Abstract
• Background and Aims Overwintering buds (turions) of the monocot aquatic pondweed species (Potamogeton distinctus) are highly tolerant to anoxic stress. Sucrose metabolism accompanied by enhanced activity of sucrose synthase (SuSy) operates actively during anaerobic elongation of pondweed turions. The aim of this study is to isolate SuSy genes from the turions and to investigate their transcriptional changes in response to anoxia and other stimuli.
• Methods SuSy genes were isolated from pondweed turions by PCR methods and transcript levels of SuSy genes were examined in response to anoxia, sugars and plant hormones. In addition, the effects of anoxia on SuSy activity were examined both in the soluble fraction and in the microsomal fraction.
• Key Results cDNAs of two SuSy genes (PdSUS1 and PdSUS2) were cloned from pondweed turions. The levels of PdSUS1 transcripts increased under anoxia but did not with sugar treatments. Anoxia-stimulated elongation of turions was further enhanced by 2,4-dichlorophenoxyacetic acid (2,4-D) and suppressed by treatments with sorbitol, 2-deoxyglucose (2-dGlc) and abscisic acid (ABA). The levels of PdSUS1 transcripts were increased by 2,4-D and decreased by sorbitol under anoxia. The levels of PdSUS2 transcripts were not significantly affected by anoxia and any other treatments. SuSy activity of turions under anoxia was enhanced in the soluble fraction, but not in the microsomal fraction.
• Conclusions Up-regulation of PdSUS1 transcription under anoxia may not be attributed to sugar starvation under anoxia. A positive correlation between stem elongation and the level of PdSUS1 transcripts was observed in turions treated with anoxic conditions, 2,4-D and sorbitol. The increase in SuSy activity in the cytosol may contribute to sugar metabolism and sustain stem elongation under anoxia.
Keywords: Anaerobic elongation, anoxia, gene expression, plant hormone, pondweed, Potamogeton distinctus, sucrose, sucrose synthase
INTRODUCTION
Plant anaerobiosis has been extensively studied, mostly in major crops with an emphasis on various aspects including energy metabolism, maintenance of homeostasis in cells, and the regulation of gene and protein expression (Gibbs and Greenway, 2003; Greenway and Gibbs, 2003). Selection of appropriate plant species and tissues as experimental materials that have notable characteristics such as high tolerance to lack of oxygen is one way to contribute to the development of this field. Many plants exposed to oxygen-deprived conditions cannot grow and will die within a few days. In contrast, Potamogeton distinctus (pondweed) can elongate its stem under anoxia (Ishizawa et al., 1999). This anaerobic growth is supported by an active glycolytic pathway (Sato et al., 2002). Potamogeton pectinatus has similar characteristics (Summers and Jackson, 1994). Its tubers remain sensitive to auxin and abscisic acid, although not to gibberellin, despite the absence of oxygen (Summers and Jackson, 1996).
Pondweed is a floating-leaved plant and forms turions (overwintering buds) in soil. Rapid starch degradation is accompanied with anoxic growth of turions (Sato et al., 2002; Harada and Ishizawa, 2003). Changes in enzymatic activity of starch degradation, such as amylolytic activity, α-amylase activity and starch phosphorylase activity have not been detected during anoxic growth, but activities of certain isozymes of amylases and starch phosphorylase are induced under anoxia in P. distinctus (Harada and Ishizawa, 2003). The incorporation into sucrose of 14C-glucose fed exogenously to turions was enhanced under anoxia, and it was concluded that more sucrose is synthesized under anoxia than is synthesized in air (Sato et al., 2002). The content of sucrose in the stems of turions decreases rapidly within 1 d of anoxia, but remains constant thereafter. These results indicate that turnover of sucrose is activated by anoxia. The activities of invertase, sucrose phosphate-synthase and sucrose synthase have been shown to increase under anoxia, the increase in the activity of sucrose synthase (SuSy; EC 2.4.1.13) being especially notable (Harada and Ishizawa, 2003). From these results, it was proposed that sucrose metabolism following rapid starch degradation plays an important role in energy production and growth of pondweed turions under anoxia.
SuSy can catalyse both synthesis and degradation of sucrose (Giegenberger and Stitt, 1993), but the degradation dominates in vivo. SuSy in the cytosol is thought to supply UDP-glucose and fructose produced by sucrose cleavage for glycolysis, and possibly starch synthesis. In addition, SuSy associated with the plasma membrane is considered to contribute to growth by supplying sugars for cellulose synthesis (Amor et al., 1995; Carlson and Chourey, 1996; Winter et al., 1997; Haigler et al., 2001). On the other hand, SuSy is one of the anaerobic proteins (ANPs) that are expressed in maize roots under anaerobic conditions (Sachs et al., 1980; Sachs et al., 1996). Two enzymes, invertase and SuSy, degrade sucrose in plants and the action of SuSy may be more energy-efficient by requiring less ATP. Accordingly, SuSy activation is thought to be a metabolic adaptation to anaerobic conditions (Guglielminetti et al., 1995; Bologa et al., 2003). Regulation of sucrose metabolism under anoxia has been studied in rice, which is the most tolerant among monocotyledonous species in major crops. Starch degradation and sucrose cleavage occur in anoxic rice and SuSy is mainly involved in sucrose degradation (Guglielminetti et al., 1995). Sucrose synthesis mediated by sucrose-phosphate synthase is particularly active in rice seedlings under anoxia, but not in barley, an anoxia-intolerant species (Guglielminetti et al., 1999). In the case of P. pectinatus, sucrose accumulates in the stem, which is the major elongating tissue under anoxia (Summers et al., 2000).
In the present study, two different SuSy cDNAs were isolated from pondweed turions in order to explore roles of SuSy in sucrose metabolism and stem elongation under anoxia. Their gene expression was examined in response to various kinds of stimuli including not only anaerobic conditions, but also sugars and plant hormones. It was found that the transcripts of one gene encoding SuSy increased during anaerobic growth in pondweed turions.
MATERIALS AND METHODS
Incubation of pondweed turions and growth analysis
Turions of pondweed were collected in a greenhouse of our laboratory and stored at 4 °C. Stem segments, 15 mm in length, were prepared by cutting off the base and the apex from turions with a razor blade after stripping off scaly leaves. Intact turions and stem segments were placed on moist filter paper and pre-incubated at 25 °C in the dark for 1 d. Then, these segments or intact turions were stood in a small vial (10 mm in diameter) containing 500 µL of incubation medium. Distilled water was the usual incubation medium, but for treatments with sugars, sorbitol, 2,4-dichlorophenoxyacetic acid (2,4-D), abscisic acid (ABA) and 2-deoxyglucose (2-dGlc), a 10 mm MES buffer (pH 7·5) was used as the incubation medium. Twenty-five μg of carbenicillin and 250 µg of vancomycin were added to the incubation medium in the case of sugar treatments to inhibit microbial growth. The vials were placed in a 100 mL glass vessel that was connected to a continuous supply of water-saturated air or pure N2 gas (Ishizawa et al., 1999). After incubation in aerobic or anaerobic conditions at 25 °C in the dark, stem segments were plunged into liquid nitrogen for extraction of RNA immediately after length measurement.
RNA isolation
Frozen turion segments (100 mg) were ground with liquid nitrogen in a pestle and mortar. The frozen powder was thoroughly mixed with 500 µL of extraction buffer containing 1 % SDS, 50 mm Tris/HCl (pH 7·5), 50 mM EDTA and 500 µL of water-saturated phenol. The mixture was centrifuged at 1300 g at 4 °C for 15 min. The supernatant was thoroughly mixed with 250 µL of water-saturated phenol and 250 µL of chloroform. The mixture was then centrifuged at 1300 g at 4 °C for 10 min, the supernatant was thoroughly mixed with the same amount of chloroform and, after centrifugation at 1300 g at 4 °C for 10 min, the supernatant pH was adjusted to below 6·0. RNA was then precipitated with isopropanol at −20 °C overnight. Following centrifugation at 7000 g at 4 °C for 15 min, the RNA was dried with 70 % ethanol and solubilized in 50 µL of water. RNA was further purified by precipitation with 2 m LiCl at 4 °C for 2 h, collected by centrifugation at 7000 g at 4 °C for 15 min, dried with 70 % ethanol, and solubilized in 30 µL of water. An amount of total RNA was determined by absorbance at 260 nm.
Subcloning and sequencing of cDNAs
For 3′RACE, first strand cDNAs were synthesized from total RNA isolated from stem segments incubated in various conditions with Oligo (dT)20-M4 Adaptor primer (Takara, Shiga, Japan) and SuperScript II (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Then RT–PCR was conducted with an oligonucleotide primer (5′-CCC AAG TTC AAC ATY GTS TCT CCT GG-3′), which was designed from a highly conserved region of nucleotide sequences of plant SuSy genes, M13 primer M4 (Takara) and Taq DNA Polymerase (Fermentas, Vilnius, Lithuania). Amplified DNA fragments were ligated into pGEM-T Easy Vectors (Promega, Madison, WI, USA) according to the manufacturer's instructions. Plasmids were introduced to competent cells by heat shock of 42 °C for 50 s. After checking insertion of gene fragments to plasmids by PCR, the plasmids for determination of nucleotide sequences were extracted by the alkali-SDS method. For 5′RACE, SMART™ Oligo cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) was used. First-strand cDNAs were synthesized with SMART II A Oligonucleotide, 5′RACE cDNA Synthesis Primer and Power Script Reverse Transcriptase (Clontech). Then RT–PCR was conducted with gene-specific primers (5′-CAG CTC AGC CTG TTC TTC-3′ for PdSUS1 and 5′-GGA GTG CAG TAA GTC TC-3′ for PdSUS2) designed from the partial nucleotide sequences of the genes, Universal Primer A Mix, and Advantage 2 Polymerase (Clontech). Amplified fragments were subcloned by the same methods as 3′RACE. Nucleotide sequences of DNA were determined by the dideoxynucleotide chain termination method using DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Biosciences, Little Chalfont, UK). Sequence analysis was performed with 373A DNA Sequencing System (Applied Biosystems, Foster City, CA, USA), GENETIX-Mac Ver 8.0. and ClustalW.
RT–PCR analysis
First-strand cDNAs were synthesized from RNA isolated from stem segments with SuperScript II or SuperScript III (Invitrogen). The synthesized cDNAs were diluted 10-fold and then used for RT–PCR. The PCR mixture contained 75 mm Tris/HCl buffer with 20 mm (NH4)2SO4, 200 nm dNTP, 250 µm MgCl2, 200 nm each two different primers, 5′-GCA GTT CGT GCT TCT AGT GAA CGA TCG-3′, which was located in the beginning of 3′-UTR region in the predicted mRNA, and 5′-GTC GGT GCT TCT TTA TTC ACT TGC CC-3′, which correspond to the complementary sequence of the ending part in the 3′-UTR, for PdSUS1 or 5′-GGT GCT AAT GGT GCA GGC CAT CAC TG-3′, which was located at the end of the ORF, and 5′-CAC GGA AAT TTG GCA TAT CGG CC-3′, which corresponds to the complementary sequence of the ending part in the 3′-UTR, for PdSUS2, 0·2 units of Taq DNA Polymerase (Fermentas) and 2 µL of diluted first strand cDNA in a final volume of 10 µL. PCR was performed for various cycles to determine a logarithmic phase of DNA amplification. The most suitable reaction for PCR analysis was selected as pre-heating at 94 °C for 4 min and then 21 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 2 min. The expression of 26S rRNA gene was used as an internal control to determine the efficiency of RT–PCR among different samples. Eight µL of each PCR product were used for 1 % agarose gel electrophoresis analysis. DNA was stained with 0·5 µg mL−1 ethidium bromide solution.
Northern blot analysis
Ten µg of total RNA was analysed by electrophoresis in 1·0 % agarose gel containing 2·2 m formaldehyde and 20 mm MOPS (pH 7·0), 8 mm sodium acetate and 1 mm EDTA (pH 8·0). The gel was blotted onto Nylon Membranes, Positively Charged (Roche, Basel, Switzerland). The digoxigenin (DIG)-labelled RNA probe was made from the 3′-UTR of PdSUS1 using the same primer set as used for RT–PCR according to the manufacturer's instructions (Roche). The membrane was hybridized with the probe at 68 °C overnight. The membrane was washed twice with a solution containing 2× SSC and 0·1 % SDS for 5 min at room temperature and then twice with a solution containing 0·1× SSC and 0·1 % SDS for 15 min at 68 °C. The DIG Luminescent Detection Kit (Roche) was used for signal detection.
Enzyme extraction and activity measurement
After incubation under various conditions, intact turions were ground in an ice-cold mortar and pestle with extraction buffer containing 100 mm Tris/HCl (pH 7·5), 1 mm EDTA, 10 mm dithiothreitol, 150 mm NaCl, and 10 mm phenylmethylsulfonyl fluoride. The homogenate was centrifuged at 7000 g at 4 °C for 20 min to remove debris. The supernatant was centrifuged at 95 000 g at 4 °C for 1 h. The microsomal fraction was resuspended in a small amount of the extraction buffer. The soluble fraction and microsomal fraction were dialysed twice against dialysation buffer containing 20 mm Tris/HCl (pH 7·5), 150 mm NaCl, and 10 mm β-mercaptoethanol for 2 h for each. Activity of SuSy in the soluble fraction and microsomal fraction was determined by the same method as described by Harada and Ishizawa (2003). Protein contents were measured by BCA regent (Pierce, Rockford, IL, USA).
RESULTS
Isolation of two distinct SuSy cDNAs from pondweed
Two different cDNA fragments of SuSy were isolated from pondweed tissues. One was from etiolated stem segments incubated under anoxia in the dark and the other was from green leaves of turions grown under aerobic condition in the light. cDNA from the former tissues was 2842 bp long and consisted of a 5′-untranslated region (UTR) of 108 bp, an open reading frame (ORF) of 2442 bp and a 3′-UTR of 292 bp. cDNA from the latter tissues was 2771 bp long and consisted of a 5′-UTR of 22 bp, an ORF of 2526 bp and a 3′-UTR of 223 bp. Figure 1 shows the amino acid sequences predicted from those two pondweed SuSy genes (PdSUS1 and PdSUS2). The nucleotide sequences revealed that PdSUS1 and PdSUS2 encode ORFs of 814 and 842 amino acid residues, respectively, with predicted molecular masses of 92·1 and 95·0 kDa, respectively. The amino acid sequences of PdSUS1 showed 74·5 % identity to those of PdSUS2. Both the amino acid sequences showed from 70–82 % identity with plant SuSys reported by others. PdSUS1 and PdSUS2 had a serine (Ser) residue in the N-terminal region (Ser 11). The Ser residue in the N-terminal region corresponding to Ser 11 of PdSUS1 and PdSUS2 is conserved in most SuSys from various species of plants (Winter and Huber, 2000) and is reported to be phosphorylated in maize (Ser 15, Huber et al., 1996). PdSUS1 and PdSUS2 consisted of two common parts, the characteristic N-terminal extension of sucrose synthase and glycosyl transferases group 1 domain. PdSUS2 had an extended alanine-rich region in the C-terminal region.
Fig. 1.

Predicted amino acid sequences of PdSUS1 and PdSUS2. The alignment was made using GENETIX-MAC Ver. 8.0. Identical amino acids are shaded and gaps are indicated by dashes. An arrowhead shows a conserved serine residue (see text). The characteristic N-terminal extension of sucrose synthase (thin underline) and a glycosyl transferases group 1 domain (broken underline) were identified by NCBI Conserved Domain Search. PdSUS2 has an alanine-rich region at the C-terminal region (bold underline). A primer for 3′ RACE of SuSy genes was designed from nucleotides encoding amino acid residues, indicated by an arrow.
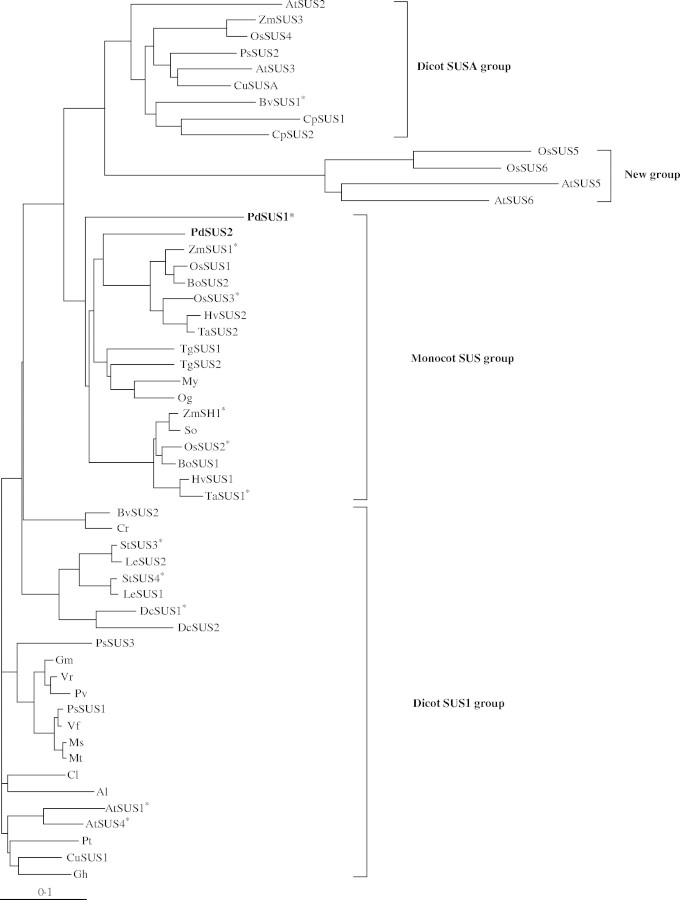
Figure 2 shows a phylogenetic tree generated with amino acid sequences of SuSys from various plants, PdSUS1 and PdSUS2. They are divided into four groups, the dicot SUS1 group, the dicot SUSA group, the monocot SUS group and the new group according to classification by Komatsu et al. (2002). Recently, a new SuSy gene named ZmSUS3 was found in maize (Carlson et al., 2002) and it belongs to the dicot SUSA group. Until now, three SuSy genes have been reported in rice (Wang et al., 1992; Huang et al., 1996). The databases of the TIGR Rice Genome Project (http://www.tigr.org/tdb/e2k1/osa1/) and the Rice Genome Annotation Database (http://ricegaas.dna.affrc.go.jp) were searched to identify SuSy genes in the rice genomes. Three new SuSy genes were found and named OsSUS4, OsSUS5 and OsSUS6. OsSUS4 is very similar to ZmSUS3 and the remaining pair (OsSUS5 and OsSUS6) are placed in the new group, including AtSUS5 and AtSUS6. PdSUS1 and PdSUS2 were placed in the monocot SUS group. PdSUS1 most resembled TgSUS2 from tulip (75·5 % identity, Balk and Boer, 1999) and PdSUS2 had highest identity to ZmSUS1 from maize (82·5 % identity, Huang et al., 1994). SuSys from Poaceae are apparently divided into two groups, one containing ZmSH1, the other including ZmSUS1. PdSUS2 was closer to the latter group than PdSUS1. However, more information is required about amino acid sequences of SuSy genes in monocotyledonous plants. Moreover, comparison between structures of exons and introns of their genes (Komatsu et al., 2002) is necessary for phylogenetic analysis of PdSUS1 and PdSUS2.
Fig. 2.
Phylogenetic tree of plant SuSy proteins. Predicted amino acid sequences were aligned using Clustal W. SuSys from Arabidopsis were named according to Baud et al. (2004). EMBL accession numbers (AGI ID for SuSys from Arabidopsis thaliana and gene number assigned by RiceGAAS for OsSUS4, OsSUS5 and OsSUS6) are given in parentheses. Al (X92378) from Alnus glutinosa; AtSUS1 (At5g20830), AtSUS2 (At5g49190), AtSUS3 (At4g02280), AtSUS4 (At3g43190), AtSUS5 (At5g37180) and AtSUS6 (At1g73370) from Arabidopsis thaliana; BoSUS1 (AF412039) and BoSUS2 (AF412037) from Bambusa oldhamii; BvSUS1 (X81974) and BvSUS2 (AY457173) from Beta vulgaris; Cl (AB018561) from Citrullus lanatus; CpSUS1 (AJ131999) and CpSUS2 (AJ132000) from Craterostigma plantagineum; Cr (X82504) from Chenopodium rubrum; CuSUS1 (AB022092) and CuSUSA (AB022091) from Citrus unshiu; DcSUS1 (X75332) and DcSUS2 (Y16091) from Daucus carota; Gh (U73588) from Gossypium hirsutum; Gm (AF030231) from Glycine max; HvSUS1 (X65871) and HvSUS2 (X69931) from Hordeum vulgare; LeSUS1 (L19762) and LeSUS2 (AJ011319) from Lycopersicon esculentum; Ms (AF049487) from Medicago sativa; Mt (AJ131943) from Medicago truncatula; My (AF530568) from Mokara ‘Yellow’; Og (AF530567) from Oncidium ‘goldiana’; OsSUS1 (X59046), OsSUS2 (X64770), OsSUS3 (L03366), OsSUS4 (OSJNBb0050D18_1·01), OsSUS5 (OSJNBb0026I12·04) and OsSUS6 (OJ1149_C12·08) from Oryza sativa; PdSUS1 (AB193515) and PdSUS2 (AB193516) from Potamogeton distinctus (this study); Pt (AY341026) from Populus tremuloides; Pv (AF315375) from Phaseolus vulgaris; PsSUS1 (AJ012080), PsSUS2 (AJ001071) and PsSUS3 (AJ311496) from Pisum sativum; So (AY118266) from Saccharum officinarum; StSUS3 (U24088) and StSUS4 (U24087) from Solanum tuberosum; TaSUS1 (AJ001117) and TaSUS2 (AJ000153) from Triticum aestivum; TgSUS1 (X96938) and TgSUS2 (X96939) from Tulipa gesneriana; Vf (X69773) from Vicia faba; Vr (D10266) from Vigna radiata; ZmSH1 (X02400), ZmSUS1 (L22296) and ZmSUS3 (AY124703) from Zea mays. SuSys that are reported to be enhanced in gene or protein expression at low oxygen concentrations are indicated by asterisks according to Baud et al. (2004), except for OsSUS2 and OsSUS3 in Huang et al. (1996) and StSUS3 and StSUS4 in Bologa et al. (2003).
Levels of PdSUS1 and PdSUS2 transcripts under anoxia
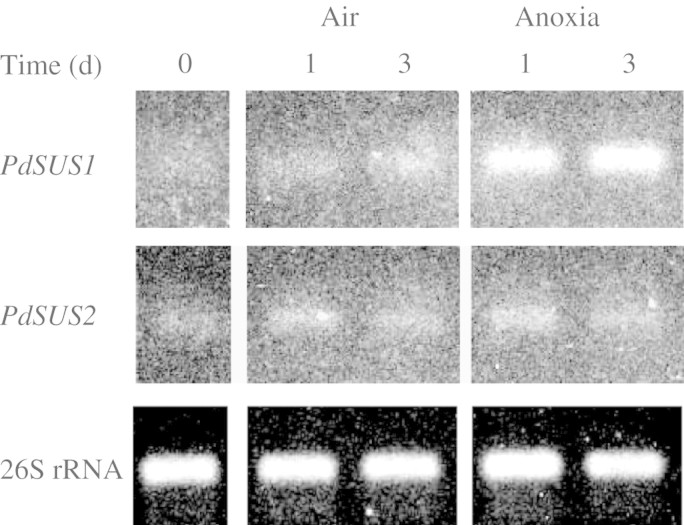
Levels of PdSUS1 and PdSUS2 transcripts in stem segments incubated in air or under anoxia were examined by RT–PCR (Fig. 3). Transcripts of PdSUS1 remained at a low level during a 3-d incubation in air, whereas it increased 1 d after anoxic incubation and then remained at the same increased level by 3 d. Enzymatic activity of SuSy in pondweed turions was not enhanced until the third day after the start of anoxic incubation (Harada and Ishizawa, 2003). These results indicate that enhancement of PdSUS1 expression under anoxia precedes the increase in SuSy enzyme activity. In contrast to PdSUS1, the transcript levels of PdSUS2 changed little during incubation either in air or under anoxia. Thus, unlike PdSUS1, PdSUS2 is not up-regulated by anoxia.
Fig. 3.
Effects of anoxia on the levels of PdSUS1 and PdSUS2 transcripts. RNA was extracted from turion segments incubated in air or under anoxia at 25 °C in the dark for 0 d, 1 d and 3 d. The amount of transcript was determined by RT–PCR. The level of 26S rRNA transcript was used as an internal control. A typical example in three independent experiments is shown.
Effects of exogenous sugars on stem elongation and levels of PdSUS1 and PdSUS2 transcripts
In many plant tissues, the expression of SuSy genes is responsive to sugars, especially glucose and sucrose (Koch et al., 1992; Koch, 1996). We examined whether exogenous glucose and sucrose affect the elongation of turion segments (Fig. 4A) and transcript levels of PdSUS1 and PdSUS2 (Fig 4B, C). Neither glucose nor sucrose at 50 mm or 100 mm significantly affected the elongation both in air and under anoxia (Fig. 4A). Figure 4B shows effects of glucose and sucrose on the levels of PdSUS1 and PdSUS2 transcripts analysed by RT–PCR. Anoxia increased the levels of PdSUS1 transcripts, but neither glucose nor sucrose had clear effects on it. The level of PdSUS2 transcripts was not much affected by anoxia, or glucose or sucrose. The conclusion was that glucose and sucrose had little effect on the level of PdSUS1 and PdSUS2 transcripts in air and under anoxia. Stem segments were also treated with sorbitol to examine effects of osmotic stress. Sorbitol at 100 mm did not affect elongation, but sorbitol at 300 mm strongly inhibited the elongation both in air and under anoxia (Fig. 4A). Sorbitol at 300 mm also showed an inhibitory effect on the level of PdSUS1 transcripts under anoxia, but it did not have clear effects on the level of PdSUS2 transcripts (Fig. 4B). On the other hand, 2-deoxyglucose (2-dGlc), a glucose analogue, slightly stimulated the elongation in air but significantly suppressed the elongation under anoxia (Fig. 4A). 2-dGlc increased the level of PdSUS1 transcripts remarkably in air but not under anoxia (Fig. 4B). 2-dGlc had little effect on the level of PdSUS2 transcripts although 2-dGlc seemed to slightly increase the PdSUS2 transcripts in air.
Fig. 4.
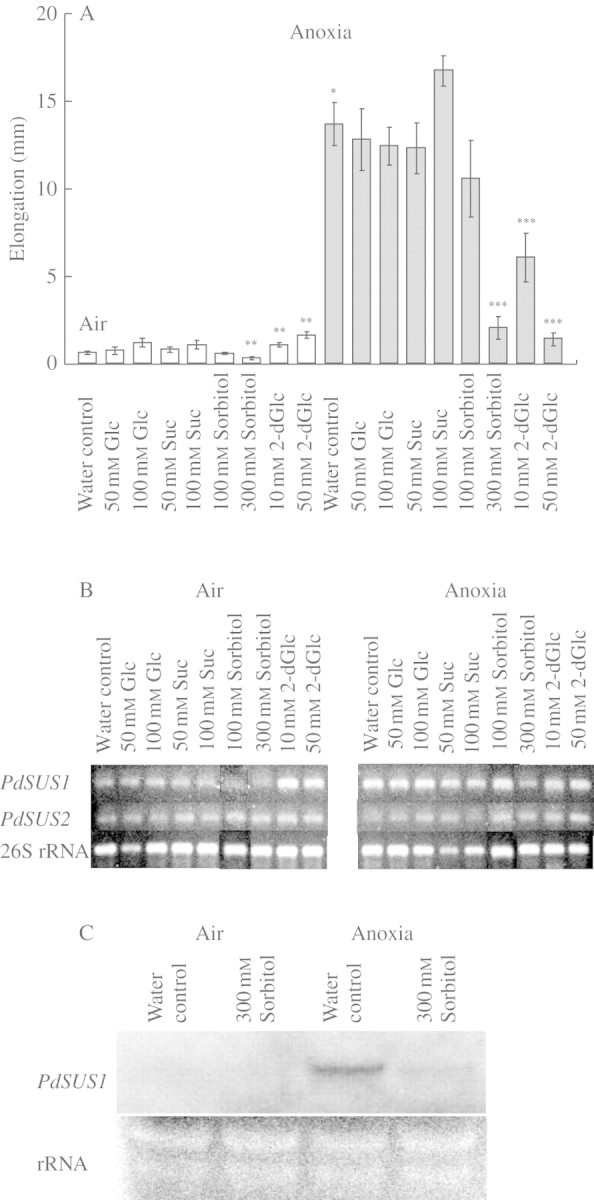
Effects of exogenous sugars on stem elongation and on the levels of PdSUS1 and PdSUS2 transcripts in turion segments. (A) Stem elongation was determined after 3 d of incubation in air (open bars) or under anoxia (shaded bars). Data for elongation are means ± s.e. of more than six turion segments. (B) The level of transcripts was determined at 3 d of incubation by RT–PCR and an example in three independent experiments is shown. (C) Northern blot analysis of PdSUS1 transcripts. Total RNA was extracted from turion segments incubated under each condition for 3 d. Ten µg of total RNA were applied to each lane and hybridized with RNA probes for PdSUS1. rRNA stained with ethidium bromide is also shown as a loading control. Significantly different from water control in air at: *P < 0·0001, **P < 0·01. Significantly different from water control in anoxia at: ***P < 0·05 (Mann–Whitney test).
We also examined the effects of anoxia and 300 mm sorbitol on the level of PdSUS1 transcripts by Northern blot analysis (Fig. 4C). An intense signal for PdSUS1 was detected after incubation in anoxia for 3 d, but not after the same period in air. Three hundred mm sorbitol depressed the increased level of PdSUS1 transcripts under anoxia.
Effects of 2,4-D and ABA on stem elongation and levels of PdSUS1 and PdSUS2 transcripts
We examined whether plant hormones, which influence elongation of pondweed stem segments, also affect the expression of SuSy genes. 2,4-D at 1 and 10 µm stimulated stem elongation in air, but only 2,4-D at 10 µm stimulated elongation in the absence of oxygen (Fig. 5A). 2,4-D also slightly enhanced the level of PdSUS1 transcripts under anoxia when the transcriptional levels were examined by RT–PCR (Fig. 5B). Northern blot analysis also showed the promotion of the level of PdSUS1 transcripts by 2,4-D under anoxia (Fig. 5C). The level of PdSUS2 transcripts was not affected by 2,4-D both in air and under anoxia (Fig. 5B). On the other hand, ABA significantly inhibited the elongation under anoxia (Fig. 5A) and slightly decreased the level of PdSUS1 transcripts under anoxia (Fig. 5B). The amount of PdSUS2 transcripts was not much affected by ABA (Fig. 5B). Then, the effects of ABA on the transcription of PdSUS1 were examined by Northern blot analysis, but none were apparent (Fig. 5C).
Fig. 5.
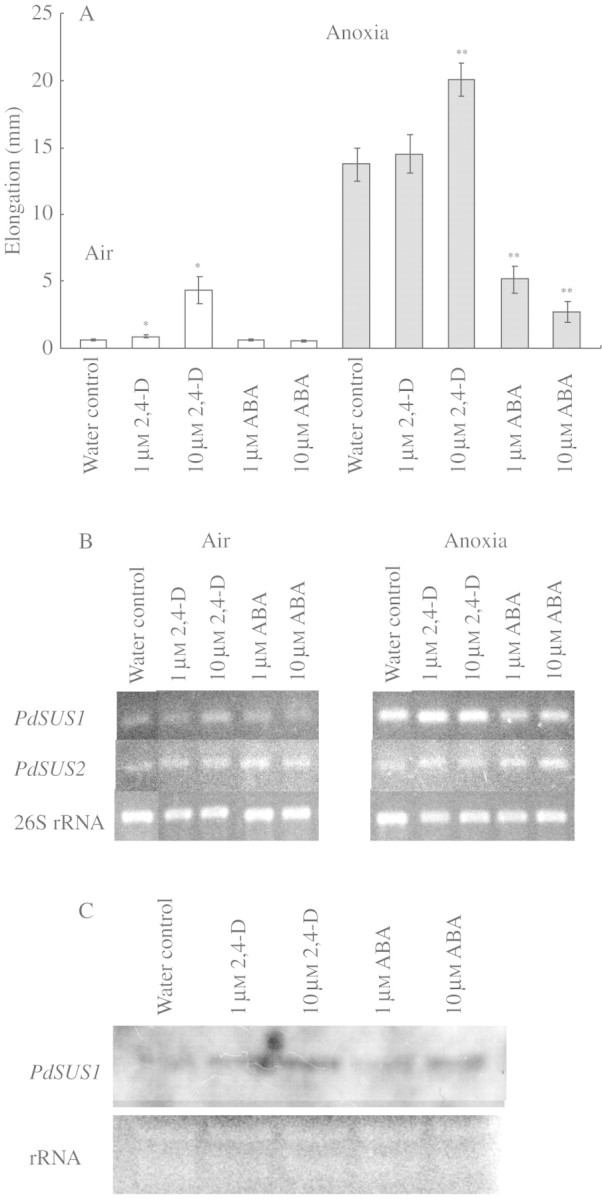
Effects of auxin and abscisic acid on stem elongation and on the levels of PdSUS1 and PdSUS2 transcripts in turion segments. (A) Stem elongation was determined after 3 d of incubation in air (open bars) or under anoxia (shaded bars). Data for elongation are means ± s.e. of more than 18 turion segments. (B) The level of transcripts was determined at 3 d of incubation by RT–PCR and an example in three independent experiments is shown. (C) Northern blot analysis of PdSUS1 transcripts. Total RNA was extracted from turion segments incubated under each condition for 3 d under anoxia. Ten µg of total RNA were applied to each lane and hybridized with RNA probes for PdSUS1. rRNA stained with ethidium bromide is also shown as a loading control. Significantly different from water control in air at: *P < 0·05. Significantly different from water control in anoxia at: **P < 0·05 (Mann–Whitney test).
Localization of SuSy activity in cell fractions in turions incubated under anoxia
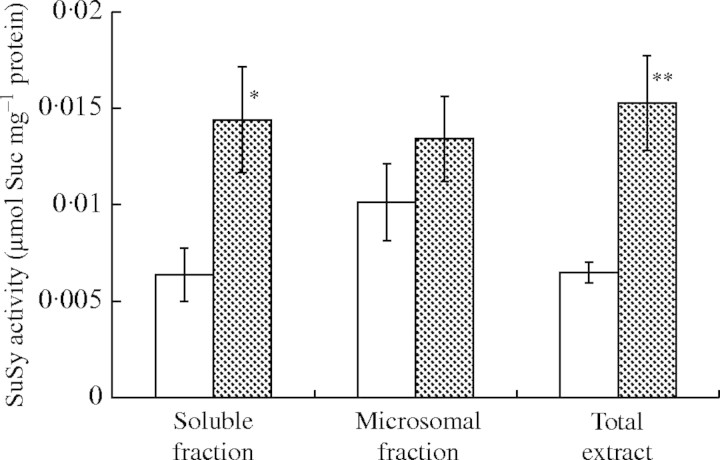
SuSy can be active in the cytosol or in a membrane-bound form. The plasma membrane-bound form has been linked to cellulose synthesis in numerous species and organs (Amor et al., 1995; Carlson and Chourey, 1996; Winter et al., 1997; Haigler et al., 2001). We examined whether soluble SuSy and membrane-bound SuSy were differentially affected by anoxia (Fig. 6). Total activity of SuSy extracted from turions under anoxia was significantly higher than that in air. SuSy activity in the soluble fraction was twice as high under anoxia as in air. However, there was no significant difference between SuSy activity in the microsomal fraction separated from pondweed turions in air and that under anoxia. These results suggest that enhancement of SuSy activity under anoxia is due to activation of SuSy in the cytosol.
Fig. 6.
Effects of anoxic conditions on SuSy activity in soluble and membrane fractions extracted from pondweed turions. SuSy was extracted from pondweed turions incubated at 25 °C in the dark in air (open bars) and under anoxia (shaded bars) for 3 d. Total activity of SuSy was determined in the supernatant after centrifugation of the extract at 7000 g for 20 min. The activity of soluble SuSy and membrane-bound SuSy was determined in the soluble fraction and in the microsomal fraction separated by ultracentrifugation at 95 000 g for 1 h. SuSy activity was measured by the method described in Materials and Methods. Data are means ± s.e. of three separate replicates. Significantly different from the activity in air at: *P < 0·1, **P < 0·05 (t-test).
DISCUSSION
We isolated two cDNAs encoding SuSy from pondweed turions (Fig. 1). In many cases, two or three SuSy genes have been isolated from one plant species. But six SuSy genes were recently found in the Arabidopsis thaliana genome (Komatsu et al., 2002; Baud et al., 2004). In the process of this study, three new genes were found from the rice genome database in addition to the three already reported (Wang et al., 1992; Huang et al., 1996). These newly discovered rice genes are unique in that they locate remotely from other rice SuSy genes in the phylogenetic tree (Fig. 2). These findings suggest a wider divergence of SuSy genes in mono- and dicotyledonous plants than was hitherto recognized. Expression of SuSy genes has been reported to be enhanced under hypoxic or anoxic conditions in several plant species including rice (Ricard et al., 1991; Guglielminetti et al., 1995), maize (Springer et al., 1986; Sachs et al., 1996), A. thaliana (Martin et al., 1993; Baud et al., 2004) and potato (Bologa et al., 2003), which differ in their tolerance to anoxia. Such genes are identified by asterisks in Fig. 2 and appear scattered in the phylogenetic tree. Thus, phylogenetic relationships based on amino acid sequences are not important so far as which SuSy gene is responsive to hypoxic or anoxic stress.
Only PdSUS1 transcripts increased under anoxia (Fig. 3) and the increase was detected 1 d after anoxic incubation, preceding a rise in activity of SuSy by 2 d (Harada and Ishizawa, 2003), suggesting that the increase in PdSUS1 transcripts contributes to the increase in the activity of SuSy. Since only two genes of SuSy were isolated from pondweed turions in this study, a possibility of involvement of other SuSy genes is not excluded in the enhancement of SuSy activity by oxygen exclusion.
There are reports concerning the mechanism of transcriptional regulation in SuSy gene expression. Arabidopsis ADH1 encoding alcohol dehydrogenase has anaerobic responsive elements (ARE) in its promoter region (Hoeren et al., 1998). The induction of ADH1 under hypoxic conditions is triggered by binding of a transcription factor, AtMYB2, to the ARE. A similar mechanism operates in the induction of ASUS1 encoding an arabidopsis SuSy (Hoeren et al., 1998). On the other hand, SuSy genes and other genes encoding enzymes that are involved in carbohydrate metabolism are reported to be sugar-responsive (Koch et al., 1992; Koch, 1996). For example, the induction of rice α-amylase in germinating rice occurs in response to sugar starvation brought about under anoxic conditions (Loreti et al., 2003). The contents of soluble sugars and sucrose in pondweed turions rapidly decrease under anoxic conditions (Sato et al., 2002; Harada and Ishizawa, 2003). Therefore, sugar starvation could be a trigger to increase the level of PdSUS1 transcripts under anoxic conditions. However, Fig. 4 shows that glucose and sucrose have no repressing effect on the level of PdSUS1 transcripts, which increases under anoxic conditions regardless of sugar additions. These results suggest that sugar starvation is not involved in the regulation of the PdSUS1 gene under anoxic conditions. Further study is necessary to explore the mechanisms regulating the expression of the PdSUS1 gene and how anoxia is sensed in pondweed turions.
SuSy is believed to play an important role in cell growth by providing UDP-glucose and fructose through sucrose cleavage for energy metabolism and for cell wall biosynthesis. The correlations between stem elongation of pondweed turions and the levels of SuSy transcripts were examined (Figs 4 and 5). Anoxic conditions enhanced shoot elongation and also increased the level of PdSUS1 transcripts. Ten μm of 2,4-D also enhanced both of them under anoxia. Three hundred mm sorbitol inhibited not only the shoot elongation but also the level of PdSUS1 transcripts under anoxic conditions. 2-dGlc stimulated stem elongation in air and enhanced the level of PdSUS1 transcripts. These cases showed a positive correlation between the level of PdSUS1 transcripts and shoot elongation. However, such a positive correlation between them was invalid in two cases. ABA significantly inhibited the anoxic elongation but it did not have clear effects on the level of PdSUS1 transcripts. In the case of 2-dGlc, stem elongation was severely restricted under anoxia but the level of PdSUS1 transcripts under anoxia was not affected. One possible explanation regarding the two results is that breakdown of the PdSUS1 transcripts was inhibited by ABA and 2-dGlc, and it resulted in hiding the inhibitory effects on the transcriptional level of the PdSUS1. Alternatively, these results may reflect a different contribution of SuSy to the regulation of stem elongation. At present, it is difficult to explain the reason why 2-dGlc stimulates shoot elongation in air but inhibits it under anoxia and how 2-dGlc increases the level of PdSUS1 transcripts. 2-dGlc may disturb carbohydrate metabolism as an analogue of glucose (Herold and Lewis, 1977) or act as an elicitor of sugar-sensing through hexokinase (Jang et al., 1997). Use of sugar analogues such as 2-dGlc as an experimental tool may be useful to investigate roles of SuSys in the control of plant growth.
SuSys in plant cells exist in the cytosol in soluble form and in association with the plasma membrane, Golgi body, tonoplast and actin filaments as a bound form (Koch, 2004). Figure 6 shows that the activity of SuSy in the soluble fraction is enhanced under anoxic conditions. SuSy in the cytosol serves to supply sugars to glycolysis under O2-deprived conditions (Guglielminetti et al., 1995). There is a strong possibility that sucrose degradation mediated by SuSy consumes less ATP than degradation by invertase (Bologa et al., 2003). These workers showed that O2-deprived conditions enhance SuSy activity in the cytosol of potato tubers but inhibit one associated with the plasma membrane. Bologa et al. (2003) suggest that an energy crisis under O2-deprived conditions is diminished by activating the SuSy pathway in glycolysis and by reducing the activity of membrane-bound SuSy engaging in biosynthesis demanding high energy costs. Ricard et al. (1998) reported that the expression of SuSy is essential for tolerance to anaerobic stress in maize, a moderately tolerant species. In pondweed turions under anoxic conditions, adenylate energy charge remains as high as in air (Ishizawa et al., 1999). It is likely that SuSy in the cytosol serves to maintain high energy status by supplying the necessary amounts of substrates to glycolysis activated in pondweed turions under anoxia with the minimum of ATP consumption. Much starch is stored in the amyloplasts of pondweed turion cells and it is the prime energy source for growth without oxygen. Enhancement of sucrose synthesis and degradation is accompanied by starch degradation in the cells (Sato et al., 2002; Harada and Ishizawa, 2003). We propose that sucrose cleavage mediated by SuSy is an important step regulating sugar metabolism of pondweed turion cells under anoxic conditions.
SuSy associated with the plasma membrane is believed to supply substrates for cell wall formation (Haigler et al., 2001). Callose formation in anoxic maize root tips is accompanied by increases in the activity of SuSy associated with the cell membrane (Subbaiah and Sachs, 2001). The induction of callose formation leads to rapid cell death of root tips, which is closely related to the tolerance of whole maize seedlings to anoxia. Moreover, hypoxia activates not only the activity of SuSy but also cellulose synthesis in wheat roots (Albrecht and Mustroph, 2003). The unexpected increase in cellulose synthesis under hypoxia is understood to enhance the mechanical strength of roots forming aerenchyma. However, no significant increases were found in the activity of SuSy in the microsomal fraction extracted from pondweed turions under anoxia, the activity remaining similar to that in air (Fig. 6). Rapid elongation of pondweed turion cells under anoxic conditions is presumably accompanied by cell wall biosynthesis. Substrates for cellulose biosynthesis are thought to be supplied from the products of sucrose cleavage mediated by SuSy associated with the plasma membrane. We cannot completely exclude a possibility that SuSy associated with the plasma membrane is activated in pondweed turion cells under anoxic conditions. A membrane-associated SuSy in maize roots (SH1) and its activity are enhanced under anoxic conditions (Subbaiah and Sachs, 2001) and its localization is regulated by phosphorylation, and thus it can exchange between the cytosol and the plasma membrane. Phosphorylation of Ser 11 residue conserved in the N-terminal regions of PdSUS1 and PdSUS2 may be involved in changes in the localization in cells. Further study of the localization of SuSys, especially PdSUS1, is necessary to discuss the role of SuSys in the accelerated site of cell elongation and vigorous anaerobic energy metabolism that characterizes pondweed turions when deprived of all oxygen.
Acknowledgments
We thank Mr K. Satoh for help with the cultivation of pondweed. This study was supported in part by Grant-in-Aid (14658165 to S.S.) for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
LITERATURE CITED
- Albrecht G, Mustroph A. 2003. Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217: 252–260. [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. 1995. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proceedings of the National Academy of Sciences, USA 92: 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk PA, de Boer AD. 1999. Rapid stalk elongation in tulip (Tulipa gesneriana L. cv. Apeldoorn) and the combined action of cold-induced invertase and the water-channel protein γTIP. Planta 209: 346–354. [DOI] [PubMed] [Google Scholar]
- Baud S, Vaultier MN, Rochat C. 2004. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis Journal of Experimental Botany 55: 397–409. [DOI] [PubMed] [Google Scholar]
- Bologa KL, Fernie AR, Leisse A, Loureiro ME, Geigenberger P. 2003. A bypass of sucrose synthase leads to low internal oxygen and impaired metabolic performance in growing potato tubers. Plant Physiology 132: 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS. 1996. Evidence for plasma membrane-associated forms of sucrose synthase in maize. Molecular and General Genetics 252: 303–310. [DOI] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS, Helentjaris T, Datta R. 2002. Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene. Plant Molecular Biology 49: 15–29. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. 1993. Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189: 329–339. [DOI] [PubMed] [Google Scholar]
- Gibbs J, Greenway H. 2003. Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Functional Plant Biology 30: 1–47. [DOI] [PubMed] [Google Scholar]
- Greenway H, Gibbs J. 2003. Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Functional Plant Biology 30: 999–1036. [DOI] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A. 1995. Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiology 108: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Loreti E, Perata P, Alpi A. 1999. Sucrose synthesis in cereal grains under oxygen deprivation. Journal of Plant Research 112: 353–359. [Google Scholar]
- Harada T, Ishizawa K. 2003. Starch degradation and sucrose metabolism during anaerobic growth of pondweed (Potamogeton distinctus A. Benn.) turions. Plant and Soil 253: 125–135. [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP. 2001. Carbon partitioning to cellulose synthesis. Plant Molecular Biology 47: 29–51. [PubMed] [Google Scholar]
- Herold A, Lewis DH. 1977. Mannose and green plants: occurrence, physiology and metabolism, and use as a tool to study the role of orthophosphate. New Phytologist 79: 1–40. [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. 1998. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-W, Chen J-T, Yu W-P, Shyur L-F, Wang A-Y, Sung H-Y, Lee P-D, Su J-C. 1996. Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Bioscience, Biotechnology and Biochemistry 60: 233–239. [DOI] [PubMed] [Google Scholar]
- Huang X-F, Nguyen-Quoc B, Chourey PS, Yelle S. 1994. Complete nucleotide sequence of the maize (Zea mays L.) sucrose synthase 2 cDNA. Plant Physiology 104: 293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL, Liao P-C, Gage DA, McMichael RW Jr, Chourey PS, Hannah LC, Koch K. 1996. Phosphorylation of serine-15 of maize leaf sucrose synthase. Plant Physiology 112: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawa K, Murakami S, Kawakami Y, Kuramochi H. 1999. Growth and energy status of arrowhead tubers, pondweed turions and rice seedlings under anoxic conditions. Plant, Cell and Environment 22: 505–514. [Google Scholar]
- Jang J-C, León P, Zhou L, Sheen J. 1997. Hexokinase as a sugar sensor in higher plants. The Plant Cell 9: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. 1996. Carbohydrate-modulated gene expression in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47: 509–540. [DOI] [PubMed] [Google Scholar]
- Koch K. 2004. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinions in Plant Biology 7: 235–246. [DOI] [PubMed] [Google Scholar]
- Koch KE, Nolte KD, Duke ER, McCarty DR, Avigne WT. 1992. Sugar levels modulate differential expression of maize sucrose synthase genes. The Plant Cell 4: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T. 2002. Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. Journal of Experimental Botany 53: 61–71. [PubMed] [Google Scholar]
- Loreti E, Alpi A, Perata P. 2003. α-Amylase expression under anoxia in rice seedlings: an update. Russian Journal of Plant Physiology 50: 737–742. [Google Scholar]
- Martin T, Frommer WB, Salanoubat M, Willmitzer L. 1993. Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. The Plant Journal 4: 367–377. [DOI] [PubMed] [Google Scholar]
- Ricard B, Rivoal J, Spiteri A, Pradet A. 1991. Anaerobic stress induces the transcription and translation of sucrose synthase in rice. Plant Physiology 95: 669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard B, Van Toai T, Chourey P, Saglio P. 1998. Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiology 116: 1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R. 1980. The anaerobic proteins of maize. Cell, 20: 761–767. [DOI] [PubMed] [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN. 1996. Anaerobic gene expression and flooding tolerance in maize. Journal of Experimental Botany 47: 1–15. [Google Scholar]
- Sato T, Harada T, Ishizawa K. 2002. Stimulation of glycolysis in anaerobic elongation of pondweed (Potamogeton distinctus) turions. Journal of Experimental Botany 53: 1847–1856. [DOI] [PubMed] [Google Scholar]
- Springer B, Werr W, Starlinger P, Bennett DC, Zokolica M, Freeling M. 1986. The Shrunken gene on chromosome 9 of Zea mays L. is expressed in various plant tissues and encodes an anaerobic protein. Molecular and General Genetics 205: 461–468. [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM. 2001. Altered patterns of sucrose synthase phosphorylation and localization precede callose induction and root tip death in anoxic maize seedlings. Plant Physiology 125: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers JE, Jackson MB. 1994. Anaerobic conditions strongly promote extension by stems of overwintering tubers of Potamogeton pectinatus L. Journal of Experimental Botany 45: 1309–1318. [Google Scholar]
- Summers JE, Jackson MB. 1996. Anaerobic promotion of stem extension in Potamogeton pectinatus Roles for carbon dioxide, acidification and hormones. Physiologia Plantarum 96: 615–622. [Google Scholar]
- Summers JE, Ratcliffe RG, Jackson MB. 2000. Anoxia tolerance in the aquatic monocot Potamogeton pectinatus: absence of oxygen stimulates elongation in association with an unusually large Pasteur effect. Journal of Experimental Botany 51: 1413–1422. [PubMed] [Google Scholar]
- Wang A-Y, Yu W-P, Juang R-H, Huang J-W, Sung H-Y, Su J-C. 1992. Presence of three rice sucrose synthase genes as revealed by cloning and sequencing of cDNA. Plant Molecular Biology 18: 1191–1194. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC. 2000. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Critical Reviews in Plant Sciences 19: 31–67. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC. 1997. Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Letters 420: 151–155. [DOI] [PubMed] [Google Scholar]