Significance
The C-C chemokine receptor type 5 (CCR5) G protein-coupled receptor (GPCR) is a prime target for preventing HIV invasion. A major difficulty in developing effective therapeutics is that the CCR5 exhibits an ensemble of ∼10–20 distinct low-energy conformations, each of which might favor binding to different ligands and/or lead to different downstream functions. X-ray structures generally provide only one of these conformations. We applied the GEnSeMBLE methodology to predict this ensemble, and we designed and carried out 11 experiments to validate the ability of this ensemble to predict binding of an HIV therapeutic to CCR5. We found that each of the mutations changes the binding site. The predicted effects of mutations on binding are in excellent agreement with experiments, providing CCR5 structures for designing new ligands.
Keywords: protein structure prediction, ligand–protein binding prediction, conformational ensemble, functional selectivity, GEnSeMBLE
Abstract
We predicted the structural basis for pleiotropic signaling of the C-C chemokine type 5 (CCR5) G protein-coupled receptor (GPCR) by predicting the binding of several ligands to the lower-energy conformations of the CCR5 receptor and 11 mutants. For each case, we predicted the ∼20 most stable conformations for the receptor along with the binding sites for four anti-HIV ligands. We found that none of the ligands bind to the lowest-energy apo-receptor conformation. The three ligands with a similar pharmacophore (Maraviroc, PF-232798, and Aplaviroc) bind to a specific higher-energy receptor conformation whereas TAK-779 (with a different pharmacophore) binds to a different high-energy conformation. This result is in agreement with the very different binding-site profiles for these ligands obtained by us and others. The predicted Maraviroc binding site agrees with the recent structure of CCR5 receptor cocrystallized with Maraviroc. We performed 11 site-directed mutagenesis experiments to validate the predicted binding sites. Here, we independently predicted the lowest 10 mutant protein conformations for each of the 11 mutants and then docked the ligands to these lowest conformations. We found the predicted binding energies to be in excellent agreement with our mutagenesis experiments. These results show that, for GPCRs, each ligand can stabilize a different protein conformation, complicating the use of cocrystallized structures for ligand screening. Moreover, these results show that a single-point mutation in a GPCR can dramatically alter the available low-energy conformations, which in turn alters the binding site, potentially altering downstream signaling events. These studies validate the conformational selection paradigm for the pleiotropic function and structural plasticity of GPCRs.
Since the finding that individuals lacking the C-C chemokine receptor type 5 (CCR5) gene are resistant to HIV, the CCR5 receptor has been established as a coreceptor for macrophage-tropic viruses, including HIV, to enter host cells (1). Several drugs aimed at disrupting the HIV–CCR5 coupling have been developed. To understand the structural mechanism of ligand binding to G protein-coupled receptors (GPCRs) and how these CCR5 targeting drugs work, we predicted the low-energy structures of the apo CCR5 receptor, which in turn were used to predict the binding sites for various ligands. The focus was on ligands already known to inhibit CCR5: Maraviroc (MVC) (2), PF-232798 (PF) (3), Aplaviroc (APL) (4), and TAK-779 (TAK) (5) because there are abundant experimental data on their binding to numerous CCR5 mutants, to test the predicted structures. The ligand structures are shown in Scheme S1. For three ligands (MVC, PF, and APL), we predict a similar binding site pharmacophore centered on a protonated nitrogen, whereas the other ligand (TAK) containing a quaternary nitrogen leads to a very different binding site pharmacophore. To validate our predictions, we identified 11 singly mutated CCR5 receptor forms selected to test the predicted binding region for the ligands. The predicted structures of ligand-bound CCR5 complexes presented here have already been used in two recent studies to provide a structural basis for (i) biophysical measurements (6) mapping the binding mode of MVC to CCR5 using genetically encoded photo–cross-linkers and (ii) viral entry experiments (7) probing the basis for G protein-coupled and -uncoupled CCR5 receptors by MVC-resistant and -sensitive HIV-1 viruses.
While this work was being finalized for submission, the CCR5 receptor was crystallized with MVC (8), allowing a direct structural validation of the predicted structures as discussed in Prediction of Ligand–CCR5 Structures and Comparison with the Crystal Structure.
Results
Prediction of the CCR5 Structure.
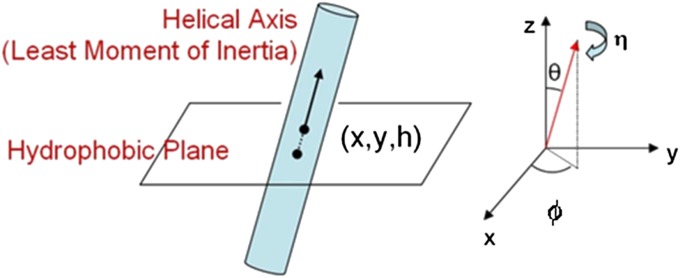
To predict the ensemble of low-energy structures (conformations) for CCR5, we used the GEnSeMBLE method (9), which performs a complete conformational sampling (∼11 billion) of transmembrane (TM) helix conformations accessible in the membrane. This method starts with templates for positioning the seven TM helices based on available experimental/predicted structures and then samples a large number of helix orientation angles as defined in Fig. 1.
Fig. 1.
Coordinate frame for the TM helix orientation in a membrane (reprinted with permission from ref. 28). The x-y plane defined as the middle plane of the membrane bilayer. The helical axis, defined as the least moment of inertia axis, intersects this plane at position x,y with residue “h.” The helical axis is tilted by an angle θ with respect to the z axis that is perpendicular to the membrane. The azimuthal angle ϕ of this tilt in the x-y plane and the rotation angle of the helix around this axis is the angle η.
During conformational sampling, we first sampled only the helix rotation angles (η) (on a 15° grid), and the BiHelix energy (10) was used to evaluate the energy of all (24)7 ∼ 4.6 billion TM helix bundle conformations. For the human CCR5 receptor structure prediction, this sampling was done starting with each of the following four X-ray–derived templates: bovine rhodopsin (PDB ID code 1u19) (11), human β2 adrenergic receptor (PDB ID code 2rh1) (12), human adenosine A2A receptor (PDB ID code 3eml) (13), and turkey β1 adrenergic receptor (PDB ID code 2vt4) (14). The full seven-helix bundle was built for the top 1,000 conformations from each template, and the side chains were optimized using the SideChain Rotamer Energy Analysis Method (SCREAM) side-chain placement protocol (15), followed by minimization (10 steps using Dreiding force field) (16). The β2 receptor template-based conformational ensemble led to the lowest energies (most stable conformations). We then selected the most stable 16 CCR5 conformations, for each of which we simultaneously sampled all three helix orientation angles (θ, ϕ, and η) allowing −10°, 0°, and +10° for the θ tilt angle and the −15°, 0° , and +15° range for both ϕ azimuthal and η rotation angles. This procedure led to a total of (27)7 ∼11 billion TM bundle conformations, for each of which we evaluated the energy rapidly using the SuperBiHelix method (17). Then the lowest-energy 2,000 conformations were built into seven-helix bundles and optimized, from which we selected the 20 lowest-energy conformations (labeled WT1 to WT20) as shown in Table S1. From these conformations, we selected eight structurally diverse seven-helix structures (highlighted rows in Table S1) for further analysis and ligand docking. The lowest-energy conformation WT1 corresponds to the predicted apo conformation of the receptor. None of the experimentally obtained GPCR structures (with the exception of opsin) has been ligand-free so it remains to be confirmed whether WT1 resembles the apo conformation of the CCR5 receptor.
These best eight diverse structures were then used to predict the binding of CCR5 ligands. The next section shows that MVC binds most strongly to the WT7 conformation rather than the lowest-energy WT1 conformation.
Prediction of Ligand–CCR5 Structures and Comparison with the Crystal Structure.
The four ligands (MVC, PF, APL, and TAK) were minimized using the B3LYP flavor of density functional theory (DFT) (with the 6–311G** basis set) using the Jaguar software package (Jaguar, version 7.8; Schrödinger, LLC). A conformational search was performed over the rotatable bonds for each ligand, and ∼20–30 conformations were selected (based on energy and diversity) (SI Materials and Methods) for each ligand. Then, each of these conformations was docked to the previously selected eight CCR5 conformations using the DarwinDock/GenDock method (18). This method for predicting ligand–protein structures samples a complete set (∼50,000) of ligand poses in the potential binding regions, which are then clustered based on root-mean-squared (rms) distances to obtain ∼2,000 family heads whose energies are then evaluated to select ∼200 families, for each of which the energies of all members are evaluated. The top 100 poses from this process are optimized to finally select the best protein–ligand structure. These ligand–CCR5 structure predictions were performed independently for each of the eight CCR5 structures using the DREIDING force field (16).
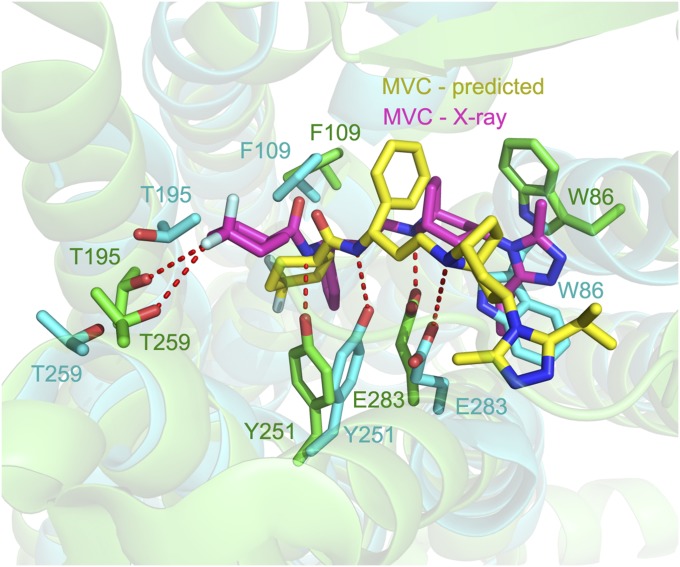
The final predicted most stable structure for the MVC/CCR5 complex is shown in Fig. 2, overlaid with the complex structure from the recent crystal structure (8) and showing the most important ligand–residue interactions. Binding of MVC stabilized the WT7 structure of CCR5, which was also stabilized by PF and APL (Fig. 3 A and B) whereas TAK selected the WT10 conformation (Fig. 3C). Similar results have been obtained earlier for the adenosine A3AR receptor, where all four selective agonists stabilized WT15 whereas all four selective antagonists stabilized WT2 or WT3 (19) conformations; for none of these eight ligands was the complex with WT1 of lowest energy. These results emphasize the importance of having the ensemble of low-energy structures, and they suggest that ligands can control the final conformation and perhaps downstream function of CCR5 as well as GPCRs in general.
Fig. 2.
Comparison of the predicted MVC binding site (MVC in yellow and binding site residues in cyan) with the experimental X-ray MVC binding site (MVC in purple and binding site residues in green).
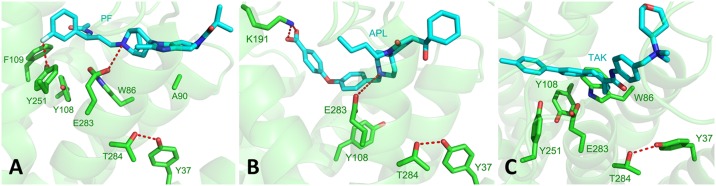
Fig. 3.
The predicted binding sites for (A) PF-232798 (PF), (B) Aplaviroc (APL), and (C) TAK-779 (TAK).
The biggest changes in the CCR5 structure upon binding MVC (WT1 to WT7) were in the η rotation angles of helices 2 and 7 (compare the binding site residues in a 2D representation in Fig. S1 A and D). The 20-degree rotation of helix 2 completely changes the interhelical hydrogen bond network of CCR5. In the wild-type protein, the structure of CCR5 is stabilized by several interactions, including a conserved N48 (TM1)–D76 (TM2) hydrogen bond, S63 (TM2)–R126 (TM3) hydrogen bond, and finally a 1-2-7 hydrogen bond network Y37 (TM1)–W86 (TM2)–T284 (TM7). The 1-2-7 network and the W86–T284 hydrogen bond, in particular, result in a compact structure for the protein that is not suitable to develop strong interactions with large ligands having a tertiary nitrogen atom, such as MVC. This is because the interaction with T284 forces W86 to be close to E283, the crucial residue for interacting with the basic part of the ligand. To overcome this problem, the W86-T284 hydrogen bond must break, which leads to a slight destabilization of the structure, which forces helices 2 and 7 to rotate to find favorable interactions with other residues from other helices, such as π–π stacking with Y108. The existence of a similar receptor activation network was proposed recently (20), but it did not include T284; instead, the network consisting of Y37, W86, Y108, and E283 residues was proposed.
Thus, binding of MVC causes helix 2 to rotate by 20 degrees, which opens up the binding pocket formed by helices 1, 2, and 7. This rotation also allows the W86 residue to find an interaction with Y108 on helix 3, while interacting with the ligand, whereas T284 still forms a hydrogen bond with Y37 on TM1. Additionally these rotations aid the hydrogen bonding between N71 (TM2)–N293 (TM7) due to a more favorable geometry of these two residues. Moreover, the 15-degree rotation of H7 allows the E283 residue to find a better orientation, optimizing the interaction between the carboxylic group and the basic nitrogen from the tropane ring of MVC, as well as the hydroxyl group of Y251 on helix 6.
The binding pocket of the predicted complex model is in very good agreement with the crystal structure of the MVC:CCR5 complex published just recently (8). Fig. 2 also shows the MVC–CCR5 interactions observed in the crystal structure of the complex for comparison with the predicted structure. Fig. S1 A and B shows these interactions using a 2D representation. All important interactions between the MVC and CCR5 residues were predicted, including (i) the salt bridge between the tropane nitrogen atom and Glu283, (ii) the hydrogen bond between the carboxyamide nitrogen and Tyr251, and (iii) the hydrophobic interactions of the phenyl group with Tyr108, Phe109, and Phe112. The predicted structure is not perfect and missed some interactions: (i) the hydrogen bond between the amine moiety of the triazole group and Tyr37 and hydrogen bonds between one of the fluorines on the cyclohexane ring with Thr195 and Thr259.
The accuracy of the predicted CCR5 structure (obtained using the human β2 adrenergic receptor template) shows the strength of the BiHelix and SuperBihelix methodology. This approach allows sampling of a complete set of TM rotations and local tilts, going well beyond simple homology modeling. This method also allows the residues crucial for ligand binding to be correctly positioned in the binding pocket, leading to high-quality protein structures starting from relatively low-homology templates (25% sequence identity between β2AR and CCR5 in the TM region).
The MVC ligand does not select the lowest-energy receptor conformation (WT1) for binding but a higher-energy one (WT7). The helix rotation η angles change from [0, −120, 0, 0, 15, 0, 0] for the WT1 conformation to [15, 0, 0, 105, 15, 0, 0] for the WT7 conformation, which is very close to the [15, 0, 10, 105, −15, 0, 0] angles calculated for the crystal conformation of CCR5 bound to MVC (8). The predicted TM domains are also in good agreement with the crystal structure (crystal TMs/predicted TMs): TM1, 26–57/26–57; TM2, 64–92/64–93; TM3, 97–132/98–131; TM4, 142–166/142–166; TM5, 187–222/192–224; TM6, 229–261/229–259; and TM7, 269–297/276–299.
The predicted PF binding mode (Fig. 3A) is similar to that of MVC, based also on the same protein configuration WT7. This result is reasonable because the ligand structures are rather similar. Thus, the mechanism of PF binding to CCR5 protein is similar to that of MVC binding, involving also the rotation of helices 2 and 7, which opens up the 1-2-7 binding pocket to allow the ligand to bind to E283. Fig. S1C shows the PF binding site in a 2D representation.
Although APL has a slightly different molecular scaffold, it shares with MVC and PF a strongly basic nitrogen atom positioned in the center of the molecule (see central N atom in the ligand structures shown in Scheme S1). Indeed, it interacts strongly with the E283 anchor point (Fig. 3B), similar to the MVC and PF cases. Thus, the preferred CCR5 configuration is WT7, just as for MVC and PF. However, the benzoic acid group of APL provides an additional anchoring point for the ligand, far from helix 7 and E283. It interacts with the K191 residue located on the verge of helix 5 and extracellular loop 2, leading to a large reduction of binding upon K191A mutation. This result is consistent with the experimental >100-fold decrease in binding affinity of APL upon K191A mutation (21, 22).
The mechanism of binding of APL also involves breaking or weakening the 1-2-7 hydrogen bond network (Y37-W86-T284), with E283 providing the anchor for the middle part of the ligand. Interestingly, the benzoic acid group also interacts with R168 on extracellular loop 2, also shown to be important for ligand binding in experimental assays (23).
The TAK antagonist is based on a completely different scaffold containing a quaternary amine group in place of the protonated nitrogen for the other three ligands, and giving the whole ligand a formal charge of +1. We predict that TAK prefers to bind to the WT10 conformation (Fig. 3C). The overall mechanism of opening the binding site via disruption of the W86-T284 hydrogen bond is preserved. As in the previous cases, the 1–2-7 hydrogen bond network is broken or weakened due to the 20-degree rotation of helix 2 and formation of a π-π stacking interaction between W86 and Y108. The T284-Y37 hydrogen bond stabilizes the ligand-bound structure, in agreement with the experimental data that show lower binding affinity of TAK upon the Y37A mutation (21). Additionally, the 15-degree rotation of helix 5 allows the T195 residue to rotate away from the toluyl group of the ligand, making more space available in the 3-5-6 pocket. On the other hand, the relatively large rotation of weakly interacting H4 does not seem to have any impact on the structure of the protein or ligand binding.
A detailed analysis of the TAK binding site shows that W86 is the strongest interacting residue with this ligand, with a −3.7 kcal/mol contribution to the overall interaction energy, which agrees well with the experimental data. This favorable interaction comes from an imperfect π–π stacking between the tryptophan residue and the benzo[7]annulene part of the ligand. We predict that the contribution of E283 residue is the second strongest one (−3.3 kcal/mol). On the other hand, the interaction of the ligand with Y108 is very weak so that the relatively large effect of the Y108A mutation seen experimentally results because WT22 becomes the most stable TAK bound conformation in the Y108A receptor mutant. Thus, the large effect of the mutation is indirect through stabilizing a different protein conformation rather than a change in direct interaction with the ligand. All other residues investigated experimentally show very weak interactions with TAK, consistent with the experimental mutation data.
Mutation Experiments.
Based on the binding structure for MVC to WT CCR5, we selected 15 single mutants to validate the predicted structure: F109Y, F112A, Q194A, Y251F, D276A, Q277A, W86A, A90H, T105A, Y108A, F109A, I198A, Y251A, Q280A, and E283A. For 11 of these mutants, binding studies of MVC and PF were carried out. The predicted results were dramatically inconsistent with experiments for the A90H and F109A mutants. Also, the experiments suggested a bigger change in binding energy for PF vs. MVC in the A90H mutant. A fundamental assumption in our selection of mutations was that the structure of the binding site would not change significantly (the established paradigm for assessing the effect of mutations). These experimental results suggested that this assumption might be invalid for CCR5 and potentially other GPCRs. This result is also consistent with our earlier studies on binding of the ligand BX471 to CCR1 receptor, where we found that a single mutation of human CCR1 to mouse and rat CCR1 dramatically changed the binding (24).
Rather than starting all over to predict the hierarchy of CCR5 structures for each of the 15 mutant receptors, we simplified the procedure as follows. We went back to the 100 lowest-energy structures from the conformational hierarchy predicted for WT CCR5; we mutated the single residue in question for each of those structures, and then we reassigned side chains, using SCREAM (15), and minimized the structures. The net result is often a dramatic reordering in the ranking of these 100 receptor structures. We then selected the 20 lowest-energy structures for docking the four ligands.
This approach leads to dramatically different hierarchies, as shown in Table S2. For example, the W86A mutant makes the WT30 conformation fourth best. This outcome is easy to understand from the structures. WT30 conformation has the W86 residue up against TM7 (Fig. S2), but replacing this residue by Ala in the W86A mutant removes this steric clash, dramatically improving the rank of the WT30 conformation to fourth.
Next, we docked the 20–30 conformations for each of the four ligands to each of the ∼10 receptor conformations for each of the mutated proteins using the DarwinDock/GenDock methods (18). The results are shown in Table 1. For the top-scoring poses, a water displacement penalty was applied if a polar residue was mutated to a nonpolar residue and could have accommodated a water molecule in the apo protein conformation. This penalty affected only the Q194A mutant.
Table 1.
Predicted lowest-energy protein structures for wild-type and mutant receptors in the apo and ligand-bound forms
| Mutations | Lowest-energy CCR5 conformations |
||||
| Apo | MVC | PF | APL | TAK | |
| Wild-type | wt1 | wt7 | wt7 | wt7 | wt10 |
| F109Y | wt3 | wt5 | wt5 | wt5 | wt3 |
| F112A | wt1 | wt5 | wt5 | wt5 | wt3 |
| Q194A | wt1 | wt5 | wt5 | wt5 | wt4 |
| Y251F | wt1 | wt2 | wt22 | wt4 | wt3 |
| D276A | wt1 | wt2 | wt7 | wt3 | wt2 |
| Q277A | wt1 | wt5 | wt5 | wt5 | wt3 |
| W86A | wt1 | wt7 | wt7 | wt7 | wt3 |
| A90H | wt3 | wt10 | wt5 | wt10 | wt3 |
| T105A | wt1 | wt2 | wt6 | wt6 | wt3 |
| Y108A | wt3 | wt2 | wt22 | wt5 | wt22 |
| F109A | wt1 | wt5 | wt7 | wt7 | wt3 |
| I198A | wt1 | wt5 | wt5 | wt5 | wt25 |
| Y251A | wt2 | wt3 | wt5 | wt5 | wt3 |
| Q280A | wt3 | wt5 | wt7 | wt7 | wt7 |
| E283A | wt3 | wt3 | wt22 | wt22 | wt3 |
Ligands used are Maraviroc (MVC), PF-232798 (PF), Aplaviroc (APL), and Tak-779 (TAK). This shows that a single mutation can dramatically affect the binding site.
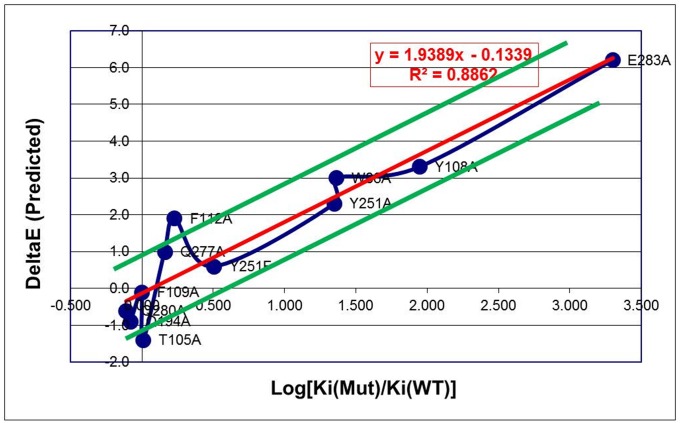
The results in Table 1 show the pronounced effect of mutations in the active site. Thus, the WT7 configuration stabilized by MVC binding to WT CCR5 is not the best binding configuration for any of the 11 mutants, all of which were aimed at modifying the binding! Instead, WT5 conformation appears seven times, WT2 four times, WT3 three times, and WT10 once. These mutants were generated experimentally (as described in SI Materials and Methods, Experimental Generation of CCR5 Mutants) and used to measure Ki values for the binding of different ligands to CCR5 WT and mutant receptors (SI Materials and Methods, Ligand-Binding Experiments). Comparing relative Ki values to relative predicted binding energies for each case led to the results in Fig. 4, which show excellent agreement with experimentally obtained Ki values for MVC binding to the mutants relative to the WT receptors, with the binding constants varying over a range larger than 3 logs. The trend for the other three ligands is also correct, but the correlation is weaker than that for MVC, due in part to the fact that the experimental changes in binding upon mutations span less than three orders of magnitude (less than 3 logs) for these ligands, which is only slightly larger than the best expected resolution from the calculations of ∼2 logs [Fig. S3 (PF), Fig. S4 (APL), and Fig. S5 (TAK)]. These results overall confirm the prediction that different conformations can be stabilized under different conditions (mutants and/or ligands).
Fig. 4.
Comparison of the predicted effect of Maraviroc binding to mutants vs. experimental data for the 11 cases studied experimentally. The red line is the fit (R2 = 0.89) whereas the green lines indicate a deviation of 1 kcal/mol.
Discussion
Analysis of the Ligand–CCR5 Binding Site in the Context of Mutations and Ligand Structure–Activity Relationship Series.
The experimental assays confirmed the predictions that the binding modes of MVC and PF232798 are rather similar, but with one exception. The W86A mutation gives a 629-fold decrease in binding of PF and a 29.8-fold decrease in binding for MVC. [Kondru et al. reported a 10-fold decrease in MVC binding upon W86A mutation (22) in slightly different conditions.] Indeed, the calculations find that for W86A, both MVC and PF bind to WT7, but with predicted energy changes of 3.1 kcal/mol and 3.5 kcal/mol.
The difference in impact of the W86A mutation on MVC/PF binding comes in part from structural differences between those two ligands. Although they share a similar scaffold, they have a different stereochemistry for the five-membered nitrogen-rich ring that forces the boat conformation of the tropane ring for PF whereas MVC retains the favorable chair conformation. Also there is an additional cyclohexane ring in the terminal part of MVC that affects the pKa value of the N3 atom, making it more basic. The quantum mechanical calculations predict the pKa value of the nitrogen in the five-membered ring to be 5.0–6.0 for MVC, but 6.0–7.0 for PF, suggesting that MVC may exist in a doubly protonated state in the experimental environment (the tropane N atom is very basic for both MVC and PF, with a predicted pKa of ∼9). Because, for WT, the residue W86 is close to the five-membered ring of both MVC and PF, this change in the protonation state could have been responsible for the predicted different interaction with tryptophan through a strong cation–π interaction (25).
However, our predicted structure makes this hypothesis unlikely because W86 is oriented in a way not allowing for the π–cation interaction in the PF. Despite a similar orientation of W86 in the MVC-bound and PF-bound models, we predict a difference in the interaction energy between those two ligands and the tryptophan residue. The binding site energy analysis allows the ligand–protein interaction to be partitioned into contributions from each residue. For the MVC binding site, we found a ligand interaction energy of −2.0 kcal/mol with W86 (which is less than the E283 contribution of −3.2 kcal/mol) whereas for PF it is −4.2 kcal/mol (almost identical to −4.3 kcal/mol contribution of E283). This difference can be understood in terms of the geometries of the ligands (the different stereochemistry of the tropane ring-five membered ring connections) and the different position of the phenyl ring with respect to the tropane ring for MVC and PF, leading to different interaction energies.
A second important difference in the experimental mutation data for these two ligands occurs for the A90H mutation, which changes the PF binding significantly, but has almost no effect on MVC binding. PF prefers the WT5 conformation for the A90H mutant whereas MVC prefers WT10. Even though both MVC and PF are anchored by E283 and occupy a similar position in the protein, they are structurally different in the part that interacts with helix 2. The bulkier pyridyl group of PF positions the ligand closer to the A90 residue, which does not affect the binding unless this residue is mutated to a larger moiety: i.e., the histidine residue. Indeed, MVC is relatively far from the A90 residue (which is on top of helix 2); as a result, replacing A90 by the bulky histidine does not affect its binding. However, the ligand PF is positioned closer to A90 so that the mutation to histidine causes a steric clash, decreasing binding. This difference arises from the difference in the receptor conformation for the A90H mutant preferred by MVC (WT10) versus PF (WT5).
On the other hand, all 11 mutations investigated force the TAK-bound CCR5 system to switch to a different conformation. This conformational change has, however, little impact on the computational mutation results. For all six mutations studied experimentally for TAK, the results from both structures were in good agreement with experimental data.
Unfortunately, the structure–activity relationship (SAR) series for the PF compound showed (3) little structural diversity. The experimental data presented in ref. 3 agrees well with the PF232798 binding pose predicted here. All 3-Substituted 1,4,6,7-Tetrahydro-imidazo[4,5-c]pyridines that were tested experimentally (compounds 39 and 41a–h from ref. 3) show a very high affinity, on the level of the PF232798 system. In the predicted docking pose, this part of the ligand interacts with the flexible N terminus of the protein and there is enough space to incorporate any substituent suggested in the original work. The modification of the ligand yielding 3-substituted derivatives (compounds 20a–h and 29–32 from ref. 3) positions the terminal part of the ligand closer to TM2, but avoiding steric clashes with any protein residues. This short SAR series further supports the predicted PF232798 binding pose.
APL (Aplaviroc) has been developed from a series of spirodiketopiperazine derivatives showing a relatively high CCR5 activity (5). In that study (5), four different spirodiketopiperazine derivatives were evaluated for anti–HIV-1 activity, and molecule E913 was selected as the lead compound for a newer round of optimizations. Our predicted docking pose of APL is in agreement with the experimental data for this series of compounds. The main difference among the earlier spirodiketopiperazine derivatives E910, E913, E916, and E917 is the length of the molecule's terminal part targeting the TM3-TM4-TM5 region of CCR5. According to the predicted pose, there is enough space to incorporate even the longest, heptylbenzene tail present in E910. This region is also highly hydrophobic, with a number of residues (F109, F112, F168, and I198) able to make relatively strong interactions with the phenyl ring present at the end of the hydrophobic tail of all these ligands. The difference responsible for the strong binding of APL is the addition of the benzoic acid group to the end of this tail, which is the most important difference between APL and E913. In APL, the phenoxy linker is able to position the new carboxylic group to promote a strong interaction with K191, known to be important in APL binding from the experimental mutation data.
For TAK-779, the original paper (4) does not disclose any SAR series. Other Takeda compounds with known CCR5 activity (like TAK-220) are completely different.
Implications for the Conformational Ensemble View of GPCRs.
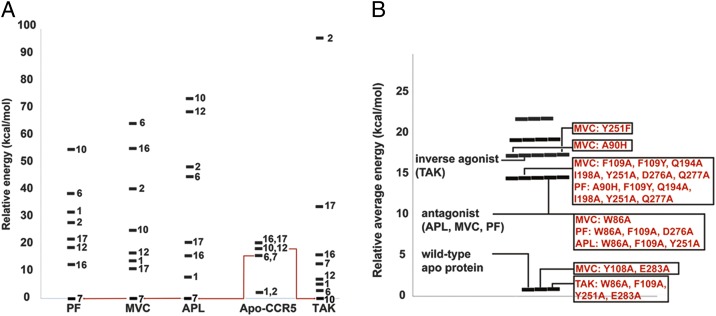
Fig. 5 summarizes the results for all ligands and mutants. Fig. 5A shows that MVC, PF, and APL prefer to bind to the WT7 receptor conformation, not to the lowest-energy WT1 conformation of the apo-protein. TAK prefers to bind to the WT10 conformation. This difference between TAK and other ligands arises because TAK has a quaternary nitrogen group in place of the tertiary nitrogen of the other ligands. Fig. 5B shows how different mutants of the receptor stabilize distinct conformations and indicates which conformation is preferred by ligands for different mutants. These changes in conformation of the mutated apo-protein explain the differential mutational data for W86A and A90H mutants on binding of MVC vs. PF compounds. This result has direct implications for the functional selectivity of GPCRs because it shows that different receptor conformations are stabilized under different conditions (ligand binding and/or mutations), which potentially can have distinct downstream effects on signaling.
Fig. 5.
(A) Conformational shuffling upon binding of different ligands. (B) CCR5 conformations preferred by different ligands and mutants.
These results show that CCR5 is a relatively plastic protein with distinctly different protein conformations stabilized by different ligands. A single mutation can considerably alter the energetics of the low-lying conformations to alter the preferred binding site. This outcome reinforces the conformational selection basis of the GPCR pleiotropic function. These results suggest that the paradigm of designing a ligand for a particular protein structure will not be sufficient for developing new therapeutics to CCR5 (and likely other GPCRs). One must consider all of the low-energy protein conformations in the design and in determining structure–activity relationships. Moreover, one must be cautious about interpreting mutation data. Thus, we consider that methods presented in this work for predicting the effect of mutations on GPCR structure will be essential to developing highly selective and active ligands targeting these receptors.
Conclusions
We developed an ensemble-based method that samples billions of conformations to eventually select an ensemble of ∼20 low-energy protein conformations for the CCR5 receptor and showed how this conformational ensemble changes upon binding of four ligands and how it changes because of mutations. To assess the validity of the binding sites, 15 mutations were predicted in CCR5 that should significantly modify the binding. Experimental binding studies were carried out for 11 of these mutants, and several cases were found that disagreed significantly from the expectations based on the predicted pose for binding of the ligand MVC to the WT protein. This result motivated us to predict the ensemble of low-energy protein structures for each of the 15 mutants, some of which showed major changes in the conformations available for ligand binding. We predicted the optimum ligand pose for the ensemble of low-energy CCR5 conformations for each of 11 mutants and compared with our experimental binding results. We found that ligand binding to the predicting mutated receptor conformations agrees well with the experimental mutation data for all four ligands. This work leads to several important general conclusions. (i) Different ligands can stabilize dramatically different seven-helix bundle conformations, complicating the use of experimental structures for designing new strongly interacting ligands. (ii) For CCR5 (and likely other GPCRs), even a single mutation can dramatically change the binding site, complicating experimental validation of ligand-binding structures. This result invalidates the standard paradigm of using directly the GPCR structure bound to the ligand to explain mutation-binding data. (iii) The changes in protein conformation stabilized by different ligands and/or mutants might lead to changes in functionality (a structural basis for GPCR pleiotropy). Indeed functional differences between agonists and inverse agonists (or antagonists) to GPCRs are well established. For example, recent studies show that single, double, and triple mutant forms of the CB1 receptor exhibit dramatic changes in function, switching from full inactivity to high constitutive activity and back for sequential mutations (26, 27). (iv) The GEnSeMBLE approach for predicting the ensemble of thermally accessible protein–ligand complexes provides a tool for accurate GPCR–ligand predictions consistent with the GPCR pleiotropy paradigm. The validation by both the mutation data and the subsequently published crystal structure for the MVC:CCR5 complex indicates that this approach may be trusted for new GPCRs for which there are little or no data.
Materials and Methods
The computational methods used in this study for the structure prediction of CCR5 receptor in the WT and mutant forms as well as ligand docking were described briefly in Results. Key aspects of the method for predicting receptor structure are that we do a very complete sampling of helix tilts and rotations (∼11 billion conformations) and use energy as the sole criterion to select a conformational ensemble for docking the ligands. Key aspects of the method for predicting the binding site are that for each of the ligands' multiple torsional conformations, we docked ∼50,000 poses to the receptor's full conformational ensemble, from which we eventually select the best pose solely on the basis of energy. SI Materials and Methods contains detailed descriptions of those computational methods along with the experimental methods used in this study for the receptor mutant generation and ligand-binding assays.
Supplementary Material
Acknowledgments
We thank Soo-Kyung Kim and Andrea Kirkpatrick for useful discussions. A portion of this research was funded by a gift from Accelerator/PharmSelex. The balance was from gifts to the Materials and Process Simulation Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413216111/-/DCSupplemental.
References
- 1.Dragic T, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 2.Dorr P, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49(11):4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupple PA, et al. An imidazopiperidine series of CCR5 antagonists for the treatment of HIV: The discovery of N-(1S)-1-(3-fluorophenyl)-3-[(3-endo)-3-(5-isobutyryl-2-methyl-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridin-1-yl)-8-azabicyclo[3.2.1]oct-8-yl]propylacetamide (PF-232798) J Med Chem. 2011;54(1):67–77. doi: 10.1021/jm100978n. [DOI] [PubMed] [Google Scholar]
- 4.Baba M, et al. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96(10):5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeda K, et al. Novel low molecular weight spirodiketopiperazine derivatives potently inhibit R5 HIV-1 infection through their antagonistic effects on CCR5. J Biol Chem. 2001;276(37):35194–35200. doi: 10.1074/jbc.M105670200. [DOI] [PubMed] [Google Scholar]
- 6.Grunbeck A, et al. Genetically encoded photo-cross-linkers map the binding site of an allosteric drug on a G protein-coupled receptor. ACS Chem Biol. 2012;7(6):967–972. doi: 10.1021/cb300059z. [DOI] [PubMed] [Google Scholar]
- 7.Berro R, et al. Use of G-protein-coupled and -uncoupled CCR5 receptors by CCR5 inhibitor-resistant and -sensitive human immunodeficiency virus type 1 variants. J Virol. 2013;87(12):6569–6581. doi: 10.1128/JVI.00099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341(6152):1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrol R, Griffith AR, Bray JK, Goddard WA., III . Structure prediction of G protein-coupled receptors and their ensemble of functionally important conformations. In: Vaidehi N, Klein-Seetharaman J, editors. Membrane Protein Structure and Dynamics: Methods and Protocols, Methods in Molecular Biology. Vol 914. New York: Humana; 2012. pp. 237–254. [DOI] [PubMed] [Google Scholar]
- 10.Abrol R, Bray JK, Goddard WA., III Bihelix: Towards de novo structure prediction of an ensemble of G-protein coupled receptor conformations. Proteins. 2011;80(2):505–518. doi: 10.1002/prot.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342(2):571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 12.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam VWT, Goddard WA., III Flat-bottom strategy for improved accuracy in protein side-chain placements. J Chem Theory Comput. 2008;4(12):2160–2169. doi: 10.1021/ct800196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayo SL, Olafson BD, Goddard WA., III Dreiding: A generic force-field for molecular simulations. J Phys Chem-Us. 1990;94(26):8897–8909. [Google Scholar]
- 17.Bray JK, Abrol R, Goddard WA, III, Trzaskowski B, Scott CE. SuperBiHelix method for predicting the pleiotropic ensemble of G-protein–coupled receptor conformations. Proc Natl Acad Sci USA. 2014;111(1):E72–E78. doi: 10.1073/pnas.1321233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goddard WA, III, et al. Predicted 3D structures for adenosine receptors bound to ligands: comparison to the crystal structure. J Struct Biol. 2010;170(1):10–20. doi: 10.1016/j.jsb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Kim S-K, Riley L, Abrol R, Jacobson KA, Goddard WA., III Predicted structures of agonist and antagonist bound complexes of adenosine A3 receptor. Proteins. 2011;79(6):1878–1897. doi: 10.1002/prot.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall SE, et al. Elucidation of binding sites of dual antagonists in the human chemokine receptors CCR2 and CCR5. Mol Pharmacol. 2009;75(6):1325–1336. doi: 10.1124/mol.108.053470. [DOI] [PubMed] [Google Scholar]
- 21.Maeda K, et al. Structural and molecular interactions of CCR5 inhibitors with CCR5. J Biol Chem. 2006;281(18):12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 22.Kondru R, et al. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol. 2008;73(3):789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- 23.Berro R, et al. Multiple CCR5 conformations on the cell surface are used differentially by human immunodeficiency viruses resistant or sensitive to CCR5 inhibitors. J Virol. 2011;85(16):8227–8240. doi: 10.1128/JVI.00767-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaidehi N, et al. Predictions of CCR1 chemokine receptor structure and BX 471 antagonist binding followed by experimental validation. J Biol Chem. 2006;281(37):27613–27620. doi: 10.1074/jbc.M601389200. [DOI] [PubMed] [Google Scholar]
- 25.Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271(5246):163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 26.Ahn KH, Scott CE, Abrol R, Goddard WA., III Kendall DA. Computationally-predicted CB1 cannabinoid receptor mutants show distinct patterns of salt-bridges that correlate with their level of constitutive activity reflected in G protein coupling levels, thermal stability, and ligand binding. Proteins. 2013;81(8):1304–1317. doi: 10.1002/prot.24264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott CE, Abrol R, Ahn KH, Kendall DA, Goddard WA., III Molecular basis for dramatic changes in cannabinoid CB1 G protein-coupled receptor activation upon single and double point mutations. Protein Sci. 2013;22(1):101–113. doi: 10.1002/pro.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrol R, Kim S-K, Bray JK, Griffith AR, Goddard WA., III Characterizing and predicting the functional and conformational diversity of seven-transmembrane proteins. Methods. 2011;55(4):405–414. doi: 10.1016/j.ymeth.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.