Significance
The neocortex is an intricate and diverse cellular network in the brain, generating complex thought and voluntary motor behavior. Although recent attention has focused on the genome and transcriptome, our goal is to study the role of posttranscriptional processing and mRNA translation in neocortical development. In this work, we show that the protein components of actively translating ribosomes and their mRNA cargo in the developing neocortex depend on the temporally specific action of an RNA-binding protein, Hu antigen R (HuR). We further show that HuR is required for the development of neocortical neurons and structure. This study contributes to our overall understanding of how the regulation of functional gene expression influences neocortical development.
Keywords: ribosome, posttranscriptional regulation, profiling, GCN2, Elav
Abstract
Precise spatiotemporal control of mRNA translation machinery is essential to the development of highly complex systems like the neocortex. However, spatiotemporal regulation of translation machinery in the developing neocortex remains poorly understood. Here, we show that an RNA-binding protein, Hu antigen R (HuR), regulates both neocorticogenesis and specificity of neocortical translation machinery in a developmental stage-dependent manner in mice. Neocortical absence of HuR alters the phosphorylation states of initiation and elongation factors in the core translation machinery. In addition, HuR regulates the temporally specific positioning of functionally related mRNAs into the active translation sites, the polysomes. HuR also determines the specificity of neocortical polysomes by defining their combinatorial composition of ribosomal proteins and initiation and elongation factors. For some HuR-dependent proteins, the association with polysomes likewise depends on the eukaryotic initiation factor 2 alpha kinase 4, which associates with HuR in prenatal developing neocortices. Finally, we found that deletion of HuR before embryonic day 10 disrupts both neocortical lamination and formation of the main neocortical commissure, the corpus callosum. Our study identifies a crucial role for HuR in neocortical development as a translational gatekeeper for functionally related mRNA subgroups and polysomal protein specificity.
Development of the mammalian neocortex follows a precise sequence of events and generates a remarkably diverse network of local and long-range circuits that mediate complex cognitive and motor functions (1–5). First, neuroepithelial stem cells lining the lateral ventricles of the nascent dorsal telencephalon undergo symmetrical cell division to expand the pool of stem cells before neurogenesis. The neurogenic phase begins when these neuroepithelial cells develop into a glial-like progenitor called radial glia (RG) and express the Paired box protein 6 (Pax6) transcription factor. Next, RG shift from symmetrical cell divisions to asymmetrical divisions and directly generate either T-box brain protein 2 (Tbr2)-expressing intermediate progenitor cells (IPCs) or doublecortin-expressing neuroblasts that populate cortical plate (CP) layers. The first neuroblasts generated from RG or IPCs populate the lower CP layers, express characteristic transcription factors (e.g., Bcl11b and Tle4 transcription factors), and differentiate to project subcortically via tracts to the thalamus, brainstem, or spinal cord. The neuroblasts generated later migrate past the first ones, express different transcription factors (e.g., Cdp and Satb2), and differentiate into upper-layer neurons that project intracortically, either to the contralateral hemisphere that forms the corpus callosum or ipsilaterally. Therefore, neocortical layers develop from RG progenitors to projection neurons in an inside-out fashion, which depends on transcriptional control (1, 4, 6, 7). Posttranscriptional mRNA regulation is also involved in determining the remarkable diversity of cellular subtypes and unique circuits, but this regulatory mechanism is poorly understood in complex systems like the neocortex (5, 8–12).
Because of the temporal delay between mRNA transcript processing and the production of functional proteins, mRNA transcript levels may not accurately reflect protein levels, and vice versa. This discrepancy has been documented globally in mammalian cells (13) and specifically in the neocortex (5, 14), where there can be a long time lag between the appearance of an mRNA and the production of its protein. Key to the spatiotemporal orchestration of mRNA fate is regulation by RNA-binding proteins (RBPs), which mediate transcript splicing, transport, stability, and ultimately several distinct and tightly controlled steps of translation (5, 12, 15–18). Translation is a particularly complex and highly regulated process (17, 19). Briefly, mRNA is first activated by binding of its 5′ untranslated region to the eukaryotic initiation factor 4F eukaryotic initiation cap complex. The activated mRNA then joins the 43S preinitiation complex, which contains a small 40S ribosomal subunit and a eukaryotic initiation factor 2 (eIF2)–GTP–tRNAMet ternary complex. During initiation, the ternary complex is removed from the 40S subunit, and the 60S ribosomal subunit is recruited to form the 80S ribosome. Actively translated mRNAs accumulate multiple 80S ribosomes per transcript, called polysomes, where active elongation occurs. Peptide elongation occurs via eukaryotic elongation factor 2 (eEF2)-dependent translocation. Eventually, translation is terminated, and ribosomes are recycled. The 40S and 60S subunits contain distinct ribosomal proteins, designated as ribosomal proteins small (e.g., RPS26 and RPS27) or ribosomal proteins large (e.g., RPL5 and RPL7), respectively. The regulators of 80S assembly, polysome accumulation, and ribosomal protein components are largely unknown in complex, developing systems such as the neocortex.
RBP-regulated posttranscriptional processing, including translation, is crucial for proper formation of the central nervous system (5). The RBP, Hu antigen R (HuR; also called ELAVL1), is estimated to have 26,000 transcriptome-wide targets in different cell lines and is implicated in multiple steps of posttranscriptional processing, including splicing, stability, and translation (20–22). However, the molecular mechanisms of HuR-mediated posttranscriptional mRNA processing, polysome specificity, and its specific role in neocortical development are currently unknown.
Here, we reveal HuR's role in the temporally specific association of functionally related mRNAs in polysomes of the developing neocortex. HuR regulates the translation of numerous mRNA transcripts that encode members of transcriptional and translational regulatory pathways. HuR associates with the eIF2-alpha kinase 4 (eIF2ak4, also known as GCN2), influencing the presence of initiation and elongation factors and determining the specificity of ribosomal proteins in neocortical polysomes. These results suggest that RBPs dynamically regulate ribosome assembly, polysome specificity, and the temporal translation of functionally related mRNA subgroups in complex developing systems such as the neocortex.
Results
HuR Is Expressed in Neurogenic RG, IPCs, and Postmitotic Projection Neurons.
Previous work using microarray analyses coupled with bioinformatics revealed that neocortical HuR mRNA was strongly expressed in the developing mice and human neocortices (5, 23, 24). In mice, HuR was found to be expressed at the following key neurogenic stages in the developing neocortex: embryonic day 11 (E11), onset of projection neuron neurogenesis; E13, predominantly lower layer projection neuron genesis; E15, predominantly upper-layer projection neuron genesis; and E18, the termination of projection neuron neurogenesis. Distinct cell types and layers of the developing neocortex have compartmentalized functions (1, 2, 4, 5, 25); therefore, it was necessary to determine the cell-type-specific expression of HuR.
We performed coimmunohistochemical staining for HuR and distinct cell-type-specific markers at stages E13, E16, and postnatal day 0 (P0) (Fig. 1A). We identified HuR protein in the neocortical ventricular zone (VZ) at E13 and E16, where it was highly enriched in cycling RG as determined by colocalization with Pax6 (specific for cycling RG) and phosphorylated histone 3 (pH3; M-phase cells) (Fig. 1A). These results suggest that HuR may be required for neocortical RG proliferation and/or differentiation, consistent with previously described roles for HuR in the cell cycle of tumor stem cells and in facilitating the exposure of neuronal precursors to Delta/Notch signals (26, 27). In the embryonic CP at E16 and later at P0, strong HuR expression was observed in differentiating deep-layer subcortically projecting neurons, as determined by colocalization with Bcl11b, and in upper-layer intracortically projecting neurons, as determined by colocalization with Satb2 and Cdp/Cux1 (1, 4, 5) (Fig. 1A). This expression pattern suggests that HuR may be involved in the specification of postmitotic identity of different projection-neuron subpopulations. Taken together, these developmental expression patterns suggest a key role for HuR as an RBP in early proliferative neocortical RG and postmitotic neocortical neurons, and possibly in neocortical circuit formation.
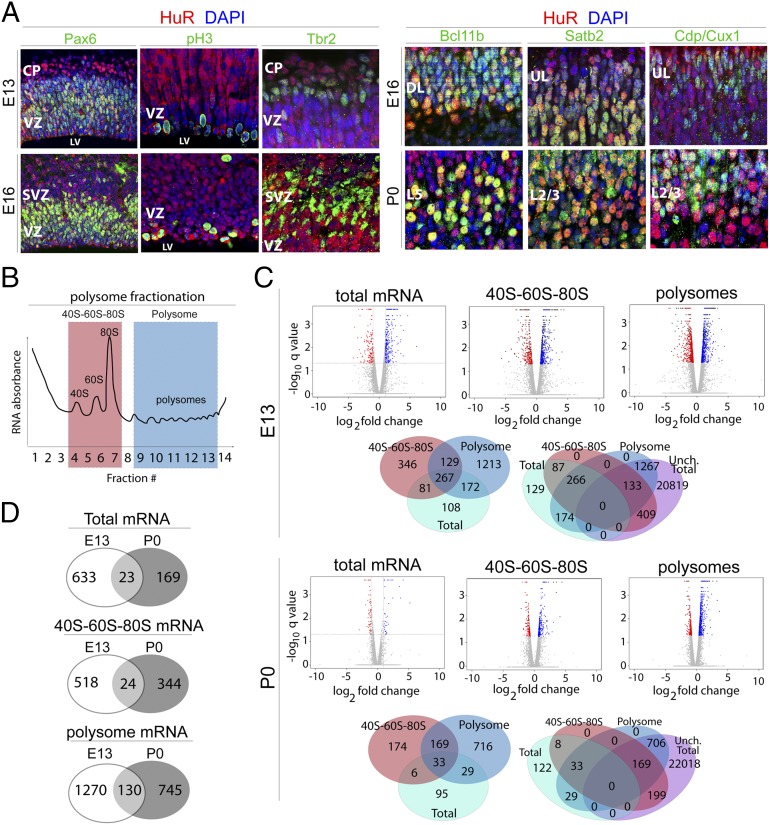
Fig. 1.
HuR regulates mRNA translation in mitotic neural stem cells and differentiating projection neurons of the developing neocortex. (A) Representative coronal confocal images of immunostained developing neocortices. HuR immunohistochemistry (red) shows that HuR is expressed in RG neural progenitors colabeled with Pax6 (green) and pH3 (green) at E13 and E16 in the ventricular zone (VZ). HuR is also expressed in postmitotic differentiated Bcl11b-positive lower-layer neurons (green) and in upper-layer neurons labeled with Satb2 and Cdp/Cux1 (green) at E16 and P0. CP, cortical plate; DL, deep layers; L2/3, layer 2/3; L5, layer 5; LV, lateral ventricle; SVZ, subventricular zone; UL, upper layers. (B) Schematic of sucrose density gradient fractionation and isolation of 40S–60S–80S and polysome cytoplasmic components for analysis. (C) E13 and P0 WT and Emx1–HuR-cKO neocortices were fractionated into 40S–60S–80S and polysomes, then subjected to RNAseq coupled with bioinformatics analysis. Volcano plots show gene-expression levels relative to WT; blue dots represent higher expression in HuR-cKO; red dots represent lower expression in HuR-cKO; and gray dots represent unchanged levels at a false-discovery rate ≤5%. Venn diagrams show the number of genes that change with HuR-cKO in 40S–60S–80S and polysomal fractions, with respect to total mRNA expression levels (whether changed or unchanged), analyzed by RNAseq at E13 and P0. (D) Venn diagrams show total, 40S–60S–80S, and polysome-associated mRNAs that change in abundance in response to HuR deletion. The mRNAs are unique to E13, unique to P0, or present at both developmental stages.
HuR Determines Temporally Distinct mRNA Enrichment in 40S–60S–80S and Polysomal Fractions of Developing Neocortices.
To identify candidate mRNAs regulated at the translational level by HuR in developing neocortices in an unbiased fashion, we performed sucrose density-gradient (10−50%) ultracentrifugation and fractionation (28, 29) coupled to RNA sequencing (RNAseq) and bioinformatics analysis (30) of combined fractions representing 40S–60S–80S and polysomes at E13 and P0 from wild-type (WT) and Emx1–Cre × HuRf/f conditional-knockout (HuR-cKO) mice (31, 32) (Fig. 1 B–D and Fig. S1). This cKO line harbors a selective deletion of HuR at approximately E11 in RG, resulting in HuR depletion in all primary projection neurons in the neocortex. The mRNA identity and levels in 40S–60S–80S and polysomal fractions were measured against total levels by RNAseq coupled to bioinformatics, where 40S–60S–80S and polysomal fractions were determined by using an RNA absorbance curve monitored during fractionation (Fig. 1B and Fig. S1A). First, results indicated that a small fraction of mRNAs’ total levels were affected in HuR-cKOs at E13 and P0 compared with WT mRNAs (Fig. 1C). At E13, we detected a total of 656 genes that were affected (2.83% of expressed genes); the levels of 433 mRNAs were lower in HuR-cKO and 223 mRNAs were higher in HuR-cKO compared with those of WT mice. At P0, we detected a total of 192 genes that were affected (0.83% of expressed genes); the levels of 56 mRNAs were lower in HuR-cKO and 136 mRNAs were higher in HuR-cKO compared with those of WT. Most of the mRNAs affected by HuR-cKO at E13 differed from those that were affected at P0 (Fig. 1D), suggesting that the absence of HuR differentially alters mRNA transcripts at different developmental stages.
Although the total levels of many mRNAs did not change, a significant number had altered levels in the distinct 40S–60S–80S and polysomal fractions at both E13 and P0 (Fig. 1C). This result suggests that HuR regulates the translation of numerous neocortical mRNAs. At E13, the total levels of 22,628 mRNAs did not significantly differ between WT and HuR-cKO; however, 542 of these were significantly different in the 40S–60S–80S fraction, and 1,400 were significantly different in the polysomal fraction of HuR-cKO compared with those of WT. At P0, the total levels of 23,092 mRNAs did not significantly differ between WT and HuR-cKO; however, 368 of these were significantly different in the 40S–60S–80S fraction, and 875 were significantly different in the polysomal fraction of HuR-cKO compared with those of WT.
Only a small number of mRNAs were regulated by HuR at both E13 and P0 in these ribosomal fractions (Fig. 1D). In the 40S–60S–80S fraction, 24 genes were regulated by HuR at both E13 and P0 (4.4% of genes affected at E13; 6.5% of genes affected at P0). In the polysomal fraction, 130 genes were regulated by HuR at both E13 and P0 (9.3% of genes affected at E13; 21.5% of genes affected at P0), suggesting a greater consistency in HuR regulation of polysomal mRNAs. Collectively, these results suggest that HuR is required for appropriate translation of a large number of distinct mRNAs in a temporally specific manner.
HuR Regulates mRNAs Encoding Distinct Members of Transcriptional, Translational, and Layer-Specific Pathways at E13 and P0.
To determine whether particular pathways are regulated by HuR in the 40S–60S–80S and polysomal fractions of developing neocortices, we performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (33, 34) of mRNAs that were identified to be differentially distributed in 40S–60S–80S and/or polysomal fractions at E13 and P0 (Fig. 2A and Fig. S1B). All distinct mRNAs are represented as colored dots on the volcano plots in Fig. 1C, which illustrates their relationship to other expressed genes. By highlighting distinct functional groups or pathways, the analyses revealed that HuR regulates subgroups of neocortical mRNAs with similar functions. At E13, GO analysis revealed that, among transcripts unchanged in total levels, mRNAs encoding proteins regulating transcription were down-regulated in polysomal fractions of HuR-cKO compared with those of WT (Fig. 2A and Fig. S1B), whereas mRNAs encoding proteins involved in translation regulation were up-regulated in polysomal fractions of HuR-cKO compared with those of WT. KEGG pathway analysis indicated that a large number of mRNAs encoding ribosomal proteins and those associated with the cell cycle were affected in HuR-cKO compared with those of WT (Fig. 2A and Fig. S1B). These data suggest that HuR regulates the active translation of mRNAs in specific pathways in the polysomal fraction at E13.
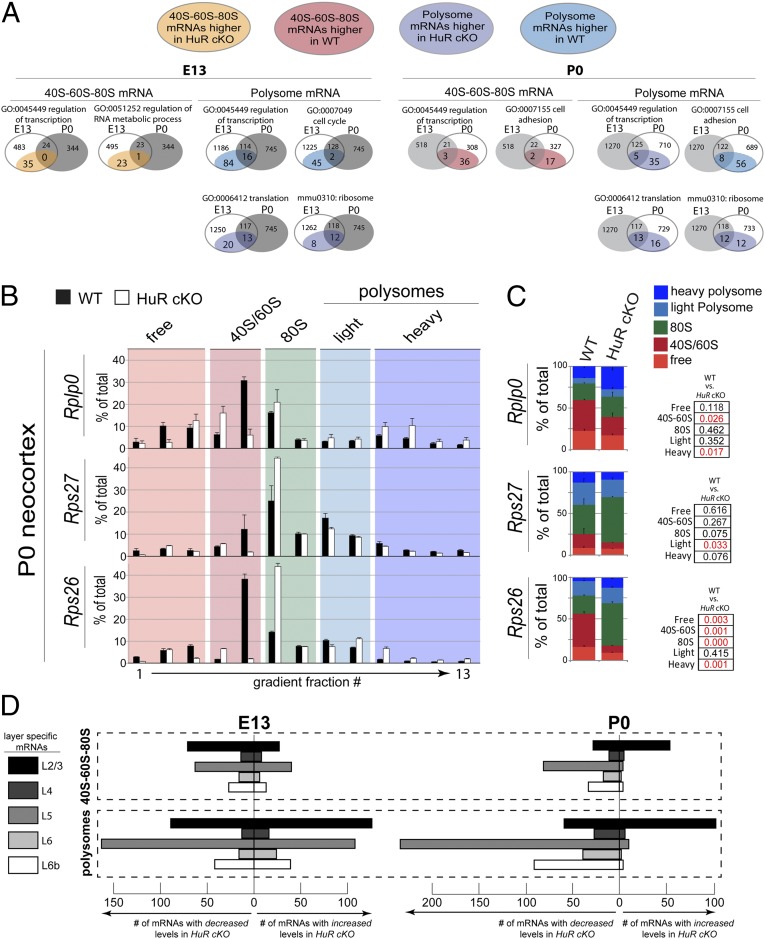
Fig. 2.
HuR regulates the polysomal positioning of functionally related mRNAs in the developing neocortex. (A) KEGG and GO bioinformatics analyses indicate that, among mRNAs stable in total levels with HuR-cKO, those altered in 40S–60S–80S and polysomal fractions are functionally related transcripts. Venn diagrams present the number of mRNAs within each functional group (e.g., regulation of transcription) that differ in either 40S–60S–80S or polysomal fractions at E13 (Left) and P0 (Right) in HuR-cKO. Developmental-stage-dependent changes in each functional group are shown as a comparison between E13 and P0. Each Venn circle sums to the total number of mRNAs that undergo changes in abundance; the changes for each functional group are highlighted as subsets of WT and HuR-cKO. (B) The mRNA candidates revealed by RNAseq were confirmed with qRT-PCR of WT (filled bars) and HuR-cKO (open bars). Fractions corresponding to the nontranslating free, 40S/60S, and 80S vs. translating light and heavy polysomes are highlighted. (C) Quantification and statistical analysis of the nontranslating and translating fractions are shown comparing WT and HuR-cKO (n = 4 cortices in two fractionations; qRT-PCRs were performed in duplicate for each fraction). Statistical significance between WT and cKO for each category with t test is indicated in red text in Right (P < 0.05). (D) Bioinformatic analysis of the number of neocortical layer-specific mRNAs increased or decreased by HuR-cKO in the 40S–60S–80S and polysome at E13 and P0, showing a particularly strong effect on layer-2/3 and -5 polysome mRNAs at both ages.
We observed that HuR plays a substantial role in positioning the mRNAs localized to the 40S–60S–80S fraction, which represents the late stage of the translation initiation/preelongation phase. Control at this level may rapidly move mRNAs into or out of a position poised for translation, whereas the total mRNA levels are unchanged. Among the mRNAs that did not change in total levels, those found in the 40S–60S–80S fraction that encoded transcriptional regulators had higher levels in HuR-cKO at E13 compared with those in WT (Fig. 2A and Fig. S1B). However, the polysome-depleted mRNAs encoding transcriptional regulators at E13 were not the same as those that were enriched in 40S–60S–80S at E13 in HuR-cKO, suggesting specificity within subgroups of HuR-regulated mRNAs in developing neocortices. These data indicate that HuR influences polysomal translation of large subsets of functionally related cytoplasmic mRNAs in developing neocortices at E13, whereas it prevents others from becoming positioned for translation within the 40S–60S–80S fraction.
GO analysis of functionally related neocortical mRNAs at P0 identified substantial changes in HuR-influenced mRNA partitioning in the 40S–60S–80S and polysomal fractions among transcripts unchanged in total levels (Fig. 2A and Fig. S1B). At P0, HuR still regulates mRNAs encoding members of transcriptional and translational control pathways, although the number is somewhat lower, and the subsets differ from those at E13. At P0, a large number of mRNAs encoding proteins involved in cell adhesion were regulated by HuR. KEGG analysis indicated that ribosomal mRNAs were affected by conditional HuR knockout, but the HuR-regulated mRNAs differed at E13 and P0. Together, these results suggest that HuR differentially regulates distinct mRNA subgroups at the 40S–60S–80S and polysomal levels of translation during development, but has a comparatively smaller effect on total mRNA levels. The particular role of HuR in influencing translational and ribosomal genes suggests an autoregulatory process at the level of translation.
We next performed quantitative RT-PCR (qRT-PCR) on fractionated WT and HuR-cKO neocortices at P0 to confirm candidates identified in Figs. 1C and 2A that did not change in total mRNA levels but exhibited differential distributions among cytoplasmic (i.e., free), 40S–60S–80S, and polysomal fractions (Fig. 2 B and C and Fig. S2). Satb2 is an example of an unaffected transcript (Fig. S2). For example, ribosomal proteins Rplp0 and Rps26 mRNAs became substantially enriched in polysome fractions of the HuR-cKO compared to WT (Fig. 2 B and C). The deep-layer neocortical transcription factor Bcl11b mRNA displayed redistribution into the 40S–60S fraction in the HuR-cKO compared with that of WT (Fig. S2). We further confirmed these results with more stringent statistical testing via simulation. Monte Carlo-based tests of significance for pairwise differences showed that Rps26 had significant shifts (P < 0.05) in free, 40S–60S, 80S, and heavy polysomes, whereas Rps27 had a significant pairwise difference in the 80S fraction in HuR cKOs. These results reinforced our previous observation that mRNAs display HuR-influenced shifts into and out of 40S–60S, 80S, and polysomal fractions in developing neocortices, whereas the total levels of many of these HuR-regulated mRNAs do not change significantly at E13 or P0.
Finally, we extended our bioinformatic analysis to assess whether HuR-regulated mRNAs are associated with distinct neocortical layers in development (35). We found that HuR-cKO had a particular influence on a large number of layer 2/3- and 5-specific mRNAs by either increasing or decreasing their association with polysomes at E13 and P0 (Fig. 2D). In this way, HuR may potentially play a role in the transition from specifying subcortically projecting lower-layer neurons to later-born intracortically projecting upper-layer neurons. Together, our data suggest that there may be temporally specific cofactors for HuR in developing neocortices, which modulate HuR-dependent translation of functionally related mRNAs.
HuR Knockout Disrupts eIF2-Alpha and eEF2 Phosphorylation.
To further investigate how HuR influences mRNA translation, we examined the integrity of the core translational components in HuR-cKO neocortices at E13 and P0. Consistent with translational dysregulation in HuR-cKO neocortices at E13, we detected increased phosphorylation of the translation initiation factor eIF2-alpha (eIF2a), in contrast to decreased eEF2 phosphorylation in the cKO compared with WT (Fig. 3A). At P0, we observed increased eIF2a phosphorylation in HuR-cKO neocortices, but eEF2 phosphorylation was also increased at this stage (Fig. 3A). However, the overall levels of eIF2a and eEF2 were largely unaffected at both stages. The observed HuR-dependent effects on core translation machinery and mRNA translation suggest that HuR specifically and temporally interacts with members of the translational machinery.
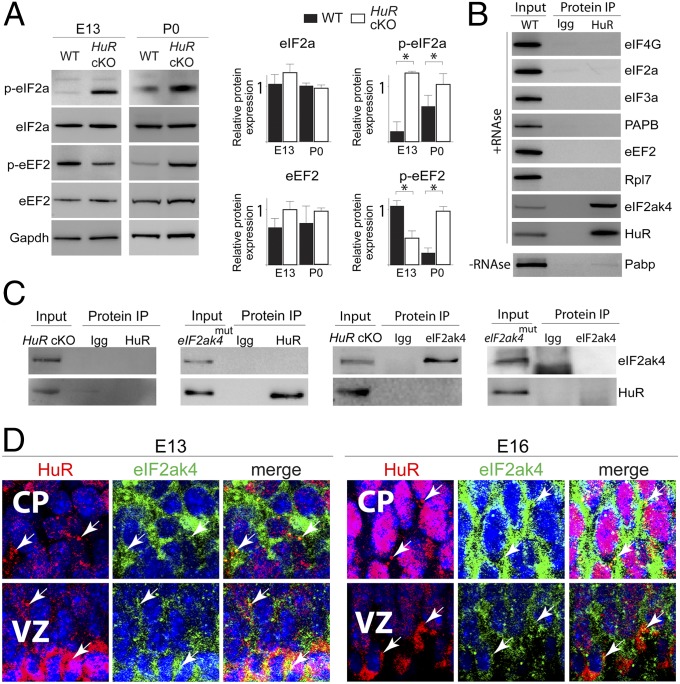
Fig. 3.
HuR associates with eIF2ak4 and influences translation factor phosphorylation in the developing neocortex. (A, Left) Western blot analysis of total neocortical lysates collected from WT and HuR-cKO at E13 and P0. (Right) Quantification is shown (n = 3). *P < 0.05 (t test normalized with respect to GAPDH loading control). (B) HuR was immunoprecipitated (IP) from E18 neocortical lysates and analyzed by Western blot for interaction with translation factor candidates. HuR was used as the positive control. (C) HuR and eIF2ak4 reverse coimmunoprecipitation from HuR-cKO and eIF2ak4 kinase-domain mutant (eIF2ak4mut). (D) Immunohistochemistry for HuR (red) and eIF2ak4 (green) in E13 and E16 neocortical coronal sections showed their colocalization in cytoplasmic puncta (white arrows) of VZ RG and differentiating CP neurons. DAPI is shown in blue.
HuR Associates with eIF2ak4 in Developing Neocortices.
To test for temporally distinct interactions of HuR with members regulating neocortical translation machinery, we performed protein coimmunoprecipitation (co-IP) with HuR and corresponding IgG antibodies from E12 and E18 neocortices, then analyzed the precipitates using mass spectrometry (Fig. S3). We considered proteins to be candidate HuR-interacting partners if the log(e) was ≤80, and spectral count was >12 for HuR co-IP and <1 for IgG. We detected a particularly high level of HuR interaction with eIF2ak4 at E18, but less at E12. Therefore, eIF2ak4 was identified as a candidate translation factor that interacts with HuR in a temporally dependent manner during neocortical development and a putative target involved in HuR-dependent translational regulation. HuR co-IP lysates were subjected to RNase treatment (to exclude RNA-mediated binding) and Western blotting, which confirmed that HuR directly interacted with eIF2ak4 but did not interact with eIF4G, eIF2a, eIF3, Pabp, eEF2, or Rpl7 at E18 (Fig. 3B).
Reverse co-IPs from P0 HuR-cKO and eIF2ak4 kinase-domain mutant (eIF2ak4mut) neocortices confirmed the specificity of interaction between HuR and eIF2ak4 (Fig. 3C) and suggested that the eIF2ak4 kinase domain is crucial for the HuR–eIF2ak4 interaction in developing neocortices. Immunostaining analysis determined that HuR and eIF2ak4 colocalize in cytoplasmic puncta (white arrows) of neural stem cells and postmitotic cells of developing neocortices at E13 and E16 (Fig. 3D). Collectively, these data suggest that HuR and eIF2ak4 are positioned to dynamically interact in differentiating neural stem cells during neocortical development and influence neocortical mRNA translation in a spatiotemporally dependent manner.
HuR and eIF2ak4 Regulate the Specificity of Initiation and Elongation Factors and Ribosomal Proteins in Neocortical 40S–60S–80S and Polysomal Fractions.
The phosphorylation of initiation and elongation factors affects their position within polysomes (36), and eIF2ak4 directly associates with the 60S subunit in yeast (37). Therefore, we hypothesized that HuR knockout would disrupt the constituent proteins of neocortical 40S–60S–80S and polysomal fractions (Fig. 4A). To identify proteins in an unbiased fashion, we performed mass spectrometry coupled with bioinformatics analysis of 40S–60S–80S and polysomal fractions isolated from HuR-cKO and WT neocortices at E13 and P0 (Fig. S4A). Mass spectrometry bioinformatic analysis (33, 34) indicated that similar targets were disrupted at E13 and P0 in HuR-cKO 40S–60S–80S and polysomal fractions, which included a number of initiation (e.g., eIF5) and elongation (e.g., eEF1A1) factors. The partitioning of numerous ribosomal proteins in both fractions was also disrupted (e.g., Rpl5 and Rpl7). The total levels of translation factors eIF5, eEF1A1, Rpl5, and Rpl7 in HuR-cKO neocortical lysates were not significantly altered at P0 compared with WT as determined by Western blot (Fig. 4B). In contrast, the fractionated HuR-cKO and WT neocortices did display changes in the protein levels of specific fractions as determined by mass spectrometry and Western blot analysis of neocortical lysates at P0 and E13 (Fig. 4 B and C and Fig. S4). In HuR-cKO neocortices at P0, we detected a dramatic decrease in the levels of eIF5 and eEF1A1 associated with the 40S–60S–80S fraction and a reduction of Rpl5 and Rpl7 associated with the polysomal fraction (Fig. 4C). Deletion of HuR did not affect eIF2a at P0; however, eIF2a prematurely entered into polysomal fractions in the HuR-cKO at E13 (Fig. S4B). These data suggest that proper neocortical polysome assembly is disrupted by HuR deletion in developing neocortices.
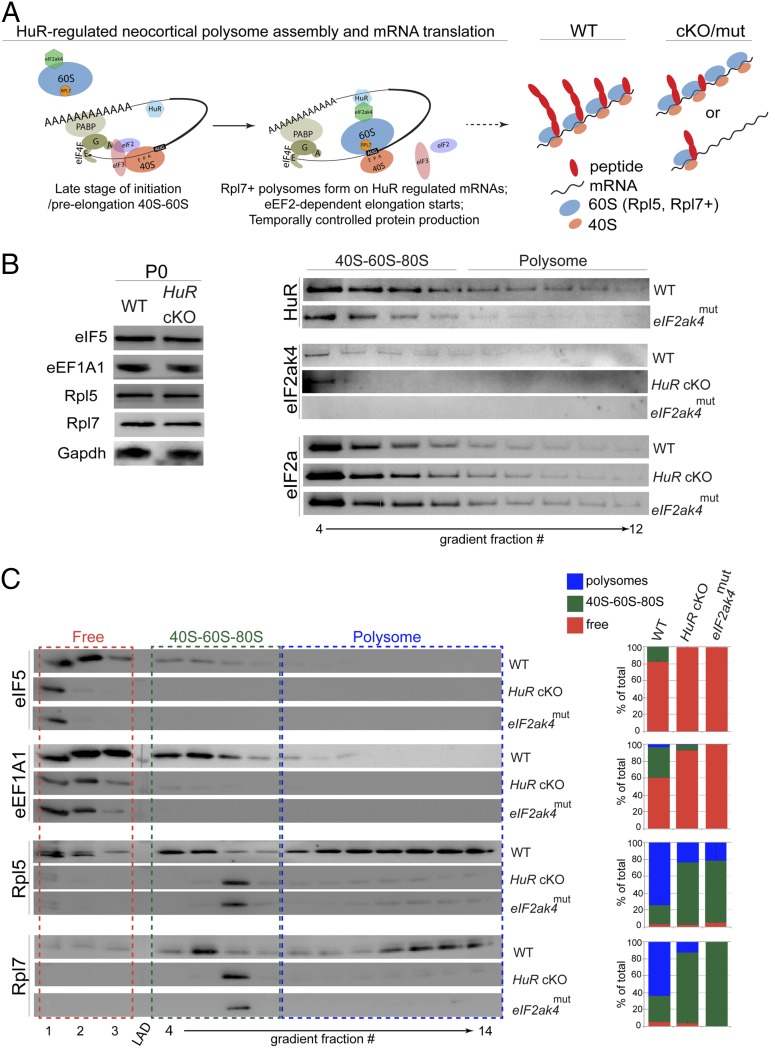
Fig. 4.
HuR and eIF2ak4 regulate specificity of translation factors and ribosomal proteins in neocortical polysomes. (A) Schematic of our model for how HuR and eIF2ak4 may interact in polysomes to influence mRNA translation. (B, Left) Western blot analysis of P0 neocortical lysates (n = 3 cortices) from WT and HuR-cKO shows unchanged total levels of translation factors and ribosomal proteins. (Right) Western blot analysis of P0 neocortical density-gradient fractionations shows that eIF2ak4mut disrupts HuR polysome enrichment, and both HuR-cKO and eIF2ak4mut disrupt eIF2ak4 polysome enrichment. The levels of eIF2a remain stable. (C) Western blot analysis (Left) and quantification (Right) of P0 WT, HuR-cKO, and eIF2ak4mut neocortices to measure the association of eIF5, eEF1A1, Rpl5, and Rpl7 with 40S-60S-80S and polysomal fractions.
To determine whether eIF2ak4 function is required for polysome assembly similar to the requirement for HuR, we performed Western blot analysis on fractionated eIF2ak4mut neocortices at P0 (Fig. 4 B and C). We observed that HuR expression decreased in polysomal fractions isolated from eIF2ak4mut neocortices, and the polysomal positioning of eIF2ak4 was severely compromised in both HuR-cKO and eIF2ak4mut (Fig. 4B). We observed that mutating the eIF2ak4 kinase domain mimicked HuR knockout with respect to the partitioning of eIF5, eEF1A1, Rpl5, and Rpl7 into the neocortical polysome (Fig. 4C), whereas the overall level of eIF2a was unaffected (Fig. 4B). These data further corroborate the preceding observation that HuR–eIF2ak4 interaction is necessary for proper assembly of the neocortical polysome. At P0, eIF2ak4 and HuR appear in both 40S–60S–80S and polysomal fractions (Fig. 4B), further suggesting a mutually interacting role in mRNA translation.
Embryonic HuR Deletion Disrupts Neocortical Lamination and Corpus Callosum Formation.
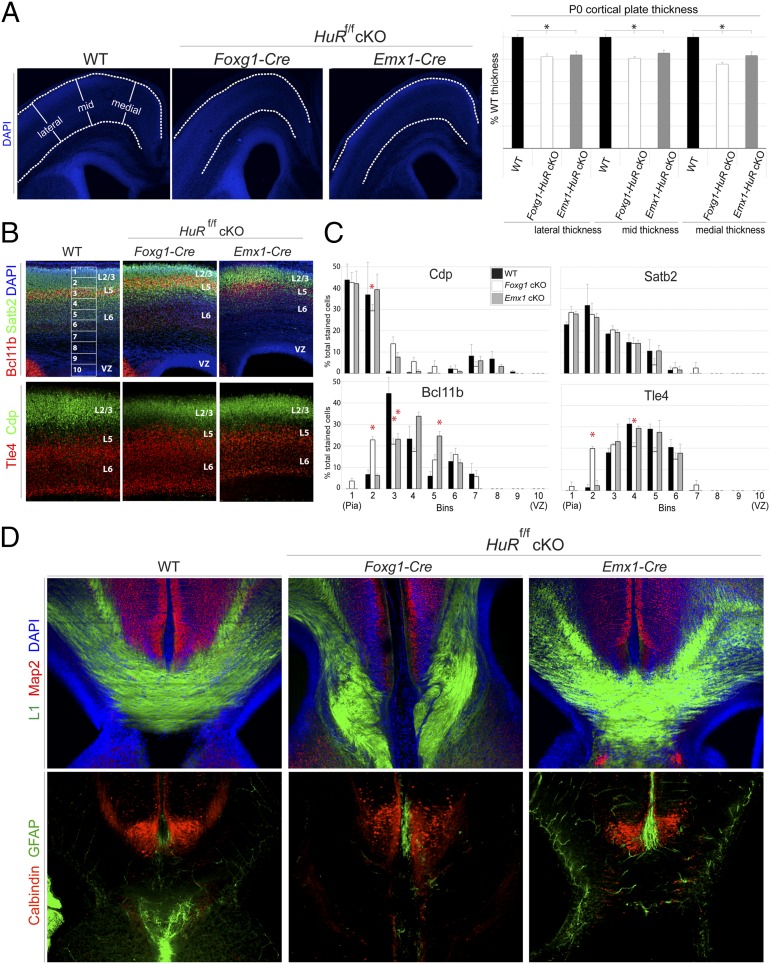
HuR is expressed in cycling RG and postmitotic differentiating projection neurons of developing neocortices (Fig. 1A). HuR knockout disrupts the polysomal association of cell-cycle mRNAs at E13, cell-adhesion mRNAs at P0, and transcription/translation-factor mRNAs at both E13 and P0 (Fig. 2 A–C). Furthermore, HuR may play a particular role in the translation of mRNAs specific to layer-2/3 and -5 neocortical neurons (Fig. 2D). Therefore, we hypothesized that loss of HuR in the developing neocortex would result in abnormal lamination and disrupt neuronal differentiation assessed in the postnatal neocortex. HuR was conditionally deleted at two different time points during neocortical development: HuRf/f × Foxg1–Cre targets neuroepithelial cells at E9, and HuRf/f × Emx1–Cre targets RG neural progenitors at E11 (38). WT and HuR-cKO brains were isolated at P0, and coronal sections were analyzed by using immunohistochemistry followed by confocal microscopy and software-based measurement. When CP thickness was measured in 4′,6′-diamidino-2-phenylindole (DAPI)-stained images, both Foxg1–HuR-cKO and Emx1–HuR-cKO animals had significantly thinner cortices than those of WT (Fig. 5A). This result suggests that HuR deletion in the developing neocortex disrupts cycling neural progenitors that generate postmitotic neurons.
Fig. 5.
HuR deletion in neuroepithelial cells and RG disrupts postnatal neocortical lamination and corpus callosum formation. (A) Immunohistochemistry on P0 coronal sections from WT, Foxg1–HuR-cKO (E9 HuR knockout, neuroepithelial stage), and Emx1–HuR-cKO (E11 HuR knockout, RG stage)-representative images (Left) and quantification (Right) of lateral, mid, and medial CP thickness (n ≥ 20 hemispheres/condition). *P ≤ 0.005 [MANOVA and ANOVA, followed by post hoc Tukey’s honest significant difference (HSD)]. (B) Immunohistochemical analysis for markers Cdp and Satb2 (green) in upper-layer differentiated neurons compared to markers Tle4 and Bcl11b (red) in lower-layer differentiated neurons of P0 coronal sections from WT, Foxg1–HuR-cKO, and Emx1–HuR-cKO. Confocal images were divided into 10 equal bins spanning from the VZ to superficial layer 2/3 (L2/3) and quantified for the presence of these markers. (C) Quantification of the distribution of each marker in WT, Foxg1-HuR-cKO, and Emx1-HuR-cKO neocortices, determined as a percentage of the total number of marker-positive cells (n = 2−4 biological replicates and 6−12 technical replicates). *P < 0.05 (MANOVA and ANOVA, followed by Tukey’s HSD post hoc if Levene statistic P ≥ 0.05 or by Games–Howell post hoc if Levene statistic P < 0.05). (D) Serial coronal sections matched for anterior-posterior level at the corpus callosum were immunostained and imaged for axonal marker L1 (green) and dendritic marker Map2 (red), showing agenesis of the corpus callosum in P0 Foxg1–HuR-cKO, but not in Emx1–HuR-cKO. Immunohistochemical analysis for calbindin-positive interneurons (red) and GFAP (green) show disruption of the glial wedge in P0 Foxg1–HuR-cKO coronal sections. DAPI is in blue.
We next assessed the distribution of postmitotic neurons within distinct neocortical layer subpopulations in Foxg1–HuR-cKO, Emx1–HuR-cKO, and WT mice at P0. We observed that Bcl11b- and Tle4-positive lower-layer neurons were ectopically redistributed into upper neocortical layers in Foxg1–HuR-cKOs, with a concurrent decrease in Cdp-positive neurons typically localized to upper layers (Fig. 5 B and C). Later deletion of HuR in Emx1–HuR-cKO RG resulted in significant redistribution of Bcl11b-positive neurons into deeper layers (Fig. 5 B and C). These data suggest that differentiation of neural stem cells and lamination of the neocortex into functionally distinct layers are disrupted in a temporally dependent manner by embryonic HuR deletion.
Upper-layer-2/3 neocortical neurons project solely intracortically, either within the ipsilateral hemisphere or to the contralateral hemisphere forming the corpus callosum. In contrast, lower-layer neurons project to subcortical targets. Because we observed significant disruption in the placement and/or identity of both upper- and lower-layer neurons in HuR-cKO, we assessed whether embryonic knockout of HuR disrupted neocortical projections at P0. Strikingly, Foxg1–HuR-cKO animals completely lacked a corpus callosum, as determined by immunostaining for L1 neural cell-adhesion molecule (L1-CAM) (Fig. 5D; serial sections matched for anterior-posterior level). In contrast, Emx1–HuR-cKO animals, which deplete HuR later in neural progenitors, maintained the intercortical connections. Although interneurons appeared to appropriately target to this midline structure in HuR-cKO, the glial wedge was largely absent in Foxg1–HuR-cKO (Fig. 5D) (39, 40). These data corroborate our finding that HuR deletion has a pronounced effect on layer-2/3-associated mRNAs in polysomes (Fig. 2D) and strongly suggest that HuR acts in a temporally specific manner to influence neocortical projection neuron differentiation, lamination, and circuit formation.
Discussion
This study indicates that absence of a neocortical RBP, HuR, alters the association of functionally related mRNAs and proteins in actively translating polysomes and influences neocorticogenesis in a stage-specific manner. We observed that HuR knockout disrupts phosphorylation of the translation factors eIF2a and eEF2, the 40S-60S-80S positioning of initiation and elongation factors, and the specificity of ribosomal proteins in polysomes. These perturbations prevent proper polysome formation and result in abnormal mRNA localization. The magnitude of the HuR-regulated transcript population suggests that HuR orchestrates a highly complex set of pathways involved in neocortical development.
The data show that HuR directly associates with eIF2ak4, which likewise influences neocortical polysome assembly. The association of eIF2ak4 with ribosomes determines the phosphorylation status of eIF2 (37). eIF2 phosphorylation, in turn, modulates the translation rate because of its association with Met–tRNA in a ternary complex and defines the 40S subunit position with respect to the initiation codon (16, 17). We found that HuR knockout increases eIF2a phosphorylation. Mutation of the eIF2ak4 kinase domain mimics HuR deletion-mediated disruption of polysome assembly, including the elimination of Rpl7- and Rpl5-positive 80S polysomes. Although eIF2 phosphorylation is generally believed to decrease translation, it promotes the translation of some transcription factors (41, 42). Our results are consistent with these reports, because HuR knockout shifted some mRNA subsets out of polysomes, whereas others were shifted into polysomes. Together, these results suggest that HuR and eIF2ak4 cooperatively determine ribosome specificity, which influences mRNA translation in developing neocortices.
Our data show that HuR regulates multiple stages of neocortical development, including the lamination of projection neurons and formation of the corpus callosum. These results suggest a mechanism for HuR-mediated regulation of a complex developing system at the level of posttranscriptional control, where temporally sensitive HuR–eIF2ak4 interaction influences polysome assembly and translation of distinct, but functionally related, mRNA members (Fig. 6). We propose a developmental model in which an RBP regulates the rapid and coordinated translation of specific sets of functionally related mRNAs, which is essential for the formation of complex neocortical circuits. These developmental changes at the level of mRNA translation in dynamic polysomes may occur with the temporal control of intrinsic interactions, possibly driven by temporally determined extracellular signaling molecules yet to be determined.
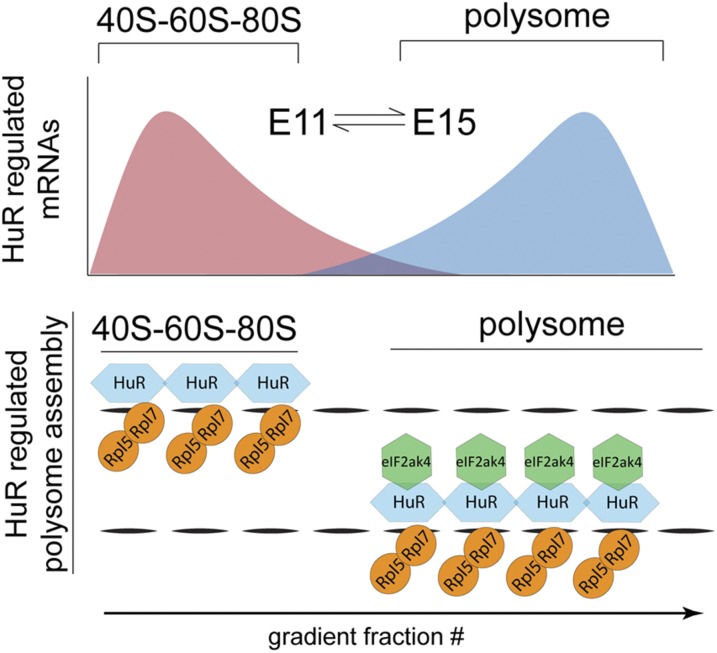
Fig. 6.
Model for dynamic RBP regulation of polysome specificity in neocortical development. HuR differentially binds distinct mRNA subsets in early vs. late neocortical neurogenesis, influencing their active polysomal translation in a temporally dependent manner. The HuR–eIF2ak4 interaction depends on the eIF2ak4 kinase domain and determines the combinatorial composition of polysomal proteins required for the temporally dependent translation of transcripts that specify neocortical circuits.
Materials and Methods
Animals.
All procedures and mouse husbandry were performed according to the Rutgers–RWJMS Institutional Animal Care and Use Committee guidelines (protocols I09-065 and I12-065-10). Generation of HuR conditional-deletion and WT littermate control animals was accomplished by using Jackson Laboratory Emx1–Cre mice [strain name: B6.129S2-Emx1tm1(cre)Krj/J; stock no. 005628] or Foxg1-Cre mice [strain name: B6.129P2(Cg)-Foxg1tm1(cre)Skm/J; stock no. 006084], crossed with HuRf/f mice (32). HuR protein depletion in genotyped HuR-cKOs was confirmed by HuR immunohistochemistry (Fig. S5). For generation of embryonic HuR deletion mice, we performed timed pregnancies in which plugs found the next day were considered E1. CD1 WT mice (Charles River) were also used for Western blot polysome analysis, along with Emx1–HuR WT littermates, compared with HuR cKOs. eIF2ak4 kinase-domain mutants (eIF2ak4mut) were obtained from the Jackson Laboratory (Strain name: B6.129S6-Eif2ak4tm1.2Dron/J; stock no. 008240).
Immunohistochemistry.
Embryonic and postnatal brains were dissected in 1× PBS and postfixed in 4% (wt/vol) paraformaldehyde (pH 7.4) overnight at 4 °C. Fixed brains were coronally sectioned on a Leica vibratome at 70 μm and prepared for immunohistochemistry as described (3, 43). Primary antibodies and concentrations used are shown in Fig. S6, and all secondary antibodies were used at 1:250 dilution in probing solution (Jackson ImmunoResearch; cy2, cy3, and cy5). Confocal imaging was performed with an FV1000MPE microscope (Olympus) by using 4×, 10×, 20×, and 60× objectives.
Polysome Fractionation.
One day before performing fractionation, sucrose density gradient columns were prepared in 11- or 2-mL ultracentrifuge polyallomer tubes (Beckman Coulter; no. 331372 or 347357). The 10–50% gradients were constructed by underlaying 10%, 20%, 30%, 40%, and 50% sucrose solutions (20 mM Tris⋅HCl, 100 mM NaCl, 10 mM MgCl2) supplemented with EDTA-free protease inhibitor (Santa Cruz Biotechnology; no. sc-29131), RNase inhibitor (Invitrogen; no. 100000840), 20 mM DTT (Invitrogen; no. NP0009), and 0.1 mg/mL cyclohexamide (Santa Cruz Biotechnology; no. sc-3508A). Columns were stored at 4 °C overnight.
E13, E16, and P0 neocortices (n ≥ 2 biological replicates per fractionation) were previously dissected and flash-frozen on dry ice. For fractionation, samples were resuspended for 10 min on ice with continuous pipetting in 250 μL of polysome extraction buffer (PEB, pH 7.4) consisting of 20 mM Tris⋅HCl, 100 mM KCl, 10 mM MgCl2, and 0.3% Igepal (Sigma-Aldrich; no. CA-630) supplemented with EDTA-free protease inhibitor (Santa Cruz Biotechnology; no. sc-29131), RNase inhibitor (Invitrogen; no. 100000840), 20 mM DTT (Invitrogen; no. NP0009), and 0.1 mg/mL cyclohexamide (Santa Cruz Biotechnology; no. sc-3508A) to homogenize the tissue. Lysates were then cleared by centrifugation (Sorvall Biofuge fresco) for 10 min at 4 °C. Total lysate RNA level was determined by using a spectrophotometer (NanoDrop ND-1000) by loading 250 or 50 μg of total RNA weight onto 11- or 2-mL columns, respectively.
Ultracentrifugation was performed at 39,000 rpm for 90 min (Sorvall Discovery 100 with Beckman Coulter SW41 rotor, UNSPSC#41103909, and buckets, #333790) for 11-mL tubes or 39,000 rpm for 50 min (Sorvall Discovery M120SE with Sorvall S-55-S rotor and buckets, #18507) for 2-mL tubes. Samples were then inserted into a tube piercer (Brandel; no. 621140007) connected to a syringe pump (Brandel) and fractionated into 14 equal-volume fractions. Total RNA absorbance was recorded throughout the fractionation (Brandel UA-6). Samples were then frozen at −80 °C. RNA was isolated from fractions by TRIzol-LS (Life Technologies; no. 10296028) extraction according to the manufacturer’s protocol, or protein analysis was performed directly from fractionated lysates with Western blot.
RNAseq and Bioinformatics.
RNA was isolated from fractionation input (total) and polysome fractions by using TRIzol LS (Life Technologies; no. 10296028) as described above. Next, equal volumes of RNA extracted from fractions 4–7 were pooled together for analysis of 40S–60S–80S-associated cytoplasmic RNA, whereas equal volumes of RNA extracted from fractions 9–12 were pooled together for polysome-associated cytoplasmic RNA. Two to three biological replicates for total, 40S–60S–80S, and polysome RNA at E13 and P0 in WT and Emx1–HuR cKO littermates, totaling 15 samples, were analyzed. Sequencing libraries were prepared by using the Illumina TruSeq RNA Sample Preparation Kit v2 according to the manufacturer's protocol. Libraries were quantified by using the Library Quantification Kit Illumina/Universal (KAPA Biosystems) and then diluted and symmetrically pooled. We performed 2 × 100-bp paired-end sequencing using the Illumina Hiseq2500 in rapid-run mode. Sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50809).
Results were aligned with the mm10 mouse genome using the University of California Santa Cruz transcript map (Illumina iGenomes) using TopHat, and comparisons between groups were made in Cufflinks and cummeRbund (30). Significant differences were judged using a 5% false discovery rate. Lists of regulated genes were assessed for enrichment of functional groups or pathways by using DAVID (33, 34). For neocortical layer-specific analysis, data from ref. 35 were used.
qRT-PCR.
RNA was isolated from sucrose gradient fractions by using TRIzol-LS (Life Technologies; no. 10296028) following the manufacturer’s protocol. qRT-PCR was performed in 10-μL reactions (equivalent fraction volumes, duplicate technical replicates for each reaction) by using the Applied Biosystems StepOne Real-Time PCR system with Step-one software (Version 2.1; no. 4376373) and the RNA-Ct 1-Step Taqman kit (no. 4392653) with Taqman probes (see Fig. S6 for catalog numbers). For each probe, n ≥ 4 neocortices were analyzed in n ≥ 2 fractionations, resulting in n ≥ 4 qRT-PCR technical replicates.
Western Blot.
Neocortical protein lysates were prepared by using either tissue-protein extraction reagent (for total protein levels; T-PER; Thermo Scientific; no. 78510) with protease inhibitor (Santa Cruz Biotechnology; no. sc29131) or taken directly from isolated polysome fractionations (PEB applied to sucrose gradients). Lysates were cleared by centrifugation for 10 min at ∼13,000 × g and analyzed for total RNA/protein content by using a spectrophotometer (NanoDrop ND-1000). The Invitrogen SureLock Western blot system with Bis-Tris 4–12% gradient gels was used with transfer onto nitrocellulose membranes (BioRad; no. 162-0214). Membranes were blocked in 5% milk, 10% FBS, and 0.3% Triton-X 100 in PBS for 1-h shaking at room temperature. Membranes were then placed in probing solution (0.3% Triton-X 100 and 10% FBS in PBS) with primary antibodies shown in Fig. S6 overnight or up to two nights while shaking at 4 °C. Blots were then washed in 0.3% Triton-X 100 in PBS three times for 5 min and placed in probing solution with corresponding HRP-conjugated secondary antibody (Jackson ImmunoResearch; 1:2,500) for 1 h at room temperature. Blots were developed (Protein Simple ChemiGlow; no. 60-12596-00) and imaged and quantified with Genesnap software and a Syngene G:Box imager.
Quantification and Analysis of qRT-PCR and Western Blot.
For the statistics of polysome fractionation qRT-PCR analyses, the raw CT value for each of the individual fractions was transformed to 2−CT and normalized to the sum total for all fractions, generating a percentage of total transcript within each fraction. For Western blot quantification, the band intensity was measured above background with Genesnap software and a Syngene G:Box imager and was similarly normalized to percentage total protein within each fraction. In both cases, each fraction's values were aggregated into different categories corresponding to different phases of polysome assembly on a total RNA absorbance curve. For qRT-PCR analysis, fractions 1–3 were summed into “free”; fractions 4 and 5 were summed into “40S–60S”; fractions 6 and 7 were summed into “80S”; fractions 8 and 9 were summed into “light”; and fractions 10–13 were summed into “heavy”—corresponding to peaks on total RNA absorbance curves monitored during fractionation. For Western blot analysis, fractions 1–3 were summed into free; fractions 4–7 were summed into “40S–60S–80S”; and fractions 8–14 were summed into “polysome.” For significance testing of qRT-PCR data, t tests were conducted between WT and HuR-cKO in each category, with P < 0.05 considered significant. SEM is shown as error bars in figures.
Further, more-stringent statistical testing of qRT-PCR data via simulation occurred as follows. Overall significance of differences in fraction means was determined by first forming a test statistic of the ratio of the sums of squares across groups (WT and cKO) to the sum of squares for error (i.e., sum of squares within groups), using the Euclidean metric across the five categories (free, 40S–60S, 80S, light polysome, and heavy polysome). This statistic is analogous to the F statistic in ordinary ANOVA, but because the response is not normally distributed, the statistic does not have a simple distribution. The sample sizes were not large enough to use permutation testing to assess significance, as was done in refs. 44 and 45, so instead we used a Monte Carlo approach. As a null distribution, we began by matching the number of biological replicates with the actual data. For each biological replicate, we averaged repeated draws of multinomial random variables with probabilities matching the observed overall means. The number of draws was selected so that the expected sum of squares for error in the simulated data matched the sum of squares for error in the observed data. The resulting test statistic was recorded, and the procedure was repeated. In the case of the overall significance test, the null distribution of the test statistics was not dependent on the number of draws per biological replicate. To test for significant pairwise free, 40S–60S, 80S, light polysome, and heavy polysome differences between groups, we used the same Monte Carlo framework but recorded the largest pairwise difference across all of the categories. Differences in the observed data were considered significant if they exceeded the 95th percentile of this “maximum pairwise difference” null distribution. Importantly, this procedure adjusts for multiple comparisons of means in the experiment. The distribution was sensitive to the number of draws per biological replicate, so all significant results were verified with sensitivity analysis for the number of draws.
IP.
HuR and eIF2ak4 IP was performed by using the Pierce Crosslink IP kit according to the manufacturer’s protocol. A total of 10 µg of GtαHuR (Santa Cruz; no. sc-5483), RbαeIF2ak4 (Cell Signaling; no. 3302S), and Gt/Rb IgG controls were applied to each column. Four neocortices were resuspended in Pierce lysis buffer, and 300 μL was applied to the column in each condition. The eluate was analyzed by Western blot as described.
CP Measurements and Layer Marker Quantification.
CP thickness was measured at three points (medial, mid, and lateral) in DAPI-stained 4× confocal images of neocortical coronal sections with Neurolucida software. Layer-marker positive cells and their distribution was quantified by drawing a grid of standard width (300 pixels) between the ventricular surface and the superficial surface of neocortical layers 2/3, dividing the grid into 10 bins of equal height as shown in Fig. 5A. Multivariate ANOVA (MANOVA) was conducted with ANOVA proceeding it if MANOVA was P < 0.05. ANOVA was then conducted followed by post hoc analysis if ANOVA was P < 0.05. Post hoc analysis depended on the equality of variances. If Levene's statistic proved that a given group had equal variances (P ≥ 0.05), Tukey’s honest significant difference (HSD) was used for multiple comparisons, whereas for unequal variances (P < 0.05), Games–Howell was used. SEM is shown as the error bars in figures.
Supplementary Material
Acknowledgments
We thank all current and previous members of the M.-R.R. laboratory as well as many colleagues on the Rutgers/Robert Wood Johnson Medical School (RWJMS) campus for comments. We thank the laboratories of Darnell, Kinzy, and Copeland for support in developing the polysome protocol. This work was supported by National Institutes of Health (NIH) Grants NS064303 and NS075367 and RWJMS start-up funds (to M.-R.R.). R.P.H. was supported by NIH Grants DA032984 and DA035594. K.T. was supported by National Science Foundation Integrative Graduate Education and Research Traineeship (IGERT) Program Fellowship DGE0801620.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50809).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408305111/-/DCSupplemental.
References
- 1.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 2.Rakic P. Evolution of the neocortex: A perspective from developmental biology. Nat Rev Neurosci. 2009;10(10):724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasin M-R, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10(7):819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- 4.Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139(9):1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deboer EM, Kraushar ML, Hart RP, Rasin M-R. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience. 2013;248C:499–528. doi: 10.1016/j.neuroscience.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18(1):28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31(4):1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66(6):848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusek G, et al. Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell. 2012;11(4):505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver DL, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13(5):551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 14.Kwan KY, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 16.Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13(6):383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- 17.Mathews M, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. [Google Scholar]
- 18.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barna M. Ribosomes take control. Proc Natl Acad Sci USA. 2013;110(1):9–10. doi: 10.1073/pnas.1218764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee N, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43(3):327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Katsanou V, et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19(6):777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Ayoub AE, et al. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108(36):14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9(2):110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 26.García-Domínguez DJ, Morello D, Cisneros E, Kontoyiannis DL, Frade JM. Stabilization of Dll1 mRNA by Elavl1/HuR in neuroepithelial cells undergoing mitosis. Mol Biol Cell. 2011;22(8):1227–1239. doi: 10.1091/mbc.E10-10-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srikantan S, Gorospe M. HuR function in disease. Front Biosci (Landmark Ed) 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esposito AM, et al. Eukaryotic polyribosome profile analysis. J Vis Exp. 2010;(40):5–8. doi: 10.3791/1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–14. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsanou V, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29(10):2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Belgard TG, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27(19):6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez M, Wek RC, Hinnebusch AG. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(6):3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou S-J, Perez-Garcia CG, Kroll TT, O'Leary DDM. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niquille M, et al. Transient neuronal populations are required to guide callosal axons: A role for semaphorin 3C. PLoS Biol. 2009;7:e1000230. doi: 10.1371/journal.pbio.1000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu T, Richards LJ. Cortical axon guidance by the glial wedge during the development of the corpus callosum. J Neurosci. 2001;21:2749–2758. doi: 10.1523/JNEUROSCI.21-08-02749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101(31):11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou D, et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283(11):7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 43.DeBoer EM, et al. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J Neurosci. 2014;34(10):3674–3686. doi: 10.1523/JNEUROSCI.3703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson M. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 45.McArdle B, Anderson M. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.