Significance
Despite the well-established connection between Ras and Src, there currently is no evidence of direct interaction between these two proteins. We show here that Src binds to and phosphorylates GTP-loaded Ras on a conserved Y32 residue within the switch I region. It has been shown that Raf binds to Ras with an affinity 1,000-fold greater than that of GAP. However, it has remained unclear how GAP is able to outcompete Raf for Ras upon Raf displacement. We show here that Y32 phosphorylation inhibits Raf binding to Ras and concomitantly promotes GAP association and GTP hydrolysis, thereby ensuring unidirectionality to the Ras GTPase cycle. These findings reveal new fundamental mechanistic insight into how Src negatively regulates Ras.
Abstract
Mutations in Ras GTPase and various other components of the Ras signaling pathways are among the most common genetic alterations in human cancers and also have been identified in several familial developmental syndromes. Over the past few decades it has become clear that the activity or the oncogenic potential of Ras is dependent on the nonreceptor tyrosine kinase Src to promote the Ras/Raf/MAPK pathway essential for proliferation, differentiation, and survival of eukaryotic cells. However, no direct relationship between Ras and Src has been established. We show here that Src binds to and phosphorylates GTP-, but not GDP-, loaded Ras on a conserved Y32 residue within the switch I region in vitro and that in vivo, Ras-Y32 phosphorylation markedly reduces the binding to effector Raf and concomitantly increases binding to GTPase-activating proteins and the rate of GTP hydrolysis. These results suggest that, in the context of predetermined crystallographic structures, Ras-Y32 serves as an Src-dependent keystone regulatory residue that modulates Ras GTPase activity and ensures unidirectionality to the Ras GTPase cycle.
The pioneering work of Harvey (1) and Kirsten and Mayer (2) showed that the Harvey strain murine sarcoma virus (HaMSV) and Kirsten strain murine sarcoma virus (KiMSV) sarcoma retroviruses cause rapid tumor formation in rats. The viral oncogenes, H-Ras and K-Ras, responsible for the oncogenic properties are altered versions of rat genes that encode enzymes with intrinsic guanine nucleotide binding and GTPase activity (3). The seminal discovery of mutationally activated RAS genes in human cancer in 1982 initiated an intensive research effort to understand Ras protein structure, function, and biology that continues to this day (4).
The three human RAS oncogenes (H-RAS, N-RAS, and K-RAS) encode highly related (90% amino acid identity) 188- or 189-amino acid proteins. They are canonical members of a large superfamily consisting of more than 150 cellular members of small monomeric GTPase proteins, which function as molecular switches in a number of signaling pathways that regulate cell proliferation, differentiation, and apoptosis (1–3, 5, 6). As do other GTP-binding proteins, Ras cycles between the inactive GDP- and the active GTP-bound forms through conformational changes near the nucleotide-binding site localized in the switch I (amino acids 30–38) and switch II (amino acids 59–72) regions (7).
Activation of the cell-surface receptor leads to the activation of Ras via guanine nucleotide-exchange factor (GEF), which binds to the Ras–GDP complex, causing dissociation of the bound GDP (8). Because GTP is present in cells at a much higher concentration than GDP, GTP binds spontaneously to the “empty” Ras molecule with the release of GEF (9, 10). Hydrolysis of GTP to GDP results from intrinsic Ras GTPase activity accelerated by GTPase-activating proteins (GAPs) that bind to and stabilize the Ras catalytic machinery, supplying additional catalytic “arginine finger” residues resulting in the inactivation of Ras and attenuation of signaling (11). Mutations in codons 12, 13, or 61 convert RAS into an active oncoprotein (12) by impeding the GTPase activity of Ras (13). Thus, unlike normal Ras, oncogenic Ras-mutant proteins are predominantly in the GTP-bound form and continuously activate the downstream effectors that promote cell proliferation, consequently leading to tumor development.
In the GTP-bound conformation, Ras has high affinity for numerous effector molecules. Ras does not chemically modify the effectors but instead regulates the activity of the effectors by recruiting them to the activators localized near Ras–GTP (14). One of the best-characterized Ras effectors is the Raf kinase. Ras and Raf are key mediators in one of the major signal-transduction pathways that regulate cell proliferation, the Ras/Raf/MEK/ERK pathway (15). Raf is recruited to Ras–GTP from the cytoplasm to the plasma membrane through two Ras-binding sites, Ras-binding domain (RBD) and the Cys-rich domain. Raf subsequently changes from a closed to an open conformation, allowing membrane-associated kinases to phosphorylate Raf at multiple activating sites (16). For example, the p21-activated protein kinase (PAK) phosphorylates c-Raf at Ser338, whereas Src family kinases phosphorylate Y341 to induce Raf activity (13, 17). Raf then dissociates from Ras–GTP (16), and this dissociation is followed by the aforementioned GAP binding and GTP hydrolysis that inactivate Ras (18).
The relationship between the first confirmed oncogene Src (v-Src) and Ras is very well established. For example, elevated Src activity has been observed in human cancer cell lines that harbor oncogenic RAS mutations (19) as well as in pancreatic ductal adenocarcinomas that are characterized by a high incidence of oncogenic K-RAS mutations (20). Furthermore, v-Src has been shown to activate Ras by phosphorylating the adaptor protein Shc, which then recruits the Grb2/Sos complex for Ras activation (21). Ras also has been shown to be essential for oncogenic v-Src–stimulated cellular transformation (22). Perhaps most mechanistically revealing is the observation that v-Src phosphorylates Raf only in the presence of Ras to activate the cell proliferative MEK/ERK signaling pathway (23). Although these and many other examples of crosstalk between Src and Ras have been described, any evidence of a direct relationship has remained elusive. We show here that Src binds to and phosphorylates Ras in vitro and in vivo on a conserved Tyr residue at position 32 within the switch I region to promote Ras GTPase activity.
Results
We asked whether the interaction between Ras and Src is dependent on the effector Raf. HEK293 cells were transfected with plasmid encoding wild-type c-Src in combination with empty expression plasmid (mock), wild-type HA-tagged human N-Ras(WT), oncogenic HA–N-Ras(12D), cytosolic GTP-bound dominant-negative HA–N-Ras(12D 186S), or the dominant-negative defective in Raf-binding HA–N-Ras(17N) mutant. c-Src coprecipitated with N-Ras(WT) and N-Ras(12D), but most intriguingly, c-Src bound strongly to the N-Ras(17N) mutant, which does not bind to Raf (Fig. 1A and Fig. S1). In addition, c-Src did not interact with cytosolic N-Ras(12D 186S) (Fig. 1A). These results suggest, for the first time to our knowledge, that c-Src binds to Ras at the cell membrane independent of Raf.
Fig. 1.
Src binds to and tyrosyl phosphorylates wild-type Ras more effectively than oncogenic Ras mutants. (A and B) HEK293 cells transfected with the indicated combination of plasmids were lysed, immunoprecipitated with anti-HA antibody, and immunoblotted with the indicated antibodies. (C and D) HEK293 cells transfected with the indicated plasmids were lysed, immunoprecipitated with anti-Flag antibody, and immunoblotted with the indicated antibodies. (E) HEK293 cells transfected with the indicated combination of plasmids were lysed, immunoprecipitated with either anti-H-Ras (Left) or anti-HA (Right) antibody, and immunoblotted with the indicated antibodies. IP, immunoprecipitation; WCE, whole-cell extract.
We next asked whether c-Src phosphorylates Ras on Tyr residues. In comparison with oncogenic N-Ras(12D), N-Ras(WT) showed a higher level of tyrosyl phosphorylation in the presence of ectopic c-Src(WT) (Fig. 1B), which corresponded with increased c-Src association to N-Ras(WT) relative to N-Ras(12D) (Fig. 1 A and B). In contrast, a negligible level of tyrosyl phosphorylation was observed on N-Ras (WT or 12D) in the presence of the kinase-dead c-Src(K295R, Y527F) mutant (Fig. S2A). The constitutively kinase-active form of transforming c-Src(Y527F) markedly increased tyrosyl phosphorylation of endogenous Ras in comparison with c-Src(WT) or kinase-dead c-Src(K295R, Y527F) (Fig. S2B). Moreover, the transforming mutants N-Ras(12V), H-Ras(12V), and K-Ras(12V) as well as N-Ras(Q61L) all displayed reduced tyrosyl phosphorylation levels in comparison with their wild-type counterparts in the presence of c-Src (Fig. 1 C–E). These results suggest that c-Src binds to and tyrosyl phosphorylates wild-type Ras isoforms more effectively than their oncogenic counterparts.
Ras is activated in response to the binding of extracellular signals, such as growth factors and cytokines, to their cognate cell surface receptors. We asked whether ligand engagement of receptors promotes tyrosyl phosphorylation of Ras. EGF treatment, a well-known mediator of Ras signaling via EGF receptor activation, markedly enhanced the level of HA–N-Ras(WT) tyrosyl phosphorylation in HEK293 cells (Fig. 2A). Consistent with this observation, the stably expressed human Flag–N-Ras(WT) in murine pro-B BaF3 cells as well as endogenous N-Ras(WT) in human myeloid TF-1 cells showed increased tyrosyl phosphorylation levels following cytokine IL-3 and GM-CSF treatments, respectively (Fig. 2B and Fig. S2C). Notably, oncogenic N-Ras(12D) in HEK293 or BaF3 cells was tyrosyl phosphorylated to a lesser level than N-Ras(WT) following EGF and IL-3 stimulation, respectively (Fig. 2 A and B). Oncogenic Ras is predominately in the active GTP-bound form that interacts with effector molecules even in the absence of external stimuli. These results thus suggest a possible inverse relationship between tyrosyl phosphorylation of Ras and effector binding. Consistent with this notion, the oncogenic N-Ras(12D) mutant showed markedly increased binding to endogenous Raf as compared with its wild-type counterpart (Fig. S3), which bound to and tyrosyl phosphorylated to a greater extent via c-Src (Figs. 1 and 2 A and B and Fig. S3).
Fig. 2.
Activation of the cell-surface receptor promotes Src-dependent tyrosyl phosphorylation of wild-type Ras more effectively than oncogenic Ras. (A) HEK293 cells transfected with the indicated combination of plasmids were serum starved and treated with (+) or without (−) 1 ng/mL of EGF. Equal amounts of lysates were immunoprecipitated with anti-HA antibody and immunoblotted with the indicated antibodies. (B) BaF3-Flag–N-Ras(parental, WT or 12D) cells were serum starved and treated with 10 ng/mL of IL-3 (+) or were left untreated (−). The cells were lysed, immunoprecipitated with anti-Flag antibody, and immunoblotted with the indicated antibodies. (C) RasB8 P0 or P3 astrocytes expressing the HA–H-Ras(12V) transgene derived from GBM-prone mice were serum starved and treated with 1 ng/mL of EGF (+) or were left untreated (−). Equal amounts of lysates were immunoprecipitated with anti-HA antibody and immunoblotted with the indicated antibodies. (D) NHA or U87 human GBM astrocytes were serum starved and treated with 1 ng/mL of EGF (+) or were left untreated (−). The cells were lysed, immunoprecipitated with anti-Ras antibody or pulled down with Raf:RBD-conjugated beads, and immunoblotted with the indicated antibodies. (E) MEFs or Src/Yes/Fyn SYF−/− MEFs were serum starved and treated with 1 ng/mL of EGF (+) or were left untreated (−). Equal amounts of lysates were immunoprecipitated with anti-pTyr antibody and immunoblotted with the indicated antibodies. (F) U87 human GBM astrocytes were treated with λ-phosphatase (λ PPase) or sodium orthovanadate (Na3V04) (+) or were left untreated (−). Cells were lysed, immunoprecipitated with anti-Ras antibody, and immunoblotted with the indicated antibodies. (G) U87 astrocytes were pretreated with sodium orthovanadate (+) or were not pretreated (−) and were treated with EGF (+) or were left untreated (−), lysed and immunoprecipitated with anti-Ras antibody or pulled down with Raf:RBD-conjugated beads, and immunoblotted with the indicated antibodies. (H) Equal numbers of U87 astrocytes were plated in a 96-well plate in quadruplicate, treated with increasing concentrations of sodium orthovanadate for 18 h, and analyzed by alamarBlue assay at 570 nM. These experiments were performed three times. PD, pulldown.
We next assessed this notion using astrocytes derived from glioblastoma (GBM)-prone mice (24). Oncogenic HA–H-Ras(12V) is stably expressed in astrocytes derived from both newborn pups (RasB8 P0) and 3-mo-old (RasB8 P3) mice; however, GBM is observed only in RasB8 P3 mice (25), suggesting a postnatal accumulation of additional, as yet undefined, pro-oncogenic events that enable the H-Ras(12V) transforming activity. Notably, astrocytes isolated from tumor-bearing RasB8 P3 mice showed increased association between HA–H-Ras(12V) and the RBD of Raf (Raf:RBD) compared with HA–H-Ras(12V) derived from astrocytes isolated from a brain of non–tumor-bearing RasB8 P0 mice (Fig. 2C). In addition, HA–H-Ras(12V) derived from RasB8 P0 astrocytes showed markedly increased tyrosyl phosphorylation and binding to c-Src in the presence of EGF as compared with HA–H-Ras(12V) obtained from RasB8 P3 astrocytes (Fig. 2C). The mechanism by which RasB8 P3 astrocytes lose responsiveness to EGF currently is unknown; however, the diminished binding to c-Src and tyrosyl phosphorylation of H-Ras(12V) are consistent with markedly increased association with Raf. These observations also suggest that the oncogenic H-Ras(12V) does not necessarily have an intrinsic defect in being subjected to tyrosyl phosphorylation via Src or an unrestrained affinity for Raf, because in astrocytes of newborn RasB8 P0 pups H-Ras(12V) behaves seemingly like wild-type Ras.
The human primary GBM cell line U87 harbors hyperactive, but otherwise wild-type, Ras that is predominantly in the active GTP-bound form, whereas normal human astrocytes (NHA) harbor Ras that is predominantly in the GDP-bound state (26). In comparison with Ras obtained from NHA, the hyperactive Ras in U87 cells, similar to H-Ras(12V) in RasB8 P3 astrocytes, showed increased binding to Raf:RBD and was less responsive to EGF-induced tyrosyl phosphorylation (Fig. 2D). Notably, EGF treatment failed to promote tyrosyl phosphorylation of endogenous Ras in Src/Yes/Fyn triple-knockout mouse embryonic fibroblasts (SYF−/− MEFs) (Fig. 2E). These results support the notion that Ras is tyrosyl phosphorylated preferentially via Src when not bound to Raf. In addition, treatment with the protein tyrosine phosphatase inhibitor sodium orthovanadate (Na3V04) markedly enhanced tyrosyl phosphorylation of Ras in U87 cells, and this effect was abrogated by cotreatment with λ-phosphatase (Fig. 2F). Sodium orthovanadate attenuated EGF-induced Ras activation and downstream signaling, concomitant with increased tyrosyl phosphorylation of Ras and decreased Raf:RBD association with Ras in U87, HEK293, and RasB8 P3 cells (Fig. 2G and Fig. S4). Consistent with these observations, sodium orthovanadate decreased cellular proliferation in a dosage-dependent manner (Fig. 2H and Fig. S4). These results suggest an as yet undefined phosphatase that dephosphorylates and activates Ras and its downstream proliferative signaling.
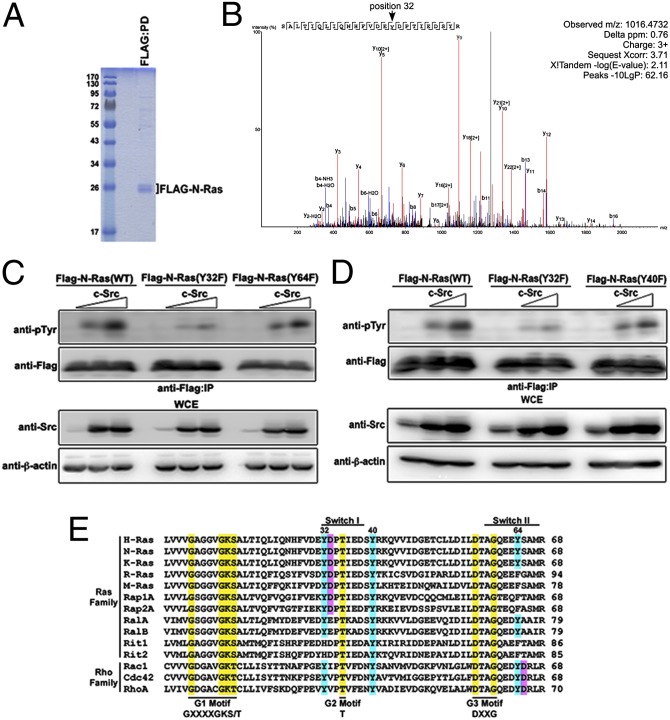
Nanoflow reversed-phase liquid chromatography-electrospray ionization-tandem mass spectrometry (nLC-ESI-MS/MS) was performed to identify tyrosyl-phosphorylated peptide fragments of H-Ras following an in vitro kinase assay using bacterially purified human GST–H-Ras and His–c-Src. Y32, Y64, Y96, and Y157 were phosphorylated by c-Src in vitro. Interestingly, a peptide array demonstrated that phosphorylated H-Ras peptides can bind to the GST–c-Abl SH2 domain, with Y32-containing peptide being the most robust (27). We next asked which N-Ras sites, if any, were phosphorylated by c-Src in vivo. FLAG–N-Ras expressed in HEK293 cells was isolated via FLAG pulldown (Fig. 3A). N-Ras was digested with trypsin, and the resulting peptides were enriched for tyrosyl-phosphorylated peptides using 4G10 platinum anti-phosphotyrosine agarose. A phosphorylated peptide consisting of amino acids 17–41 of Flag–N-Ras(WT) was detected, showing that Y32 of N-Ras is phosphorylated, but Y40 is not (Fig. 3B and Fig. S5). The spectrum of unphosphorylated N-Ras in the absence of ectopic c-Src is shown for comparison (Fig. S6). Analysis of the spectral counts for phosphorylated peptides identified by Sequest and X!Tandem with a P value greater than 0.9 identified phosphorylated Y32, Y64, and Y96. However, we did not observe any peptides containing phosphorylated Y157.
Fig. 3.
Src phosphorylates N-Ras(WT) on a conserved Tyr32 residue. (A) HEK293 cells were transfected with plasmids encoding c-Src and Flag–N-Ras. Flag pulldown was performed to isolate Flag–N-Ras, which was visualized by Coomassie staining. (B) N-Ras was affinity-captured on Flag-beads, washed, and subjected to trypsin digestion. Phospho-peptides then were captured using pTyr-specific antibodies conjugated to agarose beads. Purified peptides were eluted and subjected to LC-ESI-MS/MS. Data were analyzed using multiple database-search algorithms. Shown is an annotated representative CID spectrum for phosphorylated peptide containing Y32. β-Ions are indicated in blue, and γ-ions are indicated in red. (C and D) HEK293 cells transfected with the indicated combination of plasmids were lysed, immunoprecipitated with anti-Flag antibody, and immunoblotted with the indicated antibodies. (E) A multiple sequence alignment was performed using Clustal Omega (61). The indicated motifs, shown in yellow, have been described previously (6, 30). Ras Tyr residues that are conserved are in blue. Shown in pink are Asp residues that immediately follow a Tyr residue, making a candidate c-Src phosphorylation motif (28).
Analysis of the identified phosphotyrosine residues within the switch I and switch II regions showed that tyrosyl phosphorylation of the N-Ras(Y32F) switch I mutant is markedly attenuated in the presence of increasing concentrations of c-Src as compared with wild-type the or N-Ras(Y64F) switch II mutant (Fig. 3C). Substitution of a closely located Y40 by Phe did not affect c-Src–mediated phosphorylation of N-Ras (Fig. 3D). Unlike the cytosolic N-Ras(12D 186S) dominant-negative mutant, N-Ras(Y32F) displayed a subcellular localization profile similar to that of N-Ras(WT) (Fig. S7). These results strongly suggest that Y32 is a major Tyr residue on Ras that is phosphorylated via Src in vivo.
An alignment of a subset of Ras and Rho family members shows that Y32 is a highly conserved residue with a critical hydroxyl group that is located in the Src phosphorylation consensus motif (YD) (28) in the core effector-binding domain (Ras residues 32–40) within the switch I region (Fig. 3E), which is necessary for direct association with RBD of effectors (6, 29, 30). In addition to effector binding, Y32 has been identified as having a critical role in instigating a conformational change in Ras that modulates its GTPase activity (14, 31–36). Consistent with the notion that Y32 is important in mediating effector binding of Ras, the Flag–N-Ras(Y32F) and Flag–N-Ras(Y32E) mutants, both of which lack the critical hydroxyl group, expressed in HEK293 cells, showed reduced binding to Raf:RBD (Fig. S8 A and B). Flag–N-Ras(Y32F) also showed reduced binding to the effector Ral guanine nucleotide dissociation stimulator (RalGDS) (Fig. S8C). In addition, the introduction of the Y32F substitution in the N-Ras(12D) mutant attenuated the level of phosphorylated AKT in the presence or absence of EGF compared with N-Ras(12D)–expressing cells (Fig. S8D), suggesting that Ras-Y32 has a role in the regulation of PI3K-AKT signaling. Moreover, pharmacologic inhibition of Src kinase activity using 4-Amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2) abrogated IL-3–induced human HA–N-Ras(WT) tyrosyl phosphorylation and dramatically increased c-Raf binding to N-Ras(WT) in murine pro-B BaF3 cells (Fig. 4A). Similar results were observed in RasB8 P3 astrocytes following EGF stimulation in the presence of PP2 (Fig. S8E). Although EGF-induced phosphorylation of Ras was lost in SYF−/− MEFs (Fig. 2E), EGF treatment markedly increased the interaction between Ras and Raf:RBD in SYF−/− MEFs as compared with the parental wild-type MEFs (Fig. 4B). These results further support the notion of an inverse relationship between tyrosyl phosphorylation of Ras and effector Raf binding. However, it should be noted that in the context of Src kinase inhibition via PP2, which increases the level of the active Ras–Raf complex, the downstream signaling was paradoxically attenuated (Fig. 4A and Fig. S8E). This attenuation likely was caused by inhibition of Src-dependent Raf phosphorylation, resulting in the attenuation of Raf-dependent downstream signaling, such as ERK phosphorylation (Fig. 4 A and B and Fig. S8E). Similar observations were noted using a Ser/Thr PAK inhibitor that blocked PAK-dependent phosphorylation of Raf and its downstream signaling (Fig. S8F). Furthermore, it is well established that Src or PAK-induced Raf phosphorylation is associated with the displacement of Raf from Ras–GTP complex, an effect that is consistent with the accumulation of Ras–Raf complex upon pharmacologic inhibition of Src or PAK.
Fig. 4.
Src tyrosyl phosphorylates Ras that is GTP bound. (A) BaF3-Flag–N-Ras(WT) cells were serum starved and then were treated with 10 ng/mL of IL-3 (+) or were left untreated (−). The cells were lysed, immunoprecipitated with anti-Flag antibody, and immunoblotted with the indicated antibodies. (B) MEFs or SYF−/− MEFs were serum starved and treated with 1 ng/mL of EGF (+) or were left untreated (−). Equal amounts of lysates were pulled down using Raf:RBD-conjugated beads and immunoblotted with the indicated antibodies. (C) Bacterially purified recombinant human GST–H-Ras(WT) was preloaded with GDP, GTPγS, or GTP and subjected to an in vitro kinase assay using bacterially purified recombinant human Src kinase. A GST–pull-down assay was then performed, and cells were immunoblotted using the indicated antibodies. (D) HEK293 cells transfected with the indicated combination of plasmids were lysed, pulled down with Raf:RBD-conjugated beads or with GTP beads, and immunoblotted with the indicated antibodies. (E) HEK293 cells transfected with the indicated combination of plasmids were lysed, immunoprecipitated with anti-Flag (Left) or anti-HA (Right) antibody, and immunoblotted with the indicated antibodies.
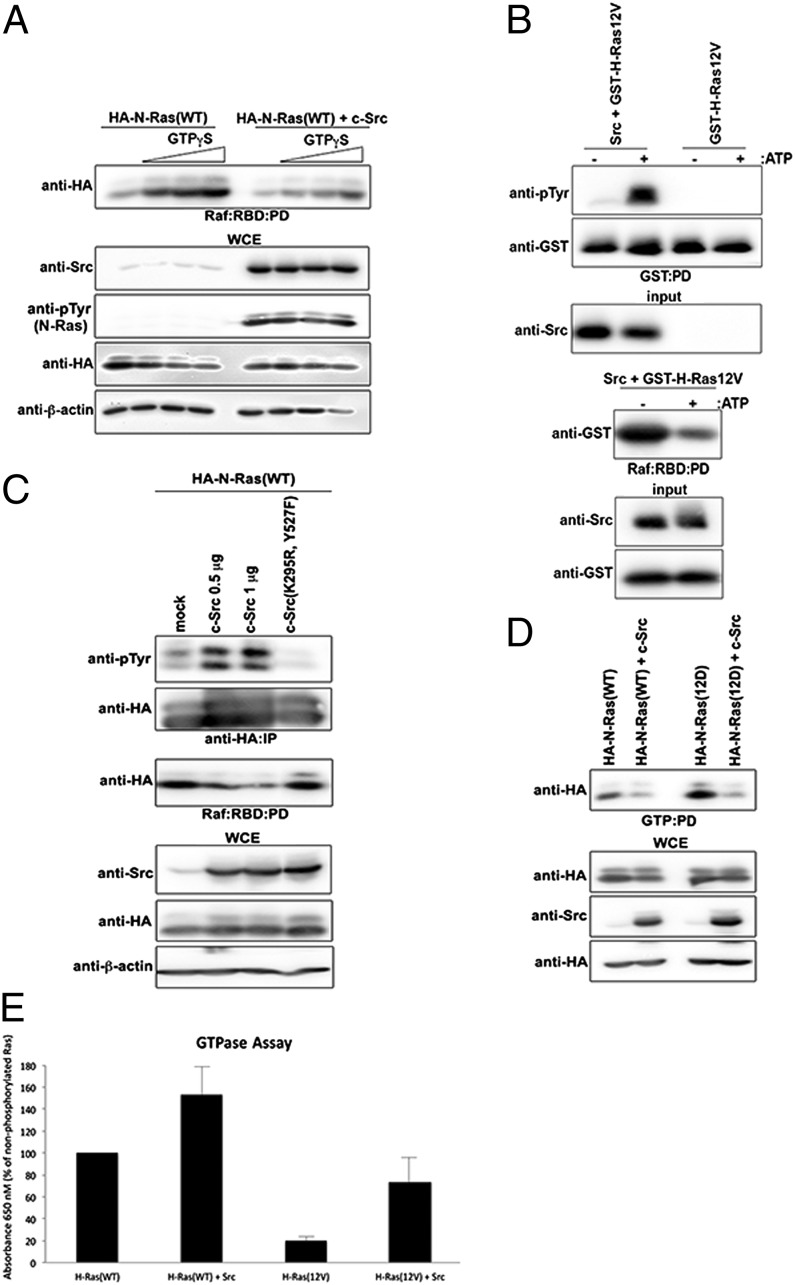
We next asked whether Ras tyrosyl phosphorylation regulates its nucleotide-binding state. The loading efficacy of purified human GST-tagged H-Ras(WT) with GDP, GTP, or nonhydrolysable GTPγS first was confirmed via pull-down analysis using Raf:RBD-conjugated beads (Fig. S9). An in vitro kinase assay using bacterially purified Src kinase on preloaded GST–H-Ras(WT) showed that GST–H-Ras(WT) loaded with GTP, but not with GTPγS or GDP, is tyrosyl phosphorylated by Src (Fig. 4C). These results suggest that Src preferentially phosphorylates Ras–GTP in vitro. Flag–N-Ras(Y116F) is a dominant-negative mutant that is defective in binding to Raf and GTP but is associated with GDP. In contrast, although Flag–N-Ras(17N) also is defective in binding to Raf, it is capable of binding to GTP (Fig. 4D). Consistent with the above in vitro data, ectopic c-Src in HEK293 cells, although coprecipitating with both Flag–N-Ras(17N) and Flag–N-Ras(Y116F), promoted tyrosyl phosphorylation of only Flag–N-Ras(17N) (Fig. 4E). These findings support the notion that Src-mediated Ras phosphorylation occurs when Ras is bound to GTP but not to GDP.
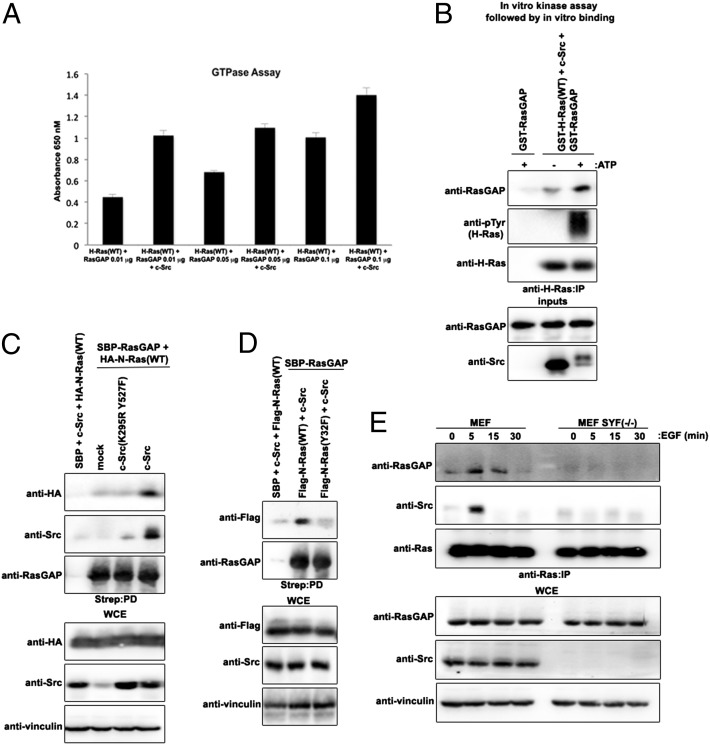
We asked whether tyrosyl phosphorylation of Ras inhibits binding to Raf. HA–N-Ras(WT) expressed in HEK293 cells showed increased binding to Raf in the presence of increasing concentrations of GTPγS (Fig. 5A). However, coexpression of c-Src, which promoted HA–N-Ras(WT) tyrosyl phosphorylation, markedly attenuated the binding to Raf (Fig. 5A). The expression of kinase-dead c-Src(K295R, Y527F) instead of wild-type c-Src rescued the binding of GTPγS-loaded HA–N-Ras(WT) to Raf:RBD (Fig. S10A). These results further suggest that Ras tyrosyl phosphorylation attenuates Raf binding to GTP-bound Ras. Considering that oncogenic Ras mutants, although displaying a reduced tyrosyl phosphorylated profile in comparison with wild-type Ras (Figs. 1 and 2 A and B), are not necessarily defective for Src-dependent tyrosyl phosphorylation (Fig. 2C), we asked whether forced phosphorylation of oncogenic Ras could lower its affinity for Raf and thereby promote GTP hydrolysis. An in vitro kinase assay was performed on bacterially purified human GST-tagged H-Ras(12V) using purified human c-Src kinase; this assay demonstrated that the recombinant human GST–H-Ras(12V) can be tyrosyl phosphorylated via c-Src (Fig. 5B). Notably, under such conditions, oncogenic GST–H-Ras(12V) had markedly reduced binding to Raf:RBD (Fig. 5B). Consistent with this observation, increasing concentrations of c-Src, but not kinase-dead c-Src(K295R Y527F), attenuated HA–N-Ras(WT or 12D) binding to Raf in HEK293 cells (Fig. 5C and Fig. S10B, respectively), and GTPγS-loaded HA–N-Ras(12D) binding to Raf:RBD was attenuated in the presence of wild-type c-Src compared with c-Src(K295R Y527F) (Fig. S10A). Notably, HA–N-Ras(WT) binding to effector RalGDS likewise was reduced in the presence of c-Src (Fig. S10C). Furthermore, ectopic expression of c-Src markedly decreased the ability of both HA–N-Ras(WT) and oncogenic HA–N-Ras(12D) to bind GTP-loaded beads (Fig. 5D), suggesting that Src-induced tyrosyl phosphorylation of Ras promotes GTP hydrolysis. Colorimetric GTP hydrolysis assay of bacterially purified GST–H-Ras(12V) confirmed the impaired GTPase activity of oncogenic Ras (Fig. S10 D and E). Most notably, tyrosyl phosphorylation of bacterially purified GST–H-Ras(12V) or GST–H-Ras(WT) using purified human c-Src kinase significantly increased the intrinsic GTP hydrolysis rate as compared with unphosphorylated counterparts (Fig. 5E). Purified GST–H-Ras(WT) in the presence of purified human c-Src increased RasGAP-stimulated GTP hydrolysis (Fig. 6A). Human recombinant GST–RasGAP bound more to tyrosyl phosphorylated bacterially purified GST–tagged H-Ras(WT) in the presence of purified c-Src (Fig. 6B). Furthermore, ectopic expression of c-Src, but not of kinase-dead c-Src(K295R Y527F), markedly increased HA–N-Ras(WT) binding to streptavidin-binding peptide (SBP)-tagged RasGAP (Fig. 6C). However, Flag–N-Ras(Y32F) showed attenuated binding to SBP-RasGAP as compared with Flag–N-Ras(WT) (Fig. 6D). Moreover, endogenous Ras showed temporal binding to RasGAP upon EGF stimulation in wild-type MEFs but not in SYF−/− MEFs (Fig. 6E). These results support the notion that Src-dependent phosphorylation of Ras promotes GAP binding and GAP-stimulated GTP hydrolysis.
Fig. 5.
Src-induced tyrosyl phosphorylation of oncogenic Ras attenuates Raf binding and promotes GTP hydrolysis. (A) HEK293 cells transfected with the indicated combination of plasmids were lysed, loaded with increasing concentrations of GTPγS, pulled down with Raf:RBD-conjugated beads, and immunoblotted with the indicated antibodies. (B) Bacterially purified recombinant human GST–H-Ras(12V) was subjected to an in vitro kinase assay with bacterially purified recombinant human Src kinase. Subsequent pull-down assays using either GST-conjugated beads (Upper) or Raf:RBD-conjugated beads (Lower) were performed, and cells were immunoblotted using the indicated antibodies. (C) HEK293 cells transfected with the indicated combination of plasmids were lysed, immunoprecipitated with anti-HA antibody or pulled down with Raf:RBD-conjugated beads, and immunoblotted with the indicated antibodies. (D) HEK293 cells transfected with the indicated combination of plasmids were lysed, pulled down with GTP-conjugated beads, and immunoblotted with the indicated antibodies. (E) An in vitro kinase assay using bacterially purified recombinant human GST–H-Ras(WT or 12V) alone or in combination with bacterially purified recombinant human Src kinase was performed and was followed by a colorimetric GTP hydrolysis assay performed in quadruplicate according to the manufacturer’s instructions. Data were normalized to the H-Ras(WT) control.
Fig. 6.
Src-induced phosphorylation of Ras increases binding to GAP and GAP-stimulated GTP hydrolysis. (A) An in vitro kinase assay using bacterially purified recombinant human GST-H-Ras(WT) alone or in combination with bacterially purified recombinant human Src kinase was performed, followed by a colorimetric GTP hydrolysis assay in the presence of increasing concentrations of bacterially purified recombinant human GST–RasGAP in quadruplicate according to the manufacturer’s instructions. (B) Bacterially purified recombinant human GST-H-Ras(WT) was subjected to an in vitro kinase assay with bacterially purified recombinant human Src kinase. Subsequent pull-down assays using either GST-conjugated beads (Upper) or Raf:RBD-conjugated beads (Lower) were performed, and cells were immunoblotted using the indicated antibodies. (C and D) HEK293 cells transfected with the indicated combination of plasmids were lysed, pulled down with streptavidin-conjugated beads, and immunoblotted with the indicated antibodies. (E) MEFs or SYF−/− MEFs were serum starved and treated with 1 ng/mL of EGF (+) or were left untreated (−) for the indicated periods of time. Equal amounts of lysates were immunoprecipitated with anti-Ras antibody and immunoblotted with the indicated antibodies.
Discussion
Studies using purified insulin receptor and Ras GTPases have suggested that Ras proteins may be phosphorylated on Tyr residues (37), and a recent report using the modified SH2 domain to phototrap potential Tyr residues identified Y32 and Y64 as possible sites of phosphorylation on H-Ras in vitro (27). However, the kinase responsible for and the functional significance of Ras phosphorylation remained unknown.
We show here, for the first time to our knowledge, that Src binds to and phosphorylates GTP-loaded Ras on Y32 in vitro and in vivo and show that this event is associated with increased Raf displacement, GAP binding, and GTP hydrolysis. Interestingly, homologous tyrosyl phosphorylation sites on RhoA, Rab24, and Ran GTPases has been described (27, 38, 39), and c-Src has been shown to phosphorylate Cdc42, R-Ras, and the G protein α subunit (40–42). Ras-Y32 is located in the switch I region within the Src phosphorylation consensus motif (YD). Previous studies have shown that Src phosphorylates Cdc42 on Y64, which is located in a candidate Src consensus motif (Fig. 3E) (40). However, we show here that Y64 in Ras is not located in the Src consensus motif and generated very few spectral counts for phosphorylated peptides in vivo.
Y32 is well conserved in the Ras GTPase superfamily and has been shown previously to play a major role in GTP hydrolysis and effector binding (14, 31–36). The effector Raf binds to Ras with an affinity that is 1,000-fold greater than that of GAPs for Ras. GAPs and Raf have overlapping binding sites on Ras (43), suggesting that the engagement of GAPs on Ras would require, at a minimum, displacement of Raf (44–46) and an as yet undefined modulation of Ras conformation that is favorable for GAP binding, because it appears highly improbable that GAPs simply can outcompete Raf at the concentrations found in cells (47). These apparent paradoxical observations in Ras biology have remained mechanistically unresolved.
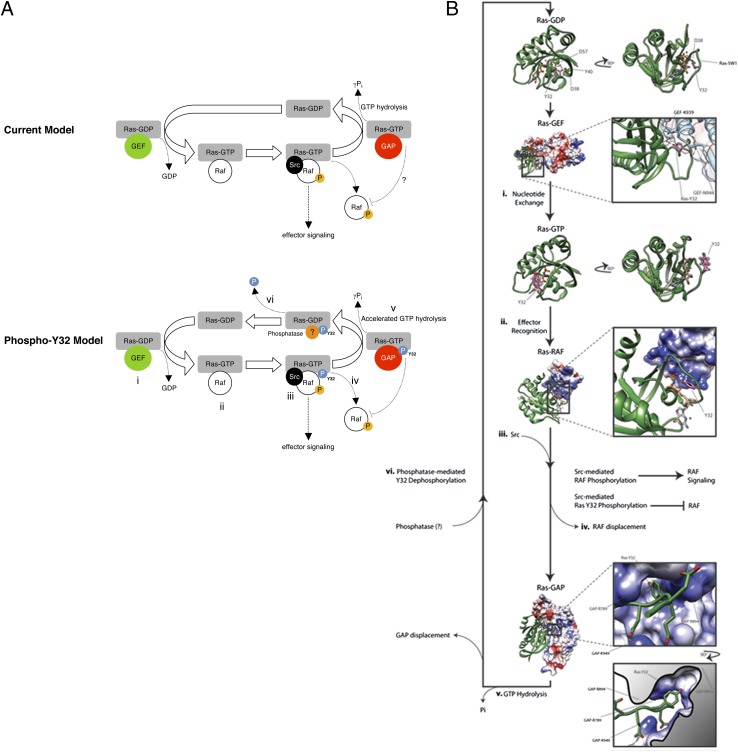
Here, we show that Src-mediated phosphorylation of GTP-loaded Ras-Y32 is associated with Raf displacement and increased GAP binding and GTP hydrolysis, revealing Y32 as an Src-dependent keystone regulatory residue that provides unidirectionality to the Ras activation cycle (see model, Fig. 7A). Consistent with this notion, effector binding of GTP-bound Ras was shown to be mediated largely through interactions with the switch I region (48). Conversely, the GDP-bound Ras has an ∼1,000-fold lower affinity for the Raf:RBD (49). The diminished affinity between GDP-bound Ras and Raf can be overcome by introducing mutations into the Raf:RBD that reinforce electrostatic interactions between Raf and the switch I region of Ras (50). These observations strongly suggest that the precise orientation of the switch I region contributes significantly to the affinity of Ras for its effectors. We propose that Y32 phosphorylation generates an electrostatic repulsion against the negatively charged D38 and D57 within the nucleotide-binding groove that alters switch I positioning or conformation so that it no longer accommodates Raf binding (Fig. 7B). This electrostatic repulsion that promotes the release of phosphorylated Y32 also is predicted to promote GTP hydrolysis by disrupting the critical hydrogen bond between the hydroxyl group of Y32 and γ-phosphate of GTP and permitting more efficient hydrolysis of γ-phosphate by the adjacent nucleophilic water molecule. Interestingly, an analogous mechanism of a bulky phosphate group destabilizing key electrostatic interactions within the active site was proposed recently for the increased GTP hydrolysis of a highly homologous plant GTPase, Toc34, upon phosphorylation (51, 52). Consistent with this notion, the cancer-associated D38N mutation in Ras was shown to cause an incomplete opening of Y32, which in turn hindered the insertion of a critical Arg (R789) required for GTP hydrolysis (53).
Fig. 7.
Structural modeling of Ras-Y32 in the Ras GTPase cycle. (A) The current and the proposed phosho-Y32 model of Ras GTPase cycle can be separated into (i) GEF-mediated nucleotide exchange, (ii) effector recognition, (iii) kinase (including Src)-mediated effector activation and Src-mediated Ras-Y32 phosphorylation, (iv) RAF displacement, (v) GTP hydrolysis, and (vi) phosphatase-mediated Ras-Y32 dephosphorylation and GEF recruitment. (B) The switch I (SW1) region and specifically Y32 make large conformational changes throughout the Ras GTPase cycle as observed through experimental atomic-resolution modeling. The phosphorylation status of Y32 is predicted to influence movement from one stage to the next. The Ras secondary structure is depicted as green ribbons, and the stick representation of Y32 is colored pink. In the atomic-resolution X-ray crystallographic model of the GDP-bound form of Ras, Y32 participates in a hydrogen bond network with D57 and a water molecule, effectively burying the side chain within the core of the Ras molecule. Furthermore, this conformation of Ras presents SW1 in the correct arrangement for GEF engagement. Pictured next is the atomic resolution model of the GEF-bound form of Ras as defined by X-ray crystallography. Representations are identical to those above; however, the GEF secondary structure is illustrated as bright blue ribbons layered beneath a molecular surface colored according to electrostatic potential: Red depicts a negative charge, and blue depicts a positive charge. A comparison of the 90° rotation of Ras-GDP and the boxed presentation of Ras–GEF clearly illustrates the conformational similarities in the SW1 orientation. In the GEF-bound form of Ras, Y32 participates in a hydrogen bond network between GEF-K939, GEF-N944, and a water molecule. Following nucleotide exchange, the side chain of Y32 does not make any intramolecular hydrogen bonds and is exposed to solvent. Shown here is the atomic resolution model of Ras–GTP as defined by NMR spectroscopy. This conformation of Ras likely could tolerate phosphorylation at Y32; however, this SW1 conformation is not suitable for Src kinase recognition. Next, the atomic resolution structure of Ras bound to the RBD of RAF illustrates the typical Ras-binding strategy for effector proteins. Heterodimerization pushes Y32 of Ras into a hydrogen bond network with the γ-phosphate of GTP and places Y32 in a more favorable position for Src binding and phosphorylation. RAF is displaced from Ras following kinase-induced activation of RAF-mediated signaling or after Y32 phosphorylation. Y32 phosphorylation generates an electrostatic repulsion against the negatively charged D38 and D57 within the nucleotide-binding groove, altering SW1 conformation so that it no longer accommodates RAF binding. The Ras–GAP heterodimer then forms upon RAF displacement. The Ras secondary structure is depicted as green ribbons, and the GAP model is represented as a molecular surface, colored by electrostatic potential: Red depicts a negative charge, and blue depicts a positive charge. The Ras-Y32–binding pocket within GAP is flanked by positively charged GAP side chains. Y32 phosphorylation increases the rate of γ-phosphate hydrolysis by accentuating the affinity of Ras-Y32 for the pocket. Following hydrolysis and GAP displacement, phosphatase-mediated Y32 dephosphorylation occurs, and Ras adopts the GDP-bound conformation observed in the crystallographic structure. Protein Data Bank ID codes are Ras–GDP: 4Q21; Ras–GEF, 1BKD; Ras–GTP, 3TGP; Ras–RAF, 4G0N; and Ras–GAP, 1WQ1. All graphics were generated using University of California San Francisco Chimera software and Adobe Illustrator CS6.
In the canonical Ras–GAP paradigm, GAP recruitment through the Ras switch I region leads to subsequent activation and increased γ-phosphate hydrolysis. Analysis of the Ras–GAP cocrystal structure indicates that Y32 docks into an amphipathic binding pocket on the surface of the GAP protein. Intriguingly, the pocket is flanked by three positively charged residues (GAP-R789, -R894, and -K949) that allow access to a mildly hydrophobic cavity. The cavity accommodates the aromatic side chain of Ras-Y32 by virtue of a Gly residue at GAP-G898. This cavity extends further and finally closes with the positively charged GAP-R903 at the cavity’s nadir (Fig. 7B). We postulate that GAP-R903 could increase the specificity of this binding pocket for phosphorylated Ras-Y32. The flanking Args likely attract the negatively charged switch I region of Ras to the GAP-binding pocket, locking Ras to GAP through electrostatic interactions between phosphorylated Ras-Y32 and GAP-R903. Consistent with this notion, we show here that Y32 phosphorylation increases not only intrinsic but also GAP-stimulated Ras GTPase activity.
We show here that oncogenic Ras mutants exhibit lower levels of tyrosyl phosphorylation, increased association with Raf, and decreased GTPase activity as compared with wild-type Ras. A previous study has shown that the aberrant positioning of Y32 in oncogenic Ras prohibited efficient GTP hydrolysis (54). The tight association to Raf with altered Y32 positioning may reduce the accessibility of Y32 to Src, explaining the observed attenuated Y32 phosphorylation in oncogenic Ras mutants. However, tyrosyl phosphorylation of oncogenic Ras can be forced by ectopic expression of c-Src, which attenuates the binding of Raf and increases GTP hydrolysis. These results support previous findings showing that the GTPase switch of mutant H-Ras is not irreversibly damaged (55). Intriguingly, we show that the lack of sensitivity toward EGF-induced tyrosyl phosphorylation of Ras in the human astrocytoma cell line U87, in which Ras is predominantly in the active GTP-bound form that is associated with Raf (26), can be rescued with the use of a general phosphotyrosine phosphatase inhibitor. These results suggest that an as yet unknown phosphatase alongside Src kinase regulates the Ras phosphorylation status to control Ras GTPase activity. Perhaps consistent with this notion, the Ras–GEF cocrystal structure indicates that Y32 is buried and participates in a hydrogen bond network with GEF-K939, GEF-N944, and a water molecule (Fig. 7B). This network promotes the switch I region of Ras to adopt a conformation amenable to GEF interaction. Notably, the structural information suggests that a dephosphorylated, but not phosphorylated, Y32 adopts the Ras–GEF crystallographic conformation, allowing GEF binding and the canonical Ras cycle to continue.
Findings from this study expand on the current understanding of the well-established codependent relationship between Src and Ras. We show that Src inactivates Ras via the phosphorylation of a conserved Y32 in the switch I region and this inactivation is associated with Raf displacement and increased GAP binding and GTP hydrolysis. These results support the notion that the phosphorylated Y32 not only would inhibit the reassociation of effector proteins, such as Raf and RalGDS, but also would augment the Ras–GAP association to accelerate GTP hydrolysis, whereas dephosphorylated Y32 would be more favorable for the formation of the Ras–GEF complex to renew the Ras GTPase cycle (Fig. 7). Results from our study also caution the use of pharmacological tyrosine kinase inhibitors that target Src in an effort to curtail the activity of Raf, because, although such inhibitors would attenuate Raf/MEK/ERK signaling, they concomitantly would promote the accumulation of active Ras–GTP–Raf and possibly other Ras–GTP–effector complexes through the suppression of Ras-Y32 phosphorylation.
Methods
Cells.
HEK293 and TF-1 cells, MEFs and MEF-SYF(−/−) cells, and U87 cells were obtained from the American Type Culture Collection. NHA and RasB8 P0 and P3 cells were generated as previously demonstrated (24, 25). HEK293, MEF, and MEF-SYF(−/−) cells and U87, NHA, and RasB8 P0 and P3 cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated FBS (Wisent) at 37 °C in a humidified 5% CO2 atmosphere. TF-1 and BaF3-Flag–N-Ras(WT or 12D) cells were maintained similarly in RPMI-1640 medium(Wisent) supplemented with 10% (vol/vol) FBS and 2 ng/mL GM-CSF or 10 ng/mL IL-3 (Invitrogen), respectively.
Generation of BaF3-Flag–N-Ras Cell Lines.
BaF3-Flag–N-Ras(WT or 12D) cells were generated using the Lonza Nucleofector kit for BaF3 cells alongside empty pcDNA3-Flag. Cell populations with stable Flag–N-Ras(WT or 12D) expression were selected with G418, sorted on a FACSAria cell sorter (BD Biosciences), and further confirmed by Western blot analysis. These stable cell lines were maintained in RPMI-1640 medium with 10% (vol/vol) FBS, 10 ng/mL IL-3, and G418.
Plasmids.
Plasmids encoding human pCGN-HA–N-Ras(WT, 12D, 17N or 12D, 186S), pCGN-HA–K-Ras(WT or 12V, 188L), and pCMV5-c-Src(WT or K295R, Y527F) were purchased from Addgene. Flag–N-Ras constructs were generated using standard protocols. Using the above plasmids as templates, Flag-N-Ras constructs were subcloned into a pcDNA3 backbone. Plasmids were verified by direct DNA sequencing.
Immunoprecipitation and Immunoblotting.
Immunoprecipitation and Western blotting were performed as described previously (56). Cells were harvested in EBC lysis buffer [50 mM Tris (pH 8), 120 mM NaCl, 0.5% Nonidet P-40] and supplemented with protease inhibitors (Roche). Lysates were immunoprecipitated using the indicated antibodies along with protein A-Sepharose (Repligen). Bound proteins were washed five times in NETN buffer [20 mM Tris (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40], eluted by boiling in sample buffer, and resolved by SDS/PAGE. Proteins were electrotransferred onto PVDF membranes (Bio-Rad), blocked, and probed with the antibodies indicated on the figures.
Mass Spectrometry Sample Preparation.
In vitro phosphorylated samples were concentrated and buffer exchanged to 50 mM ammonium bicarbonate (pH 8.2), using a 10-kDa cutoff Microcon centrifugal device (Millipore). Samples then were reduced and alkylated (as in ref. 57), digested overnight with 1 μg trypsin treated with l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK) (Promega), and lyophilized. The resulting peptides were identified using nLC-ESI-MS/MS as described below.
Purification of Ectopic Ras.
HEK293 cells were transfected with Flag–N-Ras(WT) and c-Src. Flag–N-Ras was affinity purified using the Flag M purification kit from Sigma according to the manufacturer’s recommendations. Eluted polypeptides were lyophilized and digested overnight in 50 mM ammonium bicarbonate (pH 8.2) containing 2 μg TPCK trypsin (Promega). The resulting peptides were lyophilized and reconstituted in 1 mL IAP buffer [50 mM 3-(N-morpholino)propanesulfonic acid (Mops) (pH 7.2), 10 mM sodium phosphate, 50 mM NaCl], to which 20 μL of prewashed 4G10 platinum anti-phosphotyrosine agarose beads (Millipore) were added. Samples were incubated end-over-end at 4 °C for 30 min; then beads were washed three times with IAP buffer, and peptides were eluted with 2× 100 μL 0.15% TFA (HPLC grade; Sigma). Eluted peptides were lyophilized and subjected to nLC-ESI-MS/MS.
Mass Spectrometry.
Samples were resuspended in 0.1% (MS grade) formic acid (Sigma). Analytical columns (75-µm i.d.) and 100-µm precolumns were prepared in-house from silica capillary tubing (InnovaQuartz) and were packed with 3 µm of 100-Å C18-coated silica particles (Michrom). Analytical columns were fitted with metal emitters (Thermo Proxeon) using zero dead-volume connections. Peptides were subjected to nLC-ESI-MS/MS, using a 90-min reversed-phase buffer gradient (10–30% acetonitrile, 0.1% formic acid) running at 250 nL/min on a Proxeon EASY-nLC pump in-line with a hybrid linear quadrupole ion trap (Velos LTQ) Orbitrap mass spectrometer (Thermo Fisher Scientific). A parent ion scan was performed in the Orbitrap, using a resolving power of 60,000. Simultaneously, up to the 20 most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation) using standard collision-induced dissociation (CID) fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated so that MS/MS of the same m/z (within a 10-ppm window, exclusion list size 500) detected two times within 15 s were excluded from analysis for 30 s. Thermo.RAW files were searched with the Sequest (58), X!Tandem (59), and PeaksDB (60) algorithms, using a parent mass window of 10 ppm and 0.4-Da fragment mass window, against the human_uniprot_Jan_09_2013 fasta database. Up to two missed cleavages were allowed, and oxidation of Met, deamination of Glu and Asp, and phosphorylation at Ser, Thr, and Tyr were set as variable modifications. Alkylated Cys was set as a fixed modification for the in vitro reactions.
Statistical Analyses.
An unpaired two-tailed Student t test was used for comparisons of treatment groups and cell types. All statistical analyses were performed using GraphPad PRISM 5.0 software. Statistical significance was achieved at the confidence limit indicated.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and the Canadian Cancer Society.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406559111/-/DCSupplemental.
References
- 1.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 2.Kirsten WH, Mayer LA. Morphologic responses to a murine erythroblastosis virus. J Natl Cancer Inst. 1967;39(2):311–335. [PubMed] [Google Scholar]
- 3.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1(1):2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 6.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118(Pt 5):843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 7.Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17(11 Reviews):1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 8.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 9.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 10.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Scheffzek K, et al. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 12.Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- 13.Edin ML, Juliano RL. Raf-1 serine 338 phosphorylation plays a key role in adhesion-dependent activation of extracellular signal-regulated kinase by epidermal growth factor. Mol Cell Biol. 2005;25(11):4466–4475. doi: 10.1128/MCB.25.11.4466-4475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milburn MV, et al. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 15.Avruch J, et al. Ras activation of the Raf kinase: Tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 16.Hibino K, Shibata T, Yanagida T, Sako Y. Activation kinetics of RAF protein in the ternary complex of RAF, RAS-GTP, and kinase on the plasma membrane of living cells: Single-molecule imaging analysis. J Biol Chem. 2011;286(42):36460–36468. doi: 10.1074/jbc.M111.262675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14(13):3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grewal T, Koese M, Tebar F, Enrich C. Differential Regulation of RasGAPs in Cancer. Genes Cancer. 2011;2(3):288–297. doi: 10.1177/1947601911407330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan PC, Chen HC. p120RasGAP-mediated activation of c-Src is critical for oncogenic Ras to induce tumor invasion. Cancer Res. 2012;72(9):2405–2415. doi: 10.1158/0008-5472.CAN-11-3078. [DOI] [PubMed] [Google Scholar]
- 20.Shields DJ, et al. Oncogenic Ras/Src cooperativity in pancreatic neoplasia. Oncogene. 2011;30(18):2123–2134. doi: 10.1038/onc.2010.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Geer P, Wiley S, Gish GD, Pawson T. The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions. Curr Biol. 1996;6(11):1435–1444. doi: 10.1016/s0960-9822(96)00748-8. [DOI] [PubMed] [Google Scholar]
- 22.Tokumitsu Y, Nakano S, Ueno H, Niho Y. Suppression of malignant growth potentials of v-Src-transformed human gallbladder epithelial cells by adenovirus-mediated dominant negative H-Ras. J Cell Physiol. 2000;183(2):221–227. doi: 10.1002/(SICI)1097-4652(200005)183:2<221::AID-JCP8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Williams NG, Roberts TM, Li P. Both p21ras and pp60v-src are required, but neither alone is sufficient, to activate the Raf-1 kinase. Proc Natl Acad Sci USA. 1992;89(7):2922–2926. doi: 10.1073/pnas.89.7.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding H, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61(9):3826–3836. [PubMed] [Google Scholar]
- 25.Kamnasaran D, Qian B, Hawkins C, Stanford WL, Guha A. GATA6 is an astrocytoma tumor suppressor gene identified by gene trapping of mouse glioma model. Proc Natl Acad Sci USA. 2007;104(19):8053–8058. doi: 10.1073/pnas.0611669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 27.Uezu A, et al. Modified SH2 domain to phototrap and identify phosphotyrosine proteins from subcellular sites within cells. Proc Natl Acad Sci USA. 2012;109(43):E2929–E2938. doi: 10.1073/pnas.1207358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann C. Ras-effector interactions: After one decade. Curr Opin Struct Biol. 2003;13(1):122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- 30.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 31.Wittinghofer A, Pai EF. The structure of Ras protein: A model for a universal molecular switch. Trends Biochem Sci. 1991;16(10):382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 32.Wittinghofer A, Nassar N. How Ras-related proteins talk to their effectors. Trends Biochem Sci. 1996;21(12):488–491. doi: 10.1016/s0968-0004(96)10064-5. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Karplus M. Molecular switch in signal transduction: Reaction paths of the conformational changes in ras p21. Proc Natl Acad Sci USA. 1997;94(22):11905–11910. doi: 10.1073/pnas.94.22.11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geyer M, et al. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35(32):10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- 35.Nassar N, et al. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375(6532):554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 36.Baussand J, Kleinjung J. Specific Conformational States of Ras GTPase upon Effector Binding. J Chem Theory Comput. 2013;9(1):738–749. doi: 10.1021/ct3007265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita-Yamaguchi Y, et al. In vitro tyrosine phosphorylation studies on RAS proteins and calmodulin suggest that polylysine-like basic peptides or domains may be involved in interactions between insulin receptor kinase and its substrate. Proc Natl Acad Sci USA. 1989;86(19):7306–7310. doi: 10.1073/pnas.86.19.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding J, Soule G, Overmeyer JH, Maltese WA. Tyrosine phosphorylation of the Rab24 GTPase in cultured mammalian cells. Biochem Biophys Res Commun. 2003;312(3):670–675. doi: 10.1016/j.bbrc.2003.10.171. [DOI] [PubMed] [Google Scholar]
- 39.Brucker S, Gerwert K, Kötting C. Tyr39 of ran preserves the Ran.GTP gradient by inhibiting GTP hydrolysis. J Mol Biol. 2010;401(1):1–6. doi: 10.1016/j.jmb.2010.05.068. [DOI] [PubMed] [Google Scholar]
- 40.Tu S, Wu WJ, Wang J, Cerione RA. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J Biol Chem. 2003;278(49):49293–49300. doi: 10.1074/jbc.M307021200. [DOI] [PubMed] [Google Scholar]
- 41.Zou JX, Liu Y, Pasquale EB, Ruoslahti E. Activated SRC oncogene phosphorylates R-ras and suppresses integrin activity. J Biol Chem. 2002;277(3):1824–1827. doi: 10.1074/jbc.M103133200. [DOI] [PubMed] [Google Scholar]
- 42.Hausdorff WP, et al. Tyrosine phosphorylation of G protein alpha subunits by pp60c-src. Proc Natl Acad Sci USA. 1992;89(13):5720–5724. doi: 10.1073/pnas.89.13.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moodie SA, et al. Different structural requirements within the switch II region of the Ras protein for interactions with specific downstream targets. Oncogene. 1995;11(3):447–454. [PubMed] [Google Scholar]
- 44.Minato T, et al. Quantitative analysis of mutually competitive binding of human Raf-1 and yeast adenylyl cyclase to Ras proteins. J Biol Chem. 1994;269(33):20845–20851. [PubMed] [Google Scholar]
- 45.Vogel US, et al. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- 46.Martin GA, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 47.Fujioka A, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281(13):8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 48.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103(6):931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 49.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270(7):2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 50.Filchtinski D, et al. What makes Ras an efficient molecular switch: A computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf. J Mol Biol. 2010;399(3):422–435. doi: 10.1016/j.jmb.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 51.Sveshnikova N, Soll J, Schleiff E. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc Natl Acad Sci USA. 2000;97(9):4973–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jelic M, et al. The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol Chem. 2002;383(12):1875–1883. doi: 10.1515/BC.2002.211. [DOI] [PubMed] [Google Scholar]
- 53.Gao C. Impact of Mutations on K-Ras-p120GAP Interaction. Computational Molecular Bioscience. 2013;3:8. [Google Scholar]
- 54.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmadian MR, et al. Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc Natl Acad Sci USA. 1999;96(12):7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2(7):423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen ML, et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods. 2008;5(6):459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 58.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 59.Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, et al. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics. 2012;11:M111 010587. doi: 10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.