Significance
Cellular and subcellular structures in thick biological samples typically are visualized either by genetically encoded fluorescent proteins or by antibody staining against proteins of interest. However, both approaches have drawbacks. Fluorescent proteins do not survive treatments for tissue preservation well, are available in only a few colors, and often emit weak signals. Antibody stainings are slow, do not penetrate thick samples well, and often result in considerable background staining. We have overcome these limitations by using genetically encoded chemical tags that result in rapid, even staining of thick biological samples with high-signal and low-background labeling. We introduce tools for flies and mice that drastically improve the speed and specificity for labeling genetically marked cells in biological tissues.
Keywords: immunohistochemistry, neural circuits, protein labeling, fluorescence microscopy
Abstract
Genetically encoded fluorescent proteins and immunostaining are widely used to detect cellular and subcellular structures in fixed biological samples. However, for thick or whole-mount tissue, each approach suffers from limitations, including limited spectral flexibility and lower signal or slow speed, poor penetration, and high background labeling, respectively. We have overcome these limitations by using transgenically expressed chemical tags for rapid, even, high-signal and low-background labeling of thick biological tissues. We first construct a platform of widely applicable transgenic Drosophila reporter lines, demonstrating that chemical labeling can accelerate staining of whole-mount fly brains by a factor of 100. Using viral vectors to deliver chemical tags into the mouse brain, we then demonstrate that this labeling strategy works well in mice. Thus this tag-based approach drastically improves the speed and specificity of labeling genetically marked cells in intact and/or thick biological samples.
The revolution in live imaging resulting from the use of genetically encoded fluorescent proteins (FPs) is widely appreciated (1, 2), but FPs also have had a major impact on studies of fixed, whole-mount specimens or thick sections. Processing of large or intact pieces of tissue has obvious advantages over sectioning, such as reduced tissue damage, compatibility with fast imaging modalities (e.g., light sheet microscopy), and easy subsequent 3D reconstruction. The drastic increase in imaging throughput by using whole-mount brains had a major impact on Drosophila neurobiology, in which reconstruction of neural circuits is a key requirement. Recently, several methods have been developed that allow whole-mount imaging of the mouse brain: CLARITY (3), Scale (4), SeeDB (5), and CUBIC (6) all render the brain optically transparent (although to different degrees). In such samples, imaging the native fluorescence of genetically encoded FPs offers the advantages of immediate visualization, low background, and spatially even signal. However, FP signals are easily quenched by fixation or other staining procedures, suffer from limited spectral flexibility, and often emit weak signals. Therefore, antibody detection of marker proteins remains essential in many experimental situations. Immunostaining, however, is notoriously slow, highly nonlinear, and often results in uneven labeling with high background levels. Therefore, there are undesirable tradeoffs in the antibody vs. FP labeling techniques.
These tradeoffs are a major practical issue for our research in neural circuit tracing in Drosophila (7–12). We therefore sought staining methods that combine the positive aspects of both FPs and antibody-based staining, notably fast, even, strong, and spectrally diverse signals with low background labeling. We have developed an approach based on four commercially available, orthogonal labeling chemistries (SNAP-, CLIP-, Halo- and TMP-tag) characterized by the covalent binding of a large range of fluorescent substrates to engineered enzyme tags, as described below. To use these chemistries for effective tissue labeling, we have generated the first (to our knowledge) stable transgenic reporter animals bearing these tags. We validate their use and expression in Drosophila, presenting 12 widely applicable fly strains for labeling of cells and subcellular structures. Combining these tags with the first (to our knowledge) knockin construct, we demonstrate that our approach can speed up most tissue-imaging procedures in Drosophila neurobiology from 1 wk (11, 13) to 1 h, a factor of >100, while giving more homogeneous staining and reduced background signals.
These positive results prompted us to extend the approach to mice. We have developed and validated the first (to our knowledge) viral vectors encoding these chemical tags. We show that thick brain-tissue samples can be stained rapidly with an excellent signal-to-noise ratio, allowing easy reconstruction of single neurites, critical for neural circuit mapping. We then demonstrate chemical labeling of more refined cell populations, introducing a Cre-dependent virus for intersectional labeling of genetically defined cell populations.
In conclusion, the chemical labeling reagents that we have developed and validated solve a basic but pervasive problem in tissue labeling and have immediate applications across model organisms and experimental disciplines.
Results
Expression of Chemical Tags in the Drosophila Brain.
We sought to develop a labeling system that overcomes the limitations of antibody-based immunostaining, i.e., speed (poor penetration of thick tissue samples), specificity (background staining caused by off-target binding), and complexity (number of user interactions, i.e., manual steps, in staining protocols).
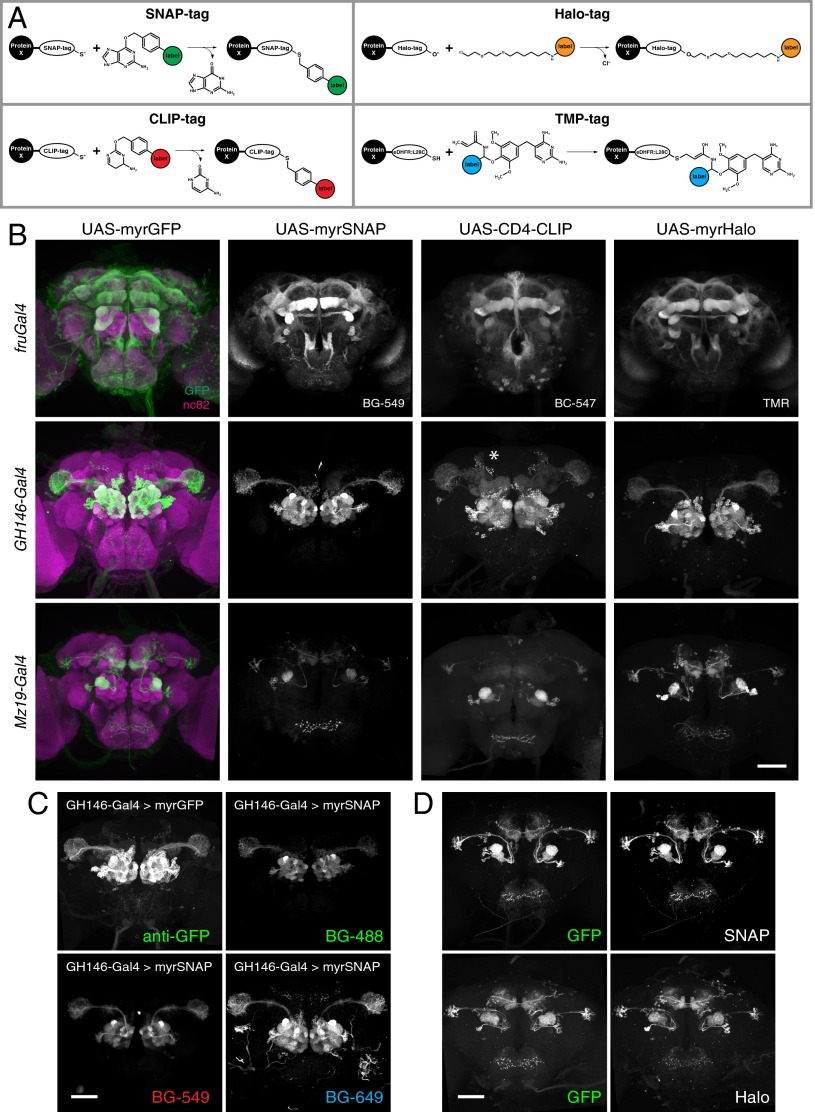
Existing chemical tagging systems were compared, and four were chosen (i) that do not require cofactors, (ii) that result in formation of a covalent bond, and (iii) for which a spectrally diverse range of fluorescent substrates is available commercially. The SNAP-tag and CLIP-tag (New England Biolabs) (14, 15), Halo-tag (Promega) (16), and TMP-tag (Active Motif) (17, 18) systems fulfilled these requirements. All four rely on the rapid formation of a covalent bond between an engineered, bio-orthogonal enzyme (tag) and a small reactive group that is fused to a reporter (substrate) (19). SNAP- and CLIP-tag are modified, 181-aa O6-alkylguanine-DNA alkyltransferases that bind benzylguanine (BG) or benzylcytosine (BC) derivatives, respectively. Halo-tag (295 aa) is an engineered bacterial haloalkane dehalogenase that binds chloroalkane groups, and TMP-tag (159 aa) is an Escherichia coli dihydrofolate reductase (eDHFR) that has been engineered to bind trimethoprim (TMP) derivatives covalently (Fig. 1A).
Fig. 1.
Expression of chemical tags in the fly brain. (A) Chemical-labeling chemistries used in this study (adapted from refs. 14–16, 18; see text for description). (B) Expression patterns of membrane-targeted tags in the Drosophila central brain. The panel is arranged in a three-row by four-column grid. Rows represent Gal4 driver lines; columns represent reporter constructs. In the far-left column, nc82 neuropil counterstaining is shown in magenta. The fluorescent substrates used are indicated for each tag. Note that although myrGFP, myrSNAP, and myrHalo are targeted attP insertions, CD4-CLIP is a P element insertion. Because of positional effects, the CD4-CLIP reporter shown here labels the bilateral anterior paired lateral neuron when crossed to GH146-Gal4 (asterisk). (C) Brains of GH146-Gal4 animals expressing either myrSNAP or myrGFP stained with GFP antibody or with fluorescent BG-488, BG-549, or BG-647 substrates, as indicated. (D) Comparison between native GFP signal and chemical labeling in brains of Mz19-Gal4 > myrGFP, myrSNAP (Upper) and Mz19-Gal4 > myrGFP, myrHalo (Lower) animals. (Scale bars: 50 µm.)
We made fusion constructs of these tags with membrane or synaptic proteins and cloned them downstream of a Gal4 upstream activating sequence (Gal4 UAS) (20). The constructs then were injected into Drosophila melanogaster, generating a total of 12 transgenic lines (Table S1).
We tested the expression of these fusion constructs using three different Gal4 driver lines: fruGal4, which drives expression in about 2,000 fruitless-positive neurons (21); GH146-Gal4, which is expressed in most olfactory projection neurons (22); and Mz19-Gal4, which is expressed in three classes of projection neurons, DA1, VA1d, and DC3 (23). As a reference, each driver line was crossed to flies expressing membrane-targeted GFP (myrGFP). When brains from the progeny of these driver–tag crosses were incubated with fluorescent SNAP, CLIP, Halo, or TMP substrates (Table S2), strong, specific labeling was observed (Fig. 1B and Fig. S1). We tested all tag–substrate combinations for potential cross-reactivity and found no signal from noncognate tag–substrate pairs, with the exception of weak binding of CLIP substrates to SNAP-tag (Fig. S2). This result is expected, because it has been shown previously that BC substrates have an ∼100-fold preference for CLIP (an engineered version of SNAP) over SNAP, whereas BG groups have an >1,000-fold preference for SNAP over CLIP (15). Because of this low cross-reactivity and the availability of spectrally diverse substrates (Fig. 1C), this labeling approach potentially allows the simultaneous visualization of up to four orthogonal channels. Importantly, because of its speed and robustness, chemical labeling is compatible with the use of both FPs and immunostaining.
Ultrafast Labeling of Thick Tissue Samples.
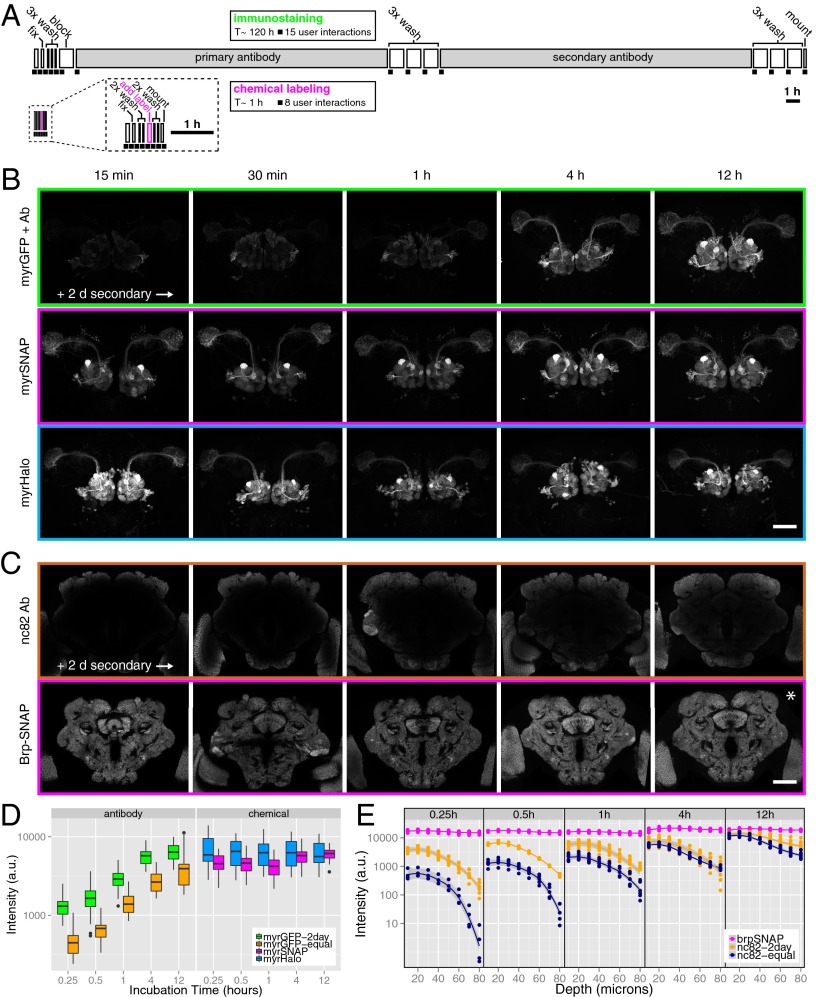
Low fluorescence after fixation is a widely appreciated limitation of genetically encoded FPs (6, 24, 25). Therefore, immunostaining is required to obtain sufficient signal from fixed samples. Antibody diffusion into tissue is a rate-limiting step for immunostaining of thick specimens. For example, homogeneous immunostaining of Drosophila brains (∼500 × 250 × 200 µm in x, y, and z planes) for the nc82/Bruchpilot (Brp) synaptic protein (26) (the standard counterstain for brain structure in Drosophila neuroanatomical studies; see below) takes 7 d (Fig. 2A) and requires >15 user interactions. This method comprises a blocking step and prolonged incubations with primary and secondary antibodies (8, 11). Similarly, homogeneous staining of 200- to 300-µm slices of mouse brain can take days to weeks, often requiring laborious cryostat reslicing of the sample. These long staining times result, in part, from the relatively large size (∼150 kDa) of antibodies, which limits their diffusion into thick tissue samples. We reasoned that the much smaller size (∼1 kDa) of fluorescent chemical labeling substrates would speed up the staining process considerably.
Fig. 2.
Ultrafast and homogeneous tag-based tissue staining. (A) Direct comparison of immunostaining and chemical-labeling protocols. In these ethograms the length of individual steps is proportional to the time required, and each black square represents a manual interaction. Chemical labeling is >100× faster (∼1 h vs. >100 h) and requires half as many (8 vs. 15) manual handling steps of the sample. (B) Staining time course of GH146 projection neurons using immunostaining against membrane-targeted GFP (Top Row), chemical labeling using SNAP-tag (Middle Row), or Halo-tag (Bottom Row). The z-maximum intensity projections from 3D confocal stacks are shown. Incubation times are indicated for primary antibody or chemical substrates (GFP antibody, SNAP BG-549, Halo-TMR, nc82-antibody). Note that incubation with secondary antibodies was for 2 d. (C) Staining time course of the nc82/Brp neuropil marker using immunostaining against Brp protein (Upper Row) or chemical labeling using SNAP-tag (Lower Row). Single coronal confocal slices through the center of the brain are shown. Note that incubation with secondary antibodies was for 2 d. Also note that background labeling increases with longer substrate incubation times (asterisk). (Scale bars in B and C: 50 µm.) (D) Quantification of signal intensity over time in GH146-Gal4 brains labeled with antibody vs. chemical labeling. Secondary antibodies for GFP immunostaining were incubated for 2 d (myrGFP-2day) or for the same duration as primary antibodies (myrGFP-equal). Fluorescence was quantified using a GH146 mask (n = 5–8 brains per condition) (Fig. S3B and SI Materials and Methods). (E) Quantification of labeling intensity (SI Materials and Methods) at different depths from the brain surface in brains labeled with nc82 antibody or chemically labeled (using Brp-SNAP). Secondary antibodies for nc82 immunostaining were incubated for 2 d (nc82-2day) or for the same duration as primary antibodies (nc82-equal). Five different time points are shown.
We directly compared the labeling speed of antibodies and chemical substrates in fly brains expressing either membrane-targeted GFP or membrane-targeted SNAP-tag in olfactory projection neurons (PNs). Brains were dissected, fixed, briefly permeabilized, and incubated with GFP antibody or fluorescent chemical substrate for 15 min, 30 min, 1 h, 4 h, or 12 h (SI Materials and Methods). After a washing step, SNAP-labeled brains were mounted. GFP-expressing brains were incubated with fluorophore-conjugated secondary antibody for 2 d, followed by washing and mounting. In a separate time-series experiment, the incubation times in primary and secondary antibodies were identical (Fig. S4A). Homogeneous staining of PNs with anti-GFP antibody required >4 h incubation with primary antibody followed by 2 d of incubation with secondary antibody (Fig. 2B, Top), whereas strong and uniform chemical labeling was visible after only 15 min (Fig. 2B, Middle). Such rapid labeling also was observed when using a membrane-targeted Halo-tag (Fig. 2B, Bottom). Quantification of staining intensity (SI Materials and Methods) revealed that antibody staining increased over the course of 12 h, whereas chemical staining reached nearly maximal levels after 15 min (Fig. 2D). These results show that the chemical labeling method leads to drastically reduced staining times, and, because no blocking step and far fewer washing steps were required, the protocol resulted in half as many user interactions (Fig. 2A).
We next assessed the uniformity and speed for labeling the nc82/Brp antigen, a widely expressed presynaptic protein (26). We compared nc82 antibody labeling of wild-type (Canton-S) brains with a SNAP-tag knockin construct inserted into the brp locus (brp-SNAP) (Fig. S5). Strikingly, nc82 immunostaining for 12 h (primary) followed by 2 d (secondary) still resulted in substantial signal drop-off in the center of the brain (Fig. 2C, Upper), but chemical labeling produced uniform staining within 15 min (Fig. 2C, Lower). Indeed we found that staining times of only 1 min were sufficient (Fig. S4D). Longer incubations did not increase neuropil labeling appreciably but instead resulted in higher background (visible in Fig. 2C, asterisk); this nonspecific labeling could be removed by extended washes (Fig. S6, asterisk). This result indicates that most of the tag molecules are occupied by substrate after minutes of incubation. We tested this finding directly by sequential labeling of the same sample with two different substrates (Fig. S6).
Using immunohistochemistry, we found that labeling the center of the brain (∼100 µm from the surface) required at least 12 h of incubation with primary antibody followed by 2 d of incubation with secondary antibody (Fig. 2E). In contrast, fluorescent SNAP substrates uniformly labeled the sample within 15 min (Fig. 2E). Therefore, the chemical labeling approach enables rapid and homogeneous staining of thick tissue samples.
Finally, we tested the effect of fixation on the labeling reaction, incubating samples for 20, 40, or 60 min in 4% (wt/vol) paraformaldehyde (PFA) dissolved in PBS before adding substrates. We found that SNAP- and CLIP-tag were largely insensitive to prolonged fixation, but Halo-tag labeling decreased and TMP-tag labeling increased with longer fixation (Fig. S7). We did not observe fixation-dependent changes in background labeling (Fig. S7).
These results show that chemical labeling of thick tissue samples is at least two orders of magnitude faster than immunostaining, requires half as many user interactions, and results in better tissue labeling (Fig. 2A and Table 1).
Table 1.
Comparison of immunostaining and chemical tag-based staining approaches
| Characteristic | Immunostaining | Chemical labeling |
| Duration* | Hours–days | Minutes–hours |
| User interactions | >15† | 8 |
| Substrate size kDa | 150 | 1 |
| Staining steps | 2‡ | 1 |
| Blocking | Yes | No |
| Background | Often high | Low |
| Epitope stability | Variable | High |
| Cell permeable | No | Yes/No |
| No. of channels | Many | 4 (currently) |
| Transgenics | No | Yes |
| Stoichiometry§ | Variable (> 1:1) | 1:1 |
Staining durations indicated are typical for thick tissue samples (hundreds of micrometers to several millimeters).
Using standard immunostaining protocol for Drosophila brains (see text for references).
Fluorophore-coupled primary antibodies are sometimes used, reducing the number of staining steps to one.
I.e., fluorescent label-to-protein tag ratio. For immunohistochemistry, this ratio cannot be easily quantified.
Applications in Drosophila Neurobiology.
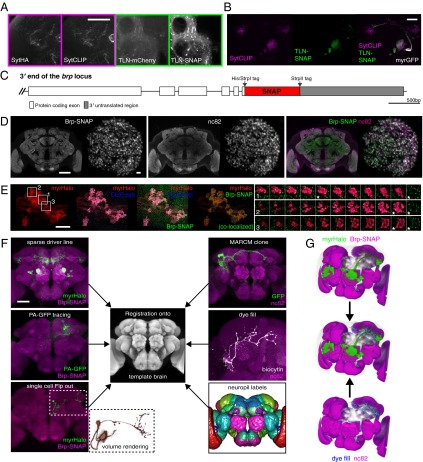
The Drosophila reagents that we have described here have immediate and widespread utility in cell and developmental biology and neurobiology. To demonstrate this utility more clearly, we optimized Drosophila transgenics for applications that are at the heart of large-scale studies of neural circuits in the fly brain. First, we constructed synaptically localized tags and compared them with previously established synaptic markers in Drosophila. Simultaneous immunostaining against the synaptotagmin-HA presynaptic marker (SytHA) (7, 27, 28) and chemical labeling with a SytCLIP construct revealed that chemical labeling consistently achieved higher signal-to-noise ratios with identical marker localization (Fig. 3A, Left). This benefit was even more pronounced when the somatodendritic marker telencephalin (TLN)-mCherry (DenMark) (29) was compared with a TLN-SNAP construct (Fig. 3A, Right). These direct comparisons point out that in some cases the chemical labeling approach can overcome one of the most common shortcomings of immunohistochemistry: high background staining caused by poor epitope specificity and/or cross-reactivity. We also used these synaptic markers simultaneously to label pre- and postsynaptic compartments of DA1 projection neurons (Fig. 3B), recapitulating previously observed localization patterns. Additional reporter constructs containing synaptic protein fusions are characterized in Fig. S1.
Fig. 3.
Key applications for chemical labeling in Drosophila neurobiology. (A) Comparison between epitope-tagged synaptic markers (HA-tag, mCherry) and synaptically targeted chemical tags (SNAP-tag, CLIP-tag). Confocal slices from the brain of a fruGal4> SytHA, SytCLIP (presynaptic markers) animal (Left) and a fruGal4> TLN-mCherry (DenMark), TLN-SNAP (somatodendritic markers) animal (Right) are shown. (Scale bar: 50 µm.) (B) Simultaneous labeling of presynaptic sites and the somatodendritic compartment of DA1 projections neurons using Mz19Gal4. (Scale bar: 50 µm.) (C) Map of the brp-SNAP knockin. (D) nc82 and Brp-SNAP signals colocalize in a brp-SNAP/+ brain simultaneously labeled with BG-549 and nc82 immunostaining. Single coronal slices through the middle of the brain (Left) and through a deconvolved image stack of the DA1 glomerulus (Right) are shown. Note the more even Brp-SNAP staining in the center of the brain. (Scale bars: 50 µm, Left; 2 µm, Right.) (E) Deconvolved, confocal z-stack images from the MB calyx region of a brp-SNAP, Mz19-Gal4 > CLIP-syb, myrHalo brain triple-labeled with fluorescent BG-488, BC-547, and Halo-SiR substrates. Colocalization analysis of SNAP-tag– and Halo-tag–labeled regions reveals presynaptic sites (Brp puncta) within PN terminals (see also Movie S1). (Scale bar: 10 µm.) Montages show three individual, triple-labeled projection neuron terminals (Right, 1–3) derived from the indicated boxed regions. One selected optical section (marked with an asterisk) is also shown with Brp-SNAP only (green), revealing that puncta are localized to the surface of each terminal. (F) Chemically labeled neurons can be registered successfully onto a template using the Brp-SNAP neuropil counterstaining, thus allowing direct comparison with imaged neurons from other sources, such as those derived from stochastic labeling (Flp-out), PA-GFP tracing, whole-cell recordings, or sparse driver lines, (e.g., Mz19-Gal4). (Scale bar: 50 µm.) (G) Overlay of DA1 projection neurons (green) from a chemically labeled brp-SNAP, Mz19-Gal4 > myrHalo brain (Left) overlaid with a dye-filled third-order olfactory neuron (cyan) from an nc82-stained brain (Right) after registration. Note the overlap between axon terminals and dendritic arbor (white arrowheads).
The nc82 monoclonal antibody is commonly used as a presynaptic marker in Drosophila and has been a key reagent in characterizing synaptic active zones (30). It also is the most widely used counterstain for neuroanatomical studies in Drosophila brain. (At present Google Scholar lists 788 publications referring to this antibody.) In computational neuroanatomy, nc82 has been used to construct standard brains for 3D atlases by image registration (7, 8, 11, 31) and is the basis of a 3D model encapsulating the recently standardized nomenclature for Drosophila brain regions (32, 33) (see also www.virtualflybrain.org). Homogeneous staining of neuropil structures throughout the brain is absolutely critical for successful registration (7, 8).
In our direct comparative studies, we found that the newly generated brp-SNAP knockin (Fig. 3C) closely recapitulated the neuropil staining obtained with nc82 antibody (Fig. 3D). However, double labeling dramatically emphasized the incomplete antibody penetration into the center of the brain. Higher-resolution imaging of the DA1 olfactory glomerulus showed complete colocalization between nc82 and Brp-SNAP puncta, but the relative labeling intensity was spatially variable; much of this variation may be caused by antibody penetration, because there was an nc82 intensity gradient from external to internal regions of the glomerulus. Next, we performed triple chemical labeling of brains expressing myrHalo, CLIP-synaptobrevin (CLIP-syb) and Brp-SNAP, and imaged DA1 PN terminals in the mushroom body (MB) (Fig. 3E). Deconvolution of these image stacks clearly revealed many discrete Brp-SNAP puncta (i.e., active zones) decorating the surface of individual PN axon terminals (Fig. 3E, 1–3). Therefore, brp-SNAP provides excellent signal-to-noise to localize key presynaptic structures. (See Movie S1 for 3D-rendered and colocalized images.)
We then confirmed that chemically labeled brains can be registered easily onto an existing nc82-immunostained template brain (IS2) (8) with 100% success (n = 84 specimens). Therefore, by using the chemical Brp-SNAP counterstain, rapidly labeled brains from different experimental sources can be registered into the same reference space reliably, enabling direct comparison of labeled structures. To demonstrate this approach, we generated a fly line bearing both an UAS-myrHalo reporter gene and brp-SNAP, crossed it to the sparse Mz19-Gal4 line, and registered the brains of progeny after 30 min of labeling (Fig. 3F). To test whether chemical labeling is sufficient for single-cell studies, we used flippase (Flp)-mediated recombination to label GH146 PNs stochastically (SI Materials and Methods). Indeed, single GH146 PNs were labeled with high signal-to-noise (Fig. 3F), showing that our approach is valuable for high-resolution neuroanatomical studies. Another powerful technique for sparse labeling of neuronal subsets is the use of photoactivatable GFP (PA-GFP). Here spatially targeted laser stimulation can identify neurons with processes in a specific region of the brain (34, 35). However, although these methods reveal single neuronal morphology, they cannot be used easily with image-registration approaches because the PA-GFP signal is substantially reduced by standard neuropil immunostaining protocols. Here we demonstrate that Brp-SNAP can be used to allow coregistration of PA-GFP specimens (Fig. 3F).
All these newly generated data can be integrated with existing resources defined by registration against nc82 template brains, such as stochastically labeled single neurons (7), lineage clones (8, 36, 37), large-scale expression screens (11), single recorded neurons (12), or the standard neuropil regions defined by the Insect Brain Name working group (33) (Fig. 3F). We demonstrated this important point by coregistration of chemically labeled Mz19-Gal4 DA1 projection neurons with a dye-filled third-order olfactory neuron (Fig. 3G) (12).
Chemical Labeling of Mouse Brain Tissue.
The positive results in flies prompted us to extend this labeling approach to a vertebrate model system. We made adeno-associated virus (AAV) constructs to deliver tags into the mouse brain. We focused on SNAP-tag with its fast labeling kinetics (14) and wide range of available substrates. In pAAV-myrSNAP, myristoylated SNAP-tag (myrSNAP) and histone-GFP (H2B-GFP) are driven by the cytomegalovirus (CMV) promoter (Fig. 4A).
Fig. 4.
Chemical labeling of the mouse brain. (A) Virus construct for chemical labeling of mouse brain. (B) A 200-µm coronal vibratome slice from a mouse brain injected with AAV-myrSNAP into hippocampus, stained with BG-549 (SI Materials and Methods). Insets show counterstaining against the neuronal marker NeuN (Left) and individual labeled neurons (Middle, arrowheads). (Scale bars: 250 µm in overview panels; 100 µm in Insets.) (C) Overview scan of a coronal vibratome slice from mouse brain unilaterally injected with AAV-myrSNAP into hippocampus and stained with BG-549 (Left). Numbered boxed areas 1–3 are expanded on the right. Single contralateral SNAP-positive projections are visible on the noninjected side (C, 2, Lower Right). (Scale bar: 500 µm.) (D) Background labeling from BG-549 is very low in zones devoid of transfected neurites (area below the dashed line; see box in B, Right). (Scale bar: 100 µm.) (E) Volume rendering of the area indicated by the dashed rectangle in C, 2, Lower Right). The high signal-to-noise ratio of BG-549–labeled processes allows facile 3D reconstruction. Four separate traced neurites are shown in different colors. (Scale bar: 200 µm.) (F) Side views of maximum-projected 3D confocal stacks of brain slices labeled with myrSNAP (Left, samples from B) or stained with anti-Tau antibody (Right) are shown with incubation times indicated. Two individual neurites are highlighted in magenta. (G) The intensity profile along the z axis for individual SNAP-labeled fibers was quantified (n = 9–11 per condition) (SI Materials and Methods). Then the normalized intensity profile was plotted for each labeling time (Left). For Tau staining, image intensity was quantified directly, because all neurons are labeled (Right) (SI Materials and Methods).
C57BL/6 mice were infected with AAV-myrSNAP in the hippocampus (SI Materials and Methods), and coronal vibratome sections were prepared. Sections were permeabilized and incubated with fluorescent SNAP substrates (SI Materials and Methods).
We observed strong and specific SNAP-tag labeling of GFP+ transfected cells in the hippocampus (Fig. 4 B and C), which were identified as neurons by costaining against NeuN (Fig. 4B, Left). Single neurons were labeled with high signal-to-noise ratio (Fig. 4B, Center) and, in contrast to intensely labeled areas containing neurites from infected cells, areas devoid of neurites exhibited no labeling (Fig. 4D). Importantly, the signal-to-noise ratio of chemically labeled brain slices was high enough to trace and reconstruct single neurites in an image stack (Fig. 4E).
Next, we wanted to assess whether mouse brain tissue could be labeled rapidly and evenly using chemical tags. We incubated permeabilized 200-µm coronal brain sections with fluorescent substrate for 30 min, 1 h, 6 h, or 12 h, acquired confocal stacks, and quantified the staining intensity of individual fibers projecting throughout the volume along the z axis (Fig. 4F and SI Materials and Methods). We found that intensity profiles of fibers were constant along the z axis after 30 min of incubation (Fig. 4G), showing that mouse brain tissue can be labeled as rapidly as whole-mount fly brains of similar thickness. Labeling of even thicker (400-µm) sections also appeared uniform after 30 min of incubation (Fig. S8D).
To compare the speed and penetration of chemical labeling in brain slices with immunohistochemistry, we performed antibody staining against the axonal marker Tau. Coronal 200-µm brain sections were incubated with anti-Tau antibody for 30 min, 1 h, 6 h, or 12 h, followed by 2-d incubation with secondary antibodies (Fig. 4F, Right). Quantification of the staining intensity along the z axis revealed a large (>50%) drop off in intensity in the center of the slice (Fig. 4G and SI Materials and Methods) even after 12 h incubation with primary antibody (2.5 d total incubation). These results mirror our findings from Drosophila brains, confirming that chemical labeling provides a significant improvement in both speed and spatial homogeneity when staining thick tissues.
We also successfully tested a range of spectrally diverse fluorescent substrates on mouse brain slices (Fig. S8 A–C). Therefore simultaneous multicolor chemical labeling should be feasible in the mouse brain.
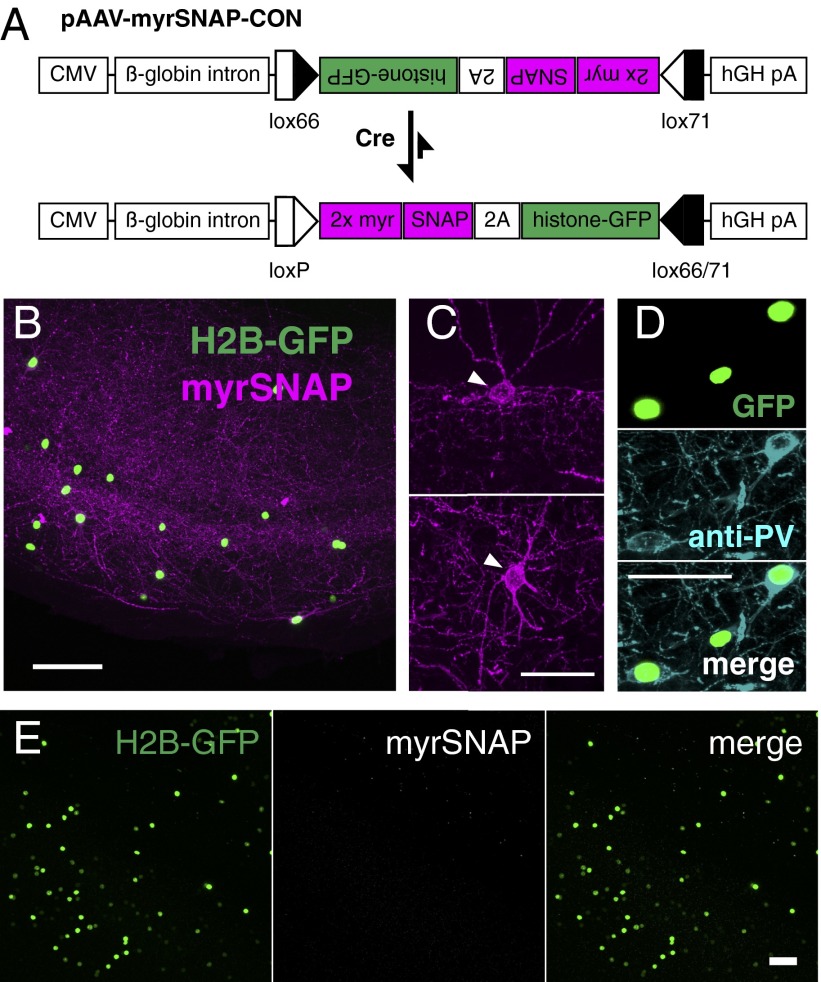
To target chemical labeling of mouse brain tissue to genetically specified neurons, we made a conditional construct, pAAV-myrSNAP-CON, in which the two markers are expressed only after Cre-mediated inversion (Fig. 5A). Adding the SNAP substrate to brain slices from parvalbumin (PV)-Cre animals injected with AAV-myrSNAP-CON (Fig. 5 B and C) specifically labeled PV+ interneurons, as confirmed by anti-PV immunostaining (Fig. 5D). Signal was completely dependent on the presence of substrate (Fig. 5E). This finding demonstrates that chemical labeling can be used in intersectional labeling strategies combining pAAV-myrSNAP-CON with the wide range of Cre-driver mouse lines and makes our approach applicable for a wide variety of studies in diverse tissues, including tracing long-range neuronal projections.
Fig. 5.
Chemical labeling of genetically defined neurons in mouse brain. (A) Virus construct for conditional (Cre-dependent) chemical labeling of mouse brain. (B) Coronal 200-µm vibratome slice from PV-Cre mouse brain injected with AAV-myrSNAP-CON, stained with BG-549. (C and D) Single neurons labeled with BG-549 (Scale bar: 250 µm) (C) and stained with anti-PV antibody (Scale bar: 50 µm) (D). (E) No-substrate control for brain injected with AAV-myrSNAP-CON (coronal vibratome slice through the hippocampus). (Scale bar: 50 µm.)
Discussion
This work addresses a fundamental need for visualizing genetically marked cellular structures in thick or intact tissue samples with high signal-to-noise ratio. It combines the advantages of immunostaining (spectral flexibility and high signal after fixation) and genetically encoded fluorophores (low background) while drastically reducing staining time. The approach presented here is practical: It uses extensively tested and commercially available reagents in a previously unreported context. Thus, little or no optimization will be required by researchers wishing to introduce this method. Because the building blocks for coupling reporter molecules to reactive groups also are available, customized substrates can be made by individual groups. The use of suitable probes can allow chemical labeling to be combined with electron or superresolution microscopy in the future. One advantage of chemical labeling in this context is its inherent linearity (i.e., one substrate molecule binds to one tag molecule), in contrast to often highly nonlinear immunostaining. Chemical labeling of appropriately tagged cellular targets therefore may allow the detection of small changes in physiological processes taking place in complex tissues.
Although our approach requires the use of transgenic animals or viral vectors and therefore cannot replace the use of antibodies against specific cellular proteins, recent advances in molecular biology (e.g., refs. 38 and 39) have greatly facilitated the generation of complex (viral) constructs. Furthermore, the clustered, regularly interspaced, short palindromic repeat (CRISPR) method promises to enable the rapid generation of transgenic animals, either for introducing reporter genes or tagging endogenous proteins (40).
We have demonstrated that chemical labeling offers many benefits for tissue staining. However, in many applications reliable, high-specificity antibodies and the endogenous fluorescence of GFP and related proteins give very satisfactory results. In such cases our reagents still may be of benefit by increasing the spectral palette or by avoiding problems with antibodies generated in the same host species. It is important to note that in our comparative analysis we have focused on GFP, which has the best characteristics among most FPs, being both bright and photostable. In contrast, other FPs fared less well when compared with SNAP-, CLIP-, or Halo-tag. For example, TLN-mCherry is significantly weaker than TLN-SNAP (Fig. 3A), and the blue and far-red FPs have not yet been adopted widely in tissue imaging. Many of the dyes used here are severalfold brighter, more photostable, and easier to separate spectrally than their FP counterparts.
Our experiments in flies and mice show that SNAP-, Halo-, and CLIP-tags are suitable for single-cell labeling (Figs. 3 E and F and 4 B and E). In contrast, using TMP-tag for this purpose will require further optimization. Additional orthogonal labeling chemistries, such as acyl carrier protein (ACP)-tag (41), could be explored in the future to allow simultaneous visualization of more than four channels. It also is likely that new tags will be engineered using the same principles as the SNAP and Halo tags. The quantitative nature of chemical labeling makes it particularly suitable for color multiplexing strategies such as Brainbow (42). We currently are working on approaches for rapid, multicolor labeling of neuronal tissue in flies and mice.
In Drosophila neurobiology, using membrane-targeted reporter constructs in combination with brp-SNAP will considerably speed up anatomical studies in which high-resolution confocal imaging requires fixed and stained specimens. Furthermore, although intensity-based image registration has been used to great effect in large-scale anatomical studies (7, 8, 10, 11), it has not yet been widely adopted, one major factor being the difficulty of obtaining successful neuropil staining. The brp-SNAP line introduced here overcomes this limitation.
We have noticed that the limiting factor for spatially even immunostaining of thick samples is the diffusion of secondary antibody into the tissue: Antibody penetration into fly brains and into ∼200-µm slices of mouse brain is incomplete after 2 h incubation with primary antibody and 12 h incubation with secondary antibody (Fig. 4F and Fig. S4B), but prolonging secondary incubation to 2 d results in more homogeneous labeling (Fig. 2C, Upper). Indeed, others have recommended extending secondary antibody incubation to 4 d (11). Arguably, incubation times are less important in a high-throughput context. However, our approach requires half as many manual handling steps, resulting in proportional labor and cost savings. In any case, reducing a standard staining time from 1 wk to 1 h results in a shortening of the experimental cycle that has major benefits: Conditions can be optimized rapidly, and experimental decisions can be made faster. Furthermore, we find that inexperienced users can obtain even and robust labeling with this method, whereas immunostaining (e.g., nc82) is subject to batch-to-batch variation, even in experienced hands.
Rapid and even staining may be of even greater significance in larger specimens. For example several recent methods can render tissue sufficiently transparent to enable whole-mount optical imaging of mouse brains (3–6). However, high-resolution, multichannel imaging of fluorescent proteins has major limitations in sensitivity. The recently developed CLARITY technique additionally promises to enable antibody staining of intact brain tissue (3). Nonetheless, the reported staining time of 4 wk for an adult mouse brain (∼5 mm across, i.e., 2.5 mm from each surface) will likely be prohibitive for many experiments, and there are still signs of significant spatial inhomogeneity. Extrapolating from our results in mouse brain slices and flies, chemical labeling could reduce this step from weeks to hours while enabling more spatially even staining. We presently are investigating transgenic reporter animals for this application.
Materials and Methods
Drosophila Stocks.
All flies used in this study were 2- to 4-d-old males with the exception of PA-GFP tracing experiments (see below). fruitlessGal4 (fruGal4) is a targeted insertion of the yeast transcription factor Gal4 into the P1 promoter of the fruitless gene (21). GH146-Gal4 (43) and Mz19-Gal4 (23) are P element-enhancer trap insertions. The generation of constructs for transgenic fly stocks is described below. UAS-DenMark (29), UAS-Syt-EGFP (27), UAS-syb-EGFP (27), and UAS-myr-GFP (44) flies were obtained from the Bloomington Stock Center. tubP-FRT-Gal80-FRT and hsFLP[86E], MKRS flies were obtained from K. Scott (University of California, Berkeley, CA) (45).
Drosophila Constructs.
pTW < Syt-CLIPm>, pTW < Syt-SNAPm>, pTW < CLIPf-Syb>, and pTW < SNAPf-Syb> constructs were made using the Multisite Gateway technology platform (Invitrogen). CLIPf and SNAPf are engineered versions of the original versions CLIP26m (CLIPm) and SNAP26m (SNAPm), respectively, that display faster labeling kinetics (46, 47). pTW is a pUASt vector containing a Gateway cassette. All CLIPm, CLIPf, SNAPm, and SNAPf coding sequences were amplified from pCLIPm, pCLIPf, pSNAPm, or pSNAPf plamids (New England Biolabs), respectively. Drosophila synaptotagmin 1 (syt1; GenBank accession no. M55048) was amplified from P{UAS-syt.eGFP}1 flies (Bloomington Stock Center) (27) using syt forward and eGFP reverse primers. Drosophila n-synaptobrevin (n-syb; GenBank accession no. S66686) was amplified from P{UAS-n-syb.eGFP}2 flies using syb forward and eGFP reverse primers. All synaptotagmin constructs described in this study are C-terminal (i.e., cytoplasmic) fusions. All synaptobrevin constructs described here are N-terminal (i.e., cytoplasmic) fusions.
UAS-TLN-CLIPm and UAS-TLN-SNAPm constructs were generated as follows: First, fusion PCR was performed on a pUAST-TLN-mCherry plasmid to remove the mCherry coding sequence and flanking linker sequences (pUAST-TLNΔcherry). A 1.5-kb fragment from the XhoI restriction site to the sequence corresponding to amino acids TVRVA of mouse Telencephalin/ICAM-5 and a 1.0-kb fragment ranging from amino acids GPWLW of Telencephalin/ICAM-5 to the MfeI restriction site were amplified. Using primers that introduced flanking XhoI and BglII sites, the full 2.7-kb fragment was cloned into pUAST-TLNΔcherry (pUAST-TLN). CLIPm and SNAPm sequences were amplified using BglII-containing primers and inserted into pUAST-TLN, thus yielding pUAST-TLN-CLIP and pUAST-TLN-SNAP.
pTW < PAT3SP-CD4-CLIPf> and pTW < PAT3SP-CD4-SNAPf> constructs were made using the Multisite Gateway technology platform. PAT3 signal peptide (SP) and CD4 sequences were amplified from UAS-CD4::spGFP1-10 flies (45).
To generate the UAS-myr-SNAPf construct, the GFP coding sequence was removed from a pJFRC-MUH-myr-GFP construct (44) using BamHI/KpnI restriction sites, and the SNAPf coding sequence was amplified from pSNAPf (New England Biolabs) and inserted.
To generate the UAS-myr-TMP construct, the GFP coding sequence was excised from a pJFRC81-L21 construct (48) using BamHI/KpnI restriction sites and was replaced by the eDHFR coding sequence (codon-optimized for Drosophila) and an N-terminal myristoylation signal.
To generate the UAS-myr-Halo construct, the TMP coding sequence was excised from UAS-myr-TMP (see above) using BamHI/KpnI restriction sites and was replaced by a Halo-tag sequence amplified from the pHT2 Halo-tag plasmid (Promega).
Generation of brp-SNAP.
We targeted the brp gene by taking advantage of a Minos-mediated integration cassette (MiMIC) (49) insertion (MI02987) 6.8 kb upstream of the stop codon present in the last coding exon of brp (Fig. S5). Because this exon is common to all nine Brp isoforms annotated in FlyBase, inserting the SNAP coding sequence immediately 5′ to the stop codon labels all Brp isoforms. The brp-SNAP targeting construct was generated by Gibson assembly (38) of the following fragments into the backbone of a pDONR221 plasmid: (i) three fragments encompassing the DNA sequence from the MiMIC insertion site to the end of the last coding exon; (ii) the SNAP coding sequence (added 5′ to the stop codon of brp); (iii) a 2.2-kb fragment of the brp 3′ untranslated region (UTR) and a cleavage site for the homing endonuclease I-CreI; and (iv) a fragment containing the 3×P3 promoter driving expression of red fluorescent protein (RFP) (used for screening of successful reduction step; see below).
The entire assembly was flanked by PhiC31 sites on both sides. Transgenic flies with the brp-SNAP construct replacing the original MiMIC insertion were made (BestGene, Inc.) and genotyped to verify the landing site and orientation of the integration. Two positive lines were identified and assayed by anti-SNAP staining. Because the integration into the brp locus generates a large duplication and an incomplete 3′ UTR (Fig. S5), one positive line was crossed to an hs-I-CreI line. The progeny were heat shocked for 30 min and subsequently crossed to balancers. Progeny from this cross then were screened for loss of the RFP eye marker, indicating successful repair of the I-CreI–induced double-strand break, resulting in reduction of the duplication and restoration of the 3′ UTR. One such fly was identified and used to generate a stock, which then was verified by PCR (Fig. S5D) and anti-SNAP staining. When crossed to OK107-Gal4, UAS-brp-RNAi, an enhancer line driving an RNAi construct against brp (30) in MBs, Brp-SNAP signal in the MBs was largely abolished (Fig. S5E), further confirming the specificity of this insertion and successful in-frame fusion with brp.
Mice.
C57BL/6 mice were obtained from Jackson Laboratory, and PV-Cre mice were obtained from Silvia Arber (Friedrich Miescher Institute for Biomedical Research, Basel). All experiments with animals were licensed under the U.K. Animals (Scientific Procedures) Act of 1986 and subject to local Animal Welfare and Ethical Review Body ethical review.
Virus Constructs, Production, and Injections.
For pAAV-CMV-myrSNAP (AAV-myrSNAP) and pAAV-myrSNAP-CON (AAV-myrSNAP-CON; “Cre-ON”) constructs, cassettes containing a tandem myristoylation signal and the SNAPf coding sequence followed by H2B-eGFP (and flanked by lox66 and lox71 sites in pAAV-myrSNAP-CO) were synthesized (Life Technologies) and inserted into a pAAV-CMV backbone using EcoRI/HindIII restriction sites. Recombination between the mutant lox66 and lox71 sites creates a wild-type loxP site and a fully mutant lox66/71 site; the latter is not well recognized by Cre recombinase, thus strongly disfavoring the reverse reaction (50).
All constructs were packaged in serotype 6. AAV-myrSNAP and AAV-myrSNAP-CON (both 6 × 1011 viral particles/mL) were unilaterally injected (1.0 µL each) into the hippocampus (bregma: −2.0 mm, midline: 1.7 mm; dorsal surface: −1.2 mm) of 2-mo-old male C57BL/6 or PV-Cre mice, respectively, using a Picospritzer III injector (Parker).
Labeling Reagents.
SNAP- and CLIP-surface labeling (i.e., cell-impermeable) substrates are abbreviated as BG- (benzylguanine) or BC- (benzylcytosine) in this study (see Table S2 for all abbreviations). Substrates were acquired either as stock solutions (e.g., Halo-TMR) or in powder form and dissolved in anhydrous dimethyl sulfoxide (DMSO) (Life Technologies) at a concentration of 1 mM (SNAP and CLIP substrates). Aliquots (5 µL) were stored at −20 °C in the presence of desiccant. We observed that using old DMSO or storing dissolved substrates in moist and/or warm conditions can lead to hydrolysis, drastically reducing labeling efficiency (SI Materials and Methods, Protocols for Chemical Labeling).
Fly Brain Staining.
Immunohistochemistry was carried out as follows: Brains were dissected in 0.1 M phosphate buffer (PB) and then fixed in 4% (wt/vol) PFA (Electron Microscopy Services) in PB at room temperature for 20 min as previously described (7). We observed that prolonged fixation (1 h at room temperature) improved signal intensity for some far-red SNAP substrates (e.g., BG-TF5) but decreased labeling of Halo-tag. For antibody staining and non–cell-permeable substrates, brains then were permeabilized by two 5-min washes in PBS + 0.3% Triton X-100 (PBS-T). Cell-permeable substrates were added without permeabilizing the brains. For immunostaining, blocking with 5% (vol/vol) goat serum was performed overnight at 4 °C. No blocking was performed for chemical staining. All following washing steps were performed twice at 10 min per wash in PBS-T (for immunostaining and chemical staining with non–cell-permeable substrates) or PBS (for cell-permeable substrates). For the blocking experiment in Fig. S4G, bromothenylpteridine (SNAP-Cell Block; S9106S; New England Biolabs) was added at 5 µM and incubated for 10 min, followed by a 20-min incubation with 0.5 µM BG-549 substrate. Fluorescent substrates (Table S2) were added at a concentration of 0.1–1 µM for 1 min–12 h on a rotating wheel, at room temperature for incubations <4 h or at 4 °C for incubations >4 h. An increase in background signal typically was observed for incubations of cell-permeable substrates for >2 h. For immunostaining, prolonged incubation of 2 d each with primary and secondary antibodies (rotating at 4 °C) was required for homogeneous staining. Primary antibodies used were mouse anti-nc82 1:20–1:40 (30) (Developmental Studies Hybridoma Bank, University of Iowa); chicken anti-GFP 1:1,000 (ab13970; Abcam); and rat anti-HA 1:200 (11 867 423 001; Roche). Secondary antibodies (all from Life Technologies) were Alexa-568 anti-mouse (A-11004) 1:1,200, Alexa-633 anti-mouse (A-21052) 1:1,200, Alexa-488 anti-chicken (A-11039) 1:1,200, and Alexa-568 anti-rabbit (A-11011) 1:1,200. Specimens were whole mounted in Vectashield (Vector Laboratories) on charged slides (Superfrost Plus, VWR) to avoid movement.
Mouse Brain Sectioning and Staining.
Coronal brain sections were prepared from mouse brains using a vibratome (SMZ 7000; Campden Instruments). Section thickness was 200 µm. Slices were transferred into glass-well plates, and excess cutting solution (ice-cold artificial cerebrospinal fluid) was removed with a pipette. Slices were fixed in 4% (wt/vol) PFA in PB for 30 min at room temperature (we found that varying fixation time from 10–30 min gave similar results) and then were washed three times for 5 min each washing in PBS. Slices were permeabilized in PBS-T for 10 min and transferred to a 24-well cell-culture plate. For chemical labeling experiments, substrates were added at a concentration of 1 µM, and the plate was incubated in a moist chamber. For experiments involving antibody staining, slices were preincubated in 1% BSA and 2% (vol/vol) goat serum in PBS-T for 1 h at room temperature before primary antibodies were added for incubation overnight at 4 °C. Slices then were washed three times for 20 min each wash in PBS-T. Secondary antibodies were added for 2 h at room temperature. After two final 20-min washes in PBS-T and one final 20-min wash in PBS, washing solution was removed, and Vectashield (Vector Laboratories) was added. Slices were mounted in Vectashield on charged slides (see above) using two sandwiched coverslips (thickness 1) to create a bridge. Primary antibodies used were mouse anti-NeuN (MAB377; Chemicon), chicken anti-GFP (ab13970; Abcam), mouse anti-Tau T49 monoclonal [a gift from G. Fraser (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK)], mouse anti-PV monoclonal [1:500; Swant; a gift from I. Greger (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK)]. Secondary antibodies (all from Life Technologies) were Alexa-488 anti-chicken (A-11039), Alexa-633 anti-mouse (A-21052), and Alexa-633 anti-rabbit (A-21071). All antibodies were used at 1:1,000 unless otherwise indicated.
In some experiments, whole brains were fixed overnight in 4% (wt/vol) PFA at 4 °C. After washing with PBS, brains were incubated in 30% (wt/vol) sucrose solution at 4 °C for 24 h before being embedded in Tissue-Tek O.C.T. (Optimal Cutting Temperature compound; Sakura Finetek) and frozen on dry ice. Cryosections (30 µm) were prepared on a Cryostat Microtome CM3050S (Leica) and stored at −20 °C in cryoprotection solution [25% (vol/vol) glycerin, 25% (vol/vol) ethylene glycol, 50% (vol/vol) PBS] until use. Staining of these cryosections was identical to the procedure described for vibratome sections.
Image Acquisition.
Confocal stacks of fly brains were acquired using a Zeiss 710 confocal microscope. Brains were imaged at 768 × 768 pixel resolution every 1 µm (voxel size 0.46 × 0.46 × 1 µm) using an EC Plan-Neofluar 40×/1.30 oil objective and 0.6 zoom factor. Images of dye-filled neurons were acquired with 2× (frame) averaging. Detailed images were taken with a Plan-Apochromat 63×/1.4 oil objective at 2–3× zoom and contained about 30 slices (each 768 × 768 pixels) with a voxel size of 0.06 × 0.06 × 0.15 µm. All images were taken using 16-bit color depths. Confocal images were registered to the IS2 template brain as previously described (8) using the computational morphometry toolkit (CMTK) available at www.nitrc.org/projects/cmtk.Triple chemical-labeled images (Fig. 3E) were acquired using a Leica TCS SP8 confocal microscope, at a 520 × 520 pixel resolution with a voxel size of 0.07 × 0.07 µm and z-step size of 0.27 µm, with a HCX Plan Apo 100 ×/1.4 Oil objective and a zoom factor of 3×.
Parameters for imaging mouse brain slices were largely identical to the parameters for fly brain imaging. An EC Plan-Neofluar 10×/0.3 objective was used for overview scans. A Plan-Neofluar 25×/0.8 multi-immersion objective was used to acquire stacks for quantifying label penetration. For reconstructions of single neurites, a stack was imaged at 0.1-µm z spacing with a Plan-Apochromat 63×/1.4 oil objective.
PA-GFP Tracing and Brp-SNAP Labeling.
Brains from 0- to 2-d-old flies were dissected in ice-cold artificial hemolymph [108 mM NaCl, 5 mM KCl, 2 mM CaCl2, 8.2 mM MgCl2, 4 mM NaHCO3, 1 mM NaH2PO4, 5 mM trehalose, 10 mM sucrose, 5 mM Hepes (pH 7.5), 265 mOsm] and allowed to adhere to a poly-d-lysine–treated coverslip within a cell-culture dish. Photoactivation was performed with a two-photon microscope as described (51). Briefly, several Kenyon cells were selected for photoactivation using baseline fluorescence at 925 nm. The somata of these cells were continuously illuminated with 710-nm light for 2 min, followed by a 30-min resting period to allow diffusion of photoactivated GFP. The brain then was fixed in 4% (wt/vol) PFA in PB for 20 min at room temperature and washed three times in PBS-T. Subsequently, 200 nM of BG-549 substrate in PBS-T was added for 20 min. Finally, brains were washed briefly with PBS-T, equilibrated in Vectashield, mounted, and imaged.
Flp-out and Brp-SNAP Labeling.
Flies of the genotype GH146-Gal4/brp-SNAP, tubP-FRT-Gal80-FRT; myrHalo/hsFLP[86E] MKRS were subjected to a single heat shock at 37 °C for 30 min between the L3 and pupal stages. Brains of adult (3–7 d old) flies were dissected, stained with BG-488 and Halo-TMR substrates, and imaged as described above.
Image Analysis and Quantification of Labeling.
For quantification of substrate penetration into fly brains, samples from GH146-Gal4 > myrSNAP or GH146-Gal4 > myrHalo or brp-SNAP animals were incubated with 1 µM BG-549 or 1 µM Halo-TMR for 15 min, 30 min, 1 h, 4 h, or 12 h. Samples from GH146-Gal4 > myrGFP or Canton-S animals were incubated with anti-GFP (1:1,000) or anti-nc82 (1:30) antibodies for 15 min, 30 min, 1 h, 4 h, or 12 h, followed by washing and incubation with fluorescent secondary antibodies (anti-chicken Alexa 568 and anti-mouse Alexa 568, respectively) for either 2 d or for the same time as primary antibodies (i.e., 15 min, 30 min, 1 h, 4 h, or 12 h).
Quantification of GH146 Labeling.
All 3D confocal stacks were registered onto a GH146-Gal4 > myrGFP template. GH146+ areas were segmented from each registered brain using a binary mask (surfaced rendered in Fig. S3B) constructed as follows: five GH146-Gal4 > myrGFP brains (stained with anti-GFP antibody for 12 h) were averaged in ImageJ. The resulting image stack was filtered (median + Gaussian) and thresholded.
Quantification of Brp-SNAP/nc82 Labeling.
All images were registered against an nc82 template brain (IS2) (8). Two binary masks were made to separate neuropil and cortex (Fig. S3A): five brp-SNAP brains labeled with BG-549 were averaged, filtered (median + Gaussian), and thresholded. To assess cortical background labeling, signal was quantified in the region resulting from subtraction of the neuropil mask from the whole brain mask (Fig. S3A). For the high-resolution three-color labeling analysis of PN terminals in the MB calyx (Fig. 3E), raw image stacks were deconvolved using Huygens Professional Deconvolution software (Scientific Volume Imaging). Further colocalization analysis of Brp puncta was performed with the same software package, and montaging was performed on deconvolved stacks in ImageJ.
Neuron Tracing.
Neuron tracing in Fig. 4 used the skeletonize module (52) in Amira (Visualization Sciences Group).
Quantification of Substrate and Antibody Penetration into Mouse Brain.
Coronal vibratome brain slices (200 µm) of animals injected with AAV-myrSNAP were incubated with 1 µM BG-549 for 30 min, 1 h, 6 h, or 12 h. Confocal stacks were resliced, and individual neurites that projected through the whole depth of the stack (n = 9–11 per condition) were traced using the ImageJ Simple Neurite Tracker tool. The tracing paths were converted to a binary mask by thresholding the result of the “Fill Out” option of Simple Neurite Tracer. Voxel intensity data then were fit as a function of z position with a gam smoothing line in R using the default options of ggplot2::stat_smooth. Antibody penetration into mouse brain sections was assessed by incubating coronal vibratome slices of the brains of PV-Cre animals injected with AAV-myrSNAP-CON with monoclonal mouse T49 anti-Tau antibody (1:2,000) for 30 min, 1 h, 6 h, or 12 h, followed by washing and 2-d incubation with secondary anti-mouse antibody (1:1,000; Alexa-Fluor 647). Data were quantified by reslicing confocal stacks in ImageJ, making a maximum intensity projection, and then using R to plot the mean image intensity at each z location, with a loess smoothing line using the default options of ggplot2::stat_smooth.
Supplementary Material
Acknowledgments
We thank K. Johnsson, V. Cornish, and I. Correa for providing labeling reagents; A. Carter and B. Hassan for DNA constructs; G. Fraser, M. Goedert, J. Krieger, and I. Greger for antibodies; K. Scott for fly stocks; S. Arber for PV-Cre mice; D. Evans for assistance with brain slice preparations; J. Manton for technical assistance including image analysis; M. Pasche for assistance with imaging and deconvolution; L. Luo, M. Landgraf, A. Ostrovsky, A. Jenett, I. Correa, and members of the G.S.X.E.J. group for comments on the manuscript; and A. Carter for our first introduction to SNAP-tag technology. This study made use of the Computational Morphometry Toolkit supported by the National Institute of Biomedical Imaging and Bioengineering. This work was supported by a European Research Council Starting Investigator grant, the European Molecular Biology Organization (EMBO) Young Investigator Programme, and Medical Research Council (MRC) Grant MC-U105188491 (to G.S.X.E.J.); an MRC Laboratory of Molecular Biology Graduate Scholarship (to J.K.); a Royal Society Dorothy Hodgkin Fellowship (to S.C.); a Boehringer Ingelheim Fonds predoctoral fellowship (to M.-J.D.); an EMBO postdoctoral fellowship (to S.F.); a Wellcome Trust/Royal Society Henry Dale award and MRC Grant MC-UP-1201/1 (to T.B.); and MRC Grant MC-UP-1201/2 (to M.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411087111/-/DCSupplemental.
References
- 1.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62(4):651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 3.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497(7449):332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hama H, et al. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14(11):1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 5.Ke MT, Fujimoto S, Imai T. SeeDB: Asimple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16(8):1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 6.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157(3):726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 7.Jefferis GSXE, et al. Comprehensive maps of Drosophila higher olfactory centers: Spatially segregated fruit and pheromone representation. Cell. 2007;128(6):1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GSXE. Sexual dimorphism in the fly brain. Curr Biol. 2010;20(18):1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu JY, Kanai MI, Demir E, Jefferis GSXE, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20(18):1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Chiang AS, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21(1):1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 11.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohl J, Ostrovsky AD, Frechter S, Jefferis GSXE. A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell. 2013;155(7):1610–1623. doi: 10.1016/j.cell.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1(4):2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 14.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21(1):86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 15.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15(2):128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 17.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: A flexible chemical tag. Nat Methods. 2005;2(4):255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW. Second-generation covalent TMP-tag for live cell imaging. J Am Chem Soc. 2012;134(33):13692–13699. doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr Opin Biotechnol. 2010;21(6):766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121(5):785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414(6860):204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 23.Jefferis GSXE, et al. Developmental origin of wiring specificity in the olfactory system of Drosophila. Development. 2004;131(1):117–130. doi: 10.1242/dev.00896. [DOI] [PubMed] [Google Scholar]
- 24.Hadjieconomou D, et al. Flybow: Genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8(3):260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 25.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 26.Kittel RJ, et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312(5776):1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- 27.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: Synaptotagmin-GFP. Genesis. 2002;34(1-2):142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 28.Chou YH, et al. Diversity and wiring variability of olfactory local interneurons in the Drosophila antennal lobe. Nat Neurosci. 2010;13(4):439–449. doi: 10.1038/nn.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolaï LJJ, et al. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci USA. 2010;107(47):20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagh DA, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49(6):833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Rein K, Zöckler M, Mader MT, Grübel C, Heisenberg M. The Drosophila standard brain. Curr Biol. 2002;12(3):227–231. doi: 10.1016/s0960-9822(02)00656-5. [DOI] [PubMed] [Google Scholar]
- 32.Milyaev N, et al. The Virtual Fly Brain browser and query interface. Bioinformatics. 2012;28(3):411–415. doi: 10.1093/bioinformatics/btr677. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, et al. Insect Brain Name Working Group A systematic nomenclature for the insect brain. Neuron. 2014;81(4):755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 34.Datta SR, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452(7186):473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- 35.Ruta V, et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468(7324):686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 36.Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr Biol. 2013;23(8):644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Yu HH, et al. Clonal development and organization of the adult Drosophila central brain. Curr Biol. 2013;23(8):633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Werling U, Edelmann W. SLiCE: A novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40(8):e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George N, Pick H, Vogel H, Johnsson N, Johnsson K. Specific labeling of cell surface proteins with chemically diverse compounds. J Am Chem Soc. 2004;126(29):8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 42.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 43.Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32(5):443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61(3):373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun X, et al. Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. ChemBioChem. 2011;12(14):2217–2226. doi: 10.1002/cbic.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellett PA, et al. Two-color STED microscopy in living cells. Biomed Opt Express. 2011;2(8):2364–2371. doi: 10.1364/BOE.2.002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109(17):6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venken KJT, et al. MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8(9):737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7(4):649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 51.Caron SJC, Ruta V, Abbott LF, Axel R. Random convergence of olfactory inputs in the Drosophila mushroom body. Nature. 2013;497(7447):113–117. doi: 10.1038/nature12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evers JF, Schmitt S, Sibila M, Duch C. Progress in functional neuroanatomy: Precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J Neurophysiol. 2005;93(4):2331–2342. doi: 10.1152/jn.00761.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.