Significance
Light-chain amyloidosis (AL) is a devastating human disease involving the clonal expansion of a plasma cell and the secretion of destabilized, amyloidogenic immunoglobulin light chains (LCs). Secreted amyloidogenic LCs aggregate extracellularly, leading to proteotoxicity on distal tissues. Available therapeutic strategies to treat AL specifically target the cancerous plasma cell population. While this approach is effective in ∼70% of patients, patients who present with substantial LC-related organ proteotoxicity are generally too sick to tolerate standard chemotherapeutics. Here, we show that stress-independent activation of unfolded protein response-associated transcription factors selectively reduces secretion of amyloidogenic LCs and decreases extracellular soluble LC aggregates associated with proteotoxicity in AL. These results identify a promising therapeutic strategy to treat AL patients unserved by current treatments.
Keywords: ER proteostasis, amyloid
Abstract
Light-chain amyloidosis (AL) is a degenerative disease characterized by the extracellular aggregation of a destabilized amyloidogenic Ig light chain (LC) secreted from a clonally expanded plasma cell. Current treatments for AL revolve around ablating the cancer plasma cell population using chemotherapy regimens. Unfortunately, this approach is limited to the ∼70% of patients who do not exhibit significant organ proteotoxicity and can tolerate chemotherapy. Thus, identifying new therapeutic strategies to alleviate LC organ proteotoxicity should allow AL patients with significant cardiac and/or renal involvement to subsequently tolerate established chemotherapy treatments. Using a small-molecule screening approach, the unfolded protein response (UPR) was identified as a cellular signaling pathway whose activation selectively attenuates secretion of amyloidogenic LC, while not affecting secretion of a nonamyloidogenic LC. Activation of the UPR-associated transcription factors XBP1s and/or ATF6 in the absence of stress recapitulates the selective decrease in amyloidogenic LC secretion by remodeling the endoplasmic reticulum proteostasis network. Stress-independent activation of XBP1s, or especially ATF6, also attenuates extracellular aggregation of amyloidogenic LC into soluble aggregates. Collectively, our results show that stress-independent activation of these adaptive UPR transcription factors offers a therapeutic strategy to reduce proteotoxicity associated with LC aggregation.
Light-chain amyloidosis (AL) afflicts 8–10 people per million per year, making it the most prominent systemic amyloid disease (1). AL is a gain-of-toxic function disease driven by a clonally expanded plasma cell that secretes amyloidogenic Ig light chains (LCs). These amyloidogenic LCs undergo extracellular misfolding and aggregation into proteotoxic soluble oligomers and amyloid fibrils that interact with distal tissues such as the kidney, heart, and gastrointestinal tract, leading to organ dysfunction and ultimately death by unknown proteotoxicity mechanism(s) (2).
The majority of AL patients must combat both a cancer (i.e., the clonally expanded plasma cell) and LC aggregation-associated proteotoxicity. The standard treatment for AL patients is chemotherapy (often combined with stem cell transplant) to eliminate the cancerous plasma cell population (3, 4). The proteasome inhibitor bortezomib, which takes advantage of the stress sensitivity of aggressively proliferating plasma cells, has transformed chemotherapy effectiveness (5–7). Regardless, ∼30% of AL patients with substantial cardiac or renal LC proteotoxicity are too ill at diagnosis to tolerate chemotherapeutics (8–10). Thus, new strategies to reduce LC organ proteotoxicity must be developed to allow more AL patients to take advantage of chemotherapy.
LC aggregation requires conformational changes and a sufficient concentration of misfolded LC in plasma, which determine the rate and extent of its concentration-dependent aggregation. Over 500 distinct LC sequences, primarily of the lambda (λ) isotype (11), have been identified in AL amyloid deposits, reflecting the significant protein heterogeneity associated with AL proteotoxicity (12). Amyloidogenic LCs generally contain an energetically destabilized variable domain that facilitates LC aggregation (2).
The amyloidogenic LC plasma concentration is determined by the extent of LC secretion from plasma cells and the rate of LC turnover in the serum. Secretion efficiency is largely dictated by the balance between folding vs. degradation, often referred to as “quality control,” in the endoplasmic reticulum (ER) (13). ER quality control is determined by the activity of the ER protein homeostasis (proteostasis) network comprising the protein folding, secretion, and degradation pathways (14–19). These pathways primarily function to prevent the secretion of destabilized, misfolding-prone proteins into the extracellular space (14–19).
Plasma cells have an evolutionarily enhanced ER and secretory pathway, enabling the proper folding and secretion of >1,000 IgGs per second (20, 21). Plasma cells normally secrete LC as IgGs [assemblies of two LCs and two heavy chains (HCs)], or as free LCs. However, in AL patients, LCs are predominantly secreted independent of HCs, sometimes as monomers, but generally as disulfide-linked homodimers called Bence−Jones proteins—the common precursor for LC aggregation (22, 23). Furthermore, LC ER quality control is compromised in AL, allowing the efficient secretion of destabilized, amyloidogenic LC sequences that can aggregate extracellularly into proteotoxic soluble oligomers and amyloid fibrils.
Herein, we sought to identify signaling pathways that can be activated to reduce the secretion of destabilized, amyloidogenic LCs, thus reducing the extracellular LC concentration available for pathologic, concentration-dependent aggregation. Using a small-molecule screening approach, we identified that unfolded protein response (UPR) (24, 25) activation selectively reduces the secretion of a destabilized, amyloidogenic LC but not a more stable, nonamyloidogenic LC sequence. The UPR transcriptionally up-regulates ER quality control in response to ER stress, primarily through the transcription factors XBP1s and/or ATF6, comprising the adaptive UPR (25). Here, we show that stress-independent activation of XBP1s and especially ATF6 is sufficient to reduce amyloidogenic LC secretion and attenuate LC aggregation. Collectively, our results demonstrate that enhancing ER quality control via adaptive UPR activation represents a therapeutic strategy to ameliorate LC aggregation-associated proteotoxicity.
Results
Screening Reveals That UPR Activation Reduces Amyloidogenic LC Secretion.
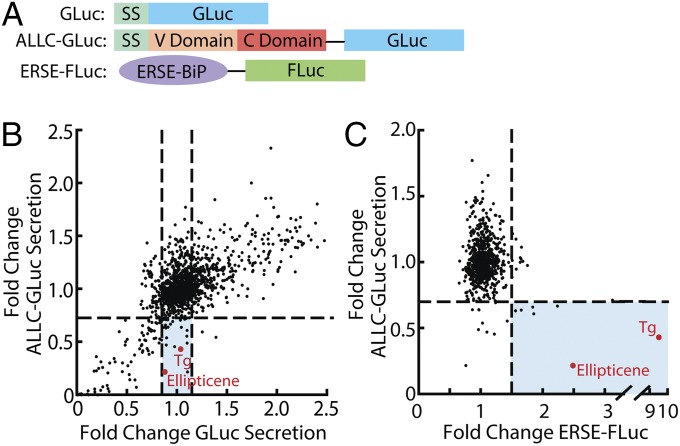
We developed a secretion reporter for LC by fusing the well-characterized AL-causing Vλ6 LC containing both constant and variable LC domains (named ALLC) (26) to an enhanced Gaussia princeps luciferase (GLuc) reporter, ALLC-GLuc (Fig. 1A) (27, 28). We used a similar GLuc fusion approach to quantify secretion of other proteins in a cell-based high-throughput screening (HTS) format (29). The secretion of ALLC-GLuc from HEK293-TRex cells was observable by immunoblotting and luminescence (Fig. S1 A and B). ALLC secretion decreased upon inhibition of protein biosynthesis using cycloheximide (CHX) or after inhibition of global protein secretion using brefeldin A (Fig. S1C), as did the secretion of GLuc, used as a control. ALLC-GLuc or GLuc was then stably incorporated into retinal pigment epithelium (ARPE-19) cells, as we previously optimized these professional secretory cells for HTS of GLuc fusion proteins such as ALLC-GLuc (29). The ALLC-GLuc secretion assay was miniaturized to a 384-well plate format, affording an HTS z-score (30) of 0.65 using cycloxheximide as a control.
Fig. 1.
ALLC-GLuc reporter screening identifies UPR modulators able to reduce secretion of amyloidogenic LC. (A) Schematic of GLuc, ALLC-GLuc, and ERSE-FLuc reporter constructs used in stable ARPE19 or HEK293T-Rex cell lines. ERSE-FLuc transcriptional reporter includes the ERSE-containing portion of the BiP promoter upstream of the firefly luciferase (FLuc) gene. SS, signal sequence. (B) Plot of LOPAC screening data comparing ALLC-GLuc secretion vs. GLuc secretion. ARPE-19 cells stably expressing ALLC-GLuc or GLuc alone were treated with compound (10 μM) for 24 h. Shaded blue section indicates compounds that reduce ALLC-GLuc secretion (>25%) and do not affect GLuc secretion (<±15% from DMSO control). (C) Plot of LOPAC screening results comparing ALLC-GLuc secretion (ARPE-19 cells, 24-h treatment) vs. UPR activation (ERSE-FLuc expressing HEK293T-Rex cells, 18-h treatment). Compounds falling below the horizontal line reduce ALLC-GLuc secretion by 25%. Compounds falling to the right of the vertical line increase ERSE-FLuc expression >1.5-fold relative to DMSO control. Compounds depicted in red are known activators of the UPR.
To identify biologic pathways that selectively reduce secretion of ALLC-GLuc, we screened the Library of Pharmacologically Active Compounds (LOPAC, Sigma Aldrich) in ARPE-19 cells secreting ALLC-GLuc or GLuc. The LOPAC was chosen because it is moderately sized (1,280 compounds), consists of compounds from major drug categories, and the biological targets for many small molecule components are known. We identified 15 molecules that significantly reduced ALLC-GLuc secretion (>25% reduction vs. vehicle control, 24-h treatment), while not affecting GLuc secretion (Fig. 1B, section shaded blue). Molecules that disrupted both ALLC-GLuc and GLuc secretion were not pursued further, as these likely reduce protein synthesis and/or disrupt global cell secretion (e.g., CHX and brefeldin A in Fig. S1C). Interestingly, the UPR activators thapsigargin (Tg), an inhibitor of sarco/endoplasmic reticulum Ca2+-ATPase (31), and ellipticene (32) selectively reduced ALLC secretion relative to GLuc (Fig. 1B), suggesting that activation of the UPR could selectively influence LC secretion. The inability of a 24-h treatment with Tg to influence GLuc secretion is consistent with previous results (27).
To further examine the relationship between UPR activation and the reduction in ALLC-GLuc secretion, we performed an additional screen using HEK293T-Rex cells stably expressing firefly luciferase under the control of the ER stress-responsive element (ERSE) promoter (ERSE-FLuc, Fig. 1A) (33). We found that 7 out of 15 molecules that selectively reduce ALLC-GLuc secretion by >25% also activated the ERSE-FLuc reporter >1.5 fold (Fig. 1C, blue), indicating that molecules that reduce ALLC-GLuc secretion are enriched for UPR activators.
Thapsigargin Selectively Reduces ALLC Secretion.
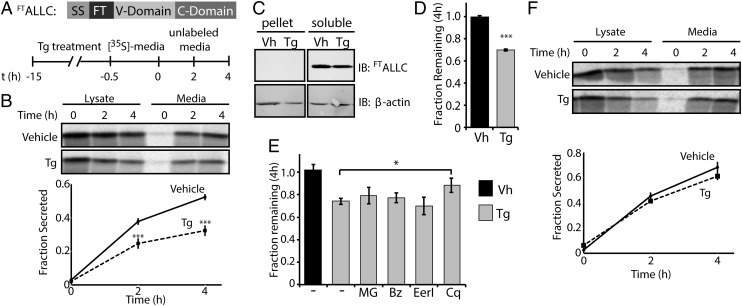
To further understand the effect of ER stress-associated UPR activation on LC secretion, we used [35S]-metabolic labeling to measure the secretion of amyloidogenic ALLC fused to an N-terminal FLAG tag (FTALLC, Fig. 2A). We preincubated cells in the absence or presence of Tg for 15 h to promote UPR-dependent remodeling of the ER proteostasis network before [35S]-metabolic labeling (Fig. 2A). FTALLC was immunopurified from media and cell lysates at specific timepoints during a 4-h chase in nonradioactive media. These experiments show that Tg pretreatment reduces FTALLC secretion by >40% (Fig. 2B). Importantly, because our Tg treatments do not drastically influence cell viability (Fig. S2 A and B), the Tg-dependent reduction in ALLC secretion cannot be attributed to cell toxicity. Furthermore the Tg-dependent reduction in ALLC secretion is also observed in cells treated with the protein kinase R-like ER kinase (PERK) inhibitor GSK2606414 (34), demonstrating that the reduced secretion does not result from PERK-dependent translational attenuation (Fig. S2C).
Fig. 2.
Thapsigargin selectively reduces the secretion of amyloidogenic LC and induces its degradation. (A) Schematic of the FTALLC construct used in pulse-chase experiments and the metabolic labeling protocol used. [35S]-labeled FTALLC was immunopurified from media and lysates collected from transfected HEK293T-Rex cells following a 15-h treatment with thapsigargin (Tg, 500 nM). (B) Representative autoradiogram and quantification of [35S]-labeled FTALLC in pulse-chase experiment described in A. Fraction secreted was calculated as described in Materials and Methods (18) (n ≥ 3). (C) Immunoblots measuring soluble and insoluble (pellet) levels of FTALLC after 15-h pretreatment with Tg. (D) Graph depicting total [35S]-labeled FTALLC (combined media and lysate protein levels as in B) remaining at 4 h in HEK293T-Rex cells following a 15-h pretreatment with 500 nM Tg (n ≥ 3). The fraction remaining was calculated as described in Materials and Methods (18). (E) Graph depicting the fraction recovery of total [35S]- labeled FTALLC at 4 h in HEK293T-Rex cells incubated in the presence of thapsigargin (Tg; 500 nM, 15 h) and the proteasome inhibitors bortezomib (Bz; 20 µM) or MG132 (MG; 20 µM), the ERAD inhibitor eeyarestatin I (EerI; 20 µM), or the autophagy inhibitor chloroquine (Cq; 100 µM). Inhibitors were incubated for 4 h before [35S] metabolic labeling and included throughout the labeling protocol shown in A. Fraction remaining was calculated as in D (n ≥ 2). (F) Representative autoradiogram and quantification of [35S]-labeled FTJTO immunopurified from media and lysates collected from transfected HEK293T-Rex cells following the same protocol and quantification as in A and B (n ≥ 3). *P < 0.05; ***P < 0.005. All error bars represent the SEM from biological replicates.
No accumulation of FTALLC was observed in cell pellets following ER stress caused by Tg treatment (Fig. 2C), indicating that ALLC does not accumulate as intracellular aggregates. Reduced FTALLC secretion was associated with a decrease in total [35S]-labeled FTALLC (media + lysate) (Fig. 2D), suggesting that Tg-associated ER stress increases FTALLC degradation. Similar results were obtained with an untagged ALLC, confirming that the FLAG tag does not significantly influence ALLC secretion and/or degradation (Fig. S2 D and E). This Tg-dependent loss in total FTALLC cannot be reversed using proteasome inhibitors (bortezomib or MG132) or the p97 inhibitor eeyarestatin I [a potent inhibitor of ER-associated degradation (ERAD) (35)], indicating that FTALLC is degraded following ER stress, at least in part through an ERAD-independent mechanism (Fig. 2E) (36). In contrast, the autophagy inhibitor chloroquine stabilized ALLC against Tg-induced degradation, indicating that Tg increases the autophagic degradation of ALLC (Fig. 2E).
Destabilized proteins are sensitive to UPR-dependent remodeling of the ER proteostasis network, whereas stable variants are generally less sensitive to this remodeling (37). Thus, we evaluated whether Tg-dependent reductions in LC secretion are also observed for a nonamyloidogenic, full-length Vλ6 LC, FTJTO (38). Tg pretreatment did not reduce the secretion of [35S]-labeled, nonamyloidogenic FTJTO (Fig. 2F), indicating that Tg-dependent UPR activation selectively reduces the secretion of an amyloidogenic, but not a nonamyloidogenic LC.
LC amyloidogenicity is highly correlated with LC stability (38, 39). Thus, we compared the in vitro relative stability of recombinant ALLC and JTO. Circular dichroism and tryptophan fluorescence measurements confirmed that the recombinant proteins have a folded, native β-sheet-rich Ig structure (Fig. S3 A and B). The relative stability of the ALLC and JTO LCs were compared by urea denaturation and monitored by tryptophan fluorescence (Fig. S3C). Although the ALLC unfolding transition is not completely reversible (Fig. S3A, black curve), the urea midpoint values for the denaturation curves (ALLC 1.8 M, JTO 2.6 M) indicate that ALLC is less stable than JTO (Fig. S3C). This suggests that the selective decrease in ALLC secretion afforded by Tg could be attributed to the inherent instability of the ALLC protein.
XBP1s and/or ATF6 Activation Reduces the Secretion of Destabilized ALLC.
Previous work has indicated that remodeling of ER proteostasis pathways selectively influences the secretion of destabilized protein variants relative to more stable protein variants (13, 40, 41). Thus, Tg-dependent UPR activation could selectively decrease the secretion of destabilized ALLC through the transcriptional remodeling of the ER proteostasis network. To differentiate between the potential effects of Tg-induced ER stress and UPR-dependent remodeling of ER proteostasis pathways on ALLC secretion, we used a HEK293T-Rex-derived cell line that expresses both a ligand-regulatable ATF6 transcription factor [DHFR-ATF6; activated by the addition of the small molecule pharmacologic chaperone trimethoprim (TMP)] and a doxycycline (Dox)-inducible XBP1s transcription factor (cells referred to as HEK293DAX) (40). In these cells, the ATF6 or XBP1s transcriptional programs can be orthogonally controlled in the absence of ER stress, which enables analysis of the functional consequences of arm-selective UPR activation independently, or in combination (Fig. S4A) (40).
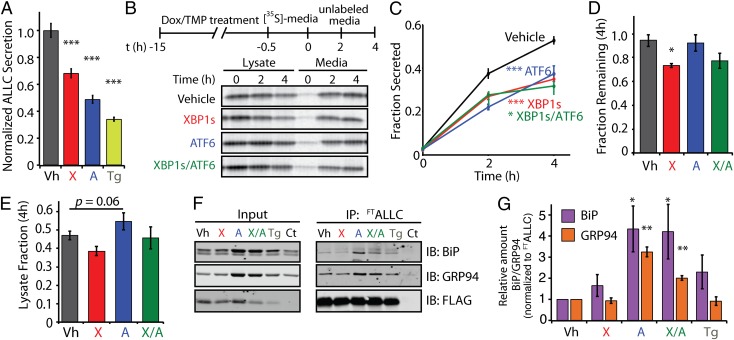
We treated HEK293DAX cells expressing FTALLC with Dox, TMP, or Tg and quantified the FTALLC levels in conditioned media by ELISA (Fig. 3A). Stress-independent activation of XBP1s or ATF6 reduced extracellular FTALLC levels to 68% and 48% of vehicle levels, respectively. Tg treatment lowered FTALLC levels to 34%. Reduced FTALLC secretion induced by XBP1s or ATF6 activation was dependent on transcription factor activity, as no reduction in FTALLC secretion was observed in TMP- or Dox-treated HEK293DYG cells—a control cell line stably expressing DHFR-YFP and Dox-inducible GFP (Fig. S4B) (40).
Fig. 3.
Stress-independent activation of XBP1s and/or ATF6 decreases the secretion of amyloidogenic LC. (A) ALLC levels determined by ELISA in conditioned media from HEK293DAX cells (40) expressing FTALLC following a 15-h preactivation of XBP1s (X; Dox, 1 μg/mL) or ATF6 (A; TMP, 10 μM) or by thapsigargin-induced stress (Tg, 500 nM). ALLC secretion levels were normalized to vehicle (Vh; DMSO) conditions (n = 3). (B) Representative autoradiogram of [35S]-labeled FTALLC immunopurified from media and lysates collected from transfected HEK293T-Rex cells following a 15-h preactivation of XBP1s (X; Dox, 1 μg/mL), ATF6 (A; TMP, 10 μM), or both (X/A). The metabolic labeling protocol used is shown. (C) Quantification of fraction secreted from autoradiograms as shown in B (n ≥ 3). (D) Graph depicting the total [35S]-labeled FTALLC remaining at 4 h in lysate and media following a 15-h preactivation of XBP1s, ATF6, or both in HEK293DAX (n ≥ 3). (E) Graph depicting the total intracellular [35S]-labeled FTALLC in lysates at 4 h in HEK293DAX cells following a 15-h preactivation of XBP1s, ATF6, or both (n ≥ 3). The lysate fraction was calculated by dividing the lysate [35S]-labeled FTALLC signal at 4 h by the total [35S]-labeled FTALLC at t = 0. (F) Immunoblot showing the recovery of the ER chaperones BiP and GRP94 in FLAG immunopurifications from cross-linked HEK293DAX cells expressing FTALLC following 15-h pretreatment with thapsigargin (Tg), XBP1s (X), ATF6 (A), or XBP1s and ATF6 (X/A), as in B. HEK293DAX cells expressing untagged ALLC is shown as a control (Ct). (G) Quantification of F achieved by comparing the signal under various conditions to vehicle, and by normalizing to the recovered FTALLC. *P < 0.05; **P < 0.01; ***P < 0.005. All error bars represent the SEM from biological replicates.
To probe the basis for the XBP1s- or ATF6-mediated reduction in ALLC secretion, we used [35S]-metabolic labeling (Fig. 3 B−E). XBP1s, ATF6, or combinatorial activation lowered the relative secreted fraction of labeled ALLC by ∼40% following a 4-h chase (Fig. 3 B and C). In contrast, stress-independent activation of XBP1s and/or ATF6 did not affect the secretion of the nonamyloidogenic FTJTO (Fig. S4C). Notably, XBP1s- and ATF6-dependent reductions in ALLC secretion proceed through distinct mechanisms. XBP1s activation resulted in a 25% decrease in total [35S]-labeled FTALLC (Fig. 3D), as observed for Tg-treatment (Fig. 2D). This XBP1s-dependent decrease in total soluble ALLC was not a result of intracellular accumulation of FTALLC in the cell pellet (Fig. S4D), implying the loss of FTALLC is due to degradation and not intracellular aggregation. ATF6 activation, in contrast, did not decrease total ALLC recovery in our [35S] metabolic labeling experiments (Fig. 3D) but instead resulted in an increase in intracellular [35S]-labeled FTALLC (Fig. 3E). Coactivation of XBP1s and ATF6 decreases FTALLC recovery to that observed for XBP1s activation. Similar results were observed with untagged ALLC (Fig. S5).
Altered processing of ALLC in the ER after XBP1s and/or ATF6 activation likely reflects distinct interactions between ALLC and ER proteostasis network components differentially induced by these transcriptional programs. We used an in situ cross-linking and immunopurification approach to sensitively measure the intracellular associations between ALLC and ER chaperones such as BiP (GRP78) and GRP94 (Fig. S6A)—two ER chaperones known to interact with LC (42, 43). As predicted, ATF6 activation, but not XBP1s activation, resulted in a significant increase in the association between ALLC and the ATF6-regulated ER chaperones BiP and GRP94 (Fig. 3 F and G), reflecting the increased levels of these two ER chaperones afforded by ATF6 activation (40). The increased association between ALLC and these ER proteostasis factors provides a mechanism to explain the ATF6-induced intracellular retention of destabilized, aggregation-prone ALLC in the ER lumen. ATF6 activation also increased interactions between FTJTO and BiP and GRP94 (Fig. S6B), albeit to a lesser extent than that observed for ALLC (Fig. 3 F and G).
XBP1s and/or ATF6 Decreases Extracellular Aggregation of Secreted ALLC.
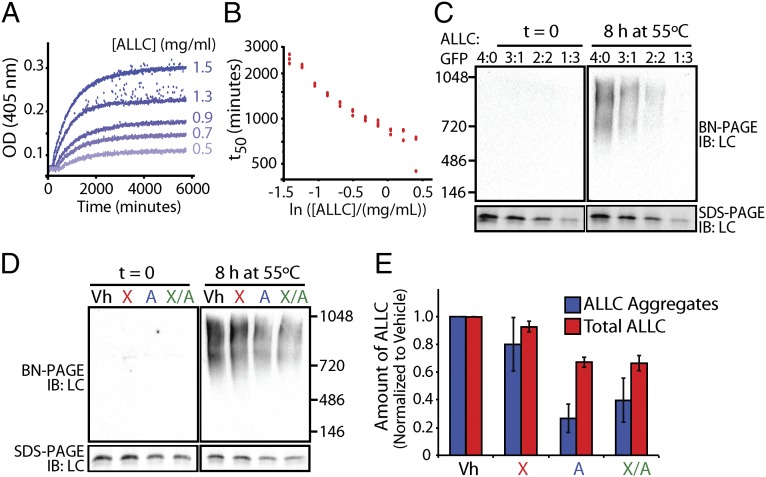
Lowering the secretion of an amyloidogenic LC should directly ameliorate AL organ proteotoxicity by reducing the extracellular concentration and therefore the extracellular aggregation of amyloidogenic LCs. To scrutinize this hypothesis, we examined the concentration-dependent aggregation of recombinant ALLC by monitoring turbidity of samples heated to 44 °C (Fig. 4 A and B). Reducing the concentration of ALLC significantly reduced the extent (Fig. 4A) and rate of aggregation (Fig. 4B), demonstrating that recombinant ALLC aggregates through a concentration-dependent mechanism.
Fig. 4.
Stress-independent activation of XBP1s and especially ATF6 reduces aggregation of cell-secreted ALLC. (A) Timecourses of recombinant ALLC aggregation at the indicated concentrations. Samples were incubated at 44 °C for the indicated time, and aggregation was measured by turbidity at 405 nm. (B) Plot of the t50 of recombinant ALLC aggregation at the indicated concentration from data as shown in A; n = 3 replicates are shown. (C) Immunoblots for BN-PAGE and SDS-PAGE of media conditioned on HEK293DAX cells expressing FTALLC for 24 h. The media was diluted with media conditioned on GFP-expressing cells, as indicated, and incubated at 55 °C for 8 h. (D) Representative immunoblots for BN-PAGE and SDS-PAGE of media conditioned on HEK293DAX cells following a 16-h preactivation of XBP1s (X), ATF6 (A), or XBP1s and ATF6 (X/A), as in Fig. 3B. FTALLC aggregation was induced by incubating the media for 8 h at 55 °C. (E) Graph depicting the quantification of soluble aggregates and total ALLC from BN-PAGE and SDS-PAGE immunoblots, as shown in D, normalized to vehicle. Error bars represent SEM from biological replicates (n = 3).
Because XBP1s and/or ATF6 activation decreases the secretion of amyloidogenic LC, we reasoned that the stress-independent activation of these transcriptional programs would similarly decrease extracellular ALLC aggregation. Heating the conditioned media of cells expressing FTALLC to 55 °C for 0–24 h produced large soluble aggregates that increase with time, as discerned by blue native polyacrylamide gel electrophoresis (BN-PAGE) (Fig. S7A) and gel filtration chromatography (Fig. S7B). Reducing the extracellular concentration of FTALLC in conditioned media by dilution with media conditioned on GFP-transfected cells shows that cell-secreted ALLC aggregation is concentration dependent (Fig. 4C), where a 1:1 [50% (vol/vol)] dilution of ALLC conditioned media nearly eliminates ALLC aggregates.
We next evaluated whether stress-independent XBP1s and/or ATF6 activation similarly attenuates ALLC aggregation. We collected conditioned media from FTALLC-expressing HEK293DAX cells following XBP1s and/or ATF6 activation and measured ALLC aggregation using BN-PAGE. ATF6 activation resulted in a 73% loss of soluble FTALLC aggregates (55 °C, 8 h incubation; Fig. 4 D and E), whereas XBP1s activation resulted in a 20% loss, and activating both XBP1s and ATF6 reduced aggregate formation by 60% (Fig. 4 D and E). As expected, the reduction in ALLC aggregation corresponds with a decrease in total extracellular ALLC (Fig. 4 D and E). Interestingly, the reduction in ALLC aggregation was more pronounced than expected from just the reduction in total extracellular ALLC, suggesting that XBP1s and/or ATF6 activation could also influence ALLC aggregation through other mechanisms. Importantly, we do not observe significant associations between ALLC and the ER chaperones BiP and GRP94 in conditioned media, indicating that the reduction in extracellular ALLC aggregation cannot be attributed to increased association with these ER chaperones secreted from cells (Fig. S8A). ALLC secretion and aggregation was not sensitive to treatment with Dox, TMP, or both in the control HEK293DYG cells (Fig. S8B), demonstrating that the reduced aggregation of ALLC observed in HEK293DAX requires XBP1s and/or ATF6 transcriptional activity.
Discussion
We show that stress-independent activation of one or both of the adaptive UPR-associated transcription factors XBP1s or especially ATF6 reduces the secretion and extracellular concentration of amyloidogenic ALLC, reducing soluble ALLC aggregate levels. This strategy has the potential to be clinically meaningful, as a >50% reduction in the amount of circulating amyloidogenic LC mediated by chemotherapeutic methods correlates with a substantial survival benefit (44). Lowering the amount of amyloidogenic LC secreted from plasma cells into the blood by UPR activation should be useful in patients with significant cardiac and/or renal involvement. This approach, with time, should dramatically reduce amyloidogenic LC oligomer levels and thus proteotoxicity, enabling the patient to tolerate established chemotherapy regimens. Arm-selective UPR activation could also be used in combination with chemotherapy approaches to further reduce the amount of circulating LC and organ proteotoxicity. Lastly, the approach could potentially be useful in cases of AL recurrence, decreasing the amyloidogenic LC secretion and organ proteotoxicity in chemotherapy-resistant AL patients.
Partitioning of amyloidogenic LCs between folding and trafficking vs. degradation pathways is determined by the interactions between the destabilized LC and ER proteostasis network components. The stoichiometry of the components of the ER proteostasis network is regulated by the activated signaling arms of the UPR (40). This in turn determines the specific ER proteostasis factors that interact with amyloidogenic LC and, thus, dictates the partitioning of LC between retention, secretion, or degradation in the ER lumen. The degradation of FTALLC upon XBP1s activation accounts for most of the observed reduction in secretion. In contrast, ATF6 activation does not induce ALLC degradation in HEK293DAX but instead leads to intracellular retention of ALLC, consistent with the increased interactions between ALLC and the ER chaperones BiP and GRP94. In AL-patient plasma cells, the retention of an amyloidogenic LC could sensitize the cells to death by subsequent chemotherapy strategies, or by itself be cytotoxic. Alternatively, degradation could eventually occur with repeated but periodic ATF6 activation, although this potential mechanism needs to be further explored. Increasing ER quality control by arm-selective UPR activation appears to be a general approach to reduce the secretion of destabilized, aggregation-prone proteins (37). Activation of ATF6 selectively reduces the secretion of destabilized, amyloidogenic transthyretin mutants from hepatocytes and can reduce intracellular accumulation of mutant rhodopsin, while not affecting secretion of the wild-type proteins (40, 41), providing strong support for this hypothesis.
Based on the results described above, we are now seeking small molecules that activate the ATF6 and/or XBP1s arms of the UPR. These small molecules would transcriptionally remodel the ER proteostasis network in plasma cells, reducing the secretion of energetically destabilized LCs irrespective of their primary structure, obviating the need for sequence- and conformation-specific AL drug design challenged by the significant sequence heterogeneity associated with LC proteotoxicity in AL. Furthermore, based on other reports demonstrating the capacity of XBP1s and/or ATF6 activation to influence aberrant ER quality control linked to disease-associated proteotoxicity (40, 41), establishing small molecules that increase ER quality control is expected to have significant potential for the treatment of additional protein aggregation diseases.
Materials and Methods
Plasmids, Cell Culture, HTS, and Recombinant Protein Production.
Detailed protocols can be found in SI Materials and Methods.
Pulse-Chase Experiments.
HEK293DAX or HEK293T-Rex cells plated on poly-d-lysine coated plates were metabolically labeled in DMEM -Cys/-Met (CellGro) supplemented with glutamine, penicillin/streptomycin, 10% dialyzed FBS, and [35S]-Translabel (MP Biomedical) for 30 min. Cells were washed with complete media and incubated in DMEM for the indicated times and harvested at the indicated times. In the case of added inhibitors, cells were pretreated for 4 h at the indicated concentrations, and inhibitors were added to the pulse and chase media. Lysates were prepared in radioimmunoprecipitation assay buffer plus protease inhibitor mixture (Roche) with 10 mM CaCl2. Proteins were immunopurified using anti-FLAG M1 agarose beads (Sigma). Protein was eluted by boiling in Laemmli buffer + 100 mM DTT, and samples were separated by SDS-PAGE. The gels were dried, exposed to phosphorimager plates (GE Healthcare), and imaged with a Typhoon imager. Band intensities were quantified in ImageQuant. Fraction secreted was calculated using the equation: fraction secreted = [extracellular [35S]-LC signal at t = t/(extracellular [35S]-LC signal at t = 0 + intracellular [35S]-LC signal at t = 0)]. Fraction remaining was calculated using the equation: [(extracellular [35S]-LC signal at t = t + intracellular [35S]-LC signal at t = t)/(extracellular [35S]-LC signal at t = 0 + intracellular [35S]-LC signal at t = 0)].
Conditioned Media Aggregation and Blue Native PAGE.
HEK293DAX cells expressing LC or GFP control were treated with vehicle, doxycycline (Dox), trimethoprim (TMP), or the combination for 16 h. The medium was removed, centrifuged at 200 × g to remove cell debris, transferred to a new tube, and protease inhibitor mixture (Roche) added. The medium was incubated at 55 °C with aliquots removed at the indicated timepoints.
Conditioned medium was added to blue native PAGE loading dye (10% glycerol, 0.5% Coomassie G-250) and then loaded onto 3–12% Bis-Tris gradient gels (Invitrogen). The cathode buffer contained 50 mM Tricene and 15 mM Bis-Tris, pH 7.0 with 0.02% Coomassie G-250. The anode buffer contained 50 mM Bis-Tris pH 7.0. The gels were transferred onto PVDF membranes, and LC was detected by polyclonal anti-human lambda LC (Bethyl Laboratories), followed by HRP-conjugated secondary antibodies. The blots were imaged using a chemiluminescence substrate (Luminata Forte Western Luminescence Substrate, Millipore) and imaged by film or with a Bio-Rad scanner.
Supplementary Material
Acknowledgments
We thank William Balch for helpful discussions. We thank Arlene and Arnold Goldstein, the National Institutes of Health (NIH) (AG046495, DK075295, and NS079882), the Ellison Medical Foundation, the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and the Scripps Research Institute for financial support. C.B.C. was supported by NIH (F32 AG042259).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406050111/-/DCSupplemental.
References
- 1.Cohen AD, Comenzo RL. Systemic light-chain amyloidosis: Advances in diagnosis, prognosis, and therapy. Hematology (Am Soc Hematol Educ Program) 2010;2010:287–294. doi: 10.1182/asheducation-2010.1.287. [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum JN, Linke RP. A molecular history of the amyloidoses. J Mol Biol. 2012;421(2-3):142–159. doi: 10.1016/j.jmb.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Jones NF, Hilton PJ, Tighe JR, Hobbs JR. Treatment of “primary” renal amyloidosis with melphalan. Lancet. 1972;2(7778):616–619. doi: 10.1016/s0140-6736(72)93014-0. [DOI] [PubMed] [Google Scholar]
- 4.Jaccard A, et al. Myélome Autogreffe (MAG) and Intergroupe Francophone du Myélome (IFM) Intergroup High-dose melphalan versus melphalan plus dexamethasone for AL amyloidosis. N Engl J Med. 2007;357(11):1083–1093. doi: 10.1056/NEJMoa070484. [DOI] [PubMed] [Google Scholar]
- 5.Kastritis E, et al. Bortezomib with or without dexamethasone in primary systemic (light chain) amyloidosis. J Clin Oncol. 2010;28(6):1031–1037. doi: 10.1200/JCO.2009.23.8220. [DOI] [PubMed] [Google Scholar]
- 6.Venner CP, et al. Cyclophosphamide, bortezomib, and dexamethasone therapy in AL amyloidosis is associated with high clonal response rates and prolonged progression-free survival. Blood. 2012;119(19):4387–4390. doi: 10.1182/blood-2011-10-388462. [DOI] [PubMed] [Google Scholar]
- 7.Reece DE, et al. Efficacy and safety of once-weekly and twice-weekly bortezomib in patients with relapsed systemic AL amyloidosis: Results of a phase 1/2 study. Blood. 2011;118(4):865–873. doi: 10.1182/blood-2011-02-334227. [DOI] [PubMed] [Google Scholar]
- 8.Comenzo RL, Gertz MA. Autologous stem cell transplantation for primary systemic amyloidosis. Blood. 2002;99(12):4276–4282. doi: 10.1182/blood.v99.12.4276. [DOI] [PubMed] [Google Scholar]
- 9.Palladini G, et al. Association of melphalan and high-dose dexamethasone is effective and well tolerated in patients with AL (primary) amyloidosis who are ineligible for stem cell transplantation. Blood. 2004;103(8):2936–2938. doi: 10.1182/blood-2003-08-2788. [DOI] [PubMed] [Google Scholar]
- 10.Merlini G, Seldin DC, Gertz MA. Amyloidosis: Pathogenesis and new therapeutic options. J Clin Oncol. 2011;29(14):1924–1933. doi: 10.1200/JCO.2010.32.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon A, Weiss DT, Pepys MB. Induction in mice of human light-chain-associated amyloidosis. Am J Pathol. 1992;140(3):629–637. [PMC free article] [PubMed] [Google Scholar]
- 12.Bodi K, et al. AL-Base: A visual platform analysis tool for the study of amyloidogenic immunoglobulin light chain sequences. Amyloid. 2009;16(1):1–8. doi: 10.1080/13506120802676781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131(4):809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 14.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 15.Smith MH, Ploegh HL, Weissman JS. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: Progress and prognosis. Cold Spring Harb Perspect Biol. 2011;3(12):a004507. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 18.Sekijima Y, et al. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121(1):73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 20.Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581(19):3652–3657. doi: 10.1016/j.febslet.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Hendershot LM. The stressful road to antibody secretion. Nat Immunol. 2003;4(4):310–311. doi: 10.1038/ni0403-310. [DOI] [PubMed] [Google Scholar]
- 22.Williamson AR, Adkonas BA. Time course of labeling of light and heavy chains of immunoglobulin G and the turnover of the free light-chain intermediates. Arch Biochem Biophys. 1968;125(2):401–409. doi: 10.1016/0003-9861(68)90596-1. [DOI] [PubMed] [Google Scholar]
- 23.Zolla S, Buxbaum J, Franklin EC, Scharff MD. Synthesis and assembly of immunoglobulins by malignant human plasmacytes. I. Myelomas producing gamma-chains and light chains. J Exp Med. 1970;132(1):148–162. doi: 10.1084/jem.132.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197(7):857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arendt BK, et al. Biologic and genetic characterization of the novel amyloidogenic lambda light chain-secreting human cell lines, ALMC-1 and ALMC-2. Blood. 2008;112(5):1931–1941. doi: 10.1182/blood-2008-03-143040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2(6):e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulleman JD, Balch WE, Kelly JW. Translational attenuation differentially alters the fate of disease-associated fibulin proteins. FASEB J. 2012;26(11):4548–4560. doi: 10.1096/fj.11-202861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulleman JD, Brown SJ, Rosen H, Kelly JW. A high-throughput cell-based Gaussia luciferase reporter assay for identifying modulators of fibulin-3 secretion. J Biomol Screen. 2013;18(6):647–658. doi: 10.1177/1087057112469405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Li WW, Alexandre S, Cao X, Lee AS. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca(2+)-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268(16):12003–12009. [PubMed] [Google Scholar]
- 32.Hägg M, et al. Induction of endoplasmic reticulum stress by ellipticine plant alkaloids. Mol Cancer Ther. 2004;3(4):489–497. [PubMed] [Google Scholar]
- 33.Hiramatsu N, Joseph VT, Lin JH. Monitoring and manipulating mammalian unfolded protein response. Methods Enzymol. 2011;491:183–198. doi: 10.1016/B978-0-12-385928-0.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Axten JM, et al. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) J Med Chem. 2012;55(16):7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem. 2008;283(12):7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruse KB, Brodsky JL, McCracken AA. Autophagy: An ER protein quality control process. Autophagy. 2006;2(2):135–137. doi: 10.4161/auto.2.2.2388. [DOI] [PubMed] [Google Scholar]
- 37.Ryno LM, Wiseman RL, Kelly JW. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr Opin Chem Biol. 2013;17(3):346–352. doi: 10.1016/j.cbpa.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wall J, et al. Thermodynamic instability of human lambda 6 light chains: Correlation with fibrillogenicity. Biochemistry. 1999;38(42):14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 39.Klimtchuk ES, et al. The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry. 2010;49(45):9848–9857. doi: 10.1021/bi101351c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoulders MD, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Reports. 2013;3(4):1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang WC, Hiramatsu N, Messah C, Kroeger H, Lin JH. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012;53(11):7159–7166. doi: 10.1167/iovs.12-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370(6488):373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 43.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. 2010;35(4):189–198. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lachmann HJ, et al. Outcome in systemic AL amyloidosis in relation to changes in concentration of circulating free immunoglobulin light chains following chemotherapy. Br J Haematol. 2003;122(1):78–84. doi: 10.1046/j.1365-2141.2003.04433.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.