Significance
Neutrophils are among the first immune cells to migrate to the site of infection and clear invading bacteria. They store large amounts of neutrophil serine proteases (NSPs) that play key roles in immune defense. Unfortunately, NSPs also contribute to tissue destruction in a variety of inflammatory disorders. In this study we discover that the pathogenic bacterium Staphylococcus aureus secretes a family of highly potent and specific NSP inhibitors that promote the pathogenicity of this bacterium in vivo. From crystallography experiments, we conclude that these proteins constitute a unique class of NSP inhibitors, which can be used to design novel treatment strategies against excessive NSP activity. Furthermore, this study significantly increases our understanding of the complex nature of S. aureus infections.
Keywords: immune evasion, bacteria, phagocytes
Abstract
Neutrophils are indispensable for clearing infections with the prominent human pathogen Staphylococcus aureus. Here, we report that S. aureus secretes a family of proteins that potently inhibits the activity of neutrophil serine proteases (NSPs): neutrophil elastase (NE), proteinase 3, and cathepsin G. The NSPs, but not related serine proteases, are specifically blocked by the extracellular adherence protein (Eap) and the functionally orphan Eap homologs EapH1 and EapH2, with inhibitory-constant values in the low-nanomolar range. Eap proteins are together essential for NSP inhibition by S. aureus in vitro and promote staphylococcal infection in vivo. The crystal structure of the EapH1/NE complex showed that Eap molecules constitute a unique class of noncovalent protease inhibitors that occlude the catalytic cleft of NSPs. These findings increase our insights into the complex pathogenesis of S. aureus infections and create opportunities to design novel treatment strategies for inflammatory conditions related to excessive NSP activity.
Infections with the human pathogen Staphylococcus aureus constitute a major risk to human health. Although this bacterium harmlessly colonizes more than 30% of the population via the nose or skin, it causes severe morbidity and mortality upon invasion of deeper tissues (1). To avert these serious infections, neutrophils play an indispensable role (2). Neutrophil serine proteases (NSPs), including neutrophil elastase (NE), proteinase 3 (PR3), and cathepsin G (CG), are important for various neutrophil functions. Active NSPs are stored within the azurophilic granules (3), but upon neutrophil activation, they either enter the nucleus to regulate extracellular trap (NET) formation (4) or they are released into the extracellular milieu to kill certain bacteria (5), cleave bacterial virulence factors (5, 6), or regulate immune responses by cleaving chemokines and receptors (7). Recently, a fourth neutrophil serine protease, denoted NSP4, was identified (8).
Given the central role of NSPs in neutrophil function, we wondered whether S. aureus had evolved mechanisms to cope with NSPs. In this study, we discover that S. aureus secretes a family of proteins that specifically and potently block NSPs: extracellular adherence protein (Eap) and the hitherto functional orphans Eap-homologue (EapH) 1 and 2. Structural studies presented here show that Eap molecules represent a unique class of noncovalent NSP inhibitors that is distinct from the well-known chelonianin class of inhibitors. These mechanistic insights can initiate development of novel, broad-range NSP inhibitors to be used in various inflammatory conditions. Furthermore, these insights increase our understanding of the pathogenicity of S. aureus and underline the exceptional capability of this pathogen to adapt to its host by modulating the immune response.
Results
Extracellular Adherence Proteins of S. aureus Inhibit NE.
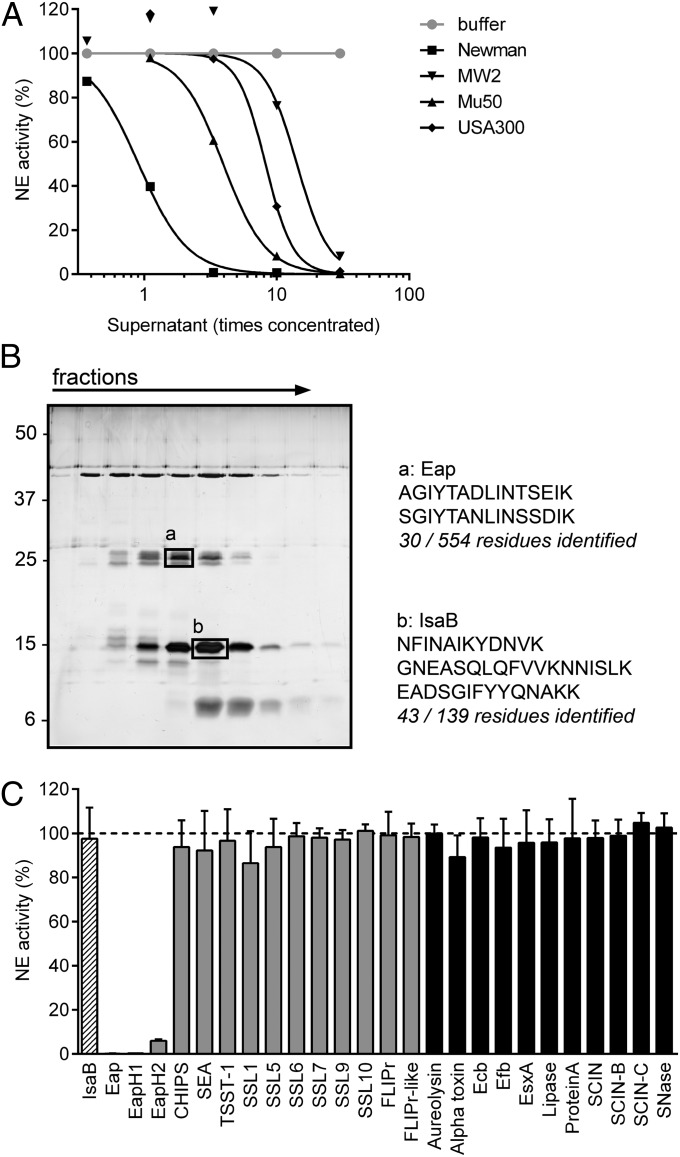
To investigate whether S. aureus secretes inhibitors of NSPs, we incubated NE with concentrated culture supernatants of different S. aureus strains and quantified residual NE activity toward a fluorescent peptide substrate. Indeed, we found that NE was inhibited by supernatants of all tested S. aureus strains (Fig. 1A). Fractionation of the supernatant of S. aureus Newman by ion-exchange and size-exclusion chromatography yielded two protein bands that corresponded with the NE inhibitory activity. These bands were identified by mass spectrometry as Eap and immunodominant surface antigen B (IsaB) (Fig. 1B). Further analysis revealed that Eap is the NE inhibitor, because NE activity was not affected by the presence of recombinant IsaB, but fully blocked by the presence of recombinant Eap (Fig. 1C). Eap is a 50- to 70-kDa protein that consists of multiple (most often four or five) repetitive EAP domains (11 kDa). The short linkers between EAP domains are susceptible to proteolysis (9), which likely explains why the band identified as Eap was only approximately 25 kDa (Fig. 1B).
Fig. 1.
Extracellular adherence protein (Eap) family of S. aureus inhibits NE activity. (A) Residual activity of 60 nM NE upon incubation with culture supernatants of multiple S. aureus strains. (B) Silver staining analysis of the fractions after gel filtration (Left). The boxes mark the protein bands analyzed by mass spectrometry and the numbers indicate the sizes of reference proteins (kilodaltons). The identified peptides are depicted next to the gel (Right). (C) Residual activity of 60 nM NE upon incubation with 100 nM of the mass-spectrometry hits Eap and IsaB (hatched bars) and other secreted proteins of S. aureus. Proteins containing a similar beta-grasp-type fold as the Eap proteins are depicted in gray, and with an unrelated structure in black. CHIPS, chemotaxis inhibitory protein of S. aureus; Ecb, extracellular complement binding protein; Efb, extracellular fibrinogen binding protein; EsxA, ESAT-6 secretion system extracellular protein A; FLIPr, formyl peptide receptor-like-1 inhibitory protein; SCIN, staphylococcal complement inhibitor; SEA, staphylococcal enterotoxin A; SNase, S. aureus nuclease; SSLs, staphylococcal superantigen-like proteins; TSST-1, toxic shock syndrome toxin-1. Data are representative of two independent experiments (A and B) or represent the mean (± SD) of three independent experiments (C). See also Fig. S1.
Eap has previously been reported to mediate bacterial agglutination, tissue adherence, and to block neutrophil migration. Although all of these functions require multiple EAP domains linked in succession (10), we found that NE inhibition is mediated by individual EAP domains (Fig. S1). Each EAP domain is characterized by a beta-grasp fold wherein an alpha-helix is positioned diagonally across a five-stranded, mixed beta-sheet (11). This fold is also found in the two S. aureus proteins that are homologous to Eap but do not share the above described functions: EapH1 (12 kDa) and EapH2 (13 kDa) (11). Likewise, we found that EapH1 and EapH2 also inhibit NE (Fig. 1C). NE inhibition is specific for the Eap protein family, because all other tested proteins of S. aureus could not inhibit NE (Fig. 1C), including molecules characterized by a similar beta-grasp–type fold as the Eap proteins (Fig. 1C, gray bars) (11, 12).
Eap Proteins Specifically Inhibit Neutrophil Serine Proteases.
NE belongs to the chymotrypsin family of serine proteases, and its amino acid sequence is most similar to that of PR3 and CG (55% and 37% amino acid identity, respectively). More distantly related chymotrypsin-like proteases include plasmin, plasma kallikrein, and thrombin. Of these six proteases, only the three NSPs appeared to be dose-dependently inhibited by the Eap proteins (Fig. 2 A–F).
Fig. 2.
Eap proteins specifically inhibit NSPs. (A) Residual NE activity toward a fluorescent-peptide substrate after incubation of 5 nM NE with variable concentrations of the three Eap proteins or CHIPS. (B–F) Residual activity of 10 nM PR3 (B), 15 nM CG (C), 10 nM plasmin (D), 1 nM plasma kallikrein (E), and 1 nM thrombin (F), determined as in A, with substrates specific for each protease. (G) Ki values (nanomolars) according to the competitive inhibition model. An accurate Ki could not be determined for EapH2/CG. ND, not determined. Data represent the mean (± SD) of three independent experiments (A–F) or represent the mean (95% CI) of at least three independent experiments (G).
Next we determined the inhibitory constant (Ki) values of each Eap protein versus the three NSPs and also measured the Ki values of the endogenous human NE/CG inhibitor SLPI (secreted leukocyte protease inhibitor) as a control. The Ki values observed for all Eap/NSP combinations were in the low-nanomolar range, which is consistent with a very potent inhibition (Fig. 2G). They are also within the same order of magnitude as the Ki of SLPI for both NE and CG. Because previous work has reported a lower Ki for SLPI/NE and SLPI/CG (0.3 nM and 10 nM, respectively) (13), our particular assay system may have even underestimated the inhibitory capacity of the Eap proteins. Importantly, the experimentally determined Ki values are all lower than the endogenous expression levels of the Eap proteins by S. aureus in culture (∼10 μg/mL or 200 nM) (14), indicating that Eap inhibition of NSPs is physiologically relevant.
Eap Proteins Are Essential for NSP Inhibition and Promote Staphylococcal Infection.
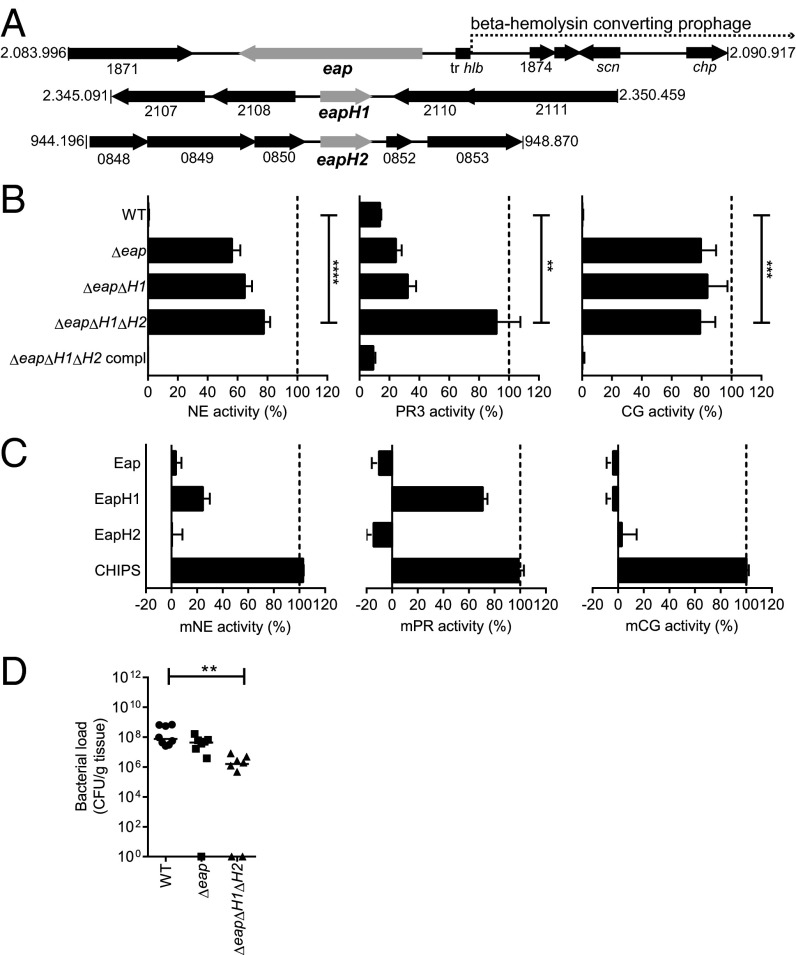
The genes for the Eap proteins lie interspersed throughout the genome, and at least two of three are present in all sequenced S. aureus strains. The eap gene is located upstream, and therefore outside, of the beta-hemolysin–converting prophage (phiNM3) that contains other immune-evasion proteins like staphylococcal complement inhibitor (SCIN) (scn) and chemotaxis inhibitory protein of S. aureus (CHIPS) (chp) (15, 16) (Fig. 3A). Neither eapH1 nor eapH2 lie in close proximity of phage-associated genes. Using sequential gene deletions by homologous recombination, we constructed a panel of three isogenic eap mutants in S. aureus strain Newman: Δeap, ΔeapΔH1, and ΔeapΔH1ΔH2 (eap-triple mutant). As a control, the eap-triple mutant was complemented with the individual genes integrated into their original genomic location (ΔeapΔH1ΔH2 compl.). All isogenic strains showed comparable growth in vitro. When incubated with the individual NSPs, stationary-phase supernatant of the WT strain could fully inhibit all three proteases, but supernatant of the eap-triple mutant had almost entirely lost this capacity (Fig. 3B). The other mutants showed that Eap is essential for NE and CG inhibition, but that all Eap proteins together are required for inhibition of PR3. Supernatant of the complemented strain showed restored NSP inhibition to levels equivalent to WT supernatant (Fig. 3B). Although S. aureus was found to be resistant to direct killing by NE and CG in vitro (17, 18), we examined whether the absence of eap genes might make S. aureus more prone to direct killing by neutrophils in vitro. Although there was a tendency toward better killing of the eap-triple mutant compared with the WT strain, this difference was not significant (Fig. S2).
Fig. 3.
Eap proteins are essential for NSP inhibition and promote staphylococcal infection. (A) Schematic overview of the genomic regions of S. aureus Newman in which eap (NWMN_1872), eapH1 (NWMN_2109), and eapH2 (NWMN_0851) are located. The location of the beta-hemolysin–converting phage is also indicated. (B) Residual activity of 5 nM NE (Left), 10 nM PR3 (Center), and 15 nM CG (Right) after incubation with culture supernatants of the isogenic mutants Δeap, ΔeapΔH1, ΔeapΔH1ΔH2 or the complemented strain ΔeapΔH1ΔH2 compl. Statistical significance was addressed with an unpaired two-tailed t test. (C) Residual activity of mouse NSPs after incubation with buffer, Eap proteins or CHIPS. (D) Murine i.v. infection model in which mice were infected retro-orbitally with 1 × 107 S. aureus Newman WT or isogenic eap mutants. Bacterial loads recovered from liver tissues 4 d after infection (n = 8). Statistical significance was addressed with a nonparametric ANOVA test. Data represent the mean (± SD) of three independent experiments (B and C) or horizontal bars indicate the median of all observations (D). See also Fig. S2.
To study the role of the Eap proteins in vivo, we compared the pathogenicity of the eap-mutant strains in a murine, liver-abscess model (19), because the bacterial burden in the liver is known to be a reliable indicator of staphylococcal virulence (20, 21). First, we confirmed that the purified Eap proteins also block murine NSPs in vitro (Fig. 3C). Then, we injected 1 × 107 bacteria retro-orbitally and determined the bacterial loads in the liver tissue after 4 d (Fig. 3D). The bacterial loads in the WT and Δeap-infected animals did not differ significantly. However, the bacterial load in the eap-triple mutant-infected mice was significantly lower than in WT-infected animals, strongly suggesting that all Eap proteins together promote S. aureus virulence in vivo.
Eap Proteins Occlude the Catalytic Site of NSPs.
EAP domains inhibit NSPs via a low nanomolar-affinity interaction, as determined by isothermal titration calorimetry (ITC) analysis of EapH1 binding to NE (Fig. S3). This complex was studied in greater detail by determining its cocrystal structure, which was refined to 1.85 Å limiting resolution (Fig. 4A and Table S1). The structure of this inhibitory complex revealed that bound EapH1 lies directly across the Asp117-His70-Ser202 active-site triad of NE (Fig. 4B). Given the size of EapH1 and the highly complementary nature of the EapH1/NE interface (surface complementarity value of 0.77; ref. 22), such an arrangement would be expected to completely impede any protein or peptide-based substrates from accessing the NE active site. Importantly, this interaction is noncovalent because contiguous electron density is not visible between the catalytic Ser202 side chain of NE and any of the EapH1 residues in its vicinity (Fig. 4C). The absence of a covalent bond strongly suggests that EapH1, and likely all EAP domains, are mechanistically distinct from a majority of physiological NSP inhibitors (e.g., alpha-1-antitrypsin), which are conventional SERPINs (7).
Fig. 4.
Eap proteins inhibit by noncovalently occluding the catalytic site of NSPs. (A) Ribbon diagram of the final refined model for the EapH1/NE complex at 1.85 Å resolution. NE (black), with the position of its Ser-His-Asp catalytic triad (orange sticks), in complex with EapH1 (cyan), with the position of its NE-contacting residues as either magenta sticks (for site 1) or purple sticks (for site 2). Cyan dotted line represents two residues, Pro67 and Glu68, that could not be accurately modeled due to poor map quality. (B) Comparison of the EapH1/NE complex with NE where both components are shown as molecular surfaces. NE is shown in black with the location of its catalytic triad in orange, whereas EapH1 is colored as in A, but depicted as a molecular surface. (C) 2Fo-Fc electron density map in the region of the NE catalytic site, contoured at 1.5 σ. The locations of the catalytic residue, Ser202, along with His60 (a component of the charge-relay system) from NE are marked, as is that of Leu59 from EapH1. Proteins are colored as in A. (D) EapH1 colored as in A, but depicted as a molecular surface (Left) or as a cartoon (Right, for reference). The image has been rotated clockwise in the plane of the page to emphasize the contiguous nature of the NE contact surface, comprised of both site 1 and site 2. (E) Comparison of the loop region comprising EapH1 site 1 in both the free (Left) and NE-bound (Center) states. The image at Right shows a superposition of this region, in both free and bound states. The locations of key residues are indicated. (F) Comparison of the EapH1/NE complex (Left, for reference) with the SLPI/NE complex (Right). Proteins are shown as ribbon diagrams with NE in black, EapH1 in cyan, and SLPI in gold. (G) Close-up view of the superposed NE catalytic site (orange sticks) in the presence of inhibitors EapH1 (cyan) and SLPI (gold). PDB ID code 2Z7F was used to render images of the NE/SLPI structure in F and G. See also Fig. S3 and Table S1.
The EapH1/NE interface buries 830 Å2 of surface area and involves 22 of the 97 EapH1 residues. A large majority of these interfacial residues (16 of 22) are found in two distinct regions of the EapH1 sequence, i.e., residues Val53-Tyr63 (hereafter denoted “site 1”) and residues Ala86-Gly90 (hereafter denoted “site 2”), and they reside in close proximity to one another in the EapH1 tertiary structure (Fig. 4D). Although site 1 and site 2 appear to work in concert to form an unbroken, pentagon-shaped surface of EapH1 that interacts with NE, the contacts formed by site 1 (628 Å2) are significantly more extensive than those formed by site 2 (176 Å2). Furthermore, the loop that comprises site 1 appears to undergo a substantial conformational rearrangement upon binding (Fig. 4E), because residues Ala58-Arg61 rotate nearly 180° in the plane of the prominent beta-sheet compared with the unbound EapH1 structure (11). Because this four-residue stretch contributes two of the three hydrogen bonds formed between site 1 and NE and accounts for almost 70% of the site-1 buried surface area (427 Å2 of 628 Å2), EapH1 most likely depends on such conformational change to tightly bind and inhibit NE.
A critical structure/function role for loop regions has also been suggested for other high-affinity protease inhibitors, most notably those of the chelonianin family that includes the endogenous, noncovalent NE inhibitors elafin (23) and SLPI (24). Besides the insertion of an inhibitory loop region into the NE active site, however, the SLPI/NE complex has little in common with the EapH1/NE complex (Fig. 4F). Furthermore, SLPI and EapH1 share no detectable sequence similarity, nor do the conformations of either inhibitor-derived loop in their NE-bound state share extensive structural identity with one another (Fig. 4G). Based on these sequence and structural differences, we propose that the Eap proteins constitute a unique class of NSP inhibitors, distinct from that of the well-known SERPINs or chelonianin class of inhibitors.
Discussion
Here we report that the human pathogen S. aureus secretes three proteins that form a unique group of NSP inhibitors. These Eap proteins are potent, highly specific, and promote S. aureus pathogenesis. From previous studies it is clear that all three genes are expressed during S. aureus infections in vivo, both in mice and humans (25, 26). The fact that S. aureus evolved three specific NSP inhibitors strongly indicates that NSPs are critical to host defense against this bacterium. However, because of the manifold physiological substrates of NSPs, the exact function of NSPs in host clearance of S. aureus is unclear. It is likely that the interpretation of previous animal models trying to address this question (5, 18, 27) has been confounded by the endogenous production of Eap proteins. In the future, these conceptual limitations can be overcome by using the bacterial deletion mutants that are presented here.
A key teleological issue remains as to why S. aureus inhibits NSPs by three distinct proteins. We speculate that the eap genes are most likely differentially expressed during an infection, creating a broader window of opportunity to block NSPs both at distinct sites of the body and times of infection. In fact, expression of both eap and eapH1 has been reported to be up-regulated in the presence of azurophilic-granule contents, whereas eapH2 expression was not affected (28). In many ways, this redundancy shows striking parallels with the diverse array of complement-evasion proteins produced by the same organism (29). The expression of multiple proteins with some level of functional redundancy may be a general principle that underlies S. aureus innate immune evasion, regardless of the specific host process that is targeted.
From a broader biological perspective, our work suggests that NSP inhibition by staphylococcal EAP domains has arisen through a distinct evolutionary trajectory from either the SERPIN or chelonianin-class inhibitors found in its human host. In contrast to the SERPINs, EAP domains do not form covalent complexes with their target(s), they do not appear to undergo large conformational changes at sites distal to their inhibitory loop, and they are relatively small (e.g., EAP domains are about one-fourth the molecular mass of the 44 kDa alpha-1-antitrypsin). Furthermore, although EAP domains are more similar in overall size to active fragments of the chelonianin-class inhibitors SLPI and elafin (∼6 kDa), their fold is entirely different and, importantly, the S. aureus molecules do not require disulfide bonds to constrain their NSP-inhibitory loop into an active conformation.
Despite the many naturally occurring NSP inhibitors found in the human body, NSPs are known to play a significant role in tissue destruction in a variety of inflammatory disorders like cystic fibrosis, chronic obstructive pulmonary disease, emphysema, and rheumatoid arthritis (7). As a consequence, targeted NSP inhibition has been considered as a plausible treatment in these diseases. However, although substantial work has been done in this area, to date no drug has been registered that targets all NSPs (7). Here we show that the Eap proteins have a high inhibitory potency and that, whereas many endogenous NSP inhibitors inhibit only two of three NSPs, they inhibit all three NSPs. Therefore, Eap proteins might serve as a template for developing a novel class of synthetic NSP inhibitors. Although drug development is clearly a long-term goal, the results we present here have already increased our understanding of the complex nature of S. aureus infections by identifying an effective mechanism through which this devastating pathogen defends itself against human innate immunity.
Materials and Methods
Screening for NE Inhibition.
S. aureus strains Newman, MW2, USA300, and Mu50 were cultured for 8 h in Icove's Modified Dulbecco's Medium (Invitrogen). Culture supernatants were concentrated (100×) on a 3-kDa Amicon-Ultra spin column (Merck Millipore). Supernatants were incubated 1:1 with 120 nM NE (Elastin Products Company) in 1 mg/mL BSA-PBS for 60 min at 37 °C. Residual NE activity was determined by the conversion of 230 μM substrate (AAPV-AMC; Calbiochem) for 60 min, and fluorescence was measured in a Flexstation fluorometer (Molecular Devices). AMC: excitation at 360 nm and emission at 460 nm. Final values are expressed as percentage of NE activity in this assay buffer.
Identification of Eap.
Supernatant of S. aureus Newman was cultured for 18 h in Todd Hewitt broth and fractionated by using the Äkta system and columns of GE Healthcare. All fractions were analyzed by 15% (wt/vol) SDS/PAGE and silver staining. Activity of the fractionated supernatant, and of the recombinant proteins (100 nM), was tested as during the screening. See SI Materials and Methods for details.
Purified S. aureus Proteins.
Toxic shock syndrome toxin-1 (TSST-1) (Bioconnect), Protein A (Sigma), and staphylococcal enterotoxin A (SEA) (Sigma) were obtained commercially. All other S. aureus proteins were produced recombinantly as His-tagged proteins in Escherichia coli and purified by using nickel affinity chromatography as described (11, 30–33).
Protease Activity Assays.
NE (5 nM), PR3 (10 nM, Elastin Products Company), CG (15 nM, Biocentrum), thrombin (1 nM, Sigma), plasmin (10 nM, Sigma), or plasma kallikrein (1 nM, Innovative Research) were incubated with inhibitors or culture supernatants for 15 min at room temperature in PBS with 0.05% (vol/vol) Igepal-Ca63 (Sigma). Residual protease activity was measured with protease-specific substrates at 37 °C in a Fluostar Omega plate reader (BMG Labtech). Data points showing linear substrate conversion were used to determine relative protease activity. Thereafter, IC50 values were determined by plotting a sigmoidal curve without constraints, or the Ki was determined according to the competitive inhibitor model by plotting the protease activity in the presence of multiple concentrations of the inhibitors Eap, EapH1, EapH2, or SLPI (R&D Systems) against the multiple concentrations of substrate used (all with maximum substrate concentrations higher than the determined Km). A list of substrates is found in Table S2.
Generation of Bacterial Mutants.
Markerless mutants of eap genes in S. aureus Newman were generated by using the pKOR1 vector (34) with minor modifications. We modified pKOR1 by replacing the lambda recombination cassette and the cat (-) and ccdB genes with a conventional multiple cloning site (cloned in ApaI/KpnI, creating the new pKOR1-mcs vector). Subsequently, we ligated the 1,000-bp regions immediately upstream and downstream of our gene of interest by using PCR overlay and cloned this product into pKOR1-mcs in E. coli DC10B (35). After electroporation into S. aureus Newman, allelic replacement was induced by temperature shift (34). The mutants were generated in a step-wise fashion: We first deleted the eap gene (resulting in MR1811, or Δeap), then eapH1 (MR1852, or ΔeapΔH1) and subsequently eapH2 (MR1860, or ΔeapΔH1ΔH2). For complementation of the ΔeapΔH1ΔH2 strain (MR1937, or ΔeapΔH1ΔH2 compl.), we also used the pKOR1 system but now cloned the upstream and downstream regions plus the gene of interest. Homologous recombination restored the presence of the eap genes at their original location. All mutants were verified by DNA sequencing and production of secreted Eap was monitored by Western blotting.
Activity of Mouse NSPs in Vitro.
Mouse bone-marrow cells were isolated as described before (36). Erythrocytes were lysed with ACK lysis buffer for 3 min at room temperature. The remaining cells were stimulated with 5 µg/mL cytochalasin B (Sigma) for 10 min at room temperature and degranulated with 1 μM fMLP (Sigma) for 15 min at 37 °C. The supernatant was used as source of murine NSPs. Diluted supernatant (1:8 for NE, 1:16 for PR3, and 1:2 for CG) was incubated with 100 nM of each Eap protein, or CHIPS. The residual activity was determined with protease-specific substrates (37). The relative mNSP activity was determined by using the slope of the linear part of the substrate-conversion graph compared with this slope in absence of inhibitors. A list of substrates is found in Table S2.
Animal Model.
The liver abscess model was performed as described, with minor modifications (19). Eight-week-old female C57/BL6 mice (purchased from Charles River) were infected with 100 µL of bacterial suspension (∼1 × 107 CFU) via retro-orbital injection. Infected mice were treated with a daily dose of caprofen (5 mg/kg; Pfizer), and 4 d after infection, mice were euthanized with pentobarbital (400 mg/kg; Merial). Their livers were removed, homogenized in PBS, and viable bacteria therein were quantified by serial dilution. The animal experiments were performed as required by the German regulations of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA), and were approved by the local governmental animal care committee (33.9-42502-04-38/2012).
Macromolecular Crystallography.
Crystals of EapH1 bound to human NE were obtained from vapor diffusion of hanging drops at 20 °C. A sample of the EapH1/NE complex was prepared by mixing a 1:1 molar ratio of each monomer, followed by centrifugal buffer exchange into 10 mM Tris at pH 7.4, 50 mM NaCl, and concentration to 5 mg/mL total protein (as judged by absorbance at 280 nM). Block-shaped crystals of various sizes appeared within 1–3 d and grew to their final dimensions over the course of a week from drops consisting of 1-μL complex mixed with 1 μL of reservoir buffer [0.1 M Hepes at pH 7.0, 1.0 M succinic acid, and 1% (wt/vol) polyethylene glycol-2000] that had been previously diluted with an equal volume of ddH2O, and that were equilibrated over 500 μL of reservoir buffer. Single crystals were harvested and flash cooled in liquid nitrogen following a brief soak in a cryopreservation solution consisting of the reservoir buffer above supplemented with 20% (wt/vol) sucrose.
X-ray diffraction data were collected at 1.22 Å wavelength by using beamline 22-ID of the Advanced Photon Source, Argonne National Laboratory. Diffraction data were indexed, integrated, and scaled by using HKL2000. Crystals of EapH1/NE grew in the space group P61 and contained a single EapH1/NE complex in the asymmetric unit. Structure solution and refinement were carried out by individual programs as implemented within the PHENIX software package (38). Initial phases were obtained by molecular replacement by using single copies of NE (PDB ID code 1HNE; ref. 39) and EapH1 (PDB ID code 1YN4; ref. 11) as sequential search models. The final model was completed after iterative cycles of manual building in COOT (40) followed by refinement using PHENIX.REFINE. In the Ramachandran plot, 97% of the residues modeled occupied favored regions, whereas the remaining 3% occupied allowed regions. Refined coordinates and structure factors (PDB ID code 4NZL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University (http://www.rscb.org/). A more complete description of the crystal properties, diffraction data quality, and characteristics of the final model may be found in Table S1.
Statistics.
Statistical analyses were performed by using GraphPad Prism 6.0. For determination of residual NSP activity in presence of various bacterial culture supernatants, normality was assumed and an unpaired, two-tailed Student t test was used. Because the number of data points in the in vivo experiment did not allow for normality testing and we had to correct for multiple comparison, we used a Krusskal–Wallis test. P < 0.05 was assumed to be statistically significant. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Supplementary Material
Acknowledgments
We thank Constantin Becker for help with animal experiments; Ted Bae, Niels Bovenschen and Rolf Urbanus for helpful discussions; Kimberly Jefferson (IsaB), Ted Bae (pKOR1), Ian Monk (DC10B) for providing reagents; the Brenkman laboratory for performing mass spectrometry; and Jos van Strijp, Erik Hack, John Lambris, and Daniel Ricklin for critical reading of the manuscript. This work was supported by a Vidi grant from the Netherlands Organization for Scientific Research (to S.H.M.R.), a Federation of European Microbiological Societies Research Fellowship grant (to D.A.C.S.), German Ministry for Education and Research Grant 01 KI 07103 Skin Staph (to M.H.), and US National Institutes of Health Grant AI071028 (to B.V.G.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team beamlines at the Advanced Photon Source, Argonne National Laboratory. A list of supporting member institutions may be found online (www.ser-cat.org/members.html).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4NZL).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407616111/-/DCSupplemental.
References
- 1.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119(9):2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol. 2011;11(8):505–518. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham CTN. Neutrophil serine proteases: Specific regulators of inflammation. Nat Rev Immunol. 2006;6(7):541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 4.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191(3):677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289(5482):1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 6.Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417(6884):91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 7.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62(4):726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera NC, et al. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc Natl Acad Sci USA. 2012;109(16):6229–6234. doi: 10.1073/pnas.1200470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammel M, Nemecek D, Keightley JA, Thomas GJJ, Jr, Geisbrecht BV. The Staphylococcus aureus extracellular adherence protein (Eap) adopts an elongated but structured conformation in solution. Protein Sci. 2007;16(12):2605–2617. doi: 10.1110/ps.073170807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain M, et al. More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infect Immun. 2008;76(12):5615–5623. doi: 10.1128/IAI.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisbrecht BV, Hamaoka BY, Perman B, Zemla A, Leahy DJ. The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens. J Biol Chem. 2005;280(17):17243–17250. doi: 10.1074/jbc.M412311200. [DOI] [PubMed] [Google Scholar]
- 12.Haas P-J, et al. The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules. J Mol Biol. 2005;353(4):859–872. doi: 10.1016/j.jmb.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Wright CD, Kennedy JA, Zitnik RJ, Kashem MA. Inhibition of murine neutrophil serine proteinases by human and murine secretory leukocyte protease inhibitor. Biochem Biophys Res Commun. 1999;254(3):614–617. doi: 10.1006/bbrc.1998.0108. [DOI] [PubMed] [Google Scholar]
- 14.Palma M, Haggar A, Flock J-I. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J Bacteriol. 1999;181(9):2840–2845. doi: 10.1128/jb.181.9.2840-2845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wamel WJB, Rooijakkers SHM, Ruyken M, van Kessel KPM, van Strijp JAG. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J Bacteriol. 2006;188(4):1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62(4):1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- 17.Belaaouaj A, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4(5):615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 18.MacIvor DM, et al. Normal neutrophil function in cathepsin G-deficient mice. Blood. 1999;94(12):4282–4293. [PubMed] [Google Scholar]
- 19.Li C, et al. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J Bacteriol. 2010;192(15):3883–3892. doi: 10.1128/JB.00237-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu SX, et al. Superantigens subvert the neutrophil response to promote abscess formation and enhance Staphylococcus aureus survival in vivo. Infect Immun. June 9, 2014 doi: 10.1128/IAI.02110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci USA. 2006;103(37):13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234(4):946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 23.Tsunemi M, Matsuura Y, Sakakibara S, Katsube Y. Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution. Biochemistry. 1996;35(36):11570–11576. doi: 10.1021/bi960900l. [DOI] [PubMed] [Google Scholar]
- 24.Koizumi M, Fujino A, Fukushima K, Kamimura T, Takimoto-Kamimura M. Complex of human neutrophil elastase with 1/2SLPI. J Synchrotron Radiat. 2008;15(Pt 3):308–311. doi: 10.1107/S0909049507060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Date SV, et al. Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J Infect Dis. 2014;209(10):1542–1550. doi: 10.1093/infdis/jit668. [DOI] [PubMed] [Google Scholar]
- 26.Joost I, et al. Transcription analysis of the extracellular adherence protein from Staphylococcus aureus in authentic human infection and in vitro. J Infect Dis. 2009;199(10):1471–1478. doi: 10.1086/598484. [DOI] [PubMed] [Google Scholar]
- 27.Reeves EP, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416(6878):291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 28.Palazzolo-Ballance AM, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180(1):500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 29.Spaan AN, Surewaard BGJ, Nijland R, van Strijp JAG. Neutrophils versus Staphylococcus aureus: A biological tug of war. Annu Rev Microbiol. 2013;67:629–650. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- 30.Xie C, et al. Suppression of experimental autoimmune encephalomyelitis by extracellular adherence protein of Staphylococcus aureus. J Exp Med. 2006;203(4):985–994. doi: 10.1084/jem.20051681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooijakkers SHM, et al. Staphylococcal complement inhibitor: Structure and active sites. J Immunol. 2007;179(5):2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- 32.de Haas CJC, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med. 2004;199(5):687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bestebroer J, et al. Functional basis for complement evasion by staphylococcal superantigen-like 7. Cell Microbiol. 2010;12(10):1506–1516. doi: 10.1111/j.1462-5822.2010.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55(1):58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012;3(2):e00277–e11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siemsen DW, Schepetkin IA, Kirpotina LN, Lei B, Quinn MT. In: Neutrophil Isolation from Nonhuman Species, Neutrophil Methods and Protocols. Quinn MT, DeLeo FR, Bokoch GM, editors. Totowa, NJ: Humana; 2007. pp. 26–27. [Google Scholar]
- 37.Kalupov T, et al. Structural characterization of mouse neutrophil serine proteases and identification of their substrate specificities: Relevance to mouse models of human inflammatory diseases. J Biol Chem. 2009;284(49):34084–34091. doi: 10.1074/jbc.M109.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwart PH, et al. In: Automated Structure Solution with the PHENIX Suite, Methods Mol Biol. Kobe B, Guss M, Huber T, editors. Vol 426. Totowa, NJ: Humana; 2008. pp. 419–435. [DOI] [PubMed] [Google Scholar]
- 39.Navia MA, et al. Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-A resolution. Proc Natl Acad Sci USA. 1989;86(1):7–11. doi: 10.1073/pnas.86.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.