Significance
The prevalence of food allergy is rising at an alarming rate; the US Centers for Disease Control and Prevention documented an 18% increase among children in the United States between 1997 and 2007. Twenty-first century environmental interventions are implicated by this dramatic generational increase. In this report we examine how alterations in the trillions of commensal bacteria that normally populate the gastrointestinal tract influence allergic responses to food. We identify a bacterial community that protects against sensitization and describe the mechanism by which these bacteria regulate epithelial permeability to food allergens. Our data support the development of novel adjunctive probiotic therapies to potentiate the induction of tolerance to dietary allergens.
Keywords: microbiome, barrier, IL-22
Abstract
Environmentally induced alterations in the commensal microbiota have been implicated in the increasing prevalence of food allergy. We show here that sensitization to a food allergen is increased in mice that have been treated with antibiotics or are devoid of a commensal microbiota. By selectively colonizing gnotobiotic mice, we demonstrate that the allergy-protective capacity is conferred by a Clostridia-containing microbiota. Microarray analysis of intestinal epithelial cells from gnotobiotic mice revealed a previously unidentified mechanism by which Clostridia regulate innate lymphoid cell function and intestinal epithelial permeability to protect against allergen sensitization. Our findings will inform the development of novel approaches to prevent or treat food allergy based on modulating the composition of the intestinal microbiota.
Life-threatening anaphylactic responses to food are an increasingly important public health problem (1). Rising disease prevalence over a short period cannot be explained by genetic variation alone, renewing interest in the role of the environment in shaping allergic sensitization to food (2, 3). First proposed more than 20 years ago, the hygiene hypothesis suggested that societal efforts to reduce exposure to infectious microbes early in life have deprived the immune system of immunoregulatory stimulation necessary for protection against allergic disease (4). As our understanding of the profound influence of commensal microbes on the maturation of the immune system has grown, more recent iterations of this hypothesis have supported the idea that alterations in the composition of the intestinal microbiota induced by environmental factors (e.g., antibiotics, diet, vaccination, sanitation) play a central role in the regulation of allergic sensitization (5–7). In particular, antibiotic use during infancy potently perturbs intestinal bacterial populations and has often been cited as a contributing factor to the rising prevalence of allergic disease (8). However, the mechanisms by which changes in the composition of the intestinal microbiota regulate allergic responses to food remain poorly understood.
The gastrointestinal tract must maintain nonresponsiveness to both an enormous variety of food antigens and the trillions of bacteria that comprise the commensal microbiota (9). Mucosal IgA and regulatory T-cell (Treg) responses induced by commensal bacteria are critical for sustaining the homeostatic host–microbe relationship and preventing intestinal inflammation (10). In addition, recent work has revealed that a heterogeneous population of innate immune cells, known collectively as innate lymphoid cells (ILCs), plays a critical role in integrating signals from the commensal microbiota to maintain homeostasis at epithelial barriers and guide adaptive immunity (11). In this report we show that sensitization to a food allergen is enhanced in mice that have been treated with antibiotics (Abx) or are devoid of commensal microbes (germ free, GF). Selective colonization of gnotobiotic mice demonstrated that the allergy-protective capacity is contained within the Clostridia, a class of anaerobic spore-forming Firmicutes that reside in close proximity to the intestinal epithelium. Reintroduction of a Clostridia-containing microbiota to Abx-treated mice blocks sensitization to a food allergen. Using microarray analysis of intestinal epithelial cells from gnotobiotic mice, we identify an innate mechanism by which Clostridia protect against sensitization to dietary antigens. Defects in intestinal permeability have been implicated in aberrant allergic responses to food, but the mechanisms governing uptake of dietary antigen have not been clear. We show here that Clostridia colonization induces IL-22 production by both RAR-related orphan receptor gamma (RORγt)+ ILCs and T cells in the intestinal lamina propria (LP) and that this cytokine acts to reduce uptake of orally administered dietary antigen into the systemic circulation, contributing, in part, to protection against sensitization.
Results
Neonatal Abx Exposure Alters the Commensal Microbiota and Enhances Food Allergen Sensitization.
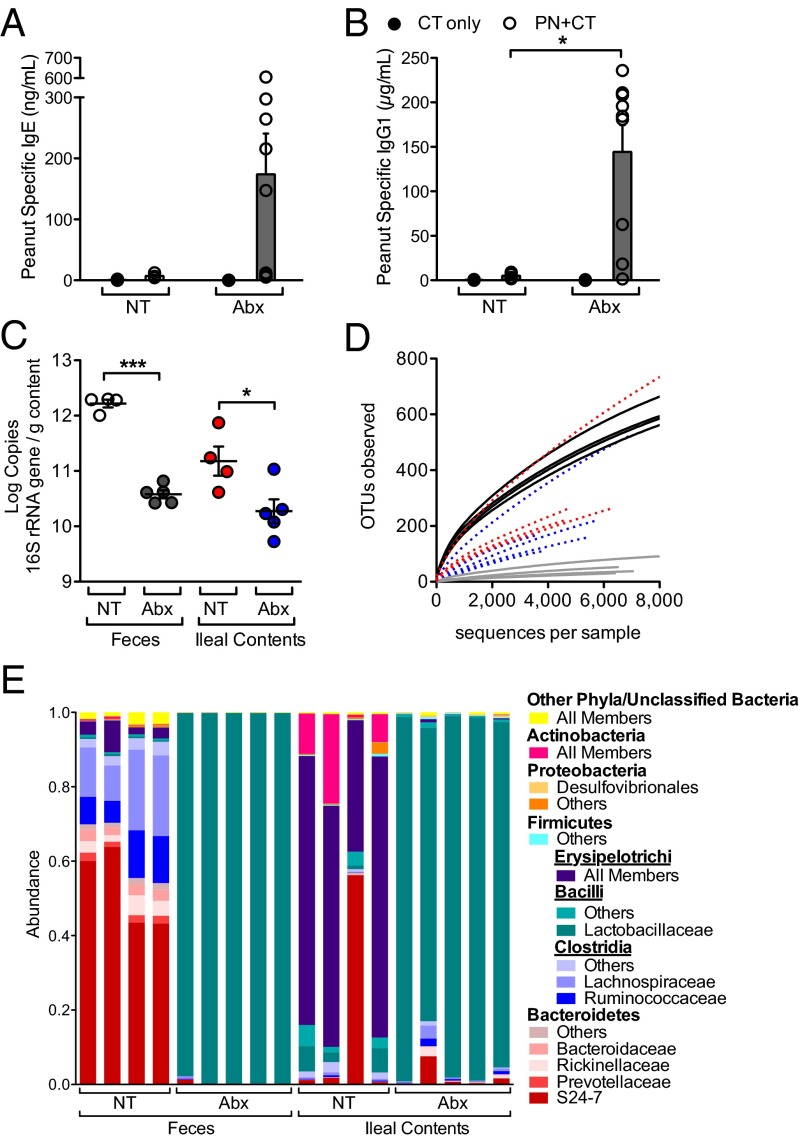
We evaluated the susceptibility of mice to food allergen sensitization by intragastric coadministration of peanut (PN) allergens and the mucosal adjuvant cholera toxin (CT), which induces PN-specific IgE, IgG1, and symptoms typical of systemic allergic hyperreactivity (12). Mice treated with Abx showed marked elevation in PN-specific IgE and IgG1 after sensitization and allergen challenge (Fig. 1 A and B). Analysis of 16S rRNA genes revealed that 6 wk of Abx treatment resulted in a significant reduction in bacterial load in both the feces and ileal contents (Fig. 1C) and altered the diversity (Fig. 1D) and composition (Fig. 1E) of the fecal and ileal microbiota. Those members of the Bacteroidetes and Firmicutes phyla most prevalent under normal conditions (Fig. 1E, no treatment, NT) were absent in fecal samples obtained from Abx-treated mice and were replaced instead by Lactobacillaceae (Fig. 1E), consistent with another recent report (13).
Fig. 1.
Neonatal Abx exposure alters the commensal microbiota and enhances food allergen sensitization. Abx treatment was initiated before weaning as described in Methods. (A and B) 3-wk-old mice were sensitized by intragastric administration of PN plus CT (PN/CT, open symbols) or CT only (closed symbols) and challenged on day 35; feces and serum were collected on day 36. Serum concentration of (A) PN-specific IgE and (B) PN-specific IgG1 was measured by ELISA (n = 4–9 mice per group from three independent experiments; each circle represents an individual mouse; bars depict mean and SEM). (C) Bacterial load in the feces or ileal contents of mice treated with Abx compared with no treatment (NT) controls. (n = 4–5 mice per group). (D) Bacterial diversity, as shown by operational taxonomic unit (97% identity) rarefaction curves in Abx-treated mice compared with NT controls: black lines, NT feces; gray lines, Abx feces; red lines, NT ileal contents; blue lines, Abx ileal contents. (E) Taxonomic classifications for the mice in C represented as proportion of total reads (Methods). *P < 0.05, **P < 0.01, ***P < 0.001 determined by Student t test (B) or one-way ANOVA with Tukey posttest (C).
A Clostridia-Containing Microbiota Protects Against Sensitization to Food Allergens.
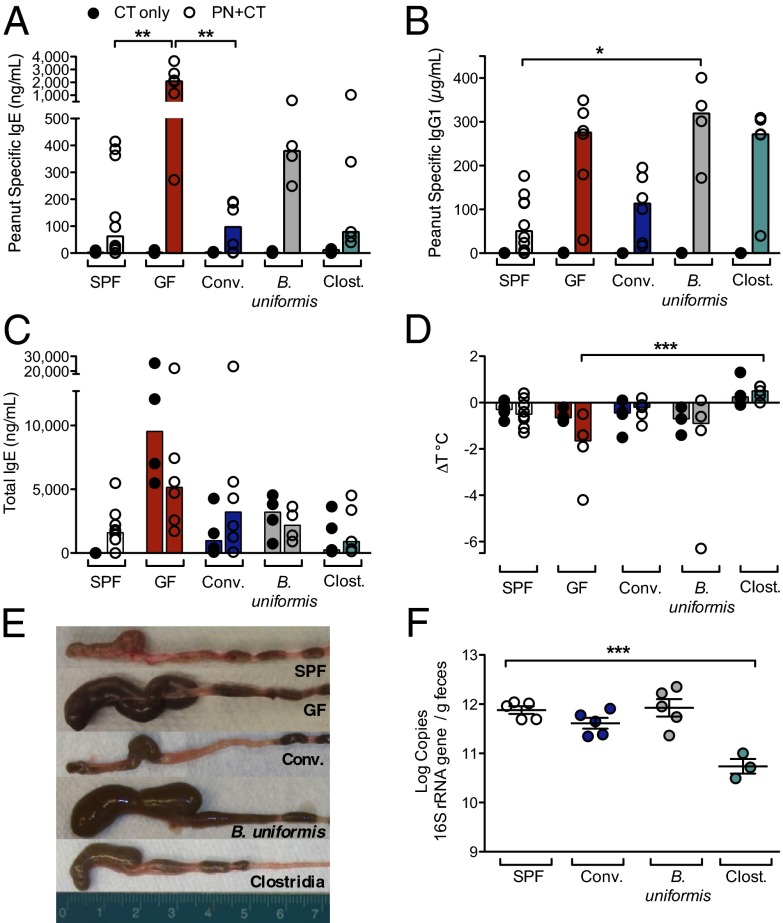
To gain insight into the populations of bacteria responsible for protection against sensitization to food allergens, we created a gnotobiotic model of food allergy. Upon sensitization with PN/CT, GF mice exhibit significantly increased levels of PN-specific IgE and IgG1 relative to mice maintained in typical specific pathogen-free (SPF) housing conditions and the reduced core body temperature at challenge characteristic of an anaphylactic response (Fig. 2 A, B, and D). In the absence of a colonizing microbiota, GF mice displayed grossly enlarged cecal size (Fig. 2E) and spontaneously higher levels of circulating IgE with increasing age (Fig. S1A). Total IgE levels in GF mice were elevated by treatment with CT or PN/CT (Fig. 2C). GF mice colonized with an SPF microbiota (conventionalized) did not show elevated levels of PN-specific IgE (Fig. 2A) or IgG1 (Fig. 2B) or a reduced core body temperature (Fig. 2D) in response to sensitization with PN/CT. The concentration of total IgE detectable in the serum of conventionalized mice was also reduced to levels similar to those seen in SPF mice (Fig. 2C). In addition, conventionalized mice displayed the normal cecal size (Fig. 2E) and bacterial load characteristic of SPF mice (Fig. 2F).
Fig. 2.
A Clostridia-containing microbiota protects against sensitization to food allergens. (A–D) Groups of SPF (white), GF (red), or gnotobiotic mice colonized with fecal/cecal material from SPF mice (Conventionalized, blue), B. uniformis (gray), or with a consortium of Clostridia (green) were sensitized with either CT only or PN/CT at weaning and challenged on day 35. (A) Concentration of PN-specific IgE, (B) IgG1, and (C) total IgE in serum of sensitized mice collected 24 h after challenge. (D) Change in core body temperature in sensitized mice (n = 4–10 mice per group from two independent experiments; closed circles, CT only; open circles, PN/CT). In A–D, each circle represents an individual mouse; bars depict median. (E) Cecal size at 13 d after colonization. (F) Bacterial load in feces collected from 5- to 10-wk-old SPF and gnotobiotic mice 14 d after colonization. n = 3–5 mice per group. F depicts mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001 determined by two-way ANOVA with the Kruskal-Wallis test (A–D) or one-way ANOVA with Tukey posttest (F).
We next examined the ability of selected members of the SPF microbiota to influence susceptibility to allergic sensitization to food. We focused on Bacteroides, Clostridium cluster XIVa, and Clostridium cluster IV, which constitute the numerically predominant taxa in the murine colon (14). Anaerobic cultures of fecal material from our SPF colony yielded Bacteroides uniformis as a representative Bacteroides species. Monocolonization of GF mice with B. uniformis resulted in a bacterial load similar to that seen in SPF and conventionalized mice (Fig. 2F) but did not reduce cecal size (Fig. 2E), rescue the drop in core body temperature in all mice (Fig. 2D), or significantly reduce the PN-specific IgE or IgG1 response seen in GF mice (Fig. 2 A and B). To colonize GF mice with Clostridia, we used chloroform-extracted spores isolated from a mixed cecal/fecal sample from a healthy SPF mouse. Sequence analysis showed that this extract was consistently and predominantly composed of members of Clostridium clusters XIVa, XIVb, and IV (Fig. S1 B–D). Colonization with this Clostridia consortium protected against sensitization to PN/CT, because levels of PN-specific and total IgE were reduced compared with GF controls (Fig. 2 A and C), and no temperature drop was seen at challenge (Fig. 2D). Cecal size in Clostridia-colonized mice was comparable to that seen in SPF mice (Fig. 2E), although the bacterial load measured in feces was significantly lower (Fig. 2F). Collectively, these data suggest that Clostridia play a role in protection against sensitization to a food allergen. We then examined whether the changes in food allergen sensitization induced by neonatal Abx administration (Fig. 1) could be reversed by selectively restoring the intestinal microbial community. In addition, we examined the response to sensitization after recovery from 1 wk of preweaning antibiotic treatment (Abx Recov.) (Fig. S2). PN-specific IgE and IgG1 and total IgE levels were reduced in serum collected at challenge from Abx-treated, PN/CT-sensitized mice that had been conventionalized (Abx conv.), Clostridia-colonized (Abx Clost.), or allowed to recover (Abx Recov.) (Fig. S2 A–C), suggesting that restoring a Clostridia-containing microbiota by either fecal gavage or removal of Abx-mediated selection is sufficient to protect against food allergen sensitization. In support of these findings, at termination, the abundance of Clostridia in fecal samples was restored to untreated levels in mice that received fecal gavage (Abx conv. or Abx Clost.) or were allowed to recover (Abx Recov., Fig. S2 D and E), although their community structures remained distinct (Fig. S2F).
Clostridia Colonization Activates Innate Immune Genes in Intestinal Epithelial Cells.
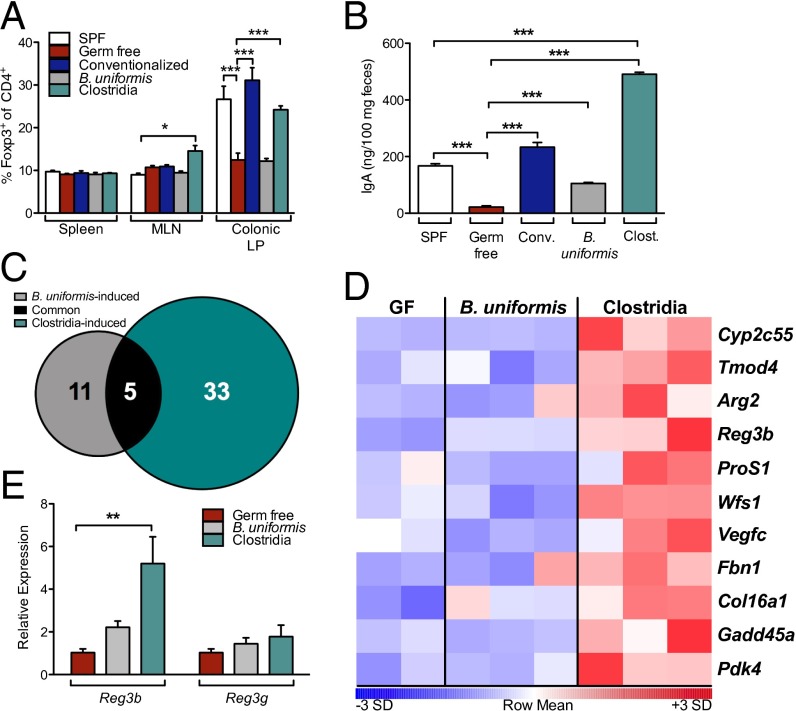
Several reports suggest that mucosa-associated Clostridia populations have a unique role in the induction of Foxp3+ Tregs and IgA, the two major arms of adaptive mucosal immunity (15–17). The ability of selected indigenous commensal bacteria to activate innate immune signaling in intestinal epithelial cells (IECs) is less well understood. We first confirmed that both conventionalized and Clostridia-colonized mice have significantly increased proportions of Foxp3+ Tregs in the colonic LP and elevated concentrations of fecal IgA compared with the baseline levels detected in GF mice (Fig. 3 A and B). Monocolonization of GF mice with B. uniformis partially restored levels of fecal IgA (Fig. 3B) but did not affect the LP Treg compartment (Fig. 3A). B. uniformis and Clostridia therefore differed in their ability both to protect against food allergen sensitization (Fig. 2) and to induce colonic Tregs and fecal IgA (Fig. 3), suggesting that they also differentially activate innate immunity. To gain insight into the role of microbial interactions with IECs in the regulation of sensitization to food allergens, we examined gene expression in IECs from GF mice and from mice colonized with B. uniformis or Clostridia. Microarray analysis showed that 38 genes in IEC from Clostridia-colonized mice and 16 from B. uniformis-colonized mice exhibited ≥1.5-fold increase in mean expression compared with GF controls (Fig. 3C). We were particularly interested in the differential up-regulation of regenerating islet-derived 3 beta (Reg3b) in Clostridia-colonized mice (Fig. 3 D and E) because it encodes an antimicrobial peptide, REG3β, which regulates the composition of the mucosa-associated microbiota (18).
Fig. 3.
Clostridia colonization activates innate immune genes in IECs. (A) Proportion of Foxp3+ Tregs among CD4+ T cells in the spleen, MLN, and colonic LP of age-matched SPF (white) and GF (red) mice and 14 d after colonization of GF mice with an SPF microbiota (Conventionalized, blue), B. uniformis (gray), or Clostridia (green) (n = 4–8 per group). (B) Concentration of IgA in feces collected from sensitized mice in Fig. 2. (C) Number of genes up-regulated in IECs relative to GF by B. uniformis (gray), Clostridia (green), or both (black) at 6 d after colonization. Genes shown exhibited significant expression above background in all samples (detection P value <0.05) and ≥1.5-fold increase in mean expression in comparison with values obtained for GF mice. (D) Heatmap depicting differential gene expression for 11 genes of interest. Samples with the highest and lowest transcript levels are red and blue, respectively. (E) Quantitative PCR verification of microarray data for selected genes. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Bonferroni posttest (A and E) or one-way ANOVA with Tukey posttest (B).
Clostridia Colonization Induces IL-22.
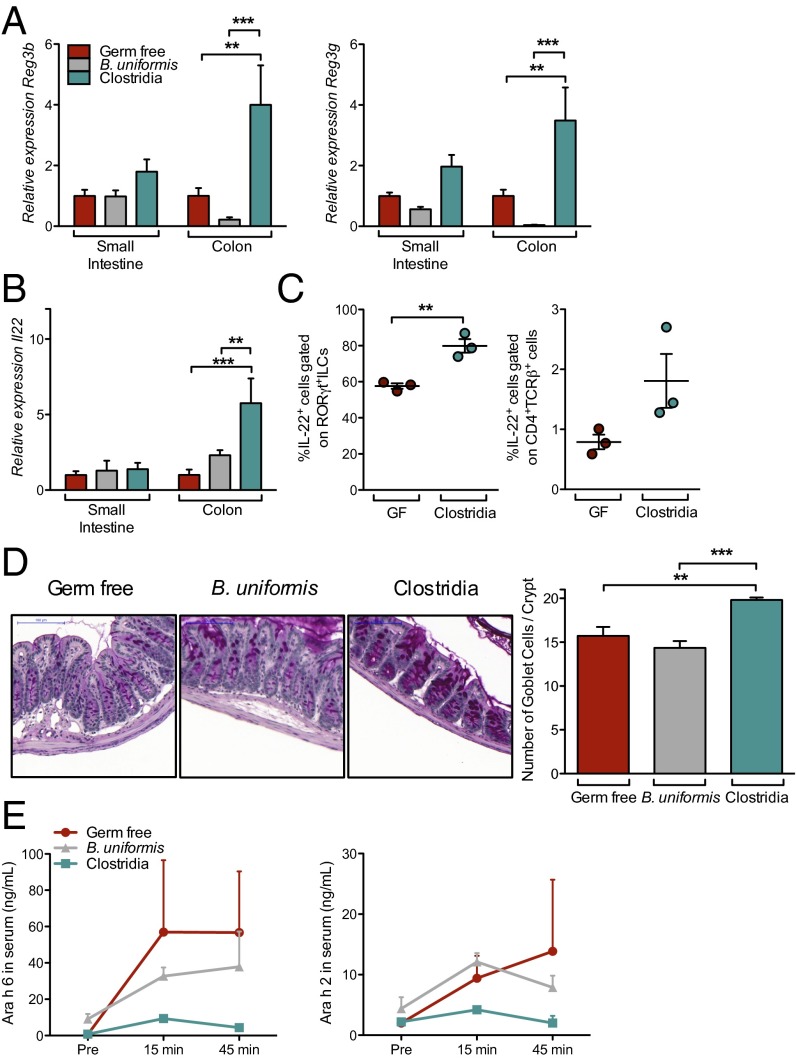
We validated our microarray results by demonstrating that Reg3b and Reg3g expression were increased in whole-tissue extracts from Clostridia, but not B. uniformis, colonized mice (Fig. 4A). IECs produce antimicrobial peptides in response to IL-22–mediated signaling (18). We found that only Clostridia colonization induced significant up-regulation of IL-22 transcripts in lamina propria lymphocyte (LPL) (Fig. 4B). Flow cytometric analysis revealed that both RORγt+ ILCs and CD4+TCRβ+ T cells produced elevated levels of IL-22 in response to Clostridia colonization (Fig. 4C and Fig. S3A). The proportion of RORγt+ ILCs within the LTi0, LTi4, and NK22 ILC3 subsets in the colonic LP was unchanged in Clostridia-colonized mice (Fig. S3B). IL-22 also protects the intestinal epithelial barrier by promoting mucus secretion by goblet cells (19); the numbers of mucus-producing goblet cells were significantly increased in mice colonized with Clostridia but not in those colonized with B. uniformis (Fig. 4D). Having identified its cellular sources and confirmed a known barrier protective functional activity in our model, we asked whether IL-22 also plays a role in regulating epithelial permeability to protein antigens. Because antigen uptake from the intestinal lumen is the first step in sensitization to a food allergen, we reasoned that Clostridia-induced IL-22 production reinforces the epithelial barrier to reduce intestinal permeability to dietary proteins. To explore this hypothesis, we developed an assay to measure the transient presence of allergen in the blood after intragastric gavage. Several Ara h proteins have been identified as the immunodominant allergens of PN (Arachis hypogaea) (20). We used sensitive capture ELISAs to measure the concentration of two of these proteins in the systemic circulation. Both Ara h 6 and Ara h 2 were readily detectable in the serum of GF mice (Fig. 4E). Colonization with Clostridia, but not B. uniformis, reduced the circulating concentrations of both proteins after gavage.
Fig. 4.
Clostridia colonization induces IL-22. (A) Reg3b and Reg3g expression from whole-tissue extracts isolated 4 d after colonization from the small intestine or colon of GF (red), B. uniformis-colonized (gray), or Clostridia-colonized (green) mice. Quantitative RT-PCR data are plotted relative to GF and normalized to Hprt (n = 8–9 mice per group from two independent experiments). (B) Il22 expression in LPL from mice in A. (C) IL-22 production by RORγt+ ILCs and T cells 6 d after colonization, determined by flow cytometric analysis of permeabilized cells (SI Methods; n = 3 mice per group representative of three independent experiments). (D) Representative images and quantification of goblet cells in distal colon of GF, B. uniformis-colonized, and Clostridia-colonized mice 6 d after colonization. n = 3–5 mice per group. (Scale bar, 100 µm.) (E) Serum Ara h 6 and Ara h 2 levels after PN gavage in GF, B. uniformis-colonized, or Clostridia-colonized mice 6 d after colonization (n = 5–12 mice per group from two independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Bonferroni posttest (A and B) or one-way ANOVA with Tukey posttest (C).
Clostridia-Induced IL-22 Regulates Allergen Access to the Bloodstream.
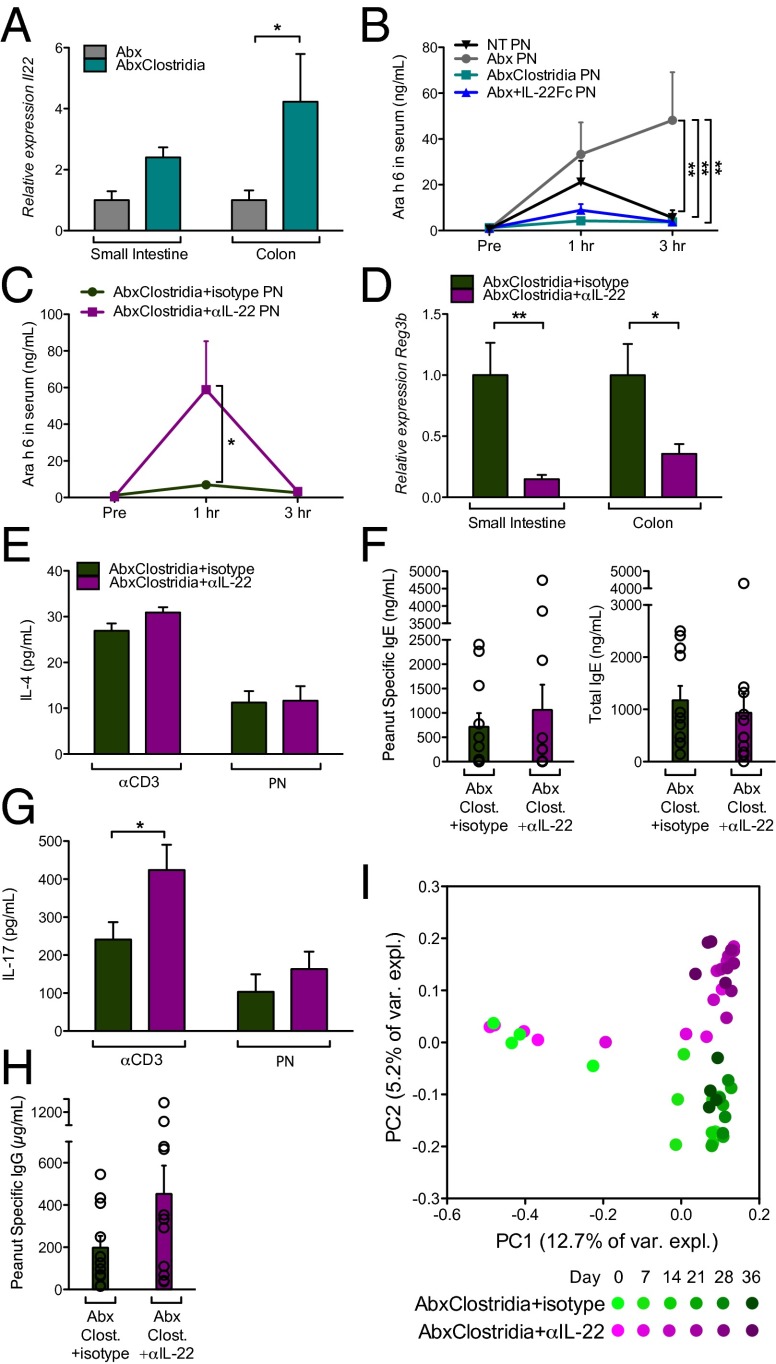
To determine whether IL-22 induced by Clostridia gavage is necessary and sufficient to reduce intestinal barrier permeability we used the Abx-depletion model. Il22 expression was significantly increased in the colon of Abx-treated Clostridia-colonized mice (Fig. 5A). Significantly higher concentrations of Ara h 6 were detected in the serum of Abx-treated mice compared with mice that received no treatment (NT; Fig. 5B); similar results were obtained for Ara h 2 (Fig. S4A). Serum Ara h 6 and Ara h 2 were reduced in Abx mice treated with an IL-22-Fc fusion protein (21) or colonized with Clostridia after 1 wk of Abx gavage (Fig. 5B and Fig. S4A), indicating that either Clostridia gavage or exogenous IL-22 is sufficient to reduce the concentration of serum allergen. To demonstrate that Clostridia-induced IL-22 regulates allergen access to the bloodstream, groups of Abx-treated Clostridia-colonized mice were given i.p. injections of a neutralizing antibody to IL-22 (22) or an isotype control before allergen challenge. Serum concentrations of Ara h 6 and Ara h 2 were significantly elevated in Clostridia-colonized mice treated with anti-IL-22 compared with mice treated with an isotype control (Fig. 5C and Fig. S4B), directly linking Clostridia-induced IL-22 production to the regulation of allergen uptake. Anti-IL-22 treatment did not affect Clostridia-mediated induction of Foxp3+ Tregs in the colonic LP (Fig. S4C). Together with the inability of IL-22-Fc to induce CD4+Foxp3+ Tregs in the colonic LP of Abx-treated mice (Fig. S4C), this result suggested that Clostridia-induced IL-22 does not expand the colonic Treg compartment. In addition, the concentration of Ara h 6 in the serum of Abx-treated mice 3 h after gavage with PN/CT was significantly higher than that detected in mice that received PN alone (Fig. S4D compared with Fig. 5B; P < 0.05), in agreement with the role of adjuvants such as CT in increasing intestinal permeability to luminal antigens (23). Serum Ara h 6 and Ara h 2 were reduced in Abx-treated Clostridia-colonized mice even when PN was administered together with CT (Fig. S4 D and E). To examine whether Clostridia-induced IL-22 production by ILCs regulates allergen uptake, we repeated the Abx treatment/Clostridia colonization in Rag−/− mice depleted of ILCs with anti-CD90 antibody (as described in ref. 24). Elevated concentrations of Ara h 6 and Ara h 2 were detectable in the serum of Abx-treated Clostridia-colonized ILC-depleted Rag−/− mice compared with mice treated with an isotype control (Fig. S4 F and G). The efficacy of anti-CD90 treatment in depleting IL-22 transcripts in the intestinal LP was confirmed by quantitative PCR (Fig. S4H).
Fig. 5.
Clostridia-induced IL-22 regulates allergen access to the bloodstream. (A) Expression of Il22 in LPL from neonatal Abx-treated mice without Clostridia colonization, or at 6 d after weaning and colonization. (B) Serum Ara h 6 at indicated time points after PN gavage in NT or Abx mice treated with or without one i.p. injection of IL-22-Fc, or by Clostridia colonization. (C) Serum Ara h 6 at indicated time points after PN gavage in Abx-treated Clostridia-colonized mice injected i.p. with neutralizing antibody to IL-22 or an isotype control. All mice in B and C received PN at 6 d after weaning, and serum levels of Ara h 6 were measured by capture ELISA (n = 5–10 mice per group, pooled from at least two experiments). (D) Expression of Reg3b in whole-tissue extracts from Abx-treated Clostridia-colonized mice treated with neutralizing antibody to IL-22 or an isotype control and sensitized with PN/CT (n = 11 mice per group, pooled from four experiments). (E) Concentration of IL-4 in culture supernatants from splenocytes of mice from D (n = 7 mice per group, representative of two experiments). (F) Concentration of PN-specific and total IgE in serum collected 24 h after challenge for mice in D (n = 11 mice per group, pooled from four experiments). (G) Concentration of IL-17 in culture supernatants from splenocytes from mice in D (n = 7 mice per group, representative of two experiments). (H) Concentration of PN-specific IgG in serum collected 24 h after challenge for mice in D (n = 11 mice per group, pooled from four experiments). (I) UniFrac analysis of fecal microbiota throughout the sensitization protocol (n = 4 mice per group). *P < 0.05, **P < 0.01 ***P < 0.001 by two-way ANOVA with Bonferroni posttest (A, B, and D) or Student t test (C and G).
Finally, we examined whether Clostridia-induced IL-22 production in the intestinal LP regulates sensitization to food allergens. Abx-treated Clostridia-colonized mice sensitized with PN/CT as in Fig. 1 and Fig. S2 were treated with anti-IL-22 or isotype control throughout the 35-d protocol. Examination at sacrifice showed that both intestinal Reg3b expression (Fig. 5D) and goblet cell numbers (Fig. S4I) were significantly reduced in mice treated with anti-IL-22 compared with isotype-treated controls, confirming that IL-22 was effectively neutralized by this treatment protocol. To assess sensitization to food, splenocytes harvested after allergen challenge were restimulated in vitro with anti-CD3 or PN as previously described (12). Oral administration of antigen with CT as a mucosal adjuvant typically induces a Th2 biased response to promote allergic sensitization (12). However, treatment of Abx-depleted Clostridia-colonized mice with anti-IL-22 throughout the course of the sensitization protocol did not result in elevated levels of IL-4 (Fig. 5E) or an increased PN-specific or total IgE response (Fig. 5F), in agreement with the absence of Th2 skewing (IL-13 and IFN-γ were also not significantly changed; Fig. S4 J and K). Instead we detected significantly elevated production of IL-17 (Fig. 5G), consistent with other reports showing that depletion of innate IL-22 promotes an adaptive Th17 response (25). PN-specific IgG increased in anti-IL-22–treated mice compared with isotype controls (P = 0.09) (Fig. 5H). Interestingly, in keeping with the antimicrobial activity of REG3β, we found that anti-IL-22 treatment altered the composition of the fecal microbiota. UniFrac analysis showed that the microbiota of anti-IL-22–treated mice increasingly diverged from that of their isotype control treated littermates during the 5 wk of treatment (Fig. 5I). Neutralization of IL-22 increased the abundance of Clostridiales throughout most of the sensitization period, whereas the abundance of Bacteroidales remained unchanged (Fig. S4L). Taken together, these data support our hypothesis that mucosa-associated Clostridia play a critical role in regulating sensitization to food allergens.
Discussion
Dietary antigens are absorbed in the small intestine and carried to the mesenteric lymph node by CD103+ dendritic cells, ultimately generating food antigen-specific Tregs that then migrate to the small intestinal LP and expand to maintain tolerance to dietary antigen (26). Our data suggest a new paradigm in which both antigen-specific tolerance and a bacteria-induced barrier protective response are required to prevent sensitization to food antigens. We identify an innate mechanism through which a predominant component of the normal mucosa-associated commensal microbiota regulates sensitization to food. Using a sensitive capture ELISA to measure the concentration of two immunodominant PN allergens in serum within hours after gavage, we show that Clostridia-induced early innate IL-22 production by RORγt+ ILCs and T cells reduces access of allergen to the bloodstream. Treatment of Abx-depleted Clostridia-colonized mice with neutralizing anti-IL-22 throughout the course of the PN/CT sensitization protocol induces enhanced production of IL-17 upon restimulation in vitro, in agreement with a role for innate IL-22 in regulating the adaptive Th17 response (25). PN-specific IgG responses increase in anti-IL-22–treated mice but, without Th2 skewing, the IgE response is unaltered. The composition of the microbiota was also transformed by treatment with anti-IL-22. The antimicrobial activity of REG3β/γ is directed against Gram-positive bacteria (18). Clostridia induce both Il22 and Reg3b/g expression and stably colonize gnotobiotic mice. In anti-IL-22–treated mice, however, increased abundance of Clostridiales correlates with reduced expression of Reg3b, suggesting that this antimicrobial peptide titrates Clostridia abundance in its colonic niche.
We also confirmed that the presence of a Clostridia-containing microbiota is associated with the adaptive expansion of the intestinal Treg compartment and class switching to IgA (16, 17), further reinforcing the immunoregulatory environment required to maintain tolerance to dietary antigen. Indeed, IgA likely contributes to immune exclusion to reduce allergen uptake; note the accelerated kinetics with which Ara h 6 and Ara h 2 reach the blood in Rag−/− mice in comparison with WT mice. Increased bacteria-induced luminal IgA and decreased systemic allergen-specific Ig in Clostridia-colonized mice may both be related to reduced systemic allergen uptake. However, Clostridia’s early induction of IL-22 may not be directly involved in the adaptive Treg and IgA phase of the Clostridia-induced protective response, because treatment with an IL-22Fc fusion protein does not result in an expansion of Tregs in the colonic LP. Instead, recent work suggests that microbial metabolites such as short chain fatty acids can regulate the proportions and functional capabilities of Foxp3+ Tregs in the colonic LP (27–29).
Direct evidence for environment-induced dysbiosis in the increasing prevalence of food allergy among children is just beginning to emerge. Studies have tied urinary levels of the commonly used antibacterial agent triclosan to food and aero-allergen sensitization (30) and prepartal or neonatal Abx use to cow’s milk allergy in infancy (31). Clostridia are enriched in the colon of both mice and humans (14). Recent work has shown that Clostridia strains isolated from healthy human feces potently induce Tregs in the colonic LP upon transfer to GF mice (17), suggesting our findings may be translatable to human disease. Oral and s.c. allergen-specific desensitization protocols are already showing promise for treating food allergy (32). Our data suggest that tolerance-inducing protocols could be effectively paired with Clostridia enrichment of gut microbiota to potentiate antigen-specific tolerance to prevent or treat food allergy.
Methods
Mice.
C57BL/6, C57BL/6Foxp3gfp, and Rag−/− mice on an inbred C57BL/6 background (33) were maintained in an SPF facility at The University of Chicago. Breeding pairs of GF C57BL/6 mice were initially provided by S. Mazmanian. C57BL/6Foxp3gfp mice were rederived GF by K. McCoy. All experiments were performed in accordance with the Institutional Biosafety and Animal Care and Use Committees.
Neonatal Abx Treatment.
C57BL/6 or C57BL/6Foxp3gfp mice were treated with a mixture of Abx, beginning at 2 wk of age, as previously described (12). For the first week, mice were given a daily intragastric gavage with 100 µL of a mixture of kanamycin (4 mg/mL), gentamicin (0.35 mg/mL), colistin (8500 U/mL), metronidazole (2.15 mg/mL), and vancomycin (0.45 mg/mL) (Sigma-Aldrich; MP Biomedicals). After weaning, the Abx were administered in the drinking water at 50-fold dilution except for vancomycin, which was maintained at 0.5 mg/mL.
Preparation of 16S rRNA-Based Amplicon Library and Data Analysis.
PCR amplicons of the V4 region of the 16S rRNA gene were sequenced on the Illumina MiSeq platform and analyzed using QIIME as described in SI Methods.
Purified PN Extract and Intragastric Sensitization.
Purified PN extract was prepared from roasted, unsalted PN by a modification of van Wijk et al., which omitted high-speed centrifugation at 10,000 × g (34). PN/CT sensitization was performed as in ref. 12 and is described in SI Methods.
Ig Detection, Isolation of Lymphocytes, and Flow Cytometry.
Methods were modified from refs. 12 and 33 and are described in SI Methods.
Microbial Isolation and Colonization of GF or Abx-Treated Mice.
B. uniformis was isolated from SPF feces. Clostridia were isolated from SPF mice by chloroform treatment. Some experimental mice were colonized from live gnotobiotic repository mice; one fecal pellet was homogenized in 1 mL sterile PBS, solids were allowed to settle, and 100 µL of the liquid phase was administered by gavage. A detailed description of bacterial colonization is given in SI Methods.
Quantitative Real-Time PCR.
RNA was prepared from freshly homogenized intestinal tissue or isolated LP cells from the small intestine and colon of GF, B. uniformis, or Clostridia-colonized mice at 4 d after colonization, Rag−/− Abx-treated Clostridia-colonized mice with or without anti-CD90.2 treatment at 6 d after colonization, or sensitized WT Abx-treated Clostridia-colonized mice with or without anti-IL-22 treatment at 24 h after challenge using the RNeasy Mini Kit (Qiagen). cDNA was produced using the iScript cDNA synthesis kit (BioRad), and quantitative real-time PCR was performed using the iQ SYBR Green supermix (Bio-Rad) on the StepOnePlus system (Applied Biosystems). Primer sequences for Il22, Reg3b, Reg3g, and Hprt are described in ref. 35. Expression of target genes was normalized to Hprt.
Microarray Analysis.
IECs were isolated from colons of GF, B. uniformis-colonized, or Clostridia-colonized mice at 6 d after colonization by shaking tissue fragments at 100 rpm for 20 min at 37 °C in 5 mM EDTA followed by vigorous vortexing and Percoll gradient centrifugation. IECs from three mice were pooled for each RNA sample in two to three independent experiments per condition. RNA was isolated as above. Samples were run on a single Illumina MouseRef-8 array at The University of Chicago Functional Genomics Facility. SI Methods provides analysis detail.
In Vivo Antibody Treatment.
SPF mice were treated with Abx by gavage for 1 wk before weaning. At weaning, mice were either placed on Abx-containing water or were colonized with Clostridia, as above. For exogenous IL-22 treatment, 20 µg of IL-22 fusion protein (IL-22-Fc, Genentech) was delivered i.p. at weaning. For depletion of IL-22, 150 µg of neutralizing antibody to IL-22 (clone 8E11, Genentech) (22) or an isotype control (GP120 10E7.1D2, Genentech) (36) was administered throughout the sensitization protocol by i.p. injection three times per week as previously described (37, 38). To deplete ILCs, 250 µg of anti-CD90.2 (clone 30H12, BioXCell) or isotype control (LTF-2, BioXCell) was administered i.p. every 3 d beginning 3 d before weaning, modified from ref. 24. The requirement for Clostridia-induced IL-22 production for the expansion of colonic Foxp3+Tregs was examined by i.p. injection of 500 μg of clone IL-22JOP (eBioscience), as previously described (39).
Assessment of Allergen Uptake.
To assess allergen uptake into serum, mice were bled before receiving 20 mg PN by gavage (±15 μg CT). Mice were bled again at indicated time points, and PN allergen concentration in serum was measured with capture ELISAs for Ara h 2 or Ara h 6 (Indoor Biotechnologies).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 5. Normally distributed data were analyzed by one-way ANOVA with Tukey posttest, two-way ANOVA with Bonferroni correction, or Student t test as appropriate to the number of comparisons to be made. Data that did not exhibit a normal distribution were analyzed using the nonparametric Kruskal-Wallis test with Dunn’s posttest.
Supplementary Material
Acknowledgments
We thank the staff of The University of Chicago Gnotobiotic Research Animal Facility for their expert technical assistance; T. Karrison (The University of Chicago Biostatistics Core) for advice on statistical analysis; S. Chervonsky and other colleagues for critical review of the manuscript; and W. Ouyang (Genentech) for providing neutralizing antibody to IL-22 (8E11), its isotype control, and an IL-22-Fc fusion protein for this study. This work was supported by Food Allergy Research and Education; a gift from the Bunning family; US National Institutes of Health Grants AI106302 (to C.R.N.), DK078938 (to S.K.M.), AI089954 (to L.Z.), AI091962 (to L.Z.), and T32AI007090-33 (to T.F.); and University of Chicago Digestive Diseases Research Core Center Grant DK42086.
Footnotes
Conflict of interest statement: A provisional US patent application (61/937952) was filed on February 10, 2014.
*This Direct Submission article had a prearranged editor.
Data deposition: The DNA sequences reported in this paper have been deposited in the MG-RAST database (project no. 7173). The Microarray data has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (series no. GSE60039).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412008111/-/DCSupplemental.
References
- 1.Berin MC, Sampson HA. Food allergy: An enigmatic epidemic. Trends Immunol. 2013;34(8):390–397. doi: 10.1016/j.it.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34(5):671–688. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao S, Feehley TJ, Nagler CR. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: Revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 6.Prioult G, Nagler-Anderson C. Mucosal immunity and allergic responses: Lack of regulation and/or lack of microbial stimulation? Immunol Rev. 2005;206:204–218. doi: 10.1111/j.0105-2896.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 7.Cho I, Blaser MJ. The human microbiome: At the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476(7361):393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 9.Nagler-Anderson C. Man the barrier! Strategic defences in the intestinal mucosa. Nat Rev Immunol. 2001;1(1):59–67. doi: 10.1038/35095573. [DOI] [PubMed] [Google Scholar]
- 10.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tait Wojno ED, Artis D. Innate lymphoid cells: Balancing immunity, inflammation, and tissue repair in the intestine. Cell Host Microbe. 2012;12(4):445–457. doi: 10.1016/j.chom.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172(11):6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 13.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagano Y, Itoh K, Honda K. The induction of Treg cells by gut-indigenous Clostridium. Curr Opin Immunol. 2012;24(4):392–397. doi: 10.1016/j.coi.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 18.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12(7):503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13(1):21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 20.Kulis M, et al. The 2S albumin allergens of Arachis hypogaea, Ara h 2 and Ara h 6, are the major elicitors of anaphylaxis and can effectively desensitize peanut-allergic mice. Clin Exp Allergy. 2012;42(2):326–336. doi: 10.1111/j.1365-2222.2011.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ota N, et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol. 2011;12(10):941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445(7128):648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 23.Lycke N, Karlsson U, Sjölander A, Magnusson KE. The adjuvant action of cholera toxin is associated with an increased intestinal permeability for luminal antigens. Scand J Immunol. 1991;33(6):691–698. doi: 10.1111/j.1365-3083.1991.tb02542.x. [DOI] [PubMed] [Google Scholar]
- 24.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336(6086):1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39(2):386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadis U, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 29.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage JH, Matsui EC, Wood RA, Keet CA. Urinary levels of triclosan and parabens are associated with aeroallergen and food sensitization. J Allergy Clin Immunol. 2012;130(2):453–460. doi: 10.1016/j.jaci.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metsälä J, et al. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology. 2013;24(2):303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 32.Henson M, Burks AW. The future of food allergy therapeutics. Semin Immunopathol. 2012;34(5):703–714. doi: 10.1007/s00281-012-0319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matharu KS, et al. Toll-like receptor 4-mediated regulation of spontaneous Helicobacter-dependent colitis in IL-10-deficient mice. Gastroenterology. 2009;137(4):1380–1390. doi: 10.1053/j.gastro.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Wijk F, et al. CTLA-4 signaling regulates the intensity of hypersensitivity responses to food antigens, but is not decisive in the induction of sensitization. J Immunol. 2005;174(1):174–179. doi: 10.4049/jimmunol.174.1.174. [DOI] [PubMed] [Google Scholar]
- 35.Upadhyay V, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13(10):947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sa SM, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178(4):2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 38.Kirchberger S, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210(5):917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mielke LA, et al. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210(6):1117–1124. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.