Significance
Protein-mediated membrane fusion is essential for eukaryotic cell biology. In several major human viral pathogens the full closure of a six-helix bundle (6HB) fusion core structure is considered imperative for the opening of a productive fusion pore. We tested this paradigm by engineering covalently restricted paramyxovirus fusion proteins that are incapable of full 6HB zippering. These proteins remained fusion competent without disruption of the restricting bonds, although fusion activity was reduced. Recombinant virions harboring the engineered fusion proteins confirmed efficient, 6HB closure-independent paramyxovirus entry. We demonstrate that full 6HB zippering does not govern paramyxovirus entry. Our results may extend to other viruses using related fusiogenic glycoproteins and affect the design of antiparamyxovirus strategies targeting virus entry.
Keywords: membrane fusion, measles virus
Abstract
Enveloped viruses such as HIV and members of the paramyxovirus family use metastable, proteinaceous fusion machineries to merge the viral envelope with cellular membranes for infection. A hallmark of the fusogenic glycoproteins of these pathogens is refolding into a thermodynamically highly stable fusion core structure composed of six antiparallel α-helices, and this structure is considered instrumental for pore opening and/or enlargement. Using a paramyxovirus fusion (F) protein, we tested this paradigm by engineering covalently restricted F proteins that are predicted to be unable to close the six-helix bundle core structure fully. Several candidate bonds formed efficiently, resulting in F trimers and higher-order complexes containing covalently linked dimers. The engineered F complexes were incorporated into recombinant virions efficiently and were capable of refolding into a postfusion conformation without temporary or permanent disruption of the disulfide bonds. They efficiently formed fusion pores based on virus replication and quantitative cell-to-cell and virus-to-cell fusion assays. Complementation of these F mutants with a monomeric, fusion-inactive F variant enriched the F oligomers for heterotrimers containing a single disulfide bond, without affecting fusion complementation profiles compared with standard F protein. Our demonstration that complete closure of the fusion core does not drive paramyxovirus entry may aid the design of strategies for inhibiting virus entry.
Membrane fusion is an essential process in eukaryotic cell biology. Examples range from fusion of lipid vesicles to sustain intracellular transport along secretory and endocytotic pathways to cell fusion in fertilization and development and to the fusion of viral with cellular membranes resulting in viral infection. Because there are several high-energy barriers (1), lipid membranes do not merge spontaneously under physiological conditions and therefore require the aid of auxiliary proteins to induce membrane stress and lower the activation energy for lipid merger. For vesicular transport, for instance, soluble N-ethylmaleimide–sensitive factor-activating protein receptor (SNARE) proteins resident in both the target and donor membrane mediate the process (2). Two fundamental differences distinguish cellular fusion machineries from viral fusogenic membrane glycoproteins (vFMGs), which mediate the entry of enveloped viruses by the fusion of the envelope with cellular membranes: (i) vFMGs do not require adapter proteins but insert a fusion peptide domain into the target membrane; and (ii) vFMGs fold into metastable prefusion conformations, obviating the dependence on external energy sources. Three distinct types of viral fusion (F) proteins have been classified: class I vFMGs are found, among others, on influenza virus, retroviruses, and paramyxoviruses; class II proteins are present on alpha- and flaviviruses; and class III proteins mediate herpes- and rhabdovirus entry (reviewed in ref. 3).
Once the fusion process is initiated, class I and II vFMGs undergo irreversible conformational rearrangements that result in refolding into thermodynamically highly stable postfusion structures. The relaxation of free energy during the transition from the pre- to the postfusion system is thought to drive membrane merger. All class I vFMGs form homotrimers, but the prefusion structures of different class representatives vary widely depending on the virus family (4–8). The stimuli for refolding also are diverse, ranging from receptor binding by an associated attachment protein to direct interaction with a receptor and/or exposure to low pH. For example, the postfusion conformations of HIV and paramyxovirus F proteins contain a characteristic fusion core structure composed of a bundle of six antiparallel α-helical heptad repeat (HR) domains known as the “six-helix bundle” (6HB). Each monomer of these F proteins features two HR domains. One, designated “HR-A or HR-1,” is located in close proximity to the fusion peptide [in the case of HIV gp41, HR-1 is separated through a 13-residue fusion peptide proximal region from the fusion peptide (9); in the case of paramyxovirus F, HR-A is directly linked to the fusion peptide]. The other, designated “HR-B or HR-2,” is in close proximity to the transmembrane domain [in the case of gp41, HR-2 is separated through an 18-residue membrane proximal external region from the transmembrane domain (9); in the case of paramyxovirus F, HR-B is separated through a seven- to nine-residue region from the transmembrane domain (10)]. In the final 6HB fusion core, the fusion peptides and transmembrane domains, and thus the target and donor membranes, are brought into immediate proximity.
Consequently, the current entry paradigm for these viruses assumes that completion of 6HB assembly is mandatory for successful infection. In a lipid-centric model of the fusion process, the approaching fusion peptide and transmembrane domains are thought to enable pore opening by introducing extreme local membrane curvature, resulting in disarray of the outer lipid monolayers followed by spontaneous merger (1, 11, 12). An alternative model assumes that the fusion peptide and transmembrane domains may assemble directly into an antiparallel bundle that extends the water-soluble 6HB core into the lipid phase, functioning as a fusion pinprick that expands into a fusion pore (13). Importantly, each of these models proposes a mechanistic link between full 6HB assembly and fusion pore opening, and, accordingly, several classes of virus-entry inhibitors, including peptides and small-molecule drug candidates, are thought to interfere with 6HB formation (14).
Therefore it was surprising that mechanistic assessment of the HIV-1 entry machinery using HR-domain–derived inhibitory peptides suggested pore opening before 6HB assembly (15–17). However, these studies also found that complete 6HB closure is required for pore stabilization and enlargement and thus for productive pore formation supporting viral entry. Recognizing that an understanding of the mechanistic basis of membrane fusion is important for the design of therapeutic strategies targeting virus entry, we explored the role of 6HB assembly in paramyxovirus entry using the measles virus (MeV) fusion machinery as an example.
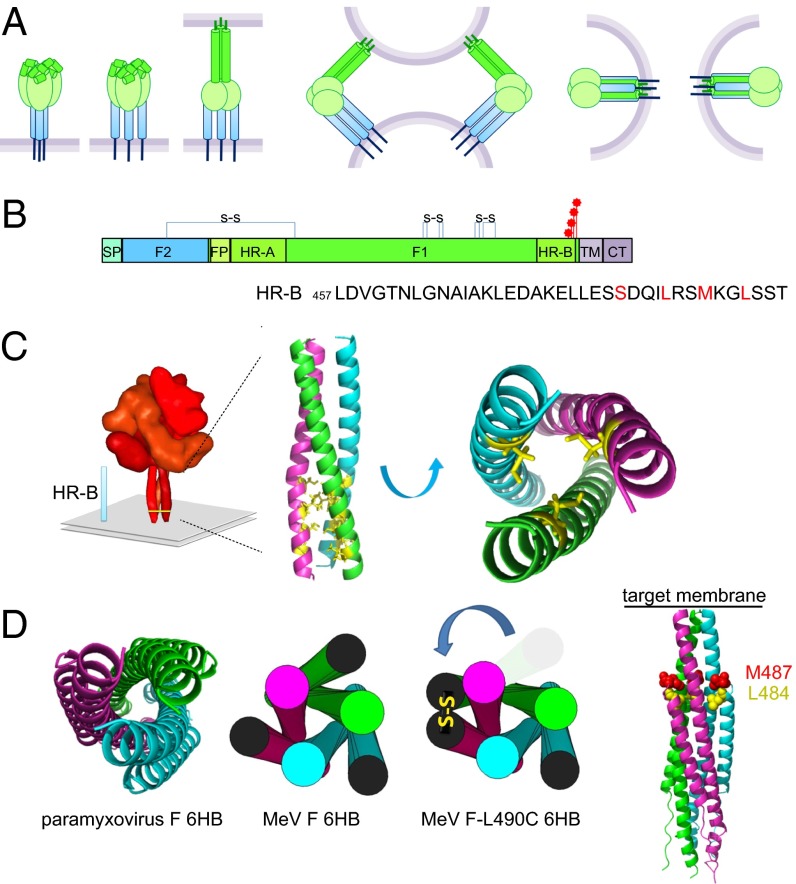
High-resolution structures of several paramyxovirus F proteins have been solved in both the prefusion (8, 18) and postfusion (19–21) conformations. In the prefusion form, the HR domain adjacent to the fusion peptide, HR-A, is broken up into multiple distinct sections forming the upper part of the prefusion F-protein head structure, and the membrane-proximal HR-B domains assemble into a short, three-helix bundle, the prefusion F-protein stalk. At least two intermediate structures are thought to guide F-protein refolding. First, the trimeric HR-B stalk dissolves, forming an open-stalk intermediate (22). Assembly of the HR-A triple helix from components of the prefusion F-protein head section then drives the fusion peptide toward the target membrane, leading to an extended prehairpin intermediate that engages the target membrane and the viral envelope simultaneously (8, 19). Hairpin formation and packing of the individual HR-B domains, which remain N-terminally connected to the F-protein ectodomain through extended chains, onto the central HR-A triple helix leads to formation of the 6HB fusion core (Fig. 1A).
Fig. 1.
Engineered cysteine bonds linking two F-protein HR-B domain monomers are predicted to prevent complete 6HB assembly. (A) Model of F protein-mediated formation of the fusion pore. (Left) Upon F-protein triggering, the HR-A triple-helix core assembles, driving the fusion peptide toward the target membrane. (Center) Hairpin formation folds the HR-B domains onto the HR-A bundle. (Right) Ultimately, this folding brings the transmembrane domains and fusion peptides into close proximity. (B) Schematic of the MeV F protein. CT, cytoplasmic tail; FP, fusion peptide; SP, signal peptide; TM, transmembrane domain. Intramonomer disulfide bonds are shown, and the cysteine substitutions are indicated by red stars. (C) Model of the prefusion MeV F protein. The HR-B region is indicated and has been enlarged (Insert) to highlight the residues that were mutated along the hydrophobic core. [MeV residues were threaded on the PIV5 structure, Protein Data Bank (PDB) ID 2B9B]. (D, Left) MeV F residues threaded on the PIV5 6HB structure (PDB ID 1SVF). (Center) Cartoon version. (Right) Model of an FL490C fusion core. One HR-B disulfide bond is shown.
To test the validity of the 6HB paradigm experimentally, we engineered covalent bonds into the MeV F-protein 6HB domains that are predicted to be incompatible with the assembly of a closed 6HB core based on the available structural information. The engineered bonds formed with high efficiency, resulting in the assembly of F-protein trimers and higher-order F-protein oligomers composed of covalently linked F-protein dimers. Having established quantitative assays to monitor the kinetics of productive cell-to-cell and virus-to-cell fusion, we examined expression assays in transit and after infection with recombinant virus to determine whether paramyxovirus entry is physically and/or thermodynamically linked to full 6HB assembly. The results of these studies advance the mechanistic understanding of paramyxovirus F protein-mediated infection, illuminate the fundamental prerequisites for productive formation of a fusion pore, and have major implications for the design and desirable drug profile of paramyxovirus-entry inhibitors. The insight gained from these experiments may extend to other viral pathogens that use class I F proteins for cell entry.
Results
To explore the feasibility of introducing covalent bonds into the prefusion HR-B domain stalk, we introduced individual cysteine substitutions at the four most membrane-proximal hydrophobic positions (L490C, M487C, L484C, and S480C) of the MeV F-protein HR-B helix (Fig. 1 B and C). Structural models of prefusion MeV F protein posit the sulfur atoms of the cysteine side chains in adjacent F-protein monomers are ∼6.1 Å apart, indicating that bond formation, although structurally conceivable in principle, requires some distortion of the prefusion stalk (Fig. S1). The remaining third thiol group may be buried in the HR-B helix interface, form a covalent bond with an adjacent F-protein trimer in a dual open-stalk configuration, or attract cellular disulfide isomerases, resulting in intracellular retention (23). Importantly, the presence of a disulfide bond between two HR-B domains is predicted to be irreconcilable with full 6HB zippering in the postfusion conformation (Fig. 1D), but prehairpin formation and the subsequent initiation of F-protein refolding still are conceivable. This approach is fundamentally distinct from the introduction of three intermonomer disulfide bonds resulting in the formation of covalently restricted prefusion F-protein trimers as previously demonstrated for MeV and parainfluenza virus 5 (PIV5) F proteins (10, 24).
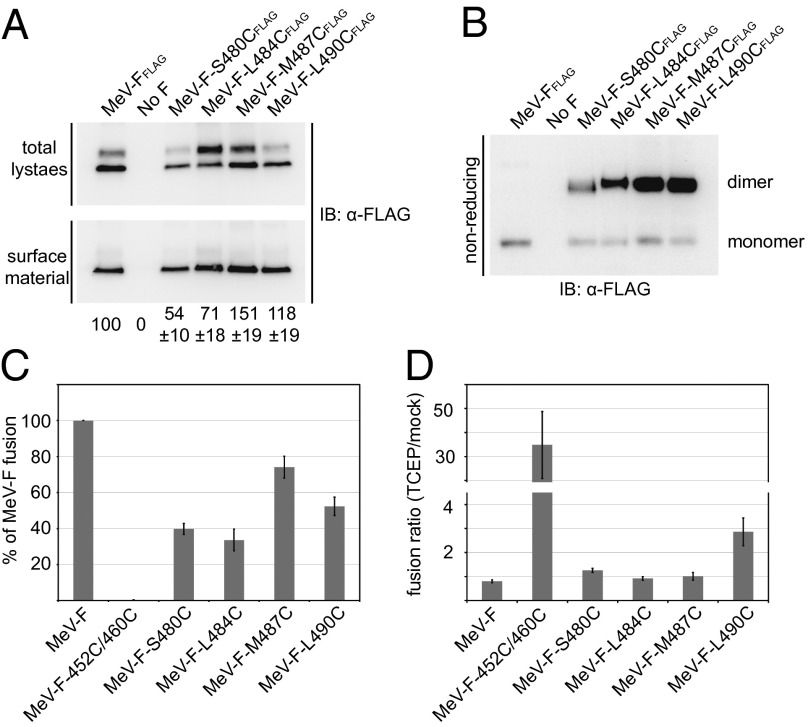
First, to assess the F-protein mutants’ competence in intracellular transport and their covalent bonding status, we transiently expressed each of the four candidates individually and subjected the transfected cells to surface biotinylation and plasma-membrane protein extraction, respectively. Although the Fcysteine constructs L490C, M487C, and L484C were surface expressed efficiently (>70% of levels observed for standard F protein), F-S480C showed an ∼50% reduction in intracellular transport competence (Fig. 2A). Gel fractionation of plasma membrane protein extracts of Fcysteine-expressing cells under nonreducing conditions demonstrated that the majority of the F-protein material is engaged in covalently linked dimers (Fig. 2B).
Fig. 2.
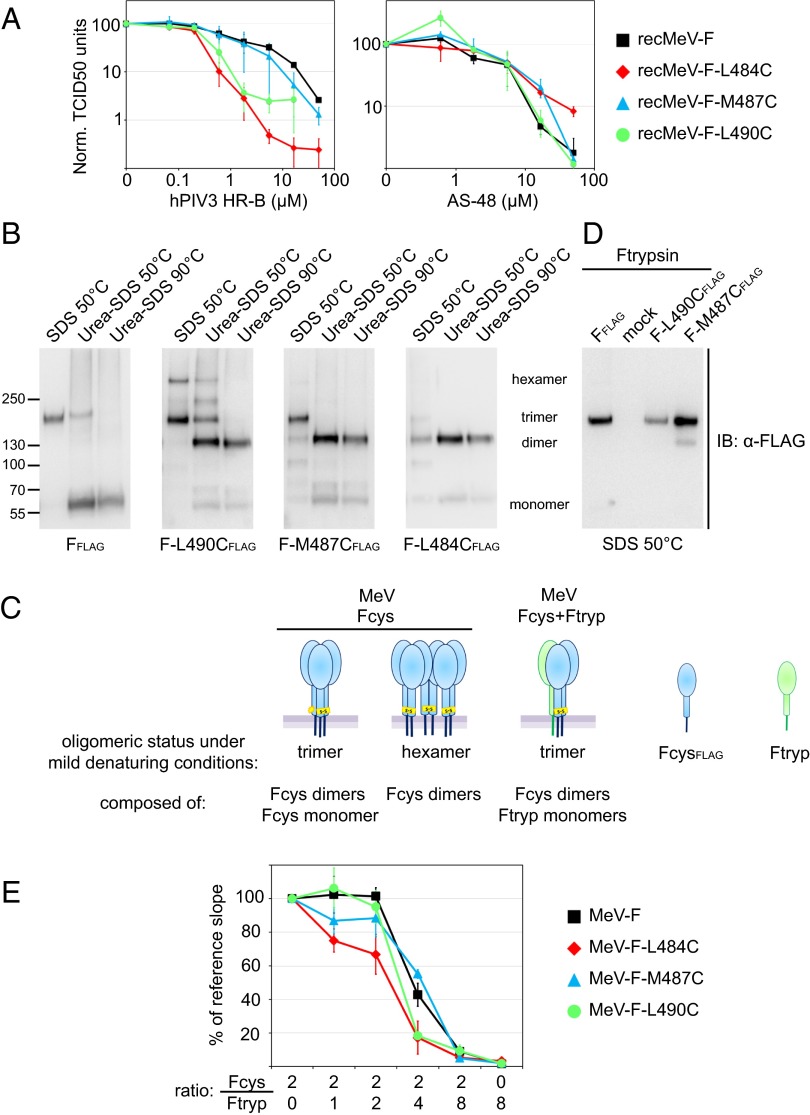
Mutant F proteins with disulfide bonds in the HR-B region predicted to prevent 6HB closure remain partially fusion active. (A) Surface expression of transiently expressed Fcysteine mutants. Densitometric quantitations show averages of three independent experiments ± SEM. (B) Intermonomer disulfide bond status of the Fcysteine mutants. Digitonin plasma membrane protein extracts were fractionated under nonreducing conditions, followed by immunoblotting using the α-FLAG antiserum. (C) Quantitative cell-to-cell fusion reporter assay. Fcysteine mutants were coexpressed with MeV H. Data shown are averages of at least four experiments ± SEM. (D) Ratios of Fcysteine fusion activities determined in the presence or absence of exogenous TCEP. Data shown are averages of at least four experiments ± SEM.
Bioactivity of F Mutants with Covalent Bonds in the HR-B Domain.
Quantitation of Fcysteine bioactivity in a luciferase reporter-based endpoint cell-to-cell fusion assay revealed fusion activities of the intracellular transport-competent mutants ranging from ∼35–75% that of the standard F protein (Fig. 2C). Only the FL490C construct showed a significant increase in bioactivity when extracellular disulfide bonds were partially reduced through limited treatment of cell monolayers with Tris(2-carboxyethyl) phosphine (TCEP), a membrane-impermeable mild reducing agent (Fig. 2D). Inclusion of an F452C/460C double mutant and nonreducing gel electrophoresis (Fig. S2) confirmed that TCEP treatment was effective. Previously, we have demonstrated that the F452C/460C mutant forms covalently linked F trimers that are permanently restricted in a prefusion conformation and require partial disulfide bond reduction to regain activity (24).
These data indicate that, of the HR-B disulfide bond candidates examined, only the intermonomer link involving HR-B residue 490 partially impairs productive F-protein refolding.
Recovery of MeV Recombinants with Disulfide Bonds in F-Protein HR-B.
A body of evidence suggests that distinct syncytia are formed by transiently expressed viral glycoproteins and viral entry (25–28). To test the bioactivity of the Fcysteines in the biologically relevant context of intact virions, we concentrated on the three most membrane-proximal constructs, L490C, M487C, and L484C, which were transported to the cell surface most efficiently, and individually replaced the F-protein–encoding ORF with the ORF of these mutants in a cDNA copy of the MeV genome. The corresponding recombinant MeV (recMeV) mutants were recovered successfully, but all showed a delayed cytopathic effect compared with unmodified recMeV (Fig. 3A). Presumably because of the resulting later breakdown of infected cell monolayers, progeny titers of standard recMeV declined more rapidly than those of the mutants in multiple-step virus growth curves (Fig. 3B). However, replication kinetics and peak titers of cell-associated and released virions of recMeV-FL490C and recMeV-FM487C revealed no significant differences from standard recMeV. Also, cell-associated progeny titers of recMeV-FL484C were affected only mildly, but yields of cell-free recMeV-FL484C were reduced by ∼90%.
Fig. 3.
recMeV incorporate covalently linked Fcysteine complexes and display growth kinetics similar to the unmodified parental strain. (A) Syncytia formation phenotype of the different Fcysteine-expressing viruses. Microphotographs were taken 36 h postinfection at a magnification of 200×. (B) Multiple-step viral growth curves of the Fcysteine recombinants. Cell-associated and released progeny viral titers are shown. Values shown are averages of three experiments ± SEM. (C) Cell-free and cell-associated viral particles contain disulfide-linked F dimers. Extracts of gradient-purified viral particle preparations were subjected to gel fractionation under nonreducing and reducing conditions, followed by immunoblotting (IB) using the α-F antiserum. (D) Protein composition and relative infectivity of the Fcysteine recombinants. Particles were normalized for equal relative amounts of F protein, and the incorporation of the viral attachment (H)-, phospho- (P), and matrix (M) proteins was assessed through immunoblotting (Left) and densitometric analysis relative to standard recMeV (Right). Mean values of three independent experiments ± SEM are shown. The absolute TCID50 units corresponding to the normalized F value are indicated.
To examine the oligomeric state of the Fcysteine proteins incorporated into viral particles, we purified cell-associated and released virions and fractionated viral proteins by SDS/PAGE under reducing and nonreducing conditions (Fig. 3C) or gradient ultra-centrifugation (Fig. S3). Resembling our initial results with transiently expressed Fcysteines, both approaches revealed that the Fcysteine complexes incorporated into recombinant virions are composed predominantly of covalently linked F dimers. However, as is consistent with the reduction in progeny titers of released particles, we could not purify enough cell-free recMeV-FM484C particles for protein detection in immunoblots. Therefore, only cell-free recMeV-FM487C and recMeV-FL490C virions could be subjected to a full protein-composition analysis (Fig. 3D). Both recombinants showed a remarkably conserved ratio of protein content to number of infectious particles, emphasizing that the engineered disulfide bonds in the F-protein complexes do not interfere with virus assembly and infectivity.
Virus-to-Cell Kinetics of the F-Modified MeV Recombinants.
Because the overall success of virus replication constitutes only an indirect readout for estimating the affect of the engineered disulfide bonds on the rate of virus entry, we established a quantitative entry assay to determine the true kinetics of virus-to-cell fusion. Two cell lines, each stably expressing complementary half-fragments of an eGFP-renilla luciferase F protein (29), were cocultured, and then virions were spin-inoculated onto the mixed-cell monolayer in the cold. A shift to physiological temperature allowed simultaneous fusion of viral particles with two adjacent cells (fusion-from-without), restoring bioactive eGFP-luciferase chimeras through cell-content mixing.
Using a live-cell luciferase substrate, we implemented this strategy to monitor the virus-to-cell fusion kinetics of all three Fcysteine recombinants in near real-time after cells were inoculated with equivalent amounts of infectious particles (Fig. 4A). In contrast to the nearly equivalent virus growth rates observed previously, only recMeV-FM487C showed a virus-to-cell fusion rate similar to that of standard recMeV in this assay. The infection kinetics of recMeV-FL490C were reduced by ∼50%, and recMeV-FL484C was incapable of inducing measurable fusion-from-without events despite robust replication of cell-associated virions in the multiple-step growth assays. These results emphasize that virus-to-cell fusion rates cannot be calculated reliably from overall replication success and further underscore our original observation that engineered disulfide bonds are less well tolerated when positioned closer to the center of HR-B and away from the transmembrane domain.
Fig. 4.
HR-B disulfide bonds remain intact during F-protein refolding into the postfusion conformation. (A) Kinetic virus-entry assay based on a fusion-from-without content-mixing assessment. The linear range of the fusion kinetics of a representative experiment is shown. (B) Immunoprecipitation of intact (Left) or lysed (Right) cell-free viral particles using specific antibodies that differentiate between the pre- (α-F pre) and postfusion (α-F trig) conformations of the F protein or antibodies directed against the FLAG epitope (M2) for specificity control (α-FLAG). Immunoblots were developed using the conformation-independent α-Ftail antiserum. (C) Cell-surface immunoprecipitation (SIP) of triggered F complexes using α-F trig antibodies. Precipitates were fractioned through nonreducing SDS/PAGE, and immunoblots were developed using the α-Ftail antiserum. Samples marked (+) were subjected to an extended incubation step at 37 °C after FIP washout. (D) Virus-to-cell fusion-from-without fusion assay in the presence of broad-spectrum PDI-inhibiting (α-PDI) antibodies (RL90), MeV-neutralizing (α-MeV H) antibodies (B5), or the small-molecule MeV entry blocker AS-48. The maximal entry rates (slopes of the linear range of the entry curves) are shown; data shown are the averages of at least four experiments ± SEM.
Effect of the Engineered Bonds on the Conformational Stability of Prefusion F Protein.
To assess the conformational state of the F proteins incorporated into released recMeV-FM487C and recMeV-FL490C virions, we used conformation-dependent anti-MeV F antibodies that differentiate between the prefusion and triggered forms of the complex (30, 31). In the absence of deliberate F-protein triggering through exposure to receptor or physical stimuli, a standard preparation of parental, unmodified recMeV particles contained a mixture of F-protein complexes in pre- and postfusion conformation (Fig. 4B, Left). Chemical lysis of the virions before immunoprecipitation unraveled the prefusion conformation, leaving only postfusion complexes detectable (Fig. 4B, Right). Remarkably, the disulfide bonds between F-protein positions 487 or 490 had opposite effects on the conformational stability of the prefusion complexes; although a higher proportion of FM487C than standard F protein was present in a postfusion conformation in intact virion preparations, the majority of FL490C was found in the prefusion form (Fig. 4B). This phenotype suggests that a disulfide bond at F-protein position 487 has a destabilizing effect on the prefusion conformation of the complex, whereas a link at position 490 has a stabilizing effect. The latter was confirmed when particle lysates were analyzed, because the FL490C complexes retained some reactivity with the prefusion F-specific antibodies even after membrane extraction.
F-Protein Refolding Does Not Require Temporary Opening of the Engineered Disulfide Bonds.
The abundance of postfusion complexes in recMeV-FM487C and recMeV-FL490C lysates combined with the predominant migration of Fcysteine extracts from purified virions as covalently linked dimers supported the notion that F-protein refolding indeed may proceed without disruption of the engineered HR-B disulfide bonds. To test this hypothesis directly, we followed a two-pronged experimental strategy. First, we determined the intermonomer covalent state of membrane-integral postfusion F complexes through surface immunoprecipitation of cells in syncytia with the postfusion F-specific antibodies, followed by gel fractionation under nonreducing conditions (Fig. 4C). Precipitated Fcysteine complexes were composed predominantly of covalently linked dimers, indicating that the engineered intermonomer disulfide bonds are present in the postfusion conformation.
Recent studies have implicated cellular protein disulfide isomerases (PDIs), which indiscriminately assist protein folding in the endoplasmic reticulum, in the specific downstream regulation of some proteins at the cell surface (32–34). To test whether refolding of the Fcysteine complexes mandates temporary opening of the engineered disulfide bonds through plasma membrane-resident PDIs, we next performed virus-to-cell fusion assays in the presence of broadly anti-PDI reactive neutralizing antibodies (35) (Fig. 4D). In parallel, we assessed the effect of the broad-spectrum PDI family inhibitor 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) on cell-to-cell fusion induced by transiently expressed mutant F proteins (Fig. S4). Neither PDI-blocking strategy reduced fusion activity significantly. In contrast, neutralizing anti-MeV antibodies and the small-molecule MeV fusion inhibitor AS-48 that was developed previously by our group (36) were highly effective in preventing MeV entry. These results indicate that the Fcysteines do not require temporary, PDI-mediated opening of the HR-B disulfide bonds to complete the refolding process.
Conformational Stability of the Postfusion Fcysteine Complexes.
The mechanistic impact of these findings depends on the validity of the modeling predictions, which suggest that the engineered covalent bonds affect the assembly of stable postfusion 6HB cores. To test this hypothesis experimentally, we determined the sensitivity of the recMeV-Fcysteines to inhibition by HR-B domain–derived synthetic peptides and, in parallel, developed a biochemical assay to monitor the stability of the paramyxovirus fusion core.
Because exogenous HR-B peptides are thought to block the entry of class I viruses by competing with the endogenous HR-B domains for binding to the HR-A triple-helix grooves (14), we hypothesized that impaired 6HB assembly should give the exogenous blocker a competitive advantage over the endogenous HR-B domains, opening a broader window of opportunity for more efficient inhibition. Dose–response curves of the peptide inhibitor generated for the Fcysteine and standard recMeVs revealed that the mutant virions were more sensitive to the blocker than the parent virus (Fig. 5A and Table S1). In comparison, the potency of the small-molecule MeV-entry inhibitor AS-48, which stabilizes a prefusion conformation of the F trimer before 6HB assembly (37), was unchanged, confirming the specificity of the assay. The enhanced potency of exogenous HR-B peptides against the Fcysteine viruses compared with standard recMeV is consistent with impaired fusion core assembly kinetics.
Fig. 5.
Fcysteine/trypsin heterotrimers are capable of opening productive fusion pores. (A) Dose–response curves of HR-B domain-derived inhibitory peptide against standard recMeV and the recMeV-Fcysteines. Progeny titers were determined by TCID50 titration after virus incubation in the presence of increasing peptide concentrations. Values shown are the averages of four experiments ± SEM. (B) Postfusion F complexes were natively extracted from the plasma membrane and exposed to increasingly stringent denaturing conditions. Samples were fractionated through nonreducing Tris-acetate-SDS/PAGE, and immunoblots were developed using specific antibodies (α-FLAG) directed against the FLAG epitope tags. The migration pattern of F monomers, dimers, trimers, and hexamers is indicated. (C) Schematic of the F transcomplementation set-up through the coexpression of Fcysteine mutants (Fcys) with bio-inactive Ftrypsin (Ftryp). Fcysteine mutants are shown in blue, and Ftrypsin mutants are shown in green; oligomer predictions are for the Fcysteine constructs only. (D) Cotransfection of cells with equal amounts of plasmid DNA encoding the specified Fcysteines and Ftrypsin. Native F complexes were extracted and analyzed under mild denaturing conditions as in B. (E) Transcomplementation fusion profiles of the Fcysteines or standard F protein with Ftrypsin. For each F construct and plasmid ratio, maximal fusion rates were determined in kinetic cell-to-cell fusion assays and normalized to the reference rate of the construct (fusion in the absence of Ftrypsin). Values shown are averages of four independent experiment sets ± SEM.
To assess the postfusion composition and stability of the three Fcysteine mutants biochemically, we heat-triggered F-protein refolding in situ and then digitonin-extracted plasma membrane proteins, subjected the extracts to increasingly stringent denaturing conditions, and determined the oligomeric state of the F complexes through nonreducing gel fractionation and immunoblotting. Standard F complexes subjected to this assay remained assembled in homotrimers at mild conditions but disintegrated rapidly into individual monomers when denaturing stringency was increased (Fig. 5B). Of note, transiently expressed prefusion F trimers are highly unstable and disintegrate into individual monomers even under mild denaturing conditions (Fig. S5), giving supporting evidence that this assay predominantly monitors the conformational stability of the 6HB fusion core.
Because we introduced single cysteine substitutions in each F monomer, we anticipated Fcysteine trimers would be composed of covalently linked homodimers and a monomer with unbonded HR-B cysteine or, alternatively, would form higher-order oligomers to balance the cysteine stoichiometry. When we applied the fusion core stability assay to the three Fcysteine mutants, each revealed a distinct oligomer composition and stability profile: FL490C assembled into extraction-resistant trimers and hexamers, the latter presumably composed of covalently linked dimers; FM487C predominantly formed trimers in situ, albeit of lower stability than standard F trimers; and FL484C complexes mostly disintegrated into covalent dimers and monomers even under mild extraction conditions (Fig. 5B). With increasing denaturing stringency, FL490C likewise dissolved into dimer and free-monomer populations. However, densitometric quantitation of the fully disintegrated samples revealed an ∼3.7- to 5.6-fold excess of the covalent dimeric over the monomeric material for all Fcysteine mutants (Table S2).
Taken together, these results confirm that positioning the disulfide bonds closer toward the center of HR-B increasingly reduces the conformational stability of the postfusion structure of the F protein. Because F-protein dimers alone are not intracellular transport competent, the dimer:monomer divergence from an hypothetical 2:1 ratio suggests that some higher-order complexes, which are composed of covalent dimers and could not be resolved in the gel, also are present in FL484C and FM487C mutants.
Enriching Fcysteine Trimers Through Transcomplementation.
To explore whether a population of mutant F-protein trimers harboring a single HR-B disulfide bond can be enriched in situ through F-protein transcomplementation (Fig. 5C), we coexpressed the three Fcysteine mutants with Ftrypsin, a previously reported F-protein construct in which the furin protease maturation site is replaced by a trypsin cleavage site (38). Although fully folding and intracellular transport competent, Ftrypsin gains fusion activity only after proteolytic activation by exogenously added trypsin. Coimmunoprecipitation of HA-tagged Ftrypsin with FLAG-tagged standard F protein or the Fcysteine variants confirmed that F heterooligomers formed readily in cells cotransfected with equal amounts of the different F-encoding plasmids (Fig. S6). When we applied the F fusion core stability assay to the cotransfected cells, only F trimers were detectable under mildly denaturing conditions (Fig. 5D). These results indicate that the higher-order Fcysteine complexes originate from the formation of inter-F trimer disulfide bonds. Coexpression of Fcysteines with Ftrypsin apparently results in the assembly of F heterotrimers containing a single HR-B disulfide bond but lacking the third free cysteine in sufficient quantity to allow intertrimer disulfide bonding.
Fusion Profile of F Heterotrimers.
Efficient heterotrimer formation and the complete lack of Ftrypsin bioactivity in the absence of exogenous trypsin allowed the functional assessment of F trimers harboring a single HR-B domain disulfide bond through the generation of transcomplementation fusion profiles. We used a transient kinetic cell-to-cell fusion assay (39, 40) that is based on dual-split eGFP-luciferase chimeras to determine maximal fusion rates for a panel of different Ftrypsin:Fcysteine ratios for each of the Fcysteine mutants (Fig. S7). For comparison, we included an Ftrypsin:standard F-protein panel. Extraction of the maximal fusion kinetics from the – in first approximation – linear sections of the individual fusion curves revealed the complementation profiles of standard F protein and the Fcysteine mutants through plotting of the normalized maximal rates relative to the complementation ratios.
Remarkably, the profiles of both FM487C and FL490C showed no significant divergence from that of standard F protein (Fig. 5E); fusion rates dropped substantially only in the presence of a twofold excess of Ftrypsin in the transfected plasmid mixture. In comparison, the decline in the FL484C profile was slightly more pronounced when Ftrypsin-encoding DNA was titered into the transfection mixture. Again, however, fusion rates were reduced substantially only when Ftrypsin-encoding plasmids were present in a twofold relative excess.
Because the fusion core stability assay revealed that F heterotrimers form efficiently at plasmid parity in the transfection mix, these matching complementation profiles of the Fcysteine variants and standard F protein indicate that preventing 6HB closure by connecting two of the HR-B domains in defined Fcysteine/trypsin heterotrimers has no detrimental effect on the opening or enlargement of a fusion pore.
Discussion
In this study we tested the prevailing model that paramyxovirus F proteins depend on the closure of the fusion core for the opening and stabilization of a fusion pore. Rather unexpectedly, we demonstrate that efficient paramyxovirus F-protein–mediated viral entry can proceed independent of complete 6HB assembly. It was suggested that, after activation of paramyxovirus F-protein refolding and insertion of the fusion peptide into the target membrane, hairpin formation and the assembly of the 6HB fusion core structure pull the viral and target membranes together (1, 41, 42). This model applies the energy released through formation of the thermodynamically stable fusion core directly to local membrane bending, and the fusion peptides and transmembrane domains function merely as anchor points that are required to transmit elastic stress into the membranes.
However, several studies have alternatively suggested that fusion peptides and transmembrane domains have an active role in the fusion process. For example, self-assembly of paramyxovirus (43, 44), influenza virus (45, 46), and HIV (47) F-protein transmembrane domains was demonstrated experimentally and is considered a determinant for proper protein folding and fusion activity (44, 47). Studies with class I vFMGs have shown that insertion of synthetic, soluble fusion peptides into the membrane can deform the lipid bilayer (48) and induce membrane fusion (49). Paramyxovirus fusion peptides derived from the PIV5 F protein were demonstrated to associate into a helical transmembrane bundle in lipid bilayers; this bundle interacts tightly with the corresponding PIV5 F transmembrane domains and forms a thermodynamically favorable coiled-coil in the lipid phase (13). In analogy to cellular SNARE proteins (50), this zippering of the F-protein helix in the bilayer —equivalent to the formation of the four-helix bundle in the SNARE transmembrane domain—may extend the water-soluble 6HB fusion core into the membrane and contribute part of the energy required to drive fusion (13). Critical functional interactions between fusion peptides and transmembrane domains also were reported for influenza virus HA (45) and HIV-1 gp41 (47).
Our study demonstrates that mutant MeV F proteins remain capable of efficiently opening, stabilizing, and enlarging fusion pores suitable for viral nucleocapsid passage, despite the presence of a covalent disulfide bond between two of the HR-B domains. Based on the available paramyxovirus fusion core structures, these bonds are considered incompatible with complete 6HB assembly. Our data can be explained by a model of paramyxovirus fusion in which partial assembly of a distorted 6HB fusion core and/or formation of 4HB or 5HB core structures is sufficient for pore opening. Alternatively, the fusion peptides and transmembrane domains could engage actively in a fusion-promoting zipper in the bilayer phase, independent of full closure of the water-soluble 6HB core (Fig. 6). Three major lines of evidence support these conclusions:
-
i)
Biochemical evidence supports the in silico predictions that the engineered disulfide bonds prevent proper 6HB assembly. Molecular modeling reveals that the membrane-proximal sections of at least one, and possibly two, F-protein HR-B domains will be unable to nest into the grooves of the HR-A triple-helix core if connected by one of the engineered disulfide bonds. Furthermore, the engineered bonds are expected to reduce the kinetics of F-protein hairpin formation, if present in prefusion F complexes and kept intact during F refolding. Our fusion core stability and peptide inhibition assays substantiate these predictions by showing that the integrity of the postfusion trimers of the fusion-active Fcysteine mutants is impaired and that the Fcysteine mutants are highly sensitized to HR-B peptide inhibition. Because the structural integrity of the postfusion paramyxovirus F trimer and the susceptibility to peptide inhibition depend strongly on the stability of the 6HB fusion core and the kinetics of F-protein refolding, respectively, these results show that the disulfide bonds are present during F-protein refolding and prevent complete zippering of the stable fusion core.
-
ii)
The opening and stabilization of productive fusion pores did not require temporary or permanent separation of the engineered inter–HR-B disulfide bonds. This notion is supported by our observation that covalently linked F dimers were readily detectable in triggered F complexes and that the extent of Fcysteine-mediated cell-to-cell fusion and viral entry kinetics, respectively, was unaffected by two distinct inhibition strategies that reportedly block plasma membrane-resident cellular disulfide isomerases (35, 51).
-
iii)
Overlapping complementation profiles of standard F protein or the Fcysteines with Ftrypsin indicate that F heterotrimers remain equally fusion competent, independent of whether they contain an Fcysteine dimer or two standard F monomers in addition to an Ftrypsin monomer. The demonstrated suppression of Fcysteine-hexamer formation in the presence of a twofold relative excess of Ftrypsin provides tangible evidence that mixed heterotrimers form efficiently under these coexpression conditions. The analysis of the complementation profiles is based on the assumption that free mixing of the coexpressed F monomers occurs and that the inactive Ftrypsin monomer does not interfere with F heterotrimer surface expression or acts functionally in a dominant-negative fashion. The Ftrypsin mutant meets these conditions, based on our F-protein coimmunoprecipitation data and the previous observation that MeV Ftrypsin is fully reactivatable by the addition of exogenous trypsin protease (38). Furthermore, uncleaved F trimers derived from the related paramyxovirus PIV5 were shown to remain competent in refolding into a postfusion conformation (19, 52), confirming that no obvious structural constraints prevent the conformational rearrangement of Ftrypsin-containing trimers in a dominant-negative manner.
Fig. 6.
Cartoon of postfusion MeV F trimers in the presence of HR-B intermonomer disulfide bonds. Assembly of the membrane-distal portion of the fusion core could be sufficient for pore opening (Lower Left), or, alternatively, a fusion peptide/transmembrane domain bundle may form in the lipid bilayer in the absence of complete assembly of the water-soluble 6HB (Lower Right). FP, fusion peptide; TM, transmembrane domain.
In addition to preventing assembly of the final fusion core, the FL490C mutation affects the rate of F-protein refolding by stabilizing a prefusion F-protein conformation. In whole-virus immunoprecipitations, prefusion F trimers of recMeV-FL490C showed a markedly higher conformational stability than those derived from recMeV-FM487C and standard recMeV. This phenotype coincided with reduced entry kinetics of recMeV-FL490C, whereas virus-to-cell fusion rates of recMeV-FM487C matched those of recMeV. The TCEP-induced boost of FL490C-mediated fusion is consistent with the notion that this most membrane-proximal disulfide bond stabilizes the prefusion conformation of the trimer. In contrast to the other mutant recMeVs, fusion rates of recMeV-FL484C particles could not be measured in the fusion-from-without assay. This phenotype is unlikely to be caused by premature FL484C trimer refolding and virion self-inactivation, because infectivity of the virus inoculum remained unchanged upon re-titering. Rather, very fast virus-to-cell fusion rates of recMeV-FL484C or an insufficient amount of functional fusion complexes displayed on recMeV-FL484C particles may prevent simultaneous engagement of and fusion with multiple target cells.
Our MeV-based data show that paramyxovirus F-protein hairpin formation releases free energy considerably in excess the amount that appears to be thermodynamically required for membrane merger. Viral entry without mandatory full 6HB zippering directly affects appropriate design strategies for antiviral therapeutics directed against paramyxovirus pathogens. Several antiviral campaigns have targeted the paramyxovirus F protein and other class I vFMGs with peptidic and small-molecule inhibitors designed to prevent viral entry by competing with the endogenous HR-B domains for binding to the HR-A triple-helix core, preventing 6HB assembly (53–55). Our study demonstrates that, in the case of paramyxovirus F proteins, complete closure of the fusion core is not mandatory for the formation and stabilization of productive fusion pores. Consequently, HR-B–competitive paramyxovirus fusion blockers may need to populate two or all three HR-A triple-helix grooves simultaneously to inhibit viral entry. This requirement for near-saturation places higher demands on target affinity and binding constants than needed from inhibitors of class I vFMGs that depend on full 6HB assembly for productive membrane merger. The need to block at least two of the HR-A docking grooves at all times is particularly challenging to reconcile with the inherent high off-rates of small-molecule therapeutics (56), diminishing the clinical value of this drug class in inhibiting paramyxovirus entry and facilitating the development of kinetic resistance through secondary escape mutations that act by accelerating F refolding.
In the context of antiviral therapies, the question of whether our paramyxovirus F-protein–based findings that viral entry can proceed independent of 6HB closure extends to other class I FMGs becomes significant. The HR-A/HR-B interface of most paramyxovirus F proteins extends at least 39 Å in length and is thus longer than that of most other class I viral F proteins (57) (Table S3). Consequently, the surface area of the buried protein–protein interface of the paramyxovirus fusion core is large in comparison with retrovirus and filovirus 6HBs. This expansive interface may underlie the ability of paramyxovirus F to form productive fusion pores very efficiently, in some cases in our study without a noticeable penalty in viral entry rates, despite being deprived of the full thermodynamic gain of complete fusion core zippering. It will be informative to test this hypothesis in future studies by subjecting a set of class I vFMGs derived from nonparamyxovirus pathogens to analogous covalent restrictions of fusion-core assembly.
Materials and Methods
Cell Lines and Transfections.
Vero (African green monkey kidney epithelial) cells (CCL-81; ATCC) and Vero cells stably expressing human SLAM [Vero-SLAM cells (58)] were maintained in DMEM supplemented with 7.5% (vol/vol) FBS at 37 °C and 5% CO2. At every third passage, Vero-SLAM cells were exposed to G418 selection to maintain SLAM expression. GeneJuice (Millipore) was used for all transient transfection reactions in adherent cells. Stably transfected dual-split protein (DSP)1 and DSP2 cells expressing DSP1–7 or DSP8–11, respectively, are derived from the human glioma cell line NP-2/CD4 (29). DSP cell lines were maintained in MEM with 7.5% (vol/vol) FBS at 37 °C and 5% CO2.
Site-Directed Mutagenesis and Epitope Tagging.
Site-directed mutagenesis was performed following the QuikChange protocol (Stratagene) using pCG-F as template, which contains the F-protein ORF derived from the MeV Edmonston (MeV-Edm) strain under the control of a CMV promoter (59). All changes were confirmed by DNA sequencing.
Quantitative Cell-to-Cell Fusion Assays.
Cell-to-cell fusion was assessed using an established luciferase reporter assay as previously described (60). In fusion experiments containing TCEP, the TCEP was added at the time of target cell overlay. To quench the reducing activity of TCEP before the completion of the experiment, 0.5 mL of FBS was added to each well 1 h after overlay with fusion-inhibitory peptide (FIP) to prevent additional fusion events.
Virus Stocks and Recovery of Recombinant MeV.
Modified vaccinia virus Ankara-expressing T7 polymerase (61) was amplified in DF-1 (chicken embryo fibroblast) cells (CRL-12203; ATCC). MeV-Edm served as the genetic basis for all MeV variants generated in this study. To produce MeV stocks, Vero-SLAM cells were infected at a multiplicity of infection (MOI) of 0.001 median tissue culture infective dose (TCID50) per cell, and cell-associated particles were harvested when virus-induced cytopathicity reached ∼90% through two freeze/thaw cycles, followed by TCID50 titration of progeny virions on Vero-SLAM cells. All Fcysteine MeV recombinants were produced by replacing the standard F-Edm ORF with the mutant F-protein–encoding sequence in the p(+)MeV-NSe plasmid, which contains a full-length cDNA copy of the MeV-Edm genome (62). Recombinant MeVs were recovered as described previously (63) with the following modifications: BHK-T7 cells were cotransfected with a plasmid carrying a cDNA copy of the recombinant MeV genome and plasmids encoding MeV-IC-B strain-derived (64) polymerase (L), nucleocapsid (N), and phospho (P) proteins. All constructs were under the control of the T7 promoter. The transfected cells were overlaid on Vero-SLAM cells 48 h after transfection, and virions in distinct, emerging infectious centers were transferred to fresh Vero-SLAM cells. The integrity of the recombinant MeVs was confirmed by RT-PCR and DNA sequencing of the entire F-protein–coding regions, using SuperScript II reverse transcriptase (Life Technologies) and random hexamer primers for first-strand synthesis. No mutations other than the engineered cysteine substitutions were found.
Virus Purification.
To purify cell-free virions, culture supernatants of infected cells showing ∼90% virus-induced syncytia formation were filtered through a 0.45-µM filter and mixed with PEG8000 (Sigma–Aldrich) [10% (wt/vol) final concentration] and NaCl [2% (wt/vol) final concentration]. The PEG-precipitated virions were pelleted (8,000 × g for 90 min at 4 °C) and then were resuspended in TNE buffer [10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA]. Recovered material was layered on a 20–60% (vol/vol) sucrose cushion and centrifuged at 100,000 × g for 90 min, and viral particles were collected from the interface of the 20–60% sucrose layers. To purify cell-associated MeV particles, infected cell monolayers were scraped in OptiMem (Life Technologies) and then were subjected to douncing (30 cycles) and low-speed centrifugation (5,000 × g, 20 min, 4 °C). The cleared material was layered on a 20–60% sucrose cushion and centrifuged at 100,000 × g for 90 min, and viral particles were collected from the interface of the 20–60% sucrose layers.
Virus Growth Kinetics.
Vero-SLAM cells (2.5 × 105 per time point) were infected at an MOI of 0.01 TCID50 per cell. At the indicated times postinfection, cell-associated and cell-free viral particles were harvested, and titers were determined by TCID50 titration on Vero-SLAM cells.
Fusion-from-Without Kinetic Dual-Split Protein Cell-Content–Mixing Assay.
DSP1 and DSP2 cells were plated in equal amounts in black-walled 96-well plates in CO2-independent medium (Gibco). EnduRen (Promega) membrane-permeable luciferase substrate was added 1 h before infection. To synchronize the initiation of infection, cells were cooled to 4 °C; then virions were spin-inoculated (MOI 20 TCID50 per cell) onto the cells for 30 min at 800 × g at 4 °C. The baseline signal was measured in a TopCount Microplate Scintillation and Luminescence Counter (PerkinElmer), followed by warming the plates to 37 °C and recording luminescence intensities at 30-min intervals over an 8-h time period. Maximal viral entry rates were calculated from the slopes of the in first approximation linear areas of the resulting luciferase activity curves, in all cases spanning the first 5 h postinfection. Where indicated, inhibitor candidates [α-PDI monoclonal blocking antibodies (RL90; Pierce Antibodies) and α-MeV H monoclonal neutralizing antibodies (B5) (65) or AS-48 (36)] were added to the cell monolayers together with the EnduRen substrate.
Surface Biotinylation, Plasma Membrane Protein Extraction, and Immunoblotting.
Cell-surface biotinylation experiments were carried out as described (63). Briefly, washed F-protein–expressing cells were biotinylated with 0.5 mg/mL sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (Pierce) and were quenched. Then biotinylated proteins were precipitated using immobilized streptavidin (GE Healthcare) after cell lysis in RIPA buffer [1% sodium deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅Cl (pH 7.2), 10 mM EDTA, 50 mM sodium fluoride, protease inhibitors (Roche), 1 mM phenylmethylsulfonyl fluoride]. Because the high-affinity biotin:streptavidin interaction impairs recovery of precipitated proteins from the matrix under nonreducing conditions, plasma-membrane protein complexes were digitonin-extracted to analyze the status of the F-protein disulfide bond. Samples were prepared using cold Native Sample Buffer [100 mM Tris⋅Cl, 10% (vol/vol) glycerol, 0.0025% Bromophenol Blue (pH 8.6), 0.1% digitonin, with 25 mM iodoacetamide] and were cleared by centrifugation (20,000 × g, 15 min, 4 °C). All protein precipitates or extracts were fractioned through SDS/PAGE under reducing or nonreducing conditions, transferred to PVDF membranes, and visualized after antibody decoration in a ChemiDoc XRS digital imaging system (Bio-Rad). Antibodies used for immunodetection were polyclonal antisera directed against the cytosolic tails of the MeV F (59) or attachment (H) (66) proteins or monoclonal antibodies directed against the viral matrix (M) or P protein (both from Millipore), the FLAG epitope (M2; Sigma), or the HA epitope (16b12; Covance). For densitometric quantitations, relative signal intensities were determined using the ImageLab software package (Bio-Rad).
Immunoprecipitation.
Immunoprecipitations were performed on intact viral particles or after particle lysis with RIPA buffer containing 25 mM iodoacetamide as described (31). Monoclonal antibodies directed against the FLAG epitope (α-FLAG), the HA epitope (α-HA), or conformation-dependent epitopes present exclusively in pre- (α-F pre) or postfusion (α-F trig) F complexes (31, 67) were used for pull-down. Cellular surface immunoprecipitations were performed on infected Vero cell monolayers. Cells were infected with the indicated recMeVs (MOI 0.4 TCID50 per cell). FIP (Bachem) was added at a final concentration of 25 μM 1 h after infection. Thirty-six hours after infection, FIP was washed out to allow fusion, and cells were cooled rapidly on ice or were cooled after 1-h incubation at 37 °C. Anti-triggered F monoclonal antibodies (1:1,500) were bound to the cells on ice for 1 h; then cells were washed in PBS to remove excess antibodies. Cells were lysed in RIPA/iodoacetamide buffer, and immunoprecipitation was performed as previously described (31).
Peptide Inhibition Assay.
Vero-SLAM cells were infected with the specified recMeV strains at an MOI of 0.02 TCID50 per cell in the presence of a parainfluenza virus type 3-derived HR-B peptide (American Peptide), which also blocks MeV F proteins (68), or AS-48. Cell-associated virus titers were determined when cytopathicity in mock-treated wells reached ∼90%. Inhibitory concentrations were calculated through four-parameter variable slope regression modeling of the resulting dose–response curves using the Prism5 (GraphPad) software package.
Fusion Core Stability Assay.
F-protein–expressing cells were placed on ice or were subjected to a 10-min heat shock at 60 °C followed by cooling on ice. Plasma-membrane proteins were harvested with cold Native Sample Buffer [100 mM Tris⋅Cl, 10% (vol/vol) glycerol, 0.0025% Bromophenol Blue (pH 8.6), 0.1% digitonin, with 25 mM iodoacetamide] and cleared by centrifugation (20,000 × g, 15 min, 4 °C). The extracted material was mixed with denaturing agents of increasing stringency [Laemmli sample buffer with 0.5% SDS or urea (2 mM)-SDS (1.25%) (wt/vol)] and were heat-denatured at the specified temperatures (50 °C or 90 °C, respectively) for 10 min. Samples were fractionated on 3–8% (wt/vol) NuPAGE Tris-acetate gradient gels (Life Technologies), followedby immunoblotting.
F-Protein Cell-to-Cell Fusion Profiles.
Vero-SLAM effector cells were transfected with plasmids encoding the DSP1–7 dual-split protein (44), MeV H, and a mixture of F-encoding plasmids at the indicated ratios. Vero-SLAM target cells received plasmid DNA encoding the DSP8–11 subunit. Premature fusion was suppressed by 75 μM FIP. Both cell populations were combined at equal ratios 24 h after transfection, and mixtures were reseeded in solid-wall 96-well plates. Reconstitution of renilla luciferase was assessed continuously at 37 °C, using the EnduRen live-cell luciferase substrate (Promega) and a Synergy H1 temperature-controlled multimode microplate reader (BioTek). Maximal fusion rates were determined as before for each F plasmid ratio examined, and values obtained for each F construct examined were normalized to those determined in the absence of Ftrypsin. For each F construct, four individual datasets were generated. The values shown are the averages of the slopes calculated individually from these datasets.
Statistical Analysis.
To assess the statistical significance of differences between sample means, unpaired two-tailed t tests were applied using the Excel (Microsoft) software package.
Supplementary Material
Acknowledgments
We thank N. Kondo and Z. Matsuda for the DSP cell lines and DSP expression plasmids; M. Takeda for the gift of B5 α-MeV H neutralizing antibodies and plasmids encoding the MeV-IC-B strain L, N, and P proteins; and B. Horvat and M. Ehnlund for providing α-F pre and α-F post monoclonal antibodies. This study was supported in part by Swiss National Science Foundation Grant 310030_153281 (to P.P.) and Public Health Service Grants AI071002 and AI083402 from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (to R.K.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403609111/-/DCSupplemental.
References
- 1.Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci. 2010;35(12):699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2(2):98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 3.Plemper RK. Cell entry of enveloped viruses. Curr Opin Virol. 2011;1(2):92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376(6535):92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 6.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci USA. 2012;109(41):16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 8.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6(5):e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zokarkar A, Connolly SA, Jardetzky TS, Lamb RA. Reversible inhibition of fusion activity of a paramyxovirus fusion protein by an engineered disulfide bond in the membrane-proximal external region. J Virol. 2012;86(22):12397–12401. doi: 10.1128/JVI.02006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15(7):675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donald JE, et al. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc Natl Acad Sci USA. 2011;108(10):3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 15.Markosyan RM, Cohen FS, Melikyan GB. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol Biol Cell. 2003;14(3):926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melikyan GB, et al. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151(2):413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markosyan RM, Leung MY, Cohen FS. The six-helix bundle of human immunodeficiency virus Env controls pore formation and enlargement and is initiated at residues proximal to the hairpin turn. J Virol. 2009;83(19):10048–10057. doi: 10.1128/JVI.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340(6136):1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102(26):9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson K, et al. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402(2):372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci USA. 2011;108(23):9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: Capture of intermediates of fusion. EMBO J. 2001;20(15):4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellgaard L, Molinari M, Helenius A. Setting the standards: Quality control in the secretory pathway. Science. 1999;286(5446):1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 24.Lee JK, Prussia A, Snyder JP, Plemper RK. Reversible inhibition of the fusion activity of measles virus F protein by an engineered intersubunit disulfide bridge. J Virol. 2007;81(16):8821–8826. doi: 10.1128/JVI.00754-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bose S, et al. Mutations in the parainfluenza virus 5 fusion protein reveal domains important for fusion triggering and metastability. J Virol. 2013;87(24):13520–13531. doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-Aguilar B, Dewispelaere K, Yi HA, Jacobs A. Significant differences in cell-cell fusion and viral entry between strains revealed by scanning mutagenesis of the C-heptad repeat of HIV gp41. Biochemistry. 2013;52(20):3552–3563. doi: 10.1021/bi400201h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence DM, et al. Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J Virol. 2000;74(4):1908–1918. doi: 10.1128/jvi.74.4.1908-1918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo N, Miyauchi K, Matsuda Z. 2011. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr Protoc Cell Biol Chapter 26:Units 26–29.
- 30.Ader N, et al. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J Virol. 2013;87(1):314–326. doi: 10.1128/JVI.01826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brindley MA, Takeda M, Plattet P, Plemper RK. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci USA. 2012;109(44):E3018–E3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prado GN, Romero JR, Rivera A. Endothelin-1 receptor antagonists regulate cell surface-associated protein disulfide isomerase in sickle cell disease. FASEB J. 2013;27(11):4619–4629. doi: 10.1096/fj.13-228577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J Cell Physiol. 2002;193(2):154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 34.Stantchev TS, et al. Cell-type specific requirements for thiol/disulfide exchange during HIV-1 entry and infection. Retrovirology. 2012;9:97–111. doi: 10.1186/1742-4690-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci USA. 1993;90(9):4112–4116. doi: 10.1073/pnas.90.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plemper RK, et al. Design of a small-molecule entry inhibitor with activity against primary measles virus strains. Antimicrob Agents Chemother. 2005;49(9):3755–3761. doi: 10.1128/AAC.49.9.3755-3761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle J, et al. Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J Virol. 2006;80(3):1524–1536. doi: 10.1128/JVI.80.3.1524-1536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maisner A, et al. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J Gen Virol. 2000;81(Pt 2):441–449. doi: 10.1099/0022-1317-81-2-441. [DOI] [PubMed] [Google Scholar]
- 39.Brindley MA, et al. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol. 2013;87(21):11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan D, et al. Dual myxovirus screen identifies a small-molecule agonist of the host antiviral response. J Virol. 2013;87(20):11076–11087. doi: 10.1128/JVI.01425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kielian M, Rey FA. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol. 2006;4(1):67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissenhorn W, Hinz A, Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581(11):2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol. 2009;386(1):14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith EC, et al. Trimeric transmembrane domain interactions in paramyxovirus fusion proteins: Roles in protein folding, stability, and function. J Biol Chem. 2013;288(50):35726–35735. doi: 10.1074/jbc.M113.514554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang DK, Cheng SF, Kantchev EA, Lin CH, Liu YT. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6:2–13. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tatulian SA, Tamm LK. Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry. 2000;39(3):496–507. doi: 10.1021/bi991594p. [DOI] [PubMed] [Google Scholar]
- 47.Reuven EM, et al. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: Implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry. 2012;51(13):2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charloteaux B, Lorin A, Brasseur R, Lins L. The “Tilted Peptide Theory” links membrane insertion properties and fusogenicity of viral fusion peptides. Protein Pept Lett. 2009;16(7):718–725. doi: 10.2174/092986609788681724. [DOI] [PubMed] [Google Scholar]
- 49.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta. 2003;1614(1):116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 50.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460(7254):525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryser HJ, Levy EM, Mandel R, DiSciullo GJ. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc Natl Acad Sci USA. 1994;91(10):4559–4563. doi: 10.1073/pnas.91.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci USA. 2006;103(47):17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porotto M, et al. Inhibition of Nipah virus infection in vivo: Targeting an early stage of paramyxovirus fusion activation during viral entry. PLoS Pathog. 2010;6(10):e1001168. doi: 10.1371/journal.ppat.1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welsch JC, et al. Fatal measles virus infection prevented by brain-penetrant fusion inhibitors. J Virol. 2013;87(24):13785–13794. doi: 10.1128/JVI.02436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roymans D, et al. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci USA. 2010;107(1):308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganellin CR, Jefferis R, Roberts SM, editors. Introduction to Biological and Small Molecule Drug Research and Development: Theory and Case Studies. Oxford, UK: Academic; 2013. p. 1. [Google Scholar]
- 57.Aydin H, Cook JD, Lee JE. Crystal structures of beta- and gammaretrovirus fusion proteins reveal a role for electrostatic stapling in viral entry. J Virol. 2014;88(1):143–153. doi: 10.1128/JVI.02023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ono N, et al. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol. 2001;75(9):4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cathomen T, Naim HY, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72(2):1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paal T, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83(20):10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh M, Billeter MA. A recombinant measles virus expressing biologically active human interleukin-12. J Gen Virol. 1999;80(Pt 1):101–106. doi: 10.1099/0022-1317-80-1-101. [DOI] [PubMed] [Google Scholar]
- 63.Plemper RK, Compans RW. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J Virol. 2003;77(7):4181–4190. doi: 10.1128/JVI.77.7.4181-4190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeda M, et al. Recovery of pathogenic measles virus from cloned cDNA. J Virol. 2000;74(14):6643–6647. doi: 10.1128/jvi.74.14.6643-6647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahara M, et al. Functional and structural characterization of neutralizing epitopes of measles virus hemagglutinin protein. J Virol. 2013;87(1):666–675. doi: 10.1128/JVI.02033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plemper RK, Hammond AL, Cattaneo R. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J Virol. 2000;74(14):6485–6493. doi: 10.1128/jvi.74.14.6485-6493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avila M, et al. Molecular determinants defining the triggering range of prefusion F complexes of canine distemper virus. J Virol. 2014;88(5):2951–2966. doi: 10.1128/JVI.03123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lambert DM, et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93(5):2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.