Abstract
• Background and Aims Shoot elongation of arrowhead tubers (Sagittaria pygmaea Miq.) is stimulated by anoxia, ethylene and CO2. The aim of this study was to characterize anoxic elongation by comparison with elongation stimulated by ethylene and CO2.
• Methods The effects of the inhibitors aminoethoxyvinylglycine (AVG) as an ethylene biosynthesis inhibitor, 1-methylcyclopropene (1-MCP) as a potent inhibitor of ethylene action, and pyrazol, an inhibitor of alcohol dehydrogenase, on shoot elongation were examined. Moreover, the effects of these gaseous factors on expression of genes possibly involved in modification of cell wall architecture were examined by polymerase chain reaction (PCR) methods.
• Key Results and Conclusions In air, promotion by 5 % CO2 and 5 µL L−1 ethylene of shoot elongation occurred. At 1% O2, ethylene also stimulated shoot elongation but CO2 did not. Pyrazol inhibited shoot elongation in hypoxia but not in normoxia, suggesting that alcohol fermentation contributes to elongation enhanced by hypoxia. AVG and 1-MCP partially prevented shoot elongation both in normoxia and in hypoxia, but they did not have significant effects in anoxia, suggesting that endogenous ethylene acts as a stimulator of shoot elongation in normoxia and in hypoxia but not in anoxia. Ethylene is not involved in anoxia-enhanced elongation. We cloned four cDNAs (SpEXPA1, 2, 3 and 4) encoding α-expansin (EXPA) and five cDNAs (SpXTH1, 2, 3, 4 and 5) encoding xyloglucan endotransglucosylase/hydrolase (XTH) from shoots of arrowhead tubers. The transcript levels of SpEXPA1 and 2 were increased by anoxia and those of SpEXPA2 were increased by 5 % CO2. Ethylene slightly elevated the level of SpEXPA4 transcripts. Anoxia enhanced the transcript levels of SpXTH1 and 4; neither ethylene nor CO2 had any effect. CO2 enhanced transcript levels of SpXTH3 and depressed those of SpXTH5. Ethylene decreased transcript levels of SpXTH5. These results suggest that four SpEXPA genes and five SpXTH genes are differently responsive to anoxia, CO2 and ethylene. Enhancement of SpEXPA1 and 2, and SpXTH1 and 4 transcript levels suggests that these gene products are involved in anoxic shoot elongation through modification of cell wall architecture.
Keywords: Anoxia, aquatic plant, arrowhead, cell wall, cell elongation, ethylene, expansin, carbon dioxide, hypoxia, Sagittaria pygmaea, submergence, xyloglucan endotransglucosylase/hydrolase
INTRODUCTION
Arrowhead (Sagittaria pygmaea) is an emergent plant and one of a number of troublesome perennial weeds in Japanese rice fields. It forms tubers at the tips of the rhizomes in summer and autumn. In natural habitats, tubers start to sprout underground in the following spring. Shoot elongation of tubers is stimulated under water, depending on water depth. Ethylene and CO2 are involved in the stimulation of shoot elongation under water (Suge and Kusanagi, 1975). Generally, ethylene usually acts as an inhibitor of cell elongation (Abeles et al., 1992), but it acts as a promoter in some cases including rice coleoptiles (Ku et al., 1970), Callitriche platycarpa and Ranunculus sceleratus (Musgrave et al., 1972) and arabidopsis in the light (Smalle et al., 1997). CO2 usually acts as an antagonist against ethylene actions (Abeles et al., 1992), but it acts as an activator of ethylene action in shoot elongation of arrowhead tubers (Suge and Kusanagi, 1975) as it does in some aquatic plants, including rice (Raskin and Kende, 1983; Ishizawa and Esashi, 1984), deepwater rice (Raskin and Kende 1984) and Sagittaria trifolia (Suge and Kusanagi, 1994). The mechanisms of such interactions between CO2 and ethylene actions on cell elongation remain unknown (Ridge, 1987).
Ishizawa et al. (1999) found that shoot elongation of arrowhead tubers is stimulated under O2-deprived conditions. The optimal concentration of O2 for shoot elongation of arrowhead tubers is approx. 1 %, similar to that in rice coleoptiles (Ishizawa and Esashi, 1984). However, unlike rice coleoptiles, shoot elongation of arrowhead tubers in the absence of O2 surpasses that in air. Such an abnormal ability has been reported in some plant species, including two species of Potamogeton (Summers and Jackson, 1994; Ishizawa et al., 1999). Acceleration of shoot elongation under O2-deprived conditions is thought to be an adaptive trait for plants under water to obtain O2 from the surface of water as soon as possible. Energy production of such plants in anoxia is attributed mainly to activation of alcoholic fermentation (Ishizawa et al., 1999; Sato et al., 2002; Harada and Ishizawa, 2003). Anoxia-enhanced shoot elongation of arrowhead tubers stems from cell elongation in the middle of leaves. Ca2+ uptake from the apoplast plays an important role in prolongation of cell elongation in anoxia (Tamura et al., 2001). However, the mechanism of anoxia-enhanced cell elongation has not been elucidated because anoxia induces plant cell death in most plants.
Cell wall extensibility is one of the key factors for regulation of plant cell growth. Various proteins in cell walls serve to modify cell wall architecture during cell growth and development (Cosgrove, 1999; Rose and Bennett, 1999). The cellulose–hemicellulose network plays a leading role in determination of the cell wall extensibility in rapidly growing cells. Expansins (Li et al., 2003) and xyloglucan endotransglucosylase/hydrolases (XTHs) (Campbell and Braam 1999; Rose et al., 2002) are attractive candidates to control wall-loosening processes during cell growth. Both expansins (Li et al., 2003) and XTHs (Yokoyama and Nishitani 2001; Yokoyama et al., 2004) have been found to be encoded by a substantial multigene family in Arabidopsis and rice genomes, suggesting that each isoform has a specific role in a certain stage of plant growth and development.
Internode elongation in deepwater rice is stimulated under submerged conditions (Kende et al., 1998). The enhancement of internodal elongation under submerged conditions is due to accumulation of ethylene in tissues, the production of which is enhanced under low O2 and high CO2 tensions (Raskin and Kende, 1984). Enhanced expression of α-expansin genes occurs in elongating tissues (Cho and Kende 1997). An interaction between ethylene and gibberellin is involved in the observed increase in cell wall extensibility of rapidly growing internodes. Like internode elongation of deepwater rice, leaf elongation of flooding-tolerant Rumex palustris is stimulated by submergence, which induces ethylene accumulation and oxygen deficiency in tissues (Voesenek et al., 2003). Submergence and ethylene treatment induce expression of expansin genes in R. palustris, but not in R. acetosa, which is flooding intolerant and does not elongate under submerged conditions, indicating that expansin has an important role in stimulated elongation by submergence (Vriezen et al., 2000).
Sachs et al. (1980) showed that normal protein synthesis in maize seedlings is suddenly repressed after exposure to anaerobic conditions. A small group of ‘transition polypeptides’ is soon produced, and another set of approx. 20 polypeptides named anaerobic polypeptides is synthesized approx. 90 min after the onset of anaerobic stress. Peschke and Sachs (1994) found that one of the genes encoding transition polypeptides, named wusl1005, is a homologue of XTH. Since the expression of the wusl1005 gene is induced by ethylene as well as anoxia, the gene product is thought to be involved in aerenchyma formation induced by flooding of maize roots (Saab and Sachs, 1996). Induction of aerenchyma formation is another adaptive trait of plants under submerged conditions.
In the present study, effects of anoxia, ethylene and CO2 on shoot elongation were examined in arrowhead tubers. Expansins and XTHs were selected as molecular markers to characterize anoxia-, ethylene- and CO2-induced elongation. We isolated four cDNAs encoding expansin and five cDNAs encoding XTH from arrowhead tubers, and found that anoxia preferentially enhanced the transcript levels of some genes.
MATERIALS AND METHODS
Plant materials and incubation
Tubers of arrowhead (S. pygmaea Miq.) were harvested from a field of the Center for Research on Wild Plants of Utsunomiya University and from a greenhouse of our department at Tohoku University, and they were stored at 4 °C in the dark (Ishizawa et al., 1999). In all experiments, tubers were used after pre-incubation at 25 °C in the dark for 1 d.
Intact tubers were used for experiments of growth analysis. A tuber having a shoot of 7 or 10 mm in length was prepared by cutting the tip of the shoot with a razor blade. Two different systems were used for aerobic treatment and anaerobic treatment. One was a closed system for growth analysis of CO2 and ethylene effects. Tubers were placed in 140 mL plastic vials that each contained 10 mL of 50 mm MES buffer (pH 7·0), in which a part of the shoot emerged from the surface of the medium. For treatments with 1 mm pyrazol, the necessary amount of the inhibitor was added to MES buffer. The vial was closed with a plastic lid and further sealed with adhesive vinyl tape. For hypoxic treatment, N2 gas was introduced into each sealed vial from one of two small holes cut in the wall of the plastic vial. Then the two holes were sealed with vinyl tape and the concentration of O2 in the vial was checked by gas chromatography as described in a previous paper (Ishizawa et al., 1999). Introduction of N2 gas into the vial was continued until the concentration of O2 in the vial reached <1 %. For treatments with CO2 and ethylene, the necessary amount of gas was introduced into a sealed vial through a hole in the side wall with a syringe. For ethylene-free and CO2-free controls, 0·5 mL of 0·25 m mercuric perchlorate and 20 % KOH, respectively, were put in the sealed vial as an absorbent. The other system used was an open system. For treatments with 50 µm aminoethoxyvinylglycine (AVG), tubers were stood in the MES buffer containing AVG in a small vial. For experiments treated with 1-methylcyclopropene (1-MCP), tubers were pre-treated with 0·1 mm 1-MCP in the MES buffer for 24 h and then washed thoroughly with distilled water immediately before the start of incubation (Sisler and Serek, 1997). Tubers were stood in the MES buffer in a small vial. Three vials with tubers were put in a 100 mL glass vessel. The glass vessel was connected to a gas through-flow system as described in a previous paper (Ishizawa et al., 1999). For induction of anoxia, hypoxia or normoxia, pure N2 gas, 1 % O2 or compressed air, respectively, was continuously introduced into the system. All incubations were done at 25 °C in the dark.
Shoot elongation was calculated as the difference between lengths before and after incubation.
RNA isolation
After incubation, shoots (approx. 0·1 g in f.w.) were cut from tubers, frozen in liquid N2, and powdered with a mortar and pestle. Frozen tissue powder collected in a microcentrifuge tube was extracted with 0·5 mL of SDS buffer solution containing 0·5 mL of 50 mm Tris–HCl (pH 7·5) with 1 % SDS and 50 mm EDTA and 0·5 mL of water-saturated phenol. After centrifugation, 0·25 mL of water-saturated phenol and 0·25 mL of chloroform were added to the supernatant. Chloroform of the same volume as the supernatant was added again to the supernatant after centrifugation, and then the upper aqueous layer was collected by centrifugation. After adjustment of the pH to below 6·0, a 0·7 vol. of isopropanol was added to precipitate total RNA. After washing with 70 % ethanol, the precipitate was dissolved in 100 µL of diethyl pyrocarbonate (DEPC)-treated water. A 50 µL aliquot of 6 m LiCl was added to precipitate RNA. RNA was washed with 500 µL of 70 % ethanol, dried, dissolved with 50 µL of DEPC-treated water, and stored at −80 °C until used.
Cloning of cDNAs for α-expansin (EXPA) and XTH genes
Reverse transcription–polymerase chain reaction (RT–PCR) was performed to isolate cDNA fragments of genes encoding EXPA and XTH from arrowhead tubers. First-strand cDNA was synthesized from total RNA (0·2 µg) extracted from arrowhead shoots using Superscript II reverse transcriptase (Invitrogen Corp., Carlsbad, CA, USA) and oligo(dT)20-M4 Adaptor primer (Takara Shuzo, Kyoto, Japan) according to the manufacturer's instructions. PCR was performed with degenerate primers corresponding to the conserved region for EXPA genes [sense primer, 5′-ATGGGIGGIGCNTGYGGNTA-3′ according to Harrison et al. (2001); antisense primer, 5′-YTGCCARTTYTGNCCCCARTT-3′ according to Rose et al.(1997)] and primers corresponding to the conserved region for XTH genes [sense primer, 5′-GARCAYGAYGARATHGAYTTYG-3′; antisense primer, M13 primer M4 (Takara Shuzo)]. The PCR protocol consisted of an initial denaturation at 94 °C for 4 min followed by 40 cycles at 94 °C for 1 min, 50 °C for 1·5 min and 72 °C for 4 min. PCR products were analysed on 1 % (w/v) agarose gel and then directly cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA). DNA sequencing was carried out using a DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and a DNA sequencer (model 373A, Applied Biosystems, Foster City, CA, USA).
To obtain full-length cDNAs, 5′ rapid amplification of cDNA ends (5′-RACE) was performed with a SMART Oligo cDNA amplification kit (Clontech, Palo Alto, CA, USA). The first-strand cDNA for 5′-RACE was synthesized from 1 µg of total RNA according to the manufacturer's protocol. cDNAs containing the 5′ end for arrowhead XTH (SpXTHs) and EXPA (SpEXPAs) were obtained by PCR amplification using an Advantage 2 PCR kit (Clontech) with 5′-specific universal primers designed from the partial cDNA sequences for SpXTHs and SpEXPAs. Gene-specific primers of 5′-RACE for SpEXPAs were 5′-CAG GAA CAG TTG CAG TGC GGC CAC-3′, 5′-CCT TTT GGG GAG TAC CGT ACA ATA GGG-3′, and 5′-GCG ACG TAA CAT CTC GTC GTC TTG TCC CC-3′ and 5′-GGG GTT CTT GAG TTG TCG TCG CCC TAA C-3′. Gene-specific primers of 5′-RACE for SpXTHs were 5′-GAG CAG CAG GAG CGT GCA GCA GTG GGC-3′, 5′-GGA TGG ATG GAT GGG GAT TGG TGG TAG-3′, 5′-CAA CAG GGG TTG TGT CGT TCT ACT TGA C-3′ and 5′-CGT GGT TAC CCC AGC CGA GCA ACC AAA-3′. The thermal cycling protocol consisted of five cycles at 94 °C for 30 s, 70 °C for 30 s and 72 °C for 3 min, followed by five cycles at 94 °C for 30 s, 68 °C for 30 s and 72 °C for 3 min, and finally followed by 25 cycles at 94 °C for 30 s, 66 °C for 30 s and 72 °C for 3 min.
Analysis of transcript levels by RT–PCR
Amounts of mRNAs transcribed from SpXTHs and SpEXPAs were estimated by RT–PCR analysis. PCR amplification was carried out using the gene-specific primers for SpXTH and SpEXPA (Table 1). RT–PCR was run with template cDNAs synthesized from 1, 2 and 4 µg of total RNA. PCR was carried out in limited cycles in which PCR products exponentially increased and reflected the initial quantity of RNA. PCR products were separated on 1 % agarose gels and stained with ethidium bromide. The actin gene was used as an internal control for RT–PCR.
Table 1.
Nucleotide sequences of primers for semi-quantitative RT–PCR
| Gene |
Sense primer |
Antisense primer |
|---|---|---|
| SpEXPA1 | 5′-GGAGGAGGATCTGTAGG-3′ | 5′-CAGGAACAGTTGCAGTGCGGCCAC-3′ |
| SpEXPA2 | 5′-TGCCTTCCAAGCCGGG-3′ | 5′-CCTTTTGGGGAGTACCGTACAATAGGG-3′ |
| SpEXPA3 | 5′-CCTTCTATCTTCATCACCG-3′ | 5′-GCGACGTAACATCTCGTCGTCTTGTCCCC-3′ |
| SpEXPA4 | 5′-AAGTCCATCACCGTGAC-3′ | 5′-GGGGTTCTTGAGTTGTCGTCGCCCTAAC-3′ |
| SpXTH1 | 5′-TGATGGGGACGACGTCT-3′ | 5′-GAGCAGCAGGAGCGTGCAGCAGTGGGC-3′ |
| SpXTH2 | 5′-GGGCAACACCAGCGG-3′ | 5′-GAGCCATCGGCTAACAAA-3′ |
| SpXTH3 | 5′-GGGGACACACACATACAT-3′ | 5′-GGATGGATGGATGGGGATTGGTGGTAG-3′ |
| SpXTH4 | 5′-AAAACCACTCCCCCCGC-3′ | 5′-CAACAGGGGTTGTGTCGTTCTACTTGAC-3′ |
| SpXTH5 | 5′-GGGGCACCAGCGAAGAA-3′ | 5′-GGGGCACCAGCGAAGAA-3′ |
| Actin | 5′-CAGAAAGATGCVTATGTTGGT-3′ | 5′-ATCAACRTCACACTTCATKAT-3′ |
Homology search for amino acid sequences
All of the sequence data were analysed using GENETYX-Mac 8·0 (SDC Software Development, Tokyo, Japan). Searches for homology of nucleotide and amino acid sequences were carried out with FASTA, BLAST and PSI-BLAST in the DNA Data Bank of Japan (DDBJ).
Statistical analysis of data
All data are represented as mean ± s.e. Data were analysed for statistical significance values by Mann–Whitney U-test using Statview version J5·0.
RESULTS
Effects of ethylene and CO2 on elongation of arrowhead tubers under hypoxic conditions
Elongation of shoots in arrowhead tubers is stimulated by three gaseous factors, ethylene, CO2 and O2-deprived conditions (Suge and Kusangi, 1975; Ishizawa et al., 1999). In preliminary experiments, we determined dose–response curves for the effects of ethylene and CO2 on shoot elongation (data not shown). Stimulation of elongation was saturated with 5 µL L−1 ethylene or 5% CO2 when each gas was applied separately to arrowhead tubers. Next, we examined the effects of either 5 % CO2 or 5 µL L−1 ethylene or both on shoot elongation of arrowhead tubers exposed to 1 % O2 or air (Table 2). Ethylene stimulated shoot elongation both in air and at 1 % O2. CO2 stimulated shoot elongation in air but not at 1 % O2. In air, a simultaneous treatment with CO2 and ethylene was more effective than a single treatment with each gas, suggesting a cooperative action of CO2 and ethylene. However, such a cooperative action of CO2 and ethylene diminished under hypoxic conditions. Maximal elongation was obtained when arrowhead tubers were simultaneously exposed to 5 % CO2 and 5 µL L−1 ethylene in 1 % oxygen.
Table 2.
Effects of 5 % CO2, 5 µL L−1 ethylene and 1 % O2 on shoot elongation in arrowhead tubers with or without 1 mm pyrazol
| O2 |
CO2 |
C2H4 |
Pyrazol |
Elongation (mm) |
|---|---|---|---|---|
| Air | − | − | − | 6·3a ± 1·4 |
| − | − | + | 5·1 ± 1·1 | |
| + | − | − | 13·1b ± 1·1 | |
| + | − | + | 10·9 ± 1·3 | |
| − | + | − | 14·3b ± 1·7 | |
| − | + | + | 11·3 ± 1·2 | |
| + | + | − | 20·0c ± 2·0 | |
| + | + | + | 11·9* ± 2·2 | |
| 1 % O2 | − | − | − | 16·4c ± 1·3 |
| − | − | + | 11·8* ± 1·4 | |
| + | − | − | 20·4c ± 1·6 | |
| + | − | + | 15·3* ± 1·4 | |
| − | + | − | 22·3d ± 1·3b | |
| − | + | + | 18·0* ± 1·5 | |
| + | + | − | 24·7d ± 1·8b | |
| + | + | + | 18·3* ± 2·1 |
Elongation was measured after incubation for 2 d in the dark. Data are the means ± s.e. of 6−22 determinations in three different experiments. Significant differences between two gaseous conditions without pyrazol are shown at P < 0·05 with different letters (a, b, c and d). Asterisks show significant differences at P < 0·05 between treatments with and without pyrazol under the same gaseous conditions.
Effects of pyrazol, AVG and 1-MCP on shoot elongation
Effects of various inhibitors were examined to determine interrelationships between CO2, ethylene and O2-deprived conditions. Most of the energy for anoxic elongation of arrowhead shoots is produced by ethanol fermentation (Ishizawa et al., 1999; Sato et al., 2002). Effects of pyrazol, an inhibitor of alcohol dehydrogenase (Dahse et al., 1994), on shoot elongation under various conditions of treatment with a combination of the three gases were examined (Table 2). Pyrazol significantly inhibited elongation at 1 % O2 regardless of the presence or absence of either CO2 or ethylene, or both. Moreover, pyrazol partially inhibited shoot elongation in anoxia (data not shown). However, pyrazol did not significantly affect shoot elongation in air except for that in the presence of both CO2 and ethylene, suggesting that pyrazol prevents the interaction between CO2 and ethylene. The results support an idea that ethanol fermentation is an important energy source to enhance shoot elongation under O2-deprived conditions.
Ethylene production is stimulated under hypoxia (Raskin and Kende, 1984). We examined the effects of AVG, a potent inhibitor of ethylene production, on shoot elongation (Table 3). AVG partially inhibited shoot elongation of arrowhead tubers in air and at 1 % O2, but it did not have significant effects on shoot elongation in anoxia. Similarly, 1-MCP, a potent inhibitor of ethylene action (Sisler and Serek, 1997), inhibited shoot elongation in air and at 1 % O2, but not in anoxia (Table 4). These results suggest that endogenous ethylene is an important stimulator of shoot elongation in normoxia and in hypoxia and that anoxia stimulates shoot elongation independently of ethylene actions.
Table 3.
Effects of AVG on shoot elongation of arrowhead tubers at different concentrations of oxygen
| Treatment |
||
|---|---|---|
| Oxygen conditions |
50 mm AVG |
Elongation (mm) |
| Air | − | 4·6 ± 0·8 |
| + | 2·2* ± 0·4 | |
| 1 % O2 | − | 10·2 ± 0·5 |
| + | 7·4* ± 0·7 | |
| N2 | – | 10·5 ± 0·8 |
| + | 9·1 ± 0·8 |
Elongation was measured after incubation for 2 d in the dark. Data are the means ± s.e. of 21–25 determinations in four different experiments. Asterisks show significant differences at P < 0·05 between treatments with and without AVG at the same oxygen concentrations.
Table 4.
Effects of 1-MCP on shoot elongation of arrowhead tubers at different concentrations of oxygen
| Treatment |
|||
|---|---|---|---|
| Oxygen conditions |
1-MCP |
Elongation (mm) |
|
| Air | – | 8·0 ± 1·1 | |
| + | 2·2* ± 0·8 | ||
| 1 % O2 | – | 12·0 ± 1·3 | |
| + | 7·7* ± 0·8 | ||
| N2 | – | 14·7 ± 2·3 | |
| + | 12·3 ± 1·3 | ||
Elongation was measured after incubation for 2 d in the dark. Data are the means ± s.e. of 6–19 determinations in two different experiments. Asterisks show significant differences at P < 0·05 between treatments with and without 1-MCP at the same oxygen concentrations.
Isolation of EXPA and XTH genes from arrowhead tubers
Growth analysis of arrowhead tubers (Tables 2–4) showed that three different factors, ethylene, CO2 and O2-deprived conditions, stimulate shoot elongation. This suggests that these gaseous factors regulate different processes of cell wall extension. We examined the effects of three gaseous factors on the expression of genes that are closely associated with cell wall extension. We selected two genes encoding EXPA and XTH as molecular markers.
cDNA fragments (each approx. 500 bp) of EXPA and XTH genes were amplified by RT–PCR with RNA, which was extracted from arrowhead shoots incubated for 2 d in air and under hypoxic conditions, and degenerate primers, which were designed from conserved amino acid sequences of EXPA and XTH. For cDNA fragments of each gene, approx. 30 positive clones were isolated and then their DNA sequences were determined. Finally, four different fragments of putative EXPA genes and five different fragments of putative XTH genes were isolated by a homology search for DNA sequences of amplified cDNA fragments using the FASTA program of the DDJB. The EXPA genes were designated SpEXPA1, 2, 3 and 4 according to nomenclature for expansin genes proposed by Kende et al. (2004), and the XTH genes were designated SpXTH1, 2, 3, 4 and 5. The nucleotide sequences containing complete open reading frames (ORFs) of these genes except for the SpXTH2 gene were determined by 5′-RACE. The DDBJ accession numbers of SpEXPA1, 2, 3 and 4 mRNAs are AB196974, AB196975, AB196976 and AB196977. The DDBJ accession numbers of SpXTH1, 2, 3, 4 and 5 mRNAs are AB196978, AB196979, AB196980, AB196981 and AB196982.
Isolated cDNAs of SpEXPA1, 2, 3 and 4 consist of 1355, 1026, 1781 and 1132 bp, respectively, with ORFs of 801, 762, 798 and 804 bp, respectively, 5′-untranslated regions (UTRs) of 146, 111, 319 and 85 bp, respectively, and 3′-UTRs of 408, 153, 664 and 243 bp, respectively. The ORFs of SpEXPA1, 2, 3 and 4 encode proteins of 267, 254, 266 and 268 amino acid residues, respectively. Figure 1 shows alignment of the amino acid sequences predicted from cDNA sequences of SpEXPA1, 2, 3 and 4. The amino acid identity between the two genes among the four SpEXPAs was between 39·7 and 63·6 %. Four SpEXPAs had the same motifs as EXPAs previously identified from various seed plants (Li et al., 2003), i.e. conserved cysteine (C) residues in the N-terminal region of the protein, a putative catalytic domain, an HFD (His-Phe-Asp) motif in the central portion of the protein, and conserved tryptophan (W) residues in the C-terminal region. From the genome sequence database, 26 and 32 genes of EXPA from Arabidopsis and rice, respectively, have been identified (Li et al., 2003). Phylogenetic analysis (Fig. 2) showed that SpEXPA1 and 4 are close to OsEXPA2 and 4, and that SpEXPA3 is close to that of AtEXPA6 and OsEXPA7.
Fig. 1.
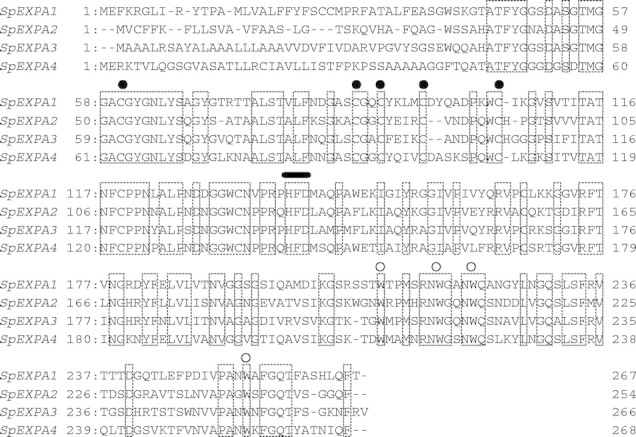
Multiple alignment of the deduced amino acid sequences of four arrowhead α-expansins. The deduced amino acid sequences were aligned by using the Clustal-W program. Positions of highly conserved Cys (C) residues, HFD and a Try (W) residue are indicated by filled circles, a black bar and open circles, respectively, above each amino acid residue. Common amino acid residues of the four α-expansins are enclosed in boxes.
Fig. 2.
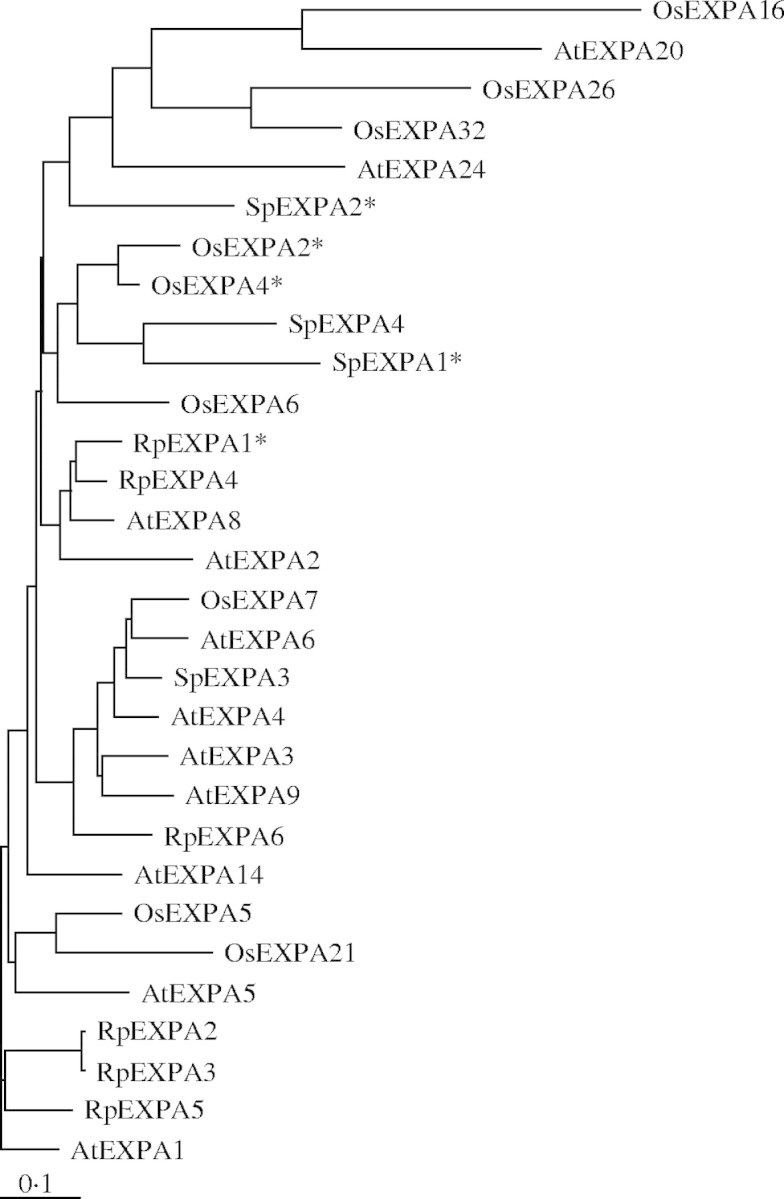
Phylogenetic analysis of α-expansin proteins. The full-length amino acid sequences of four arrowhead expansins, 11 Arabidopsis expansins, nine rice expansins and six Rumex expansins were aligned with Clustal-W and TreeViewPPC software programs. Genes marked with an asterisk show that the expression is enhanced by submergence or O2-deprived conditions. The GenBank accession numbers of Arabidopsis and rice α-expansin genes are given at the expansin web site (http://www.bio.psu.edu/expansins/). Accession numbers of Rumex palustris genes are as follows: RpEXPA1 (AF167360), RpEXPA2 (AF167361), RpEXPA3 (AF167362), RpEXPA4 (AF167363), RpEXPA5 (AF167364), RpEXPA6 (AF167356).
Isolated cDNAs of SpXTH1, 2, 3, 4 and 5 consist of 1101, 708, 1273, 1181 and 1029 bp, respectively, with ORFs of 918, 613, 1005, 912 and 852 bp, respectively, 5′-UTRs of 97, 0, 130, 198 and 94 bp, respectively, and 3′-UTRs of 86, 95, 138, 71 and 83 bp, respectively. The ORFs of SpXTH1, 2, 3, 4 and 5 encode proteins of 306, 204, 335, 304 and 284 amino acid residues, respectively. Figure 3 shows the alignment of the amino acid sequences predicted from cDNA sequences of SpXTH1, 2, 3, 4 and 5. Five SpXTHs shared between 55·8 and 63·0 % amino acid identity. All of the SpXTHs conserved the DEIDFEFLG motif, which is thought to function as a catalytic site of both hydrolase and transferase activities (Okazaki et al., 1992; Campbell and Braam, 1999), and contained one potential N-linked glycosylation signal (N-X-S) immediately following the conserved DEIDFEFLG motif (Campbell and Braam, 1999). At present, the numbers of XTH genes identified in the Arabidopsis and rice genome sequence databases are 33 and 29, respectively. Figure 4 shows a phylogenetic tree of 14 AtXTHs, seven OsXTHs, five SpXTHs and one maize XTH (ZmXTH). All five SpXTHs belong to group I/II subfamilies. SpXTH1 belongs to group I, and the other four SpXTHs belong to group II (Li et al., 2003).
Fig. 3.

Multiple alignment of the deduced amino acid sequences of five XTH cDNAs from arrowhead tubers. The deduced amino acid sequences were aligned by using the Clustal-W program. The black bar shows the predicted active centre of glucanase and the white bar shows the predicted N-linked glycosylation signal (N-X-S). Common amino acid residues of the five XTHs are enclosed in boxes.
Fig. 4.
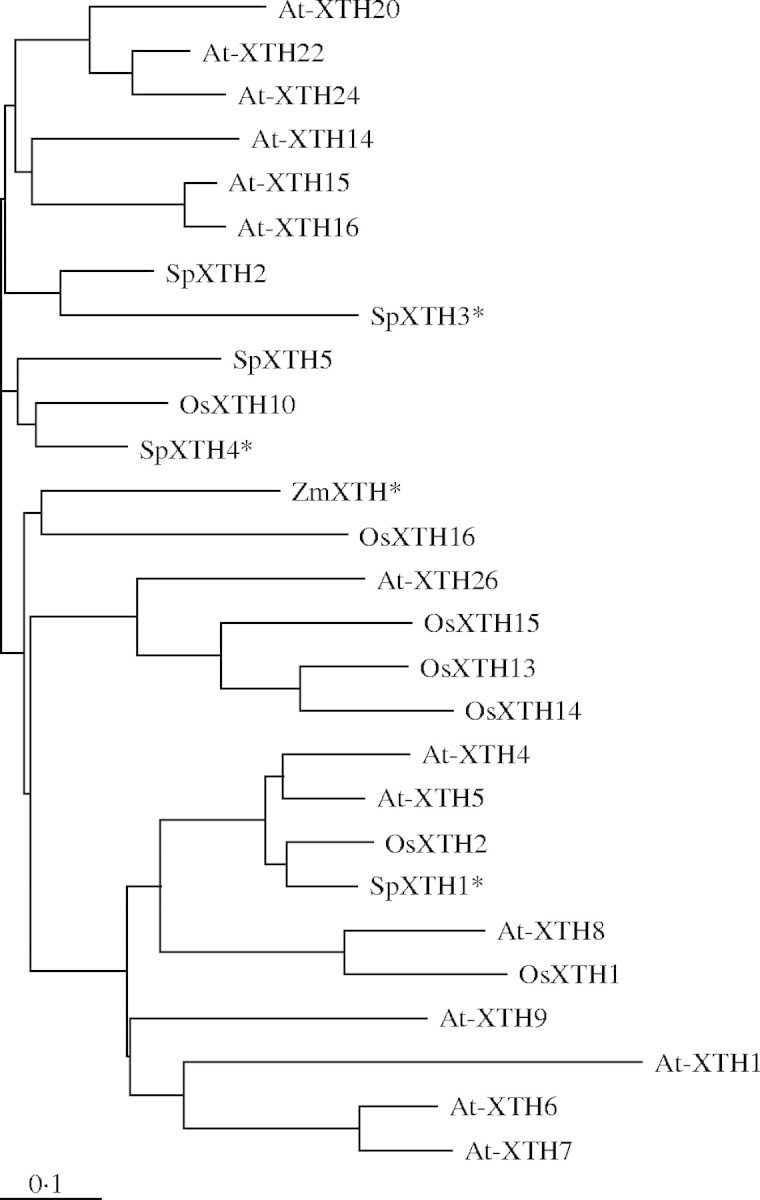
Phylogenetic analysis of XTH proteins. The full-length amino acid sequences of four arrowhead XTHs, 14 Arabidopsis XTHs, seven rice XTHs and one maize XTH were aligned by using Clustal-W and TreeViewPPC software programs. The expressions of four genes (SpXTH1, 3 and 4 and ZmXTH) marked with an asterisk is enhanced by anoxic conditions. Identification numbers (AGI ID, Arabidopsis Genome Initiative 2000) of Arabidopsis genes are summarized by Yokoyama and Nishitani (2001). The GenBank accession number of maize (Zea mays L.) XTH is U15964.
Effects of anoxia, CO2 and ethylene on levels of SpEXPA transcripts
Figure 5A shows the SpEXPAs transcripts of tuber shoots treated with anoxia, 5 % CO2 and 5 µL L−1 ethylene. In air, all of the four SpEXPA transcripts were detected at 12 and 24 h after the start of incubation. The transcript levels of SpEXPA1 and SpEXPA2 increased slightly at 12 and 24 h after the start of incubation under anoxia, whereas the levels of SpEXPA3 and SpEXPA4 did not change. The accumulation of SpEXPA2 transcripts in anoxia seemed to be more than that of SpEXPA1 transcripts. We confirmed that the transcripts of SpEXPA2 increased 6 h after incubation under anoxic conditions in a separate experiment (data not shown). The levels of SpEXPA2 transcripts seemed to be slightly increased by 24 h treatment with 5 % CO2. Ethylene slightly elevated the level of SpEXPA4 transcripts after 24 h incubation. These results suggest that four SpEXPA genes are differently responsive to anoxia, CO2 and ethylene.
Fig. 5.
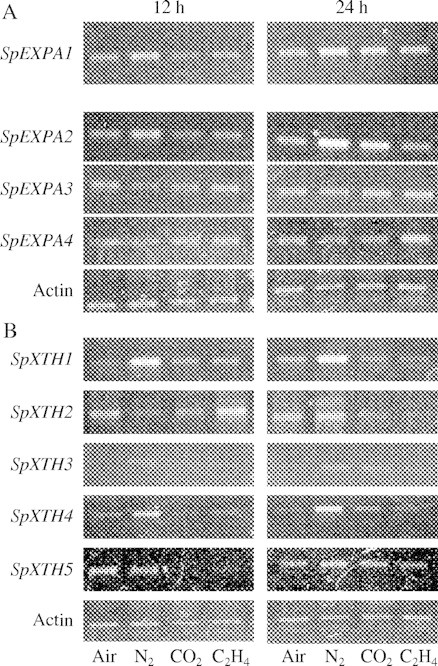
Transcript levels of four expansin genes (A) and five XTH genes (B) of arrowhead tubers incubated in normoxia, anoxia, 5 % CO2 and 5 µL L−1 ethylene at 25 °C in the dark. The incubation period was 12 or 24 h. The PCRs were subjected to 23 cycles (SpEXPA1), 30 cycles (SpEXPA2 and SpEXPA3) and 25 cycles (SpEXPA4 and actin genes) for expansin genes and to 20 cycles (SpXTH1), 30 cycles (SpXTH2), 23 cycles (SpXTH3) and 25 cycles (SpXTH4, XTH5 and actin genes) for XTH genes of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 4 min.
Effects of anoxia, CO2 and ethylene on levels of SpXTH transcripts
Figure 5B shows the transcript levels of SpXTHs in tuber shoots treated with anoxia, 5 % CO2 and 5 µL L−1 ethylene. In air, transcripts of SpXTH1, 2, 4 and 5 were detected throughout a 24 h incubation period, but SpXTH3 transcripts were not detected. Anoxia elevated the levels of SpXTH1 and 4 transcripts after 12 and 24 h incubation and slightly elevated the levels of SpXTH3 transcripts. In contrast, the transcript level of SpXTH2 was slightly lowered after 12 h incubation in anoxia. CO2 inhibited the accumulation of SpXTH5 transcripts after 12 h incubation and slightly enhanced that of SpXTH3 transcripts after 24 h incubation. Ethylene enhanced the accumulation of SpXTH2 transcripts after 12 h incubation, repressed that of SpXTH5 transcripts after 12 h incubation and decreased SpXTH2 transcripts after 24 h incubation. It is notable that only anoxia enhanced the accumulation of SpXTH1 and 4 transcripts.
DISCUSSION
Interactions among the actions of three gaseous factors on elongation have been examined in various plant tissues. In internodes of deepwater rice, hypoxia and high CO2 stimulate ethylene production and thereby cooperatively promote elongation (Raskin and Kende, 1984). On the other hand, in rice seedlings, hypoxia and high CO2 do not stimulate ethylene production (Raskin and Kende, 1983), and the three gaseous factors promote the coleoptile elongation in an additive manner, suggesting that high CO2 and low O2 sensitize seedlings to ethylene or that the three gases stimulate elongation independently (Raskin and Kende, 1983; Ishizawa and Esashi, 1984). On the other hand, in petioles of R. palustris, hypoxia-stimulated petiole elongation is accompanied by enhancement of sensitivity to ethylene and the expression level of the putative ethylene receptor gene (Vriezen et al., 1997). However, in this case, CO2 has no effects on elongation and ethylene production (Voesenek et al., 1997). In Potamogeton pectinatus stems, anoxia-stimulated elongation is not due to ethylene action because ethylene does not stimulate the stem elongation (Summers and Jackson, 1993, 1994) and P. pectinatus is not able to produce ethylene (Summers et al., 1996). In the present study, elongation of arrowhead shoots was significantly promoted by hypoxia and anoxia in addition to ethylene and CO2 (Tables 2–4). The enhanced elongation in hypoxia was inhibited by AVG and 1-MCP, suggesting that endogenous ethylene is partially involved in hypoxic elongation (Tables 3, 4). Hypoxia may increase ethylene production of arrowhead shoots as in deepwater rice or it may enhance the sensitivity of ethylene as in R. palustris. Since ethylene production from ACC requires molecular oxygen (Abeles et al., 1992), ethylene production under anoxia should be greatly suppressed. In fact, AVG and 1-MCP did not have significant effects on anoxic elongation of arrowhead tubers, suggesting that anoxic elongation is independent of ethylene action. Moreover, pyrazol, an inhibitor of alcohol dehydrogenase, was most effective in hypoxic elongation (Table 2). These results suggest that the signal transduction pathway in anoxia is different from that operating in ethylene-stimulated elongation.
Expansins are thought to play a key role in primary wall-loosening processes during plant growth and secondary wall-loosening processes during fruit ripening, pollination, germination and abscission (Cosgrove, 1999; Cosgrove et al., 2002). The Arabidopsis genome and the rice genome contain 38 and 80 ORFs, respectively, and these ORFs encode expansin-like proteins. Expansins are currently divided into four subfamilies, EXPA (α-expansin), EXPB (β-expansin), EXLA (EXPANSIN-LIKE A) and EXLB (EXPANSIN-LIKE B) (Kende et al., 2004). In this study, we isolated cDNA fragments of four EXPA genes (SpEXPA1, SpEXPA2, SpEXPA3 and SpEXPA4) from arrowhead tubers.
The transcript levels of a few expansin genes have been reported to be elevated by submergence or by plant hormones involved in submergence-induced elongation. In deepwater rice, OsEXPA2 and OsEXPA4 are induced by submergence and gibberellin treatment (Cho and Kende 1997), whereas in Japonica rice seedlings, the accumulation of OsEXPA2 and OsEXPA4 transcripts is enhanced by submergence or hypoxia but not by anoxia (Huang et al., 2000). In Rumex, the level of RpEXPA1 transcripts is greatly increased by submergence and ethylene treatment, suggesting that RpEXPA1 is involved in leaf elongation under water (Vriezen et al., 2000). Moreover, two α-expansin genes were isolated from a semiaquatic fern, Regnellidium diphyllum (Kim et al., 2000). These expansins form their own branch rather than being a member of any other expansin subfamily of seed plants in a phylogenetic tree of expansin proteins, but the levels of transcripts of the one gene, RdEXPA1, are increased by submergence or ethylene. These findings indicate that the expression of EXPA genes may play an important role in mediating elongation in submerged plants. In the case of arrowhead (Fig. 5A), anoxia increased the transcript levels of SpEXPA1 and 2, but ethylene did not affect the transcript levels of SpEXPA2. Judging from the responsiveness to anoxia and ethylene, the control of SpEXPA2 transcript levels seems to be different from that of deepwater rice OsEXPA2 and Rumex RpEXPA1. However, three genes, OsEXPA2 and OsEXPA 4 and SpEXPA1, which are responsive to submergence or hypoxia, belong to the same cluster in the phylogenetic tree (Fig. 2). The amino acid sequence of EXPA seems to be related to the function that EXPA fulfills in plant tissues under O2-deprived conditions. The rice genome has more diversity of EXPBs than of EXPA, and the expression of EXPB genes is also correlated with internode elongation of deepwater rice (Lee and Kende, 2001). EXPBs may be important in the regulation of shoot elongation in monocotyledonous plant tissues including arrowhead tubers.
XTHs are enzymes that change cell wall architecture during plant growth and development through modification of the cellulose–xyloglucan framework (Rose et al., 2002). Like expansins, XTHs are versatile in cell wall modification because they are thought to be involved in cell wall extension during growth (Rose and Bennett, 1999) as well as cell wall construction during tissue development (Campbell and Braam, 1999; Rose et al., 2002). The Arabidopsis genome and rice genome have 33 members (Yokoyama and Nishitani, 2001) and 29 members (Yokoyama et al., 2004), respectively, of the XTH gene family. In this study, we isolated cDNA fragments of five members of the XTH gene family (SpXTH1, SpXTH2, SpXTH3, SpXTH4 and SpXTH5) from arrowhead tubers, although one of them (SpXTH2) is a partial fragment. The transcript levels of SpXTH1 and 4 were increased by anoxia but not by ethylene or CO2 (Fig. 5B). Saab and Sachs (1996) found that one of the maize genes encoding XTH (wusl1005, named ZmXTH herein) is induced by flooding or ethylene treatment. Accumulation of ZmXTH transcripts is inhibited by aminooxyacetic acid (AOA), a potent inhibitor of ethylene biosynthesis, which also prevents aerenchyma formation under hypoxia, suggesting that the ZmXTH gene is associated with aerenchyma formation. However, the transcript levels of this gene are elevated by anoxia, in which aerenchyma formation does not occur without ethylene accumulation in tissues. In anoxia, AOA has no effect on the transcript levels of ZmXTH. Saab and Sachs suggested that at least two mechanisms operate for ZmXTH gene induction. One is active under hypoxia, depending on ethylene actions, and the other operates independently of ethylene actions under anoxia. In the case of arrowhead tubers, hypoxic elongation was dependent on ethylene action, but anoxic elongation was independent of ethylene action (Tables 3, 4). The accumulation of SpXTH1 and 4 transcripts was not induced by ethylene but was induced by anoxia. According to these characteristics, SpXTH1 and 4 resemble ZmXTH. Arrowhead plants develop aerenchyma in both aerobic and anaerobic conditions. Aerenchyma formation of aquatic plants is usually different from that of terrestrial plants (Schussler and Longstreth, 1996). SpXTH1 and 4 may be involved in rapid elongation in anoxia rather than aerenchyma formation through cell wall modification.
This study showed that shoot elongation of arrowhead tubers is stimulated by anoxia, ethylene and CO2. The results suggest that hypoxic elongation is partially mediated by ethylene action but that anoxic elongation is independent of ethylene action. We found EXPA genes and XTH genes.
Acknowledgments
We thank Professor H. Omokawa and Dr H. Kuramochi, the Center for Research on Wild Plants, Utsunomiya University, for proving arrowhead tubers. We also thank Mr K. Satoh for help in the cultivation of S. pygmaea. We are grateful to for Mr Banjo Mitsui (Rohm and Haas Japan K.K. AgroFresh) for kindly providing 1-MCP. This study was supported in part by a Grant-in-Aid (14658165 to S.S.) for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME Jr. 1992.Ethylene in plant biology, 2nd edn. San Diego: Academic Press. [Google Scholar]
- Campbell P, Braam J. 1999. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends in Plant Science 4: 361–366. [DOI] [PubMed] [Google Scholar]
- Cho H-T, Kende H. 1997. Expansins in deepwater rice internodes. Plant Physiology 113: 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 1999. Enzymes and other agents that enhance cell wall extensibility. Annual Review of Plant Physiology and Plant Molecular Biology 50: 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho H-T, Hoffmann-Benning S, Moore RC, Blecker D. 2002. The growing world of expansins. Plant and Cell Physiology 43: 1436–1444. [DOI] [PubMed] [Google Scholar]
- Dahse I, Petzold U, Bernstein M. 1994. Redox activity and alcohol-mediated energization of the plasma membrane of plant cells. Journal of Plant Physiology 143: 675–680. [Google Scholar]
- Harada T, Ishizawa K. 2003. Starch degradation and sucrose metabolism during anaerobic growth of pondweed (Potamogeton distinctus A. Benn.) turions. Plant and Soil 253: 125–135. [Google Scholar]
- Harrison EP, McQueen-Mason SJ, Manning K. 2001. Expression of six expansin genes in relation to extension activity in developing strawberry fruit. Journal of Experimental Botany 52: 1437–1446. [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. 2000. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.). Planta 211: 467–473. [DOI] [PubMed] [Google Scholar]
- Ishizawa K, Esashi Y. 1984. Gaseous factors involved in the enhanced elongation of rice coleoptiles under water. Plant, Cell and Environment 7: 239–245. [Google Scholar]
- Ishizawa K, Murakami S, Kawakami Y, Kuramochi H. 1999. Growth and energy status of arrowhead tubers, pondweed turions and rice seedlings under anoxic conditions. Plant, Cell and Environment 22: 505–514. [Google Scholar]
- Kende H, Bradford KJ, Brummell DA, Cho H-T, Cosgrove DJ, Fleming AJ, et al. 2004. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Molecular Biology 55: 311–314. [DOI] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho H-T. 1998. Deepwater rice: a model plant to study stem elongation. Plant Physiology 118: 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cho H-T, Kende H. 2000. α-Expansins in the semiaquatic ferns of Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation. Planta 212: 85–92. [DOI] [PubMed] [Google Scholar]
- Ku HS, Suge H, Rappaport L, Pratt HK. 1970. Stimulation of rice coleoptile growth by ethylene. Planta 90: 333–339. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H. 2001. Expansin of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiology 127: 645–654. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. 2003. Expansins and cell growth. Current Opinion in Plant Biology 6: 603–610. [DOI] [PubMed] [Google Scholar]
- Musgrave A, Jackson MB, Ling E. 1972. Callitriche stem elongaiton is controlled by ethylene and gibberellin. Nature New Biology 238: 93–96. [Google Scholar]
- Okazaki K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K. 1992. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransfease that mediates molecular grafting between matrix polysaccharides in plant cell walls. Journal of Biological Chemistry 268: 25364–25368. [PubMed] [Google Scholar]
- Peschke VM, Sachs MM. 1994. Characterization and expression of transcripts induced by oxygen deprivation in maize (Zea mays L.). Plant Physiology 104: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin I, Kende H. 1983. Regulation of growth in rice seedlings. Journal of Plant Growth Regulation 2: 193–203. [Google Scholar]
- Raskin I, Kende H. 1984. Regulation of growth in stem sections of deep-water rice. Planta 160: 66–72. [DOI] [PubMed] [Google Scholar]
- Ridge I. 1987. Ethylene and growth control in amphibious plants. In: Crawford RMM, ed. Plant life in aquatic and amphibious habitats. Oxford: Blackwell, 53–76. [Google Scholar]
- Rose JKC, Bennett AB. 1999. Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends in Plant Science 4: 176–183. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Lee HH, Bennet AB. 1997. Expression of a divergent expansion gene is fruit-specific and ripening-regulated. Proceedings of the National Academy of Sciences of the USA 94: 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. 2002. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology 43: 1421–1435. [DOI] [PubMed] [Google Scholar]
- Saab IN, Sachs MM. 1996. A flooding-induced xyloglucan endo-transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiology 112: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R. 1980. The anaerobic proteins of maize. Cell 20: 761–767. [DOI] [PubMed] [Google Scholar]
- Sato T, Harada T, Ishizawa K. 2002. Stimulation of glycolysis in anaerobic elongation of pondweed (Potamogeton distinctus A. Benn.) turions. Journal of Experimental Botany 53: 1847–1856. [DOI] [PubMed] [Google Scholar]
- Schussler EE, Longstreth DJ. 1996. Aerenchyma develops by cell lysis in roots and cell separation in leaf petioles in Sagittaria lancifolia (Alismataceae). American Journal of Botany 83: 1266–1273. [Google Scholar]
- Sisler EC, Serek M. 1997. Inhibitors of ethylene responses in plants at the receptor level: recent developments. Physiologia Plantarum 100: 577–582. [Google Scholar]
- Smalle J, Haegman M, Kurepa J, van Montagu M, van der Straeten D. 1997. Ethylene can stimulate Arabidopsis hypocotyls elongation in the light. Proceedings of the National Academy of Sciences of the USA 94: 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suge H, Kusanagi T. 1975. Ethylene and carbon dioxide: regulation of growth in two perennial aquatic plants, arrowhead and pondweed. Plant and Cell Physiology 16: 65–72. [Google Scholar]
- Suge H, Kusanagi T. 1994. Ethylene and carbon dioxide in the regulation of initial growth in Sagittaria trifolia L. Weed Research 39: 237–242. [Google Scholar]
- Summers JE, Jackson MB. 1993. Promotion of stem extension in an aquatic momocot (Potamogeton pectinatus L.) by the complete absence of oxygen, and by partial oxygen shortage. In: Jackson MB, Black CR, eds. Interacting stresses on plants in a changing climate. Heidelberg: Springer-Verlag, 315–325. [Google Scholar]
- Summers JE, Jackson MB. 1994. Anaerobic conditions strongly promote extension by stems of overwintering tubers of Potamogeton pectinatus L. Journal of Experimental Botany 45: 1309–1318. [Google Scholar]
- Summers JE, Voesenek LACJ, Blom CWPM, Lewis MJ, Jackson MB. 1996.Potamogeton pectinatus is constitutively incapable of synthesizing ethylene and lacks 1-aminocyclopropane-1-carboxylic acid oxidase. Plant Physiology 111: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Kuramochi H, Ishizawa K. 2001. Involvement of calcium ion in the stimulated shoot elongation of arrowhead tubers under anaerobic conditions. Plant and Cell Physiology 42: 717–722. [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Vriezen WH, Smekens MJE, Huitink FHM, Bögemann GM, Blom CWPM. 1997. Ethylene sensitivity and response sensor expression in petioles of Rumex species at low O2 and high CO2 concentrations. Plant Physiology 114: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LACJ, Benschop JJ, Cox BMCH, Groeneveld HW, Mellenaar FF, Vreeburg RAM, Peeters AJM. 2003. Interactions between plant hormones regulate submergence-induced shoot elongation in the flooding-tolerant dicot Rumex palustris Annals of Botany 91: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, van Rijn CPE, Voesenek LACJ, Mariani C. 1997. A homolog of the Arabidopsis thaliana ERS gene is actively regulated in Rumex palustris upon flooding. Plant Journal 11: 1265–1271. [DOI] [PubMed] [Google Scholar]
- Vriezen WH, De Graaf B, Mariani C, Voesenek LACJ. 2000. Submergence induces expansin gene expression in flooding-tolerant Rumex palustris and not in flooding-intolerant R. acetosa. Planta 210: 956–963. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. 2001. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology 42: 1025–1033. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKV, Nishitani K. 2004. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiology 134: 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]


