Significance
Sequences derived from ancient viruses have been shown to make up a substantial part of animal genomes. Bornaviruses, a genus of nonsegmented, negative-sense RNA virus, also have left their DNA copies in the genomes of a number of vertebrate lineages. Recent studies have demonstrated that some endogenous bornavirus-like elements (EBLs) may have acquired functions in their hosts as a result of exaptation. In this study, we show that protein encoded by an EBL in the genome of the thirteen-lined ground squirrel efficiently blocks infection and replication of extant bornavirus. To our knowledge, this is the first report showing that endogenous nonretroviral RNA virus elements may function in antiviral defense, providing a potential role for RNA virus endogenization in host evolution.
Keywords: endogenous nonretroviral viruses, antiviral immunity
Abstract
Animal genomes contain endogenous viral sequences, such as endogenous retroviruses and retrotransposons. Recently, we and others discovered that nonretroviral viruses also have been endogenized in many vertebrate genomes. Bornaviruses belong to the Mononegavirales and have left endogenous fragments, called “endogenous bornavirus-like elements” (EBLs), in the genomes of many mammals. The striking features of EBLs are that they contain relatively long ORFs which have high sequence homology to the extant bornavirus proteins. Furthermore, some EBLs derived from bornavirus nucleoprotein (EBLNs) have been shown to be transcribed as mRNA and probably are translated into proteins. These features lead us to speculate that EBLs may function as cellular coopted genes. An EBLN element in the genome of the thirteen-lined ground squirrel (Ictidomys tridecemlineatus), itEBLN, encodes an ORF with 77% amino acid sequence identity to the current bornavirus nucleoprotein. In this study, we cloned itEBLN from the ground squirrel genome and investigated its involvement in Borna disease virus (BDV) replication. Interestingly, itEBLN, but not a human EBLN, colocalized with the viral factory in the nucleus and appeared to affect BDV polymerase activity by being incorporated into the viral ribonucleoprotein. Our data show that, as do certain endogenous retroviruses, itEBLN potentially may inhibit infection by related exogenous viruses in vivo.
Endogenous retroviruses have accumulated in the genomes of many organisms over evolutionary time and occupy about 8% of the human genome (1). Although almost all endogenous retroviruses in animal genomes appear to lack intact ORFs and are not expressed in somatic tissues, some retain the potential to express mRNA and even produce the proteins they encode (2, 3). There is mounting evidence that the expression of human endogenous retroviruses (HERVs) is associated with some autoimmune diseases, cancers, and schizophrenia, as well as with viral infections (4–6), suggesting that the induction of HERVs may be linked to immunological aberrations in the host. In addition, it has been reported that endogenous retroviruses have the ability to recombine with exogenous or other endogenous retroviruses to produce intact infectious viruses (7, 8). These observations indicate that the expression of endogenous retroviruses may directly cause deleterious consequences to the hosts.
On the other hand, recent studies have revealed clearly that endogenous retroviruses have been coopted to play new and beneficial roles in their hosts (9, 10). For instance, in mammals, envelope genes from endogenous retroviruses are involved in the formation of the placenta during the fusion of syncytiotrophoblast cells (9, 11). In addition, a retroviral-like aspartic protease, skin aspartic protease, is known to play a key role in determining the texture of skin by modulating the degree of hydration in mammals (12, 13). Furthermore, it has been known for decades that endogenous retrovirus-derived elements, such as Friend virus susceptibility 1 (Fv1) gene and endogenous fragments from Jaagsiekte sheep retrovirus (enJSRV), protect host cells from infection by exogenous retroviruses (10, 14, 15). These lines of evidence suggest that evolution has favored the persistence of endogenous retroviral elements that have the potential to protect their hosts against related viruses.
Recent advances in the availability of genomic sequences from many animal species, as well as the development of tools for sequence comparisons, have revealed that nonretroviral viruses also have endogenized in many mammalian species (16–18). Bornavirus, an enveloped, nonsegmented, negative-strand RNA virus in the order Mononegavirales, is unique among animal RNA viruses, because it not only replicates in the cell nucleus but also readily establishes a long-lasting, persistent infection in the absence of overt cytopathogenesis (19). Bornavirus does not integrate into the host genome during its replication cycle, but, interestingly, we and others recently found that DNA sequences derived from ancient bornaviruses are endogenized in the genomes of many vertebrate species, including humans (16, 17). The endogenous fragments of bornavirus in mammalian genomes originate predominantly from the region encoding the viral nucleoprotein (N), which encapsidates viral genomic RNA to form nucleocapsids, and we therefore designated them “endogenous bornavirus-like nucleoproteins” (EBLNs) (16).
In a previous study, we showed that EBLNs from the human and the thirteen-lined ground squirrel (TLS) (Ictidomys tridecemlineatus) genomes, named “hsEBLN” and “itEBLN,” respectively, have significant sequence similarity to the N ORFs of extant mammalian bornaviruses, Borna disease virus (BDV). The elements hsEBLN-1 and -2, which are located in chromosomes 10 and 3 of the human genome, respectively, encode ORFs with an overall 41% amino acid sequence identity to BDV N (16). itEBLN also has an intact ORF and shows ∼77% amino acid sequence identity to N. Furthermore, it has been shown that some hsEBLNs, including hsEBLN-1 and -2, are expressed as RNA and that a predicted mRNA transcript of itEBLN is provided by NCBI RefSeq (XM_005342477). Interestingly, we have found that long terminal repeats (LTR-1C and LTR-21B) from endogenous retroviruses exist in close proximity and upstream of itEBLN ORF (20). In fact, we could detect the predicted transcripts of itEBLN in several tissue samples from both breeding and wild-captured TLSs by RT-PCR (Fig. S1). From these observations, it is tempting to speculate that EBLNs encode functional proteins as a consequence of exaptation or cooption by their hosts.
In this study, we cloned hsEBLN-1 and itEBLN sequences from the human and TLS genomes, respectively, to determine whether these EBLNs encode potentially functional proteins. We found that the protein encoded by itEBLN, but not hsEBLN-1, colocalizes with the viral factory in the nucleus and markedly decreases infection and the replication of exogenous BDV. Furthermore, the protein encoded by itEBLN appeared to be incorporated into the viral ribonucleoproteins (RNPs) in infected cells. These results suggest that, like some endogenous retroviruses, itEBLN has the potential as a coopted gene in the host to inhibit infection by genetically related viruses.
Results
Expression and Reconstruction of EBLN Elements from the Human and TLS Genomes.
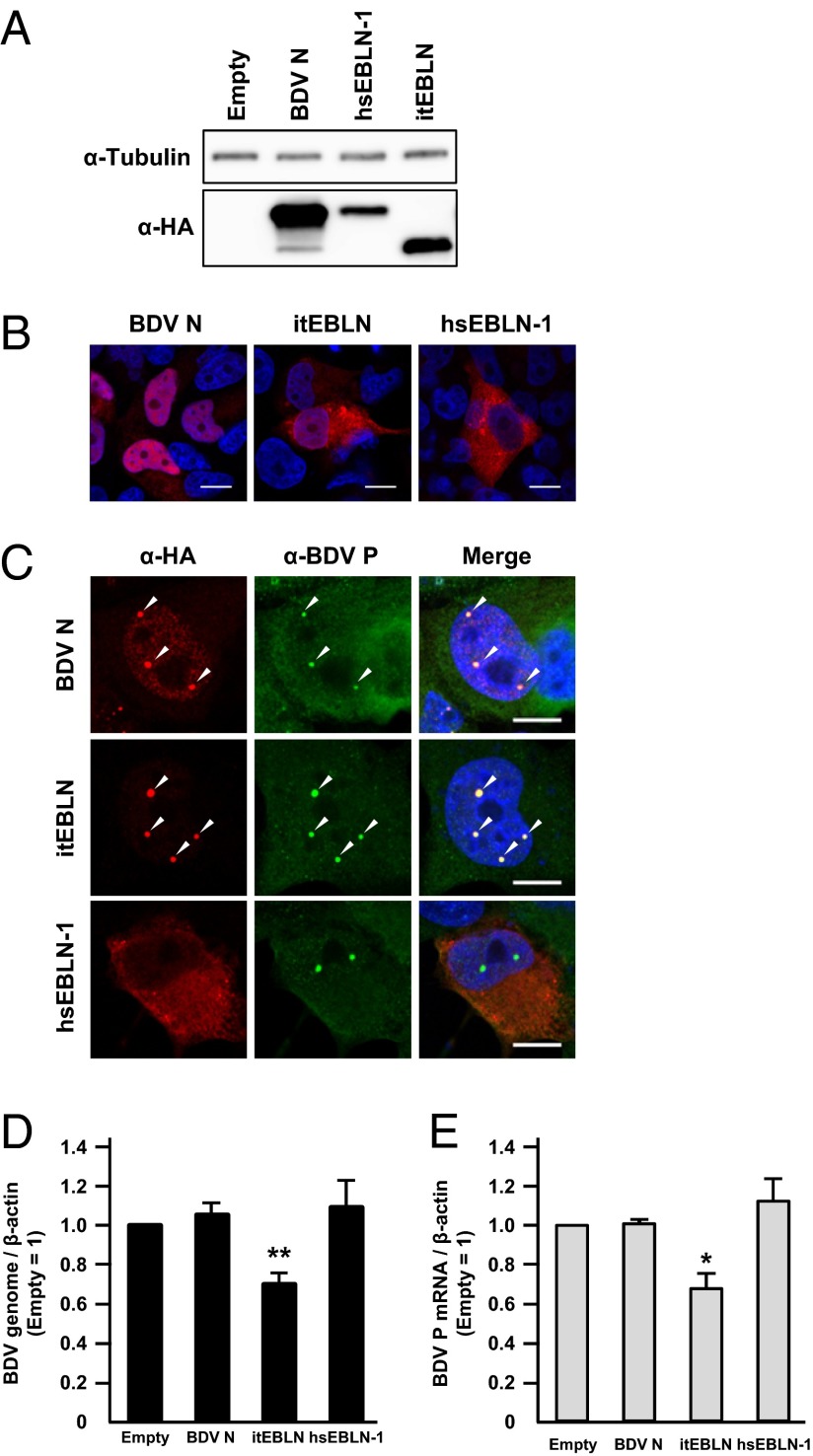
To investigate the potential roles of the EBLN elements in the human and TLS genomes, we first amplified EBLN sequences from the genomic DNAs and cloned the products into expression vectors. Although the expression of hsEBLN-1 and itEBLN has been predicted in previous studies (16, 18), the structure and transcription initiation sites of the EBLN RNAs had not been characterized. In this study, therefore, we cloned only the regions homologous to BDV N. To detect the expressed EBLN products, an HA-tag was fused to the N terminus of the coding sequences. At first, we transfected the plasmids into human oligodendroglioma (OL) cells and investigated the expression of the reconstituted proteins by Western blotting at 24 h posttransfection. As shown in Fig. 1A, expression of the recombinant proteins was detected as ∼35- to 40-kDa products, indicating that the EBLNs may have the ability to express stably the proteins in mammalian cells. In addition, we carried out immunofluorescence assays to examine the subcellular localization of the proteins. As shown in Fig. 1B, although BDV N clearly localizes in the nucleus, itEBLN and hsEBLN-1 appear to be distributed predominantly in the cytoplasm of the transfected cells, probably because these proteins lack the nuclear localization signal (NLS), which is present in the N terminus of BDV N (see Fig. 5).
Fig. 1.
Expression of itEBLN inhibits BDV replication in transfected human cells. (A) Expression of recombinant EBLN proteins in transiently transfected OL cells. The HA-fused itEBLN, hsEBLN-1, and BDV N constructs were transfected into OL cells, and the expressed proteins were detected by Western blotting using anti-HA antibody. (B and C) Subcellular localization of recombinant EBLNs in transfected cells. The expression plasmids were transfected into uninfected cells (B) and OL cells persistently infected with BDV (C), and the distributions of EBLN proteins and BDV P were visualized by immunofluorescence assay with anti-HA (red) and anti-P (green) antibodies, respectively. Cells were counterstained with DAPI. Arrowheads in C indicate vSPOTs (bornavirus viral factories). (Scale bars, 10 μm.) (D and E) Expression of itEBLN decreases the level of BDV RNAs in persistently infected cells. Forty-eight hours after transfection of the indicated constructs, the amounts of BDV genomic RNA (D) and mRNA (E) were quantified by real-time RT-PCR. The bars labeled “Empty” represent cells transfected with an empty vector. The values are presented as the mean ± SE of three independent experiments. Statistical significance was analyzed by the two-tailed t test. *P < 0.05, **P < 0.01.
Fig. 5.

Alignment of amino acid sequences in BDV N and itEBLN. Amino acids that are in BDV N and itEBLN are shown by blue shading. The sequences of the NLS and NES and two P-binding sites (PBS-1/2) are indicated. Arrowheads indicate the predicted amino acids residues essential for interaction with the viral RNA. The asterisks between the sequences indicate the residues predicted to be involved in the tetramerization of BDV N. Red and blue asterisks indicate residues interacting with the preceding and following crystallographic neighbors, respectively, to form the tetramer. The initiation site of itEBLN-M2 is indicated also.
Next, we transfected the EBLN plasmids into OL cells persistently infected with BDV. In a previous study, we demonstrated that BDV generates dot-like structures, viral speckles of transcripts (vSPOTs), in the nucleus (21). The vSPOT is the viral factory in which essential events in the BDV replication cycle take place and could be the same structured nuclear bodies and have the same function as Joest–Degen inclusion bodies in vivo (22–24). Despite the lack of nuclear localization activity, itEBLN was strongly redistributed to vSPOTs in the nucleus, as was N (Fig. 1C, arrowheads). On the other hand, hsEBLN-1 seemed not to be relocated in the infected cells. This observation suggests that itEBLN, but not hsEBLN-1, may have the potential to interact with the viral components in the infected cells, leading to colocalization with vSPOT in the nucleus.
itEBLN Inhibits BDV Replication in Persistently Infected Cells.
In a previous study, Geib et al. (25) revealed that transient expression of BDV phosphoprotein (P), but not N, inhibits viral replication in Vero cells persistently infected with BDV. Therefore, we next investigated whether expression of hsEBLN-1 and itEBLN affects BDV replication in persistently infected cells. To evaluate viral transcription/replication, we measured the level of viral RNAs in cells transfected with the EBLN plasmids. Consistent with previous reports, transient expression of N in persistently infected OL cells did not exert any effect on BDV replication (Fig. 1 D and E). In contrast, intriguingly, expression of itEBLN, but not hsEBLN-1, significantly reduced the levels of both viral genomic and mRNA at 48 h posttransfection (Fig. 1 D and E). This result suggested that, unlike N, itEBLN could inhibit both the transcription and replication of BDV.
Expression of itEBLN Confers Resistance to Exogenous BDV Infection.
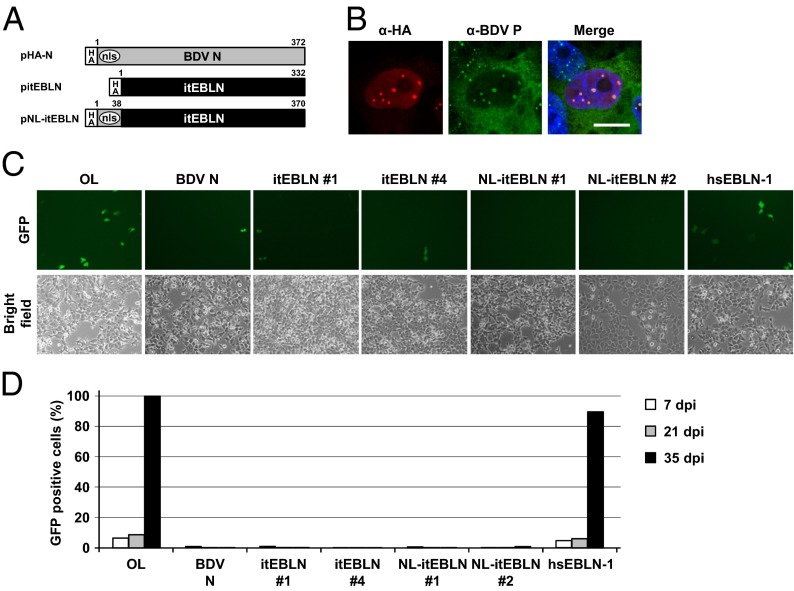
Previous studies revealed that expression of BDV N protects cells from subsequent infection by BDV (25). Although the detailed mechanism of this resistance to superinfection has not been elucidated, it is assumed that the overexpression of the viral nucleocapsid component may interfere with an early step of BDV infection, such as nuclear transport of viral RNP, intranuclear dissemination, or initiation of viral replication (25). To determine whether itEBLN also can affect BDV infection, we established OL cell lines stably expressing itEBLN or hsEBLN-1. In this experiment we also generated a plasmid, pNL-itEBLN, in which itEBLN is fused with the NLS of BDV N at the N terminus (Fig. 2A), to investigate the effect of nuclear localization of the protein. As with N, the product of pNL-itEBLN clearly localized in the nuclei of transiently transfected cells (Fig. 2B). We then infected the cell lines with cell-free virions of recombinant BDV (rBDV) expressing GFP, rBDV P/M-GFP, with a multiplicity of infection (M.O.I.) of 0.1 and monitored the propagation of GFP for at least 3 wk. As shown in Fig. 2 C and D, the expression of itEBLN and NL-itEBLN, but not hsEBLN-1, almost completely protects the cells against BDV infection for the observation period, as does N, suggesting that itEBLN protects against BDV infection in the nucleus.
Fig. 2.
Expression of itEBLN in human cells inhibits BDV infection. (A) Schematic representations of the recombinant proteins of BDV N and itEBLN. An expression plasmid, pNL-itEBLN, was generated by fusing the NLS of BDV N (amino acids 1–38) to the N terminus of the itEBLN ORF. (B) Subcellular localization of NL-itEBLN in the transiently transfected BDV/OL cells. (Scale bar, 10 μm.) (C) OL cells stably expressing pHA-N, pitEBLN, pNL-itEBLN, and phsEBLN-1 were inoculated with cell-free rBDV expressing GFP at an M.O.I. of 0.1. GFP expression was monitored by fluorescence microscopy. The cells were photographed 4 d after infection. (D) The percentage of GFP-expressing cells was monitored over a period of 35 d.
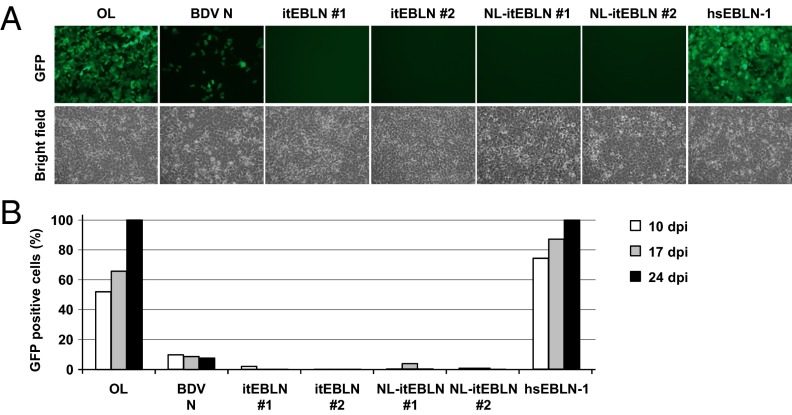
We next performed cocultivation experiments using the EBLN-expressing OL cell lines and Vero cells persistently infected with rBDV P/M-GFP, to investigate whether itEBLN also can inhibit the cell-to-cell spread of the infection, which presumably is the main route of BDV transmission. Repeated experiments revealed that, despite incomplete protection of the cells expressing N, itEBLN constructs were almost completely resistant to virus infection (Fig. 3). These results indicated that itEBLN could have a strong inhibitory effect on BDV infection, even after cell-to-cell transmission. To exclude the possibility that the NLS region (amino acids 1–38) of BDV N in the constructs has an effect on BDV replication, we also conducted the experiments using two different plasmids, pNLsv-itEBLN and pNL-DsRed, in which itEBLN and DsRed were fused with SV40 and BDV N NLS at the N terminus, respectively (Fig. S2). The results in Fig. S3 show that the NLS region of N is not involved in the inhibitory effect of NL-itEBLN on BDV replication.
Fig. 3.
Expression of itEBLN protects against cell-to-cell transmission of BDV. (A) OL cells stably expressing pHA-N, pitEBLN, pNL-itEBLN, and phsEBLN-1 were cocultured with Vero cells persistently infected with rBDV P/M-GFP. Three days after cocultivation, the cells were treated with Zeocin or G418 to eliminate the Vero cells. GFP expression was monitored by fluorescence microscopy. The cells were photographed 24 d after cocultivation. (B) The percentage of GFP-expressing cells was monitored over a period of 24 d.
Incorporation of itEBLN into BDV RNP.
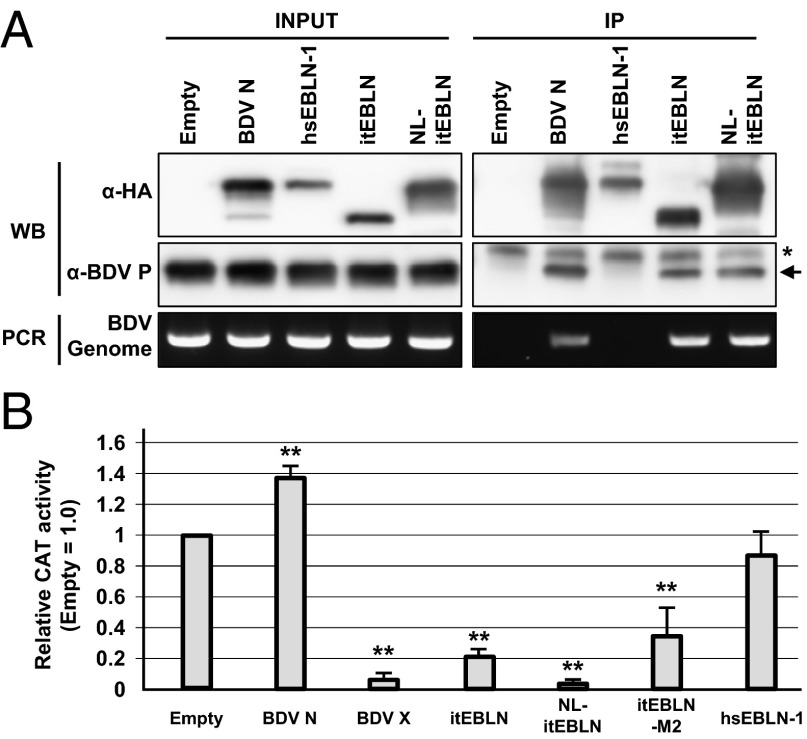
The results shown above suggest that the itEBLN protein may be incorporated directly into BDV RNP, resulting in inhibition of viral replication. To test this possibility, we performed an immunoprecipitation analysis using BDV-infected cells transfected with the EBLN plasmids. At 24 h posttransfection, the cell extracts were immunoprecipitated with anti-HA antibody, and the viral RNP components were detected by Western blotting and RT-PCR. As shown in Fig. 4A, both BDV genomic RNA and BDV P were clearly precipitated with itEBLN and N, whereas hsEBLN-1 seemed not to interact with BDV RNP in the cells. This observation revealed that itEBLN may be incorporated efficiently into the viral RNP in infected cells.
Fig. 4.
itEBLN is incorporated into viral RNPs and affects BDV polymerase activity. (A) Immunoprecipitation analysis of itEBLN in OL/BDV cells. OL cells persistently infected with BDV were transfected with the indicated constructs, lysed 24 h after transfection, and immunoprecipitated with anti-HA antibody. “Empty” indicates cells transfected with empty vector (pcDNA3). BDV P was detected by anti-BDV P antibody (arrow). The asterisk indicates nonspecific bands detected in all cells. BDV genomic RNA was detected by RT-PCR using primers specific for the genome sense RNA within the BDV P region. (B) Expression of itEBLN inhibits BDV polymerase activity in a minireplicon assay. HEK293T cells were cotransfected with a set of BDV minigenome plasmids and the expression constructs for itEBLN, hsEBLN-1, BDV N, and BDV X. Forty-eight hours after transfection, the cells were lysed and subjected to CAT assays. The CAT activities are expressed as the ratio relative to empty vector-cotransfected cells (Empty). The values are the mean ± SE of three independent experiments. Statistical significance was analyzed by the two-tailed t test; **P < 0.01.
itEBLN Inhibits BDV Polymerase Activity in a Minireplicon System.
We next determined whether itEBLNs can affect the polymerase activity of BDV directly. To this end, we used a minireplicon system of BDV, which synthesizes recombinant BDV nucleocapsids containing an artificial, minigenome reporter RNA, after transfection of expression plasmids encoding BDV N, P, RNA-dependent RNA polymerase protein (L), and the minigenome (26). We carried out the minireplicon assay in the presence or absence of plasmids expressing itEBLN. Consistent with previous observation (26), the viral polymerase activity was strongly inhibited when BDV X was cotransfected with the minireplicon constructs (Fig. 4B). Interestingly, although BDV N could not inhibit the polymerase activity of the minireplicon, all of the itEBLN constructs, including itEBLN-M2, which is translated from the AUG codon at amino acid position 132 in the itEBLN sequence (Fig. 5), efficiently decreased the polymerase activity in the system (Fig. 4B). Taken together, these results suggest that itEBLN acts as a dominant-negative inhibitor of N by being incorporated into the viral RNP.
The Putative Tetramer and RNA Interaction Domains of BDV N Are Conserved in itEBLN.
Previous studies revealed that BDV N contains several signal sequences, including the NLS and nuclear export signal (NES), and putative P-binding sites (27–29). Furthermore, structural studies of BDV N determined the amino acids essential for homotetramer formation and interaction with viral RNA (30, 31). To predict the function of itEBLN as a dominant-negative inhibitor of N, we aligned the amino acid sequences of BDV N and itEBLN. As shown in Fig. 5, BDV N contains an NLS in the N-terminal region, whereas itEBLN lacks a homologous sequence in that region. On the other hand, putative binding sites for P (PBS-1 and -2) seem to be highly conserved between itEBLN and BDV N (Fig. 5). In addition, the sequences predicted to be involved in tetramerization of BDV N also seem to be conserved in the sequence of itEBLN, with the exception of the sequence in the N terminus of BDV N (Fig. 5, asterisks). We also found that the corresponding residues essential for interaction with the viral RNA (K164, R165, K242, and R297) (Fig. 5, arrowheads) are well conserved in itEBLN. A structural model based on BDV N also revealed that the regions surrounding the RNA interaction sites are conserved in itEBLN (Fig. S4), suggesting that itEBLN may retain the ability to form nucleocapsids with BDV N.
Discussion
In this study, we showed that the protein expressed from an endogenous bornavirus fragment from the TLS genome, itEBLN, is incorporated into BDV RNPs and inhibits BDV replication. This conclusion is supported by the following observations. First, we found that the itEBLN protein was colocalized with the viral factory of BDV in the nucleus. Second, the expression of this protein markedly reduced the replication level of BDV in persistently infected cells. Furthermore, the cells stably expressing the itEBLN protein were completely resistant to infection by exogenous BDV through both the cell-free and cell-to-cell routes. Third, we showed that the itEBLN protein can precipitate BDV RNP components, including viral genomic RNA, in infected cells. Finally, coexpression of itEBLN reduced BDV polymerase activity using the minireplicon system. To our knowledge, this is the first report that an endogenous, nonretroviral virus efficiently inhibits infection by the related exogenous virus.
In this study we used an overexpression system of recombinant itEBLN with human cell culture systems. Thus, to demonstrate the host-specific exaptation of itEBLN, it would be necessary to investigate whether TLSs actually exhibit resistance to BDV infection. At present, however, we could neither find any available cultured cells of TLSs nor establish experimental infection using TLSs. On the other hand, we could detect the expression of predicted mRNA of itEBLN in tissue samples from both breeding and wild-captured TLSs by RT-PCR (Fig. S1 A and B). The immunoblot analysis using a BDV N-specific polyclonal antibody showed only a faint band at the predicted size in the heart samples (Fig. S1C). Although the expression of itEBLN protein remained obscure because of the specificity of the antibody, itEBLN may protect BDV infection in vivo if the mRNA of itEBLN is efficiently translated into the protein in the squirrel cells. We will continue to make an effort to establish infection systems of TLSs with BDV.
Our results showed that the expression of itEBLN not only reduces viral replication but also blocks de novo BDV infection. Immunofluorescence and immunoprecipitation assays indicated that the itEBLN protein interacted efficiently with BDV RNP in the infected cells. Among the RNP components, the most likely candidate is the N protein, because BDV N is known to assemble tightly as a homotetramer (30). BDV N forms the nucleocapsid, which serves as the template for RNA synthesis with the L and P proteins (31, 32), strongly suggesting that itEBLN participates in heteromultimerization with BDV N. It has been determined that the residues predicted to be important for tetramer formation by N are located in the N and C termini of the protein (Fig. 5) (30). A comparison of amino acids in the itEBLN protein and BDV N revealed that the identity between BDV N and itEBLN may be sufficient to permit heteromultimerization. It is highly likely, therefore, that the itEBLN protein efficiently coassembles with BDV N into viral nucleocapsids in infected cells. In a previous study, Geib et al. (25) demonstrated that transient expression of recombinant N in cells persistently infected with BDV does not inhibit BDV replication, even though the transduced N was colocalized with the viral factory in the nucleus. This observation also was confirmed in our experiments shown in Fig. 1, suggesting that the itEBLN may act like a dominant-negative mutant of N in the viral nucleocapsids and exert a deleterious effect on the viral replication. In fact, a number of substitutions, especially at the N terminus, were found in the itEBLN sequence (Fig. 5). Such heterogeneity might destabilize tetramer formation or the interaction with viral RNAs, leading to inhibition of viral polymerase activity.
Alternatively, it may be possible that the itEBLN protein interacts directly with P and inhibits the functions of P. In fact, the sequences corresponding to the P-binding sites in N have been shown to be well conserved in itEBLN. P plays important roles in viral replication, as a viral polymerase cofactor, and in nucleocytoplasmic shuttling of the viral nucleocapsid (32–34). Although the intracellular distribution of P seems not to be altered by overexpression of itEBLN in the cells (Fig. 1C), the interaction between P and itEBLN may affect the dynamics of P in the infected cells. Furthermore, we previously demonstrated that the expression of P regulates the translation efficiency of BDV X, which is a negative regulator of BDV polymerase (35). Thus, it also is possible that their interaction affects the translation efficiency of X, resulting in the inhibition of polymerase activity of BDV. Thus, it is conceivable that the effect of itEBLN on BDV replication may be multifaceted, via interaction with both N and P. Further experiments will be necessary to understand the mechanism of the inhibitory effect of itEBLN on BDV replication.
Our results may reveal an intriguing strategy for inhibiting exogenous virus infection by endogenous viral fragments. Previous studies clearly demonstrated that endogenous viral products are efficiently incorporated into the capsids of incoming, genetically related viruses, resulting in the inhibition of viral replication. A well-studied example is the Fv1 gene in mice. Fv1 originated from the gag gene of an ancient retrovirus, which was endogenized several million years ago into the genome of a common ancestor of mice and is known to restrict infection by specific strains of murine leukemia virus (MLV) (14, 36). Although the amino acid sequence of Fv1 is distant from the restricted MLVs, recent studies clearly indicate that direct interaction of Fv1 with the capsid protein of MLV induces the anti-MLV function of Fv1 (37). Another instance is the enJSRV. This endogenous virus is known to inhibit exogenous Jaagsiekte sheep retrovirus (JSRV) infection by two different mechanisms (38). First, the envelope protein encoded by enJSRVs is expressed and binds to the cellular receptors used by JSRV (39). The Gag protein expressed by enJSRVs also is known to have deleterious effects on the replication of exogenous JSRV (15). A misfolded Gag protein of enJSRVs coassembles with JSRV Gag and forms chimeric viral capsids, which are degraded by the proteasome system of the infected cells (40). In addition, Monde et al. (41) recently have demonstrated that the Gag proteins of an endogenous betaretrovirus coassemble with the Gag protein of a distinct retrovirus, HIV-1, to modulate the late phase of HIV-1 replication. Together with our observation that itEBLN also has the ability to form nucleocapsids with BDV N, interaction of endogenous viral products with the capsids or nucleocapsids of incoming exogenous viruses may be an effective way to regulate virus infections.
Recent studies have demonstrated that the human genome contains many intrinsic factors that prevent viral infection (10, 42). Such factors could be acquired during evolution as consequences of the battle between viruses and their hosts. There is no doubt that the arms race between the host and virus has affected the evolution of both. In addition to the sophistication of the immune system, host organisms must have acquired many genes to overcome infection by pathogens over many generations of evolution. Exaptation is the coopting of exogenous sequences as new genes, with functions distinct from their original purpose, into the genome (10, 43). The cooption of endogenous retroviruses as antiviral factors is a good example of such exaptation. Until now, retroviruses have been considered the only virus family that has been coopted as new functional genes in the host genomes. Our results strongly suggest that exaptation of nonretroviral viral genes may have occurred during the coevolution of bornaviruses and their hosts.
At present, some EBLNs from human and nonhuman primate genomes, including hsEBLN-1 and hsEBLN-2, have been shown to express RNAs that potentially encode proteins (16, 44). We could not demonstrate that the hsEBLN-1 protein had an inhibitory effect on the BDV replication, and the possibility that hsEBLN-1 plays a role in the cellular environment or acts at the level of RNA remains to be elucidated. On the other hand, hsEBLN-2 has been shown to express protein in human cells (44). A recent study also revealed that hsEBLN-2 is a candidate gene of the recurrent 3p12-p14 loss in cervical cancer and could be a tumor suppressor in cervical cancer (45). The human EBLNs are thought to have been generated 40–45 million years ago. Nevertheless, relatively long ORFs, are conserved in these elements, especially in hsEBLN-1 and -2, and have a high level of amino acid identity with the N protein of current bornaviruses (16). These observations suggest the intriguing possibility that hsEBLN-1 and -2 have been adapted during evolution with new functions in the host cells. We currently are working on understanding the coopted roles of the EBLNs, and other endogenous nonretroviral elements, in mammalian genomes. These studies could provide new insights into the coevolution and between viruses and their hosts and the cooption of new genes in hosts after infection by exogenous viruses.
Materials and Methods
Cells.
OL and Vero cell lines were cultured in DMEM-high glucose (4.5% wt/vol) supplemented with 5% (vol/vol) FBS and 4 mM glutamine. HEK293T cells were cultured in DMEM-low glucose (1.0%) supplemented with 10% (vol/vol) FBS. OL cells persistently infected with the BDV strain huP2br (OL/BDV) were cultured using the same conditions as the parental cell line. The cell lines stably expressing hsEBLN-1, itEBLN, or BDV N were established by the limiting dilution method and were maintained in culture medium with Zeocin (Invitrogen) or G418 (Invitrogen).
Virus Infection.
OL cell lines stably expressing the EBLN constructs were infected with cell-free rBDV P/M-GFP (46) virions at an M.O.I of 0.1. After absorption for 1 h, the cells were washed with PBS and passaged within 2 or 3 d. In addition, the OL cell lines were cocultured with Vero cells persistently infected with rBDV P/M-GFP. Three days after the cocultivation, the cells were treated by Zeocin or G418 to eliminate the Vero cells. The infection rates of the cells were determined by measuring GFP expression using a FACSCalibur flow cytometer (BD Biosciences) or Tali Image-Based Cytometer (Invitrogen).
Minireplicon Assay.
Minireplicon assays were carried out according to the methods of Yanai et al. (26). Briefly, HEK293T cells were seeded in 12-well plates and transfected with expression plasmids of BDV N (0.1 μg), P (0.01 μg), L (0.1 μg), and Pol II-driven minigenome plasmids (0.1 μg), with or without EBLN and BDV X expression plasmids (0.5 μg), using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, the cells were lysed, and cell lysates were prepared for chloramphenicol acetyltransferase (CAT) assay.
Supplementary Material
Acknowledgments
This study was supported in part by the Funding Program for Next Generation World-Leading Researchers and Grant-in-Aid for Scientific Research (A) 26253027 from the Japan Society for the Promotion of Science (to K.T.), by Grants-in-Aid for Scientific Research on Innovative Areas 24115709 and 25115508 (to T.H.) from the Ministry of Education, Culture, Science, Sports and Technology of Japan, and by grants from Takeda Science Foundation (to K.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407046111/-/DCSupplemental.
References
- 1.Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000;10(6):651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 2.Bannert N, Kurth R. Retroelements and the human genome: New perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 4.Krieg AM, Gourley MF, Perl A. Endogenous retroviruses: Potential etiologic agents in autoimmunity. FASEB J. 1992;6(8):2537–2544. doi: 10.1096/fasebj.6.8.1592206. [DOI] [PubMed] [Google Scholar]
- 5.Singh S, Kaye S, Gore ME, McClure MO, Bunker CB. The role of human endogenous retroviruses in melanoma. Br J Dermatol. 2009;161(6):1225–1231. doi: 10.1111/j.1365-2133.2009.09415.x. [DOI] [PubMed] [Google Scholar]
- 6.Christensen T. HERVs in neuropathogenesis. J Neuroimmune Pharmacol. 2010;5(3):326–335. doi: 10.1007/s11481-010-9214-y. [DOI] [PubMed] [Google Scholar]
- 7.Evans LH, et al. In vivo interactions of ecotropic and polytropic murine leukemia viruses in mixed retrovirus infections. J Virol. 2006;80(10):4748–4757. doi: 10.1128/JVI.80.10.4748-4757.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy-Burman P. Endogenous env elements: Partners in generation of pathogenic feline leukemia viruses. Virus Genes. 1995;11(2-3):147–161. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- 9.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 10.Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol Evol. 2012;27(11):627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard D, et al. Identification and characterization of a novel retroviral-like aspartic protease specifically expressed in human epidermis. J Invest Dermatol. 2005;125(2):278–287. doi: 10.1111/j.0022-202X.2005.23816.x. [DOI] [PubMed] [Google Scholar]
- 13.Matsui T, et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol Med. 2011;3(6):320–333. doi: 10.1002/emmm.201100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoye JP. Fv1, the mouse retrovirus resistance gene. Rev Sci Tech. 1998;17(1):269–277. doi: 10.20506/rst.17.1.1080. [DOI] [PubMed] [Google Scholar]
- 15.Mura M, et al. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci USA. 2004;101(30):11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horie M, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463(7277):84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: Multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6(7):e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6(11):e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomonaga K, Kobayashi T, Ikuta K. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 2002;4(4):491–500. doi: 10.1016/s1286-4579(02)01564-2. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki Y, Kobayashi Y, Horie M, Tomonaga K. Origin of endogenous bornavirus-like nucleoprotein elements in thirteen-lined ground squirrels. Genes Genet Syst. doi: 10.1266/ggs.89.143. in press. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto Y, et al. Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe. 2012;11(5):492–503. doi: 10.1016/j.chom.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Schneider U, Naegele M, Staeheli P, Schwemmle M. Active borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein. J Virol. 2003;77(21):11781–11789. doi: 10.1128/JVI.77.21.11781-11789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chase G, et al. Borna disease virus matrix protein is an integral component of the viral ribonucleoprotein complex that does not interfere with polymerase activity. J Virol. 2007;81(2):743–749. doi: 10.1128/JVI.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki S, Ludwig H. In borna disease virus infected rabbit neurons 100 nm particle structures accumulate at areas of Joest-Degen inclusion bodies. Zentralbl Veterinarmed B. 1993;40(4):291–297. doi: 10.1111/j.1439-0450.1993.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 25.Geib T, et al. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J Virol. 2003;77(7):4283–4290. doi: 10.1128/JVI.77.7.4283-4290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanai H, et al. Development of a novel Borna disease virus reverse genetics system using RNA polymerase II promoter and SV40 nuclear import signal. Microbes Infect. 2006;8(6):1522–1529. doi: 10.1016/j.micinf.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi T, et al. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology. 1998;243(1):188–197. doi: 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 28.Berg M, Ehrenborg C, Blomberg J, Pipkorn R, Berg AL. Two domains of the Borna disease virus p40 protein are required for interaction with the p23 protein. J Gen Virol. 1998;79(Pt 12):2957–2963. doi: 10.1099/0022-1317-79-12-2957. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi T, et al. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J Virol. 2001;75(7):3404–3412. doi: 10.1128/JVI.75.7.3404-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolph MG, et al. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11(10):1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Hock M, et al. RNA induced polymerization of the Borna disease virus nucleoprotein. Virology. 2010;397(1):64–72. doi: 10.1016/j.virol.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Schwemmle M, et al. Interactions of the borna disease virus P, N, and X proteins and their functional implications. J Biol Chem. 1998;273(15):9007–9012. doi: 10.1074/jbc.273.15.9007. [DOI] [PubMed] [Google Scholar]
- 33.Schwemmle M, Jehle C, Shoemaker T, Lipkin WI. Characterization of the major nuclear localization signal of the Borna disease virus phosphoprotein. J Gen Virol. 1999;80(Pt 1):97–100. doi: 10.1099/0022-1317-80-1-97. [DOI] [PubMed] [Google Scholar]
- 34.Yanai H, et al. A methionine-rich domain mediates CRM1-dependent nuclear export activity of Borna disease virus phosphoprotein. J Virol. 2006;80(3):1121–1129. doi: 10.1128/JVI.80.3.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe Y, Ohtaki N, Hayashi Y, Ikuta K, Tomonaga K. Autogenous translational regulation of the Borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. PLoS Pathog. 2009;5(11):e1000654. doi: 10.1371/journal.ppat.1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382(6594):826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 37.Hilditch L, et al. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci USA. 2011;108(14):5771–5776. doi: 10.1073/pnas.1100118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnaud F, Varela M, Spencer TE, Palmarini M. Coevolution of endogenous betaretroviruses of sheep and their host. Cell Mol Life Sci. 2008;65(21):3422–3432. doi: 10.1007/s00018-008-8500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer TE, Mura M, Gray CA, Griebel PJ, Palmarini M. Receptor usage and fetal expression of ovine endogenous betaretroviruses: Implications for coevolution of endogenous and exogenous retroviruses. J Virol. 2003;77(1):749–753. doi: 10.1128/JVI.77.1.749-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnaud F, Murcia PR, Palmarini M. Mechanisms of late restriction induced by an endogenous retrovirus. J Virol. 2007;81(20):11441–11451. doi: 10.1128/JVI.01214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A. Human endogenous retrovirus K Gag coassembles with HIV-1 Gag and reduces the release efficiency and infectivity of HIV-1. J Virol. 2012;86(20):11194–11208. doi: 10.1128/JVI.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bieniasz PD. Intrinsic immunity: A front-line defense against viral attack. Nat Immunol. 2004;5(11):1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 43.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33(9):663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lando M, et al. Identification of eight candidate target genes of the recurrent 3p12-p14 loss in cervical cancer by integrative genomic profiling. J Pathol. 2013;230(1):59–69. doi: 10.1002/path.4168. [DOI] [PubMed] [Google Scholar]
- 46.Daito T, et al. A novel borna disease virus vector system that stably expresses foreign proteins from an intercistronic noncoding region. J Virol. 2011;85(23):12170–12178. doi: 10.1128/JVI.05554-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.