Abstract
• Background and Aims The Laperrine's olive (Olea europaea subsp. laperrinei) is an endemic tree from Saharan massifs. Its populations have substantially regressed since the Pleistocene and are presently distributed in a fragmented habitat. Long-term persistence of this taxon is uncertain and programmes of preservation have to be urgently implemented. To define a conservation strategy, the genetic diversity and breeding system of this tree have to be investigated.
• Methods One hundred and eleven ramets were prospected in the laperrinei populations from the Tamanrasset region, southern Algeria. Genetic polymorphism was revealed at nuclear and chloroplast DNA (cpDNA) microsatellite loci allowing a comparative assessment of the genetic diversity of laperrinei and Mediterranean populations based on bi-parental and maternal markers. Additionally, nuclear microsatellite markers enabled the genotypes to be identified unambiguously.
• Key Results Based on nuclear microsatellite data, the total diversity was high (Ht = 0·61) in laperrinei populations and similar to that observed in western Mediterranean populations. A substantial cpDNA diversity (Ht = 0·19) was also observed. Genetically identical ramets originated from the same stump (which can cover >80 m2) were identified in each population. Sixteen per cent of genets exhibited more than one ramet. In addition, several cases of somatic mutations were unambiguously revealed in distinct ramets stemming from the same stump.
• Conclusions These data show that highly isolated and small laperrinei populations are able to maintain a high genetic diversity. This supports the existence of relict trees persisting for a very long time (probably since the last humid transition, 3000 years ago). It is proposed that the very long persistence associated with an asexual multiplication of highly adapted trees could be a strategy of survival in extreme conditions avoiding a mutational meltdown due to reproduction in reduced populations.
Keywords: Chloroplast DNA, clonal structure, conservation, microsatellite, olive tree, Saharan desert, threatened species, wild genetic resources
INTRODUCTION
The olive (Olea europaea subsp. europaea; Oleaceae) is an emblematic species that represents one of the most important fruit trees in the Mediterranean basin (Loumou and Giourga, 2003). Its primary genetic resources are taxonomically classified in the Olea europaea complex in which six subspecies are recognized (Green, 2002). Wild olive populations are distributed on a large area covering southern Asia, a great part of Africa and southern Europe (Green, 2002). Some populations are presently affected by several threats, due to human activities and/or climatic vicissitudes, so that programmes of evaluation and conservation should be urgently implemented (e.g. Médail et al., 2001). The genetic diversity in this complex has been analysed in a few studies (e.g. Hess et al., 2000; Besnard et al., 2001), but more specific research is needed to investigate the evolution of threatened taxa such as the subspecies maroccana (High Atlas, Morocco) or laperrinei (Saharan mountains).
The Laperrine's olive (Olea europaea subsp. Laperrinei) is a relict taxon restricted to massifs of central-southern Sahara and eastern Sahel (Wickens, 1976; Quézel, 1978; Maley, 1980; Médail et al., 2001; Green, 2002). This tree is present at high altitudes [from (1000) 1400 to 2800 m] on volcanic or eruptive rocks, generally in cliffs and banks of canyons (Fig. 1). This taxon is adapted to very dry conditions and in Hoggar (southern Algeria) it persists in areas reaching a mean rainfall of about 20–100 mm per year (Quézel, 1965). In a recent study (Besnard and Bervillé, 2002), maternally inherited genetic markers have attested that this subspecies is closely related to the eastern Mediterranean olive (O. e. europaea). However, this result was not supported by nuclear data which highlighted closer relationships between the laperrinei populations and north-western African taxa (Hess et al., 2000; Besnard et al., 2001). These genetic relationships probably indicate the existence of relatively recent exchanges between sub-tropical African and Mediterranean populations (during the Pleistocene) but the importance of such events has still to be assessed. This wild olive tree also displays traits of potential interest for the cultivated olive. It could be an important genetic resource for drought adaptation and, for instance, could be used as rootstock in dry areas.
Fig. 1.

Individuals belonging to the subspecies laperrinei in the Ti-n-Aleo canyon at Tanarouatine. The ramets are numbered 1 to 8. Based on molecular characterization, only three genets were distinguished: T-in-Aleo (1 and 2), T-in-Aleo (3) and T-in-Aleo (4–8). The stump of the last one covers about 80 m2.
Olea e. laperrinei populations have substantially regressed with desert expansion during the Pleistocene (Wickens, 1976; Quézel, 1978; Maley, 1980). As a consequence, its present distribution is highly fragmented and populations are generally composed of a few individuals (<100), and sometimes of only one isolated individual. Present populations of O. e. laperrinei endure extreme dry conditions and insufficient rainfall during several consecutive years could seriously alter their local persistence. Thus, this taxon has disappeared from Tibesti, Chad, during the Pleistocene (Wickens, 1976; Quézel, 1978). In addition, camels and cattle browse young shoots of this tree, thus limiting its development and possibilities of regeneration. A preservation programme should allow protection of present populations or reforestation of some zones where O. e. laperrinei is seriously declining or has recently disappeared (Baali-Cherif et al., 2000).
In desert conditions, the survival of O. e. laperrinei is greatly dependent on its particular traits. For instance, this taxon has the capacity to harness water from a great depth (due to an adapted root system) and its reduced leaf surface (lanceolate leaves) limits water loss. Moreover, this taxon is also considered to have great persistence ability. The natural vegetative multiplication (by development of shoots from stump) should favour locally adapted genotypes and/or regeneration of individuals after long unfavourable periods. In small populations, propagation of adapted genotypes via an asexual mechanism should limit inbreeding (which could be associated with inbreeding depression; e.g. Wang et al., 1999) and therefore confer a selective advantage in relict populations (e.g. Cupressus dupreziana; Pichot et al., 2001). Moreover, O. e. laperrinei trees rarely fructify in their natural environment and their regeneration is null (Maire, 1933; Quézel, 1965). For conservation purposes, it will be necessary to understand the reproductive biology of O. e. laperrinei. The survey of the genetic diversity in populations should help to reach this objective and to propose a strategy of preservation (e.g. El Mousadik and Petit, 1996; Pichot et al., 2001).
In this context, the genetic diversity was evaluated and a search was carried out to see if a clonal reproductive strategy occurs in populations of O. e. laperrinei from Hoggar. Genetic polymorphism was investigated at nuclear microsatellite (SSR) loci which are reported to be highly polymorphic and are suitable markers to discriminate different genotypes. These loci allowed the presence of clones in populations to be tested. Additionally, the genetic diversity in O. e. laperrinei in comparison to a few western Mediterranean populations was investigated using both nuclear and chloroplast DNA markers.
MATERIALS AND METHODS
Plant material
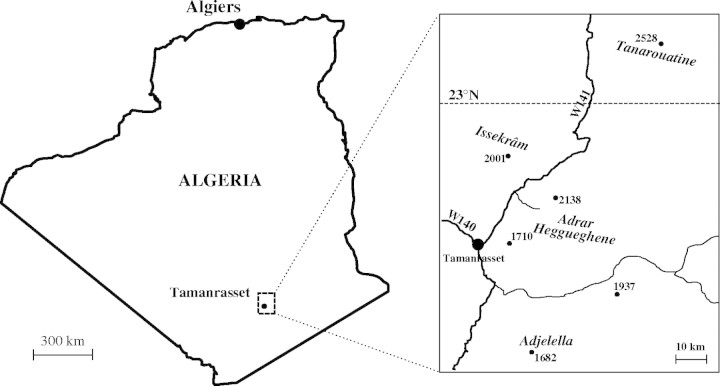
One hundred and eleven ramets of Olea europaea subsp. laperrinei (Batt. & Trab.) Ciferri were sampled in the east and north-east of Tamanrasset in four distinct mountain massifs (Adjelella, Adrar Heggueghene, Issekrâm and Tanarouatine; Table 1 and Figs 1 and 2). These mountains are located in an area covering about 2500 km2. It was attempted to prospect all ramets present in each massif but, on some massifs (i.e. Adjelella and Adrar Heggueghene), a few ramets growing in cliffs were not accessible and were not prospected. A few leaves were collected and desiccated in silica gel for each ramet sampled. Furthermore, in Issekrâm, four distinct ramets (named Ilennanene 8a, b, c and d) originated from the same old stump were sampled on an area of about 20 m2. These ramets have each a limited development and are intensively grazed by camels and cattle. They were characterized in order to test their genetic identity.
Table 1.
Geographic origin of the wild populations characterized in the present study
| Subspecies |
Geographic origin |
Altitude (m) |
Latitude |
Longitude |
NR |
NG |
|---|---|---|---|---|---|---|
| Subsp. laperrinei | Adjelella, south-east Tamanrasset, Algeria | 1530 | 22°38′N | 5°3′E | 10 | 9 |
| Adrar Heggueghene, east Tamanrasset, Algeria | 1390–1600 | 22°47′N | 5°37′E | 65 | 57 | |
| Issekrâm, north Tamanrasset, Algeria | 1525–1605 | 22°55′N | 5°33′E | 23 | 20 | |
| Tanarouatine, north-east Tamanrasset, Algeria | 1580–1600 | 23°06′N | 6°03′E | 13 | 8 | |
| Subsp. europaea | Gué de Constantine, south-east Algiers, Algeria | 30 | 36°42′N | 3°33′E | 20 | 20 |
| Birkhadem, south-east Algiers, Algeria | 30 | 36°42′N | 3°39′E | 14 | 14 | |
| Mount Belloua, Tizi Ouzou, Algeria | 790 | 36°47′N | 4°04′E | 7 | 7 | |
| Tamanar, Essaouira, Morocco | ∼25 | 32°00′N | 9°33′W | 6 | 6 |
NR corresponds to the number of ramets prospected at each site, and NG corresponds to the number of genets identified among these ramets.
Fig. 2.
Geographical location of the four mountain massifs (near Tamanrasset; 22°47′N, 5°31′E) in which populations of O. e. subsp. laperrinei were prospected in the present study.
Additionally, to compare the genetic diversity of laperrinei populations with that of populations belonging to the europaea subspecies, wild Mediterranean olives (oleasters) were characterized. Thirty-four ramets were prospected in the south-east of Algiers (on two sites separated by 10 km: Birkhadem and Gué de Constantine; Table 1). Oleasters from Tamanar, Morocco (six) and Mt Belloua, Algeria (seven), previously characterized using RAPDs and cytoplasmic markers (Besnard et al., 2001, 2002), were also considered in this study (Table 1).
All individual DNAs were extracted from leaves using a CTAB method (Besnard et al., 2000).
Genetic markers
To characterize all individuals, nuclear microsatellites [or single strand repeats (SSR)] were chosen because such genetic markers should reveal a higher level of polymorphism compared with other markers (e.g. cytoplasmic DNA, AFLP or RAPD; Belaj et al., 2003) and allow a direct estimation of the heterozygosity. Nine SSR loci were selected from the literature: DCA1, DCA3, DCA8, DCA9, DCA14, DCA15 (Sefc et al., 2000; Bandelj et al., 2004), GAPU45 (Carriero et al., 2002), PA(ATT)2 (Saumitou-Laprade et al., 2000; Khadari et al., 2003) and EMO3 (de la Rosa et al., 2002). Their choice was based on three criteria: (1) loci displaying polymorphism (from 4 to 11 alleles) were selected in the cultivated olive; (2) only loci with an observed heterozygosity (Ho) similar to the expected heterozygosity (Hs) were used to eliminate all loci with null alleles; and (3) in order to perform multiplexing of loci, allele size range was considered. For each locus, one primer was labelled with a fluorochrome. Two distinct fluorochromes were used: FAM (blue) or HEX (green). PCR amplification of each locus was done separately. The PCR reaction mixture contained 10 ng of DNA template, 1× reaction buffer, 2 mm MgCl2, 0·2 mm dNTPs, 0·2 µmol of each primer, and 0·75 unit of DNA polymerase (Invitrogen) in a total volume of 25 μL. Reaction mixtures were incubated in a thermocycler (T1; Biometra) for 4 min at 94 °C followed by 36 cycles consisting of 1 min at 94 °C, 1 min at the defined annealing temperature (50 or 55 °C) and 1 min at 72 °C. The last cycle was followed by a 10-min extension at 72 °C. Electrophoresis of PCR products was directly carried out on a denaturing 5 % polyacrylamide gel using an automated sequencer (ABI 377; Applied Biosystems).
Twenty polymorphic sites (i.e. nine microsatellite or indel sites and 11 PCR-RFLPs) of the chloroplast DNA (cpDNA) were also investigated in the laperrinei and western Mediterranean europaea populations: ccmp5, ccmp7 (Weising and Gardner, 1999), the 14 polymorphic loci developed by Besnard et al. (2003), a microsatellite locus present in the trnT–trnL intergenic spacer and three additional restriction sites in the matK spacer, which have recently been identified to be polymorphic in the olive complex (Table 2). The new loci (Table 2) were PCR amplified at an annealing temperature of 53 °C using conditions described before. For the other loci, PCR conditions reported in Weising and Gardner (1999) and Besnard et al. (2003) were used. PCR or restricted-PCR fragments were electrophoresed on a 5 % acrylamide gel as previously indicated. A sub-sample of five individuals per massif was characterized for each locus. The polymorphic loci were then used to characterize the whole sample.
Table 2.
Primer pairs for PCR amplifications of three chloroplast DNA regions displaying polymorphism in the Olea europaea complex (G. Besnard, R. Rubio de Casas and P. Vargas, unpubl. res.)
| cpDNA region |
Primer sequence (5′–3′) |
Fragment name |
T (°C) |
Polymorphism type |
Fragment size (bp) |
|---|---|---|---|---|---|
| trnT-L | F: CATTCCTCCGCTTTCATTCG | trnT-L-poly-A | 53 | LP; Poly A | 106, 107 |
| R: TATGTCTCTCTTCCTGCCAC | |||||
| matK | F: ATATCCACTTATCTTTCAGGAG | matK-2-TaqI/MseI | 53 | RS; TaqI | 101 → 101 (–) |
| R: TGGATTTATTGTCATAACCTGG | → 63 + 38 (+) | ||||
| RS; MseI | 101 → 101 (–) | ||||
| → 54 + 47 (+) | |||||
| matK | F: AGATAGTAAAATCTCATAATTTTC* | matK-3-TaqI | 53 | RS; TaqI | 102 → 102 (−) |
| R: GGGGTATTAGTATATCTAACAC | → 24 + 78 (+) |
The fragment names, size variants, polymorphism types and annealing temperatures (T) are given.
The bold nucleotide indicates a nucleotide change comparatively to the reference sequences (A→T) to create an absence–presence polymorphism of a TaqI restriction site.
LP, Length polymorphism; RS, restriction site polymorphism (nucleotide substitution); (−), absence of the restriction site; (+), presence of the restriction site.
Statistical analyses
Based on nuclear SSR data, the genetic relationships between ramets of the subspecies laperrinei were assessed. Shared allele distances, defined as 1 – proportion of shared alleles (Jin and Chakraborty, 1993), were computed between pairs of all individuals using the software Populations 1.2·28 (Langella, 1999). A phenetic tree was built using the neighbour-joining algorithm (Nei, 1987). Bootstrap values were computed using 1000 re-samplings on loci to evaluate support of the branches in both phenetic analyses.
When identical genotypes were identified on several independent ramets, it was necessary to evaluate the possibility that they represented the same genet. The probability that identical genotypes could result from independent formation of zygotes was estimated (e.g. Parks and Werth, 1993; Setsuko et al., 2004). The probability that a zygote acquires a given diploid genotype, Pgen, was calculated, following Parks and Werth (1993). Pgen = (Π pi)2h where pi is the frequency in the population of each allele represented in the genotypes and h is the number of loci that are heterozygous. The Pgen value represents the probability that two sampled ramets belonging to different genets would have the same genotype by chance.
Observed (Ho) and expected (Hs) heterozygosity was computed at each nuclear SSR locus independently in the laperrinei and western Mediterranean europaea populations using the Fstat software (Goudet, 1995). These estimations were based on the different genets and enabled the detection of a putative excess of homozygosity at a locus due to null alleles. The total genetic diversity (Ht; Nei, 1987) was also estimated.
RESULTS AND DISCUSSION
Marker data
Nuclear SSR markers showed a very good transferability from europaea to laperrinei confirming previous observations (Rallo et al., 2003). The nine nuclear SSR loci were polymorphic in both subspecies (Table 3). The total number of alleles revealed in these taxa was similar [85 for western Mediterranean (on 47 individuals) and 89 for laperrinei populations (on 94 genets from Hoggar; see below); Table 3]. The total gene diversity was also comparable in both subspecies (Ht = 0·63 and 0·61; Table 3). Only 31 alleles (21·7 %) were common to both europaea and laperrinei samples analysed indicating a great differentiation between these two taxa. Moreover, in the populations studied, the observed heterozygosity was very similar to the expected heterozygosity at each nuclear SSR locus, suggesting no excess of homozygosity in populations. In addition, the investigated loci should not display null alleles (or at a very low frequency; <2 %) and should be suitable markers for a genetic population study in wild olive relatives. Such an absence of null alleles is probably due to the choice of nuclear SSR loci which have revealed very low deviation in the observed heterozygosity from the expected heterozygosity in previous studies on cultivated olives (Sefc et al., 2000; de la Rosa et al., 2002; Khadari et al., 2003; Bandelj et al., 2004). It is recommended that this choice criterion be used to avoid loci displaying null alleles being included.
Table 3.
Number of alleles (Na), observed and expected heterozygosities (Hoand Hs), total genetic diversity (Ht) for each locus for pooled data from each of the two subspecies, europaea and laperrinei
|
europaea |
laperrinei |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus |
Allele size range |
Na |
Ho |
Hs |
Ht |
Allele size range |
Na |
Ho |
Hs |
Ht |
||||||||||
| Nuclear SSR | ||||||||||||||||||||
| DCA1 | 208–244 | 10 (5) | 0·44 | 0·45 | 0·45 | 222–280 | 24 (5) | 0·91 | 0·92 | 0·92 | ||||||||||
| DCA3 | 231–255 | 6 (3) | 0·52 | 0·45 | 0·45 | 229–253 | 8 (3) | 0·73 | 0·69 | 0·68 | ||||||||||
| DCA8 | 127–165 | 16 (5) | 0·83 | 0·83 | 0·89 | 119–147 | 11 (5) | 0·88 | 0·83 | 0·83 | ||||||||||
| DCA9 | 167–207 | 16 (9) | 0·88 | 0·82 | 0·84 | 169–193 | 11 (9) | 0·58 | 0·61 | 0·63 | ||||||||||
| DCA14 | 153–192 | 13 (0) | 0·91 | 0·85 | 0·88 | 144–165 | 12 (0) | 0·83 | 0·77 | 0·78 | ||||||||||
| DCA15 | 247–271 | 5 (1) | 0·59 | 0·57 | 0·61 | 251–258 | 2 (1) | 0·20 | 0·19 | 0·19 | ||||||||||
| PA(ATT)2 | 106–124 | 5 (2) | 0·66 | 0·61 | 0·66 | 106–109 | 2 (2) | 0·13 | 0·12 | 0·12 | ||||||||||
| GAPU45 | 183–185 | 2 (1) | 0·04 | 0·04 | 0·04 | 185–193 | 5 (1) | 0·62 | 0·58 | 0·59 | ||||||||||
| EMO3 | 212–226 | 12 (5) | 0·92 | 0·87 | 0·89 | 195–224 | 14 (5) | 0·84 | 0·79 | 0·79 | ||||||||||
| Total | – | 85 (31) | 0·64 | 0·61 | 0·63 | – | 89 (31) | 0·64 | 0·61 | 0·61 | ||||||||||
| cpDNA* | – | 5 (0) | – | – | 0·67 | – | 4 (0) | – | – | 0·19 | ||||||||||
Nuclear and cpDNA data were analysed separately.
For Na, the number of alleles shared between laperrinei and western europaea populations is in brackets.
Based on the combination of the 20 investigated characters (see Table 4).
Four cpDNA haplotypes were revealed in the subspecies laperrinei (Table 4): CL1 (86 %), CL2 (3 %), CL3 (9 %) and CL4 (2 %). The laperrinei haplotypes are related to the haplotype CE1 from the Mediterranean basin (Table 4) and all share a deletion in trnS-G-indel-1 specific to the lineage E1 (Besnard et al., 2002, 2003). This sustains the hypothesis of a single maternal origin of present populations in Hoggar probably originating in northern Africa (Besnard et al., 2002). The four haplotypes were present in the Adrar Heggueghene massif, while only one haplotype (CL1) was revealed in the Adjellela and Tanarouatine massifs. Furthermore, in the sample of the subspecies europaea, cpDNA haplotypes CE1 (4.3 %), COM1 (13·1 %), COM2 (65·2 %), CCK (15·2 %) and CCK2 (2·2 %) were identified. These haplotypes belong to three distinct cpDNA lineages (E1, E2 and E3; Table 4) as described in Besnard et al. (2002). The haplotype CCK2 is closely related to CCK and is newly described in this study (Table 4). The total cpDNA genetic diversity was higher in western Mediterranean populations (Ht = 0·67) than in laperrinei populations (Ht = 0·19), but this is probably a consequence of multiple maternal origins in the europaea populations from the western Mediterranean (Besnard et al., 2002).
Table 4.
Chloroplast DNA haplotypes detected in the populations from western Mediterranean (europaea) and Hoggar (laperrinei) based on the combination of 20 loci
| Characters |
||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype |
Lineage* |
ccmp5† |
ccmp7† |
trn T-L-poly-T |
trn T-L-poly-A |
psb K-trn S-poly-T/A |
trn G-poly-T |
trn G-R-poly T |
trn S-G-indel-1 |
trn S-G-indel-2 |
Trn T-L-TaqI |
Trn T-L-AcsI |
mat K-RsaI |
mat K-TaqI |
trn S-G-MseI |
trn S-G-NdeI |
mat K-2-TaqI |
mat K-2-MseI |
mat K-3-TaqI |
trn G-PstI |
psb K-trn S-MseI |
|||||||||||||||||||
| CE1 | E1 | 105 | 120 | 81 | 106 | 109 | 87 | 60 | 105 | 60 | + | − | + | + | + | + | − | − | + | − | − | |||||||||||||||||||
| CL1 | E1 | 105 | 120 | 81 | 106 | 109 | 87 | 60 | 105 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| CL2 | E1 | 105 | 120 | 81 | 106 | 108 | 87 | 60 | 105 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| CL3 | E1 | 105 | 120 | 82 | 106 | 109 | 87 | 60 | 105 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| CL4 | E1 | 104 | 120 | 81 | 106 | 109 | 87 | 60 | 105 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| CCK | E3 | 106 | 120 | 81 | 106 | 109 | 87 | 60 | 114 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| CCK2 | E3 | 105 | 120 | 81 | 106 | 109 | 87 | 60 | 114 | 60 | + | − | + | + | + | − | − | − | + | − | − | |||||||||||||||||||
| COM1 | E2 | 106 | 121 | 81 | 106 | 108 | 87 | 60 | 117 | 52 | + | − | + | + | − | − | − | − | + | − | − | |||||||||||||||||||
| COM2 | E2 | 105 | 121 | 81 | 106 | 108 | 87 | 60 | 117 | 52 | + | − | + | + | − | − | − | − | + | − | − | |||||||||||||||||||
The haplotypes CE1, CCK, CCK2, COM1 and COM2 were detected in europaea populations while CL1 to CL4 were only detected in laperrinei populations.
Eight polymorphic loci were observed.
For the nine first fragments, size variants in base pairs are given. For the next ones, restriction site presence (+) or absence (−) are indicated.
Named according to Besnard et al. (2002; unpubl. res.).
For ccmp5 and ccmp7, size of the variants was corrected comparatively to the previous estimation (Besnard et al., 2002).
Evidence of clones in laperrinei populations
The genetic similitude between all ramets of the subspecies laperrinei was assessed using a phenetic approach. In the phenogram, most branches were very poorly supported and there is no obvious individual cluster corresponding to the four massifs prospected in the Tamanrasset area (data not shown). Furthermore, the present results indicate that several ramets display the same genotype based on the nine loci. In all cases, genetically undistinguished ramets were sampled on the same site at a relatively small distance (<15 m; for example, see Fig. 1). Thus, on the 111 ramets analysed, only 94 genetic profiles were distinguished in the subspecies laperrinei. In contrast, based on nuclear SSR markers, the europaea individuals from the Maghreb were all distinguished. In the laperrinei populations, 12 distinct genetic profiles were each assigned to several ramets. For these genets, the probability of obtaining the same genetic profile (Pgen) was extremely low (ranging from 2·4 × 10−11 for I-n-Tôunine 9 and 10 to 5·7 × 10−7 in Ti-n-Aleo1-2) and, therefore, two different genotypes are expected to be distinguished in the present analysis. Consequently, it was considered that two undistinguished ramets correspond to a clone. There is no doubt that the undistinguished ramets have resulted from the development of shoots from an old stump because, in all cases, undistinguished ramets were sampled on the same site at small distances from each other. These genets can cover >80 m2 (Fig. 1) and should have originated from very old stumps (probably persisting for >1000 years). Clonal growth was similarly reported in the europaea subspecies based on RAPD data [Besnard, 1999; oleaster populations Al Asharinah (Syria), Torvizcon (Spain) and Mt Boron (France)]. Nevertheless, the importance of this feature was not assessed in Mediterranean populations but the observation of stumps with a surface superior to 20 m2 was exceptional, maybe due to higher competition in Mediterranean populations.
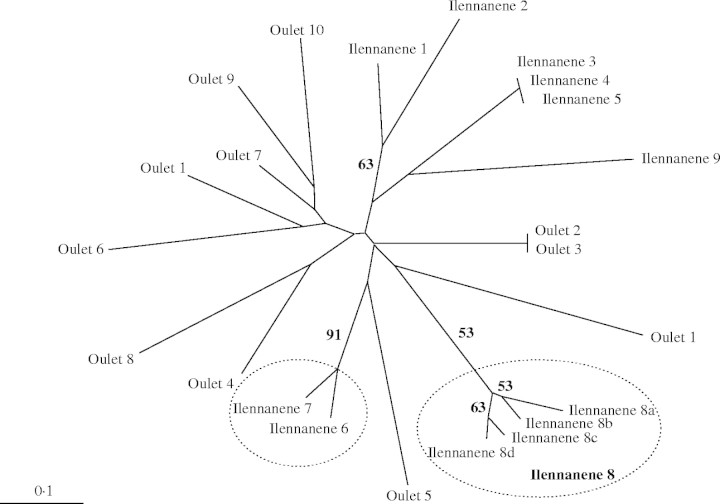
In the Adrar Heggueghene population, the three ramets of the genet I-n-Toûnine 1-2-3 each display three alleles at the DCA1 locus (e.g. allele sizes 236, 240 and 244). Such a result was found only on this genet. Similar observations have been reported on SSR loci in other vegetative-reproducing species such as grape vine (Franks et al., 2002; Riaz et al., 2002). It can be supposed that the presence of a third allele is due to a somatic mutation and that chimerism has been maintained during the development of the individual (e.g. Franks et al., 2002; Rodríguez López et al., 2004). Another interesting feature concerns the individual Ilennanene 8 for which the four ramets are genetically very similar but different (Fig. 3). Indeed, each ramet can be distinguished from each other, based on one to three alleles at the loci DCA1 (genotypes 240–272, 240–274 or 240–276), DCA14 (genotypes 151–156 or 152–156) and DCA9 (genotypes 181–189 or 181–193). These results were verified by two independent characterizations. These observations demonstrate that mutations can accumulate along time on distinct ramets originating from the same ancestral genet, as already shown in cultivated olive during prolonged periods of vegetative reproduction (Garcia-Díaz et al., 2003; Lopes et al., 2004). This propensity for clonal variation in wild-growing trees probably indicates that some stumps are very old and/or persist in environmental conditions, such as an intense grazing, favouring an emergence of mutations present in meristematic tissues. Indeed, shoot damage can unlock somatic mutations by promoting axillary growth (e.g. Marcotrigiano, 2000). Furthermore, it is suspected that other ramets from the same stump were distinguished due to mutations. Such ramets should be genetically similar and sampled on the same site and at a small distance. Among similar ramets, only Ilennanene 6 and 7 are indicative of this pattern (Fig. 3). These ramets were sampled at a distance of 3 m and were distinguished by both alleles of the locus DCA1 (genotypes 246–270 and 240–272). Excluding the locus DCA1, the probability of obtaining the same genetic profile for the eight other loci for Ilennanene 6 and Ilennanene 7 was very low (Pgen = 7·4 × 10−5). Consequently, it is suspected that these ramets originated from the same stump.
Fig. 3.
Un-rooted phenetic tree showing the genetic similitude between ramets of the Issekrâm population using nine nuclear SSR loci. This analysis is based on shared allele distances (Jin and Chakraborty, 1993) and the neighbour joining algorithm (Nei, 1987). The bootstrap values superior to 50 % are indicated on each corresponding branch. The two genets Oulet 2-3 and Ilennanene 3-4-5 display two and three ramets, respectively. The surrounded clusters of ramets correspond to two groups of genetically similar ramets thought to be distinguished based on mutations.
Conclusion and further prospects
To summarize, approx. 16 % of genets with several ramets were found in the laperrinei populations. Only four individuals displayed more than two ramets (from 3 to 5). The mean number of ramets per genet was 1·23. The long survival in combination with clonal growth should be an important feature explaining the persistence of the laperrinei populations in Saharan mountains (Honnay and Bossuyt, 2005). This probably explains why this olive subspecies is able to maintain a high genetic diversity in very small populations over a very long period. It remains to clarify if the taxon could have sexually reproduced since the last humid transition, about 3000–3500 years ago (Quézel, 1965). The future persistence of these populations should not be dependent on strong genetic erosion (e.g. genetic drift due to sexual reproduction in reduced populations) but could be affected by serious environmental changes.
The authors can advise on the strategy to use for genetic resources management and the reforestation of zones where O. e. laperrinei is seriously declining or has recently disappeared. The vegetative propagation of existing individuals in different massifs or the production of novel plants stemming from seed germination can be proposed. The first alternative has the advantage of employing individuals which are highly adapted to very dry conditions since these trees have been naturally selected over a very long period. Such trees should be preserved in collections and characterized for their drought resistance.
The analysis of other populations is still needed to give a more exhaustive inventory of the genetic resources of this subspecies. Population genetic studies should bring new insights into the reproductive strategy of this taxon. Prospects, particularly from Niger and Tassili n'Adjer, are in progress to attain these objectives.
Acknowledgments
We thank Kristina M. Sefc, Nadia Bouguedoura, Valérie Legué, Nicole Galland, Anne Streiff, Nevena Basic, Christian Parisod and Nicolas Juillet for their helpful comments on this manuscript.
LITERATURE CITED
- Baali-Cherif D, Kara-Mostefa-Khelil L, Zemit O, Meghaoui A, Sahki A, Bouguedoura N. 2000. Etude de quelques aspects biologiques de l'olivier de Laperrine (Olea laperrini) en vue de la mise en place d'une banque ex situ. In: llizi W, ed. Redecouvrir et réinventer une sylviculture en zones arides. Djanet: CRSTRA-INRF, 82–89. [Google Scholar]
- Bandelj D, Jakše J, Javornik B. 2004. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica 136: 93–102. [Google Scholar]
- Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, et al. 2003. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theoretical and Applied Genetics 107: 736–744. [DOI] [PubMed] [Google Scholar]
- Besnard G. 1999.Etude de la diversité génétique de l'olivier cultivé et de ses formes sauvages apparentées à l'aide de marqueurs moléculaires: applications pour l'identification variétale et pour la gestion des ressources génétiques. PhD Thesis, Université Montpellier II. [Google Scholar]
- Besnard G, Bervillé A. 2002. On chloroplast DNA variations in the olive (Olea europaea L.) complex: comparison of RFLP and PCR polymorphisms. Theoretical and Applied Genetics 104: 1157–1163. [DOI] [PubMed] [Google Scholar]
- Besnard G, Baradat P, Chevalier D, Tagmount A, Bervillé A. 2001. Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rRNA genes. Genetic Resources and Crop Evolution 48: 165–182. [Google Scholar]
- Besnard G, Khadari B, Baradat P, Bervillé A. 2002.Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theoretical and Applied Genetics 104: 1353–1361. [DOI] [PubMed] [Google Scholar]
- Besnard G, Khadari B, Villemur P, Bervillé A. 2000. Cytoplasmic male sterility in the olive (Olea europaea L.). Theoretical and Applied Genetics 100: 1018–1024. [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. 2003. A set of primers for length and nucleotide-substitution polymorphism in chloroplastic DNA of Olea europaea L. (Oleaceae). Molecular Ecology Notes 3: 651–653. [Google Scholar]
- Carriero F, Fontanazza G, Cellini F, Giorio G. 2002. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theoretical and Applied Genetics 104: 301–307. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics 92: 832–839. [DOI] [PubMed] [Google Scholar]
- Franks T, Botta R, Thomas MR, Franks J. 2002. Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theoretical and Applied Genetics 104: 192–199. [DOI] [PubMed] [Google Scholar]
- Garcia-Díaz A, Oya R, Sánchez A, Luque F. 2003. Effect of prolonged vegetative reproduction of olive tree cultivars (Olea europaea L.) in mitochondrial homoplasmy and heteroplasmy. Genome 46: 377–381. [DOI] [PubMed] [Google Scholar]
- Goudet J. 1995. Fstat (version 1.2): a computer programme to calculate F-statistics. Journal of Heredity 86: 485–486. [Google Scholar]
- Green PS. 2002. A revision of Olea L. (Oleaceae). Kew Bulletin 57: 91–140. [Google Scholar]
- Hess J, Kadereit JW, Vargas P. 2000. The colonization history of Olea europaea L. in Macaronesia based on internal transcribed spacer 1 (ITS-1) sequences, randomly amplified polymorphic DNAs (RAPD), and inter simple sequence repeats (ISSR). Molecular Ecology 9: 857–868. [DOI] [PubMed] [Google Scholar]
- Honnay O, Bossuyt B. 2005. Prolonged clonal growth: escape route or route to extinction? Oikos 108: 427–432. [Google Scholar]
- Jin L, Chakraborty R. 1993. Estimation of genetic distance and coefficient of gene diversity from single-probe multilocus DNA fingerprinting data. Molecular Biology and Evolution 11: 120–127. [DOI] [PubMed] [Google Scholar]
- Khadari B, Breton C, Moutier N, Roger JP, Besnard G, Bervillé A, et al. 2003. The use of molecular markers for germplasm management in a French olive collection. Theoretical and Applied Genetics 106: 521–529. [DOI] [PubMed] [Google Scholar]
- Langella O. 1999. Populations 1.2.28 software (http://www.pge.cnrs-gif.fr/bioinfo/populations). [Google Scholar]
- Lopes MS, Mendonça D, Sefc KM, Gil FS, da Câmara Machado A. 2004. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience 39: 1562–1565. [Google Scholar]
- Loumou A, Giourga C. 2003. Olive groves: “The life and identity of the Mediterranean”. Agriculture and Human Values 20: 87–95. [Google Scholar]
- Maire R. 1933. Études sur la flore et la végétation du Sahara central. Mémoires de la Société d'Histoire Naturelle de l'Afrique du Nord No. 3, Mission du Hoggar II, 166–168. [Google Scholar]
- Maley J. 1980. Les changements climatiques de la fin du Tertiaire en Afrique: leur conséquence sur l'apparition du Sahara et de sa végétation. In: Williams MAJ, Faure, H, eds. The Sahara and the Nile. Rotterdam: AA Balkema, 63–86. [Google Scholar]
- Marcotrigiano M. 2000. Herbivory could unlock mutations sequestered in stratified shoot apices of genetic mosaics. American Journal of Botany 87: 355–361. [PubMed] [Google Scholar]
- Médail F, Quézel P, Besnard G, Khadari B. 2001. Systematics, ecology and phylogeographic significance of Olea europaea L. subsp. maroccana (Greuter & Burdet) P. Vargas et al, a relictual olive tree in south-west Morocco. Botanical Journal of the Linnean Society 137: 249–266. [Google Scholar]
- Nei M. 1987.Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Parks JC, Werth CR. 1993. A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae). American Journal of Botany 80: 120–126. [DOI] [PubMed] [Google Scholar]
- Pichot C, El Maataoui M, Raddi S, Raddi P. 2001. Surrogate mother for endangered Cupressus. Nature 412: 39. [DOI] [PubMed] [Google Scholar]
- Quézel P. 1965.La végétation du Sahara. Du Tchad à la Mauritanie. Stuttgart: Gustav Fischer Verlag. [Google Scholar]
- Quézel P. 1978. Analysis of the flora of Mediterranean and Sahara Africa. Annals of the Missouri Botanical Garden 65: 479–534. [Google Scholar]
- Rallo P, Tenzer I, Gessler C, Baldoni L, Dorado G, Martín A. 2003. Transferability of olive microsatellite loci across the genus Olea Theoretical and Applied Genetics 107: 940–946. [DOI] [PubMed] [Google Scholar]
- Riaz S, Garrison KE, Dangl GS, Boursiquot JM, Meredith CP. 2002. Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. Journal of the American Society for Horticultural Science 127: 508–514. [Google Scholar]
- Rodríguez López CM, Wetten AC, Wilkinson MJ. 2004. Detection and quantification of in vitro-culture induced chimerism using simple sequence repeat (SSR) analysis in Theobroma cacao (L.). Theoretical and Applied Genetics 110: 157–166. [DOI] [PubMed] [Google Scholar]
- de la Rosa R, James CM, Tobutt KR. 2002. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Molecular Ecology Notes 2: 265–267. [Google Scholar]
- Saumitou-Laprade P, Vassiliadis C, Epplen JT, Hardt C. 2000. Isolation of microsatellite loci for paternity testing in Phillyrea angustifolia L. (Oleaceae). Molecular Ecology 9: 112–114. [DOI] [PubMed] [Google Scholar]
- Sefc KM, Lopes MS, Mendonça D, Rodrigues dos Santos M, Laimer da Câmara Machado M, da Câmara Machado A. 2000. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Molecular Ecology 9: 1171–1173. [DOI] [PubMed] [Google Scholar]
- Setsuko S, Ishida K, Tomaru N. 2004. Size distribution and genetic structure in relation to clonal growth within a population of Magnolia tomentosa Thunb. (Magnoliaceae). Molecular Ecology 13: 2645–2653. [DOI] [PubMed] [Google Scholar]
- Wang J, Hill WG, Charlesworth D, Charlesworth B. 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genetical Research 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Weising K, Gardner RC. 1999. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19. [PubMed] [Google Scholar]
- Wickens GE. 1976. The flora of Jebel Marra (Sudan) and its geographical affinities. Kew Bulletin Additional Series 5: 1–368. [Google Scholar]