK, et al. Peroxisome proliferator-activated receptor-gamma activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol Cancer Ther. 2010;9(12):3221–3232. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey JS, Rave-Harel N, McGillivray SM, Coss D, Mellon PL. Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol Endocrinol. 2004;18(5):1158–1170. doi: 10.1210/me.2003-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nerlov C, et al. A regulatory element that mediates co-operation between a PEA3-AP-1 element and an AP-1 site is required for phorbol ester induction of urokinase enhancer activity in HepG2 hepatoma cells. EMBO J. 1992;11(12):4573–4582. doi: 10.1002/j.1460-2075.1992.tb05559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berthelsen J, Vandekerkhove J, Blasi F. Purification and characterization of UEF3, a novel factor involved in the regulation of the urokinase and other AP-1 controlled promoters. J Biol Chem. 1996;271(7):3822–3830. doi: 10.1074/jbc.271.7.3822. [DOI] [PubMed] [Google Scholar]
- 38.Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering AP-1 function in tumorigenesis: Fraternizing on target promoters. Cell Cycle. 2007;6(21):2633–2639. doi: 10.4161/cc.6.21.4850. [DOI] [PubMed] [Google Scholar]
- 39.Adiseshaiah P, Lindner DJ, Kalvakolanu DV, Reddy SP. FRA-1 proto-oncogene induces lung epithelial cell invasion and anchorage-independent growth in vitro, but is insufficient to promote tumor growth in vivo. Cancer Res. 2007;67(13):6204–6211. doi: 10.1158/0008-5472.CAN-06-4687. [DOI] [PubMed] [Google Scholar]
- 40.Caramel J, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24(4):466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 41.Desmet CJ, et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci USA. 2013;110(13):5139–5144. doi: 10.1073/pnas.1222085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doehn U, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol Cell. 2009;35(4):511–522. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38(1):114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394(6696):909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 45.Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11(8):943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Ferretti E, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21(4):627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galvan A, et al. Gene expression signature of non-involved lung tissue associated with survival in lung adenocarcinoma patients. Carcinogenesis. 2013;34(12):2767–2773. doi: 10.1093/carcin/bgt294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.