Abstract
• Background and Aims In the last decades, the geographical location of the centre of origin of Quercus suber (cork oak), a strictly western Mediterranean oak species, has been the subject of controversy.
• Methods RFLP variation over the whole chloroplast DNA molecule and PCR–RFLPs over seven specific cpDNA fragments were analysed phylogeographically to reconstruct the evolutionary history of cork oak.
• Key Results Nine chlorotypes of the ‘suber’ cpDNA lineage were identified throughout the species range. Using closely related Mediterranean oak species as outgroup, the chlorotypes showed a clear phylogeographical pattern of three groups corresponding to potential glacial refuges in Italy, North Africa and Iberia. The most ancestral and recent groups were observed in populations located in the eastern and western parts of the species range, respectively. Several unrelated chlorotypes of the ‘ilex’ cpDNA lineage were also identified in specific western areas.
• Conclusions The results support a Middle-Eastern or a central Mediterranean origin for cork oak with subsequent westward colonization during the Tertiary Period, and suggest that the ‘ilex’ chlorotype variation does not reflect entirely cytoplasmic introgression by Q. ilex but originated partly in Q. suber.
Keywords: cpDNA RFLP and PCR–RFLP variation, evergreen Mediterranean oaks, phylogeography, Quercus suber, Quercus ilex
INTRODUCTION
Quercus suber (cork oak) has a quite narrow geographical range as compared with the other main evergreen Mediterranean oak species, e.g. Quercus coccifera/Q. calliprinos (holly oak) and Q. ilex (holm oak). It is restricted to several discontinuous areas located exclusively in the western part of the Mediterranean Basin and along the Atlantic coast of North Africa and of south-western Europe. As reported previously by Bellarosa (2003), the few relict areas of south-eastern Italy constitute the far eastern limit of the species. Cork oak avoids limestone substrates and usually grows in hot parts of the humid and sub-humid Mediterranean areas with at least 450 mm mean annual rainfall and >4–5 °C mean temperature for the coldest month. These physiological requirements may account predominantly for the observed natural distribution pattern of Q. suber, which belongs to subgenus Cerris (Tutin et al., 1993) and has been used since Antiquity to produce cork.
Some authors have suggested that Q. suber may have originated in the Iberian peninsula where the species has its present main range. This assessment was based on geobotanical studies (Sauvage, 1961) and on allozyme variation in the whole cork oak range, which revealed a substantially higher genetic diversity in the Iberian populations as compared with those from North Africa, Italy and Provence (France) (Toumi and Lumaret, 1998). Alternatively, fossil records from oak species of subgenus Cerris dating to the Tertiary were found in the Balkanic Peninsula, and other authors considered that Q. suber probably appeared first in more eastern countries, either in the Balkanic peninsula or, alternatively, in the Middle Eastern–Peri-Caucasian area (Palamarev, 1989; Bellarosa, 2003; Bellarosa et al., 2005, and references therein). It has been suggested that the species expanded westward during the late Miocene and was widespread throughout the Mediterranean Basin during the Pliocene, with several other Mediterranean oak species of subgenus Cerris. During the Quaternary glaciations, cork oak may have survived in scattered refugia which possessed favourable microclimate conditions, and from which post-glacial colonization occurred over recent millennia. However, reliable scientific evidence is lacking to verify this scenario. The Tertiary and early Quaternary remains (megafossils and pollen) found in several European countries did not allow taxonomic identification at the species level (Smit, 1973; Kvacek and Walther, 1989) and could be attributable, therefore, to any Mediterranean oak species of the Cerris group, most of which (e.g. Q. cerris, Q. trojana, Q. macrolepis) grow exclusively in the eastern and central parts of the Mediterranean Basin. Molecular phylogenetic reconstruction could be a promising way to elucidate the geographical origin of Q. suber by identifying the location of the most ancestral genotypes currently observed in the species. Such identification should be obtained by comparing molecular variation in Q. suber with that in related species of subgenus Cerris.
Among available genetic molecular markers, restriction-site analysis of chloroplast DNA (cpDNA), a molecule inherited maternally in oaks (Dumolin et al., 1995), has been shown to be a powerful tool for phylogenetic reconstruction at both inter- and intra-specific levels. In several (mostly regional) studies, PCR–RFLP variation of a few cpDNA fragments was analysed in Q. suber, Q. ilex and Q. coccifera populations sampled predominantly in Iberia and Morocco (Belahbib et al., 2001; Jimenez et al., 2004; Staudt et al., 2004; Lopez-de-Heredia et al., 2005). In these studies, chlorotypes belonging to two very distinct cpDNA lineages were identified in cork oak. In the first lineage, named ‘suber’, which may be considered as the original and most widely distributed lineage in cork oak, Jimenez et al. (2004) identified four related chlorotypes. Their partial geographical distribution was reported by Lopez-de-Heredia et al. (2005). In addition, in two specific geographical areas, i.e. north-eastern and southern Morocco, and eastern Iberia with adjacent French Catalonia, all the sampled cork oak populations were shown to possess chlorotypes corresponding to a second cpDNA lineage (‘ilex’) shared with holm oak. This fact was interpreted as the result of multiple and mainly unidirectional cytoplasmic introgression of Q. suber by Q. ilex. From their experimental crosses, Boavida et al. (2001) reported the occurrence of asymmetric hybridization between cork oak and holm oak. However, in several surveys of cpDNA variation in these species, evidence was provided that, in initial hybridization and in back crosses, Q. ilex is predominantly, but not exclusively, the maternal species (Belhabib et al., 2001; Staudt et al., 2004). Quercus suber and Q. ilex possess overlapping geographical distributions (Tutin et al., 1993) but they are not very closely related, as shown from both cytoplasmic and nuclear genetic analyses (Belahbib et al., 2001; Toumi and Lumaret, 2001; Petit et al., 2002; Bellarosa et al., 2005) and belong to subgenera Cerris and Schlerophyllodrys, respectively (Tutin et al., 1993).
In the present work, RFLP analysis of the whole chloroplast DNA molecule is used for the first time in Q. suber to analyse the phylogeographical variation of cpDNA over the whole species range, including the eastern part that was poorly investigated previously. Using the same technique, 25 related chlorotypes of the ‘ilex’ lineage were distinguished previously in 174 Q. ilex populations sampled over the entire species geographical distribution (Lumaret et al., 2002). These chlorotypes were mapped and treated cladistically to build a phylogram, using related Mediterranean oak species as an outgroup, and the evolutionary history of Q. ilex was reconstructed successfully (Lumaret et al., 2002). In the present study, a complementary PCR–RFLP analysis of cork oak cpDNA, using several, DNA fragment/endonuclease combinations, was performed to identify additional phylogenetically informative characters based on very small fragment changes, and usually not detected with the standard RFLP technique. The main objectives were: (a) To identify, map and conduct a phylogeographical analysis of Q. suber cpDNA variation in the ‘suber’ lineage, to infer the probable route followed by the species during its spread over the Mediterranean Basin at the end of the Tertiary period; (b) to determine putative ice-age refugia and the main post-glacial re-colonization routes; and (c) to identify the ‘ilex’ lineage chlorotypes observed in Q. suber according to those described previously in Q. ilex (Lumaret et al., 2002). Specifically, the aim of the present study was to determine whether the ‘ilex’ lineage cpDNA variation of Q. suber is entirely due to cytoplasmic introgression by Q. ilex or else originated partly in Q. suber, from novel mutational events occurring in already introgressed cork oak material.
MATERIALS AND METHODS
Plant material
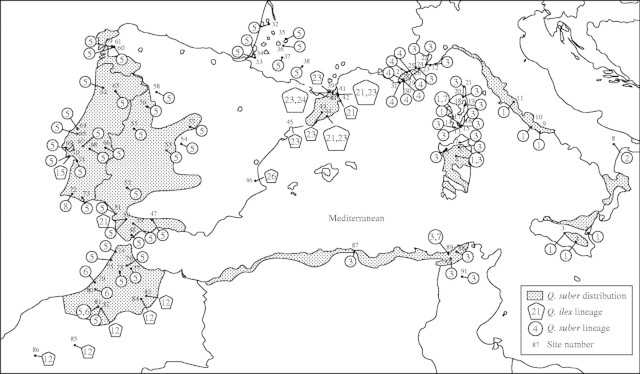
To analyse RFLP variation over the whole cpDNA molecule, the tree material used in the study was collected from 91 Q. suber populations distributed over the entire geographical range of the species (Fig. 1). Geographical coordinates and sample sizes of the localities may be obtained from the corresponding author. The populations were either pure (42 sites) or mixed with Q. ilex in 45 sites. A single Q. suber population (no. 69) was mixed with Q. coccifera and four populations were mixed with both evergreen oak species (nos 39, 41, 46 and 50). A mean of 4·02 trees per population was analysed for RFLP variation over the whole cpDNA genome. In the populations where cpDNA variation was observed, the sample size was increased to 10·1 trees per population, on average. In addition, as a complement, a sub-sample including tree material from 39 populations (nos 1, 3, 4, 6–11, 14, 16, 17, 22–38, 52, 57, 59, 69, 75, 79, 87, 88, 90 and 91) was scored for RFLP variation in seven specific cpDNA fragments amplified by using a polymerase chain reaction (PCR–RFLP) technique. Species identification was based on morphological characters as described in Flora Europaea (Tutin et al., 1993). Moreover, cpDNA variation was analysed in Q. cerris (by using the same RFLP and PCR–RFLP techniques) and Q. trojana (with PCR–RFLPs exclusively), two species closely related to Q. suber. Several Q. cerris individuals (three from two sites in France, three from one site of central Italy, two from one site of north-western Hungary, one from central Greece, and one from southern Macedonia), and two individuals of Q. trojana from southern Macedonia were used as outgroups.
Fig. 1.
Geographical distribution of the eight and six chlorotypes of the ‘suber’ and ‘ilex’ lineages identified, respectively, in 91 Q. suber populations scored for RFLP variation over the whole cpDNA molecule. The identity of sampled populations and cpDNA chlorotypes assayed through RFLP as well as affiliation to the ‘suber’ or ‘ilex’ cpDNA lineages are indicated.
Isolation and restriction endonuclease analysis of the whole cpDNA molecule
Leaf-bearing branches collected on the trees were placed in the dark for 8 d to de-starch the leaves before they were ground in liquid nitrogen and freeze-dried. Chloroplasts were isolated from 4-g aliquots of freeze-dried powder and cpDNA was extracted from chloroplasts as described by Mariac et al. (2000). Aliquots of 20 µg of chloroplast DNA were incubated for 5 h with three six-cutter (AvaI, BamHI and DraI) and one four-cutter endonucleases (HhaI), according to the recommendations of the suppliers (Boehringer-Germany, Appligene-France). These restriction enzymes provided consistent restriction patterns with a large number of fragments (usually over 45). The restriction digests were separated by electrophoresis on horizontal 0·85 % agarose-slab gels. The ‘1-kb ladder’ DNA (Eurogentec) was used as size standard. Gels were stained with ethidium bromide and photographed under UV light. For each cpDNA restriction endonuclease pattern, DNA restriction fragment sizes were determined using ‘Bande’ software (Duggleby et al., 1981).
PCR–RFLP analyses of cpDNA
Although RFLP analysis over the total cpDNA molecule (cpDNA-RFLP) is very efficient in identifying the putative variants, it can fail to differentiate between very small fragment-size changes (addition, deletions), if these concern either very large (>10 000 bp) or very small (<400 bp) fragments. Small fragment-size changes can be more easily detected with a PCR–RFLP technique, which was used as a complement to the RFLP analysis of the whole cpDNA molecule. Universal primers are only available for the large copy region of cpDNA (Grivet et al., 2001).
Total DNA was extracted from freeze-dried powder using a DNeasy Plant Minikit (QIAGEN). Chloroplast DNA was amplified using seven pairs of universal chloroplast primers corresponding to fragment TF (Taberlet et al., 1991); fragments AS, CD, DT, HK (Demesure et al., 1995), FV (Dumolin-Lapègue et al., 1997) and SR (Grivet et al., 2001). The four CD, DT, FV and HK amplified fragments and the three AS, TF and SR fragments were cut using endonucleases TaqI and HinfI, respectively. Digestion products were separated in 8 % polyacrylamide gels (18 × 28 cm) stained with ethidium bromide and photographed under UV light. The ‘Smart ladder’ (100–1000 bp) (Eurogentec-France) was used as a size standard.
Identification of cpDNA mutations
The cpDNA restriction endonuclease patterns of individual trees were scored for fragment-length differences. When possible, the cpDNA changes were identified as either length or site mutations. For instance, in the RFLP study of the whole cpDNA molecule, the detection of specific changes, each revealed from an individual oak tree by several restriction enzymes, suggests that alterations in the length of the fragments may be due to DNA length mutations rather than restriction site mutations. By scoring those length variants arbitrarily as the same mutation (same letter), counting the same insertion/deletion several times was avoided and, thereby, overestimating the number of distinct mutations.
Relationships between chlorotypes
Even if distinct site mutations (based on distinct restriction sites) were identified using either RFLP method, the possibility that both revealed, at least partly, the same length mutations cannot be ruled out and justifies separate data treatment. The different chlorotypes were scored for presence/absence and pooled to compute a similarity matrix (F-values) as proposed by Nei and Li (1979). The fraction of shared fragments was estimated by F = 2Nxy/(Nx + Ny) where Nxy is the number of bands chlorotype x and chlorotypes y have in common, and Nx and Ny are the number of bands found for chlorotypes x and y, respectively. The distance (1 – F values) was then analysed using the UPGMA option of PHYLIP 3·5 (Felsenstein, 1993). In addition, for both RFLP and PCR–RFLP analyses considered separately, using the main Q. cerris chlorotype as an outgroup, the DNA changes observed in the several chlorotypes identified in Q. suber were scored for presence/absence and were analysed cladistically by enumeration of the parsimonious trees using PAUP 4.0b10 (Swofford, 2000). A strict consensus tree was obtained and 1000 bootstrap samples were used to place confidence limits on branch points in the tree.
Geographical distribution and genetic diversity of cpDNA haplotypes
The geographical distribution of the different chlorotypes identified in the sampled plant material from RFLP variation over the whole cpDNA molecule was mapped. Nei's (1987) genetic diversity statistics, adapted to small and unequal sample sizes, were calculated for each population (hs) and over all populations (ht), and the proportion of diversity resulting from genetic differentiation among populations (Gst) was estimated using GENETIX 4.03 (Belkhir et al., 2001).
RESULTS
Of all the Q. suber individuals analysed by cpDNA-RFLP with four restriction enzymes (AvaI, BamHI, DraI and HhaI), two very divergent types of restriction patterns were distinguished and these contained, on average, 24 % of distinct fragments. One type, corresponding to the molecular lineage ‘suber’ was observed in 84 % of cork oak trees and in 100 % of the Q. cerris and Q. trojana individuals analysed in the study. A second type, the ‘ilex’ lineage, was found in 16 % of the cork oak individuals, and was composed exclusively of restriction profiles similar to those identified previously in Q. ilex populations (Lumaret et al., 2002). The restriction patterns observed in both of these cpDNA lineages were also very distinct from those observed in the European white oak species of section Quercus (R. Lumaret, J.-Ph. Ripoll, C. Mir et al., unpublished data).
The size of the chloroplast genome was estimated by adding together the size of the fragments generated by each endonuclease, particularly those produced by BamHI, which provided fewer and larger-sized fragments, and was estimated to be 141–142 kb in cork oak. In the Q. suber material, which showed cpDNA-RFLP restriction patterns corresponding to the ‘suber’ lineage, five, four, three and four distinct patterns and an average of 55·5, 47·0, 40·0 and 38 fragments were observed for HhaI, DraI, AvaI and BamHI, respectively. By comparison with the restriction profiles observed in Q. suber material from continental Italy (used as a reference because they showed the highest similarity to the patterns observed in Q. cerris), the mutations responsible for cpDNA variation were distinguished. Thirteen site mutations and three length mutations were identified (Table 1) and categorized with numbers and letters, respectively. Eight distinct haplotypes (chlorotypes) were distinguished in cork oak material from all the mutations obtained with the four restriction enzymes (Table 1). Four of these chlorotypes, S1, S3, S4 and S5, had a wide geographical distribution (Fig. 1) and were observed in 9·0, 19·9, 8·5 and 42·2 % individuals, respectively. The four other chlorotypes, S2, S6, S7 and S8, had a very restricted distribution and their frequencies were 0·8, 3·0 0·5 and 1·1 %, respectively. In Q. cerris, one additional chlorotype of the same lineage was observed which differed from S1 by a single mutation (no. 14) using HhaI. A 5300-bp fragment observed in S1 is replaced by a 5260-bp fragment in Q. cerris.
Table 1.
Restriction fragment length changes (kb) compared with the fragment size observed in the Q. suber individuals possessing chlorotype S1 (reference) in cork oak material and their occurrence in the other chlorotypes of Q. suber cpDNA lineage
| Fragment size (bp) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Restriction enzyme |
No. |
Reference |
Mutant |
Chlorotypes |
|||||
| HhaI | 1 | 5600 + X | → | 5680 | S3–S4–S7 | ||||
| 2 | 5300 + X | → | 5450 | S3–S4–S5–S6–S7–S8 | |||||
| 3 | 5300 | → | 5150 + X | S8 | |||||
| a | 4760 | → | 4650 | S5–S6 | |||||
| 4 | 4080 + X | → | 3880 | S5–S6 | |||||
| 5 | 1545 + X | → | 1560 | S3–S4–S7 | |||||
| 6 | 7730 | → | 5210 + 2520 | S7 | |||||
| DraI | a | 2680 | → | 2570 | S5–S6 | ||||
| 7 | 2045 + X | → | 2065 | S4 | |||||
| b | 2040 | → | 1980 | S5–S6–S8 | |||||
| c | 1550 | → | 1580 | S5–S6–S8 | |||||
| AvaI | 8 | 10590 + 3440 | → | 14010 | S5–S6 | ||||
| c | 5220 | → | 5250 | S5–S6 | |||||
| 9 | 3930 + X | → | 3900 | S5–S6 | |||||
| 10 | 3120 | → | 3050 + X | S8 | |||||
| b | 2220 | → | 2180 | S5–S6 | |||||
| BamHI | 11 | 9310 + X | → | 9390 | S6 | ||||
| 12 | 4520 + X | → | 4480 | S2 | |||||
| 13 | 2960 + X | → | 2970 | S5–S6 | |||||
| a | 2950 | → | 2840 | S5–S6 | |||||
Cork oak material was scored for RFLP cpDNA variation by using four restriction enzymes.
Changes attributable to site and length mutations are indicated with numbers and letters, respectively. Changes attributable to the same mutation are indexed with the same letter.
Some fragments could not be accurately sized due to small size or multiple superimposed bands (X).
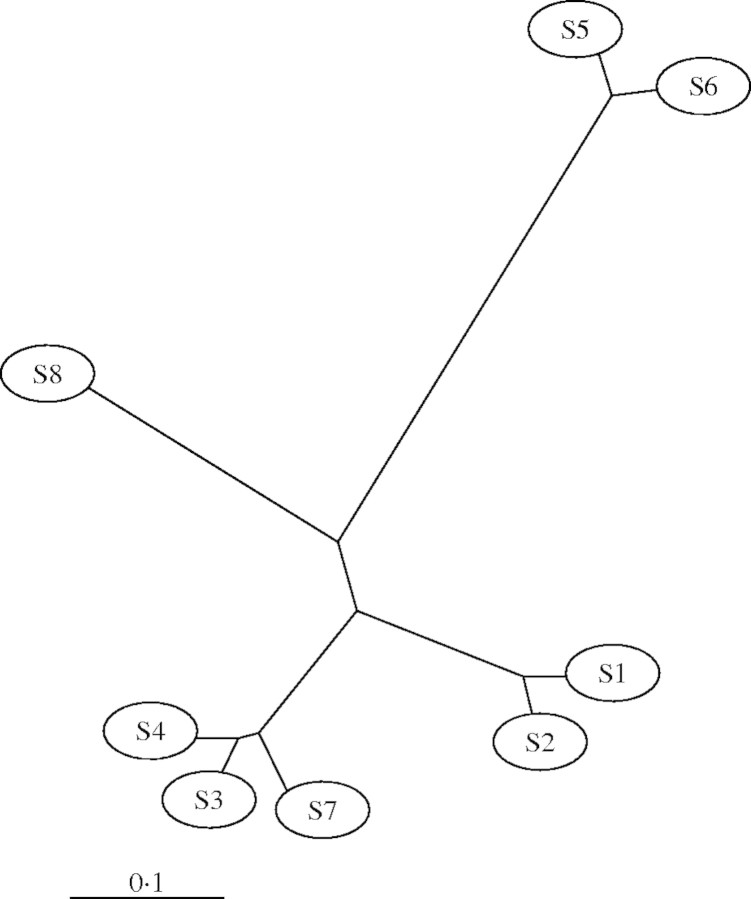
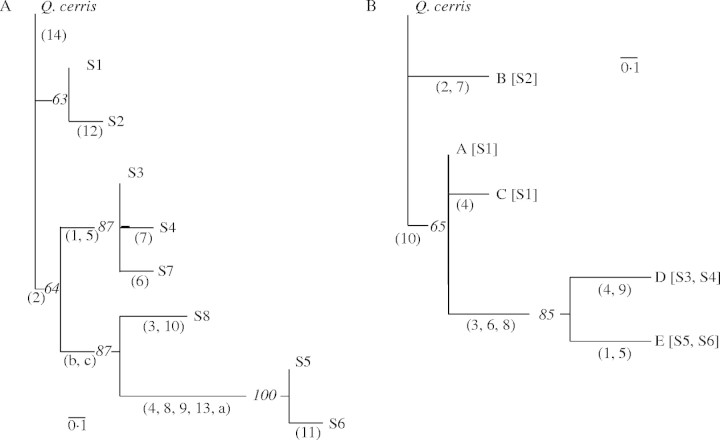
In Q. suber, the similarity index (F) values obtained from all of the mutations (site and length) ranged from 0·35 to 0·94. When the distance matrix obtained from the 1 – F values was constructed using UPGMA algorithm, four groups of chlorotypes could be distinguished (Fig. 2). Two of these groups, S1–S2 and S3–S4–S7, corresponded to cork oak material sampled exclusively in the eastern part of the species range, whereas the two other groups, S5–S6 (very distant from the other chlorotypes) and S8, possessed an Iberian-Moroccan distribution (Fig. 1). With the Dollo parsimony method, a similar general clustering pattern was obtained from the analysis of presence/absence of the 17 cpDNA mutations, of which 11 were phylogenetically informative, with Q. cerris as an outgroup (Fig. 3A). A single most-parsimonious tree was obtained (17 steps) with no character reversion. The consistency and retention indices were equal to 1·000. The consensus tree was obtained with confidence values ranging from 63 to 100 % of the major branches (Fig. 3A). Three clusters could be distinguished, each with a distinct geographical distribution (Fig. 1). A first cluster included two chlorotypes, S1 and S2, which differ from the chlorotype observed in Q. cerris by one and two mutations, respectively. S1 was found in Sicily, in a part of Sardinia and of Corsica, and in central Italy, whereas S2 was restricted to the south-eastern part of Italy (Apulia region). A second cluster is characterized by the phylogenetically informative mutations 1 and 5 and contains chlorotypes S3, S4 and S7. Chlorotype S3 was observed in Sardinia, Corsica, in the most eastern part of Provence (France), and in Tunisia and Algeria. S4, which differs from S3 by mutation 7, was only found in the western part of Provence, while S7 was observed exclusively in two individual trees collected in Tunisia (site 89) and Corsica (site 18), respectively. The third cluster was characterized by the two phylogenetically informative length mutations (b and c) and included the chlorotypes S5, S6 and S8 which are all restricted to the western part of the Q. suber geographical area. S5 was distributed widely in Morocco and in the Iberian Peninsula, whereas S6 and S8 were restricted to the centre of Morocco and to a single site located in southern Portugal, respectively. S5 and S6, which differ from each other by a single mutation (11) were very distinct from the other chlorotypes and shared five specific mutations (4, 8, 9, 13 and a).
Fig. 2.
UPGMA unrooted phenogram based on the proportion of shared site and length mutations for the eight chlorotypes observed in Q. suber populations showing a ‘suber’ lineage cpDNA molecule and scored for four endonuclease RFLPs.
Fig. 3.
Phylograms based on the consensus trees resulting from parsimony analysis of the eight RFLP chlorotypes (A) and of the five PCR–RFLP chlorotypes (B) identified in Q. suber populations showing a ‘suber’ lineage cpDNA molecule. In both data treatments, Q. cerris was used as outgroup. The number of mutations observed is indicated in parentheses for each node. For each major branch, the percentage of times that the defined group occurred in the 1000 bootstrap samples is indicated in italics. In (B), cpDNA haplotypes defined through PCR–RFLP are indicated by capital letters, and the corresponding RFLP chlorotypes in (A) are given in the square brackets.
PCR–RFLP cpDNA analyses of 39 Q. suber populations distributed over the entire species range and which possessed the main cpDNA-RFLP chlorotypes of the ‘suber’ lineage (S1, S2, S3, S4, S5 and S6) provided interesting additional data regarding the phylogeographical organization of cpDNA in Q. suber. No polymorphism was observed in using trnA-trnS and trnD-trnT. The five other cpDNA primer pairs combined with the two restriction enzymes identified five distinct chlorotypes (from A to E) containing two site mutations and eight length mutations (Table 2). Chlorotype ‘A’ was also observed in all the Q. cerris trees sampled in France, Italy and Hungary. However, the Q. cerris individuals from Greece and Macedonia and the two Macedonian Q. trojana trees possessed an additional chlorotype which differed from chlorotype A by a single mutation (no. 10) also found in the Q. suber population 8 located in south-eastern Italy (Fig. 1).This chlorotype, which was observed in the central parts of the ranges of Q. cerris and Q. trojana where Q. suber does not occur, was chosen as a reference (outgroup) for the phylogenetic analysis of cpDNA PCR–RFLP variation in cork oak. The consensus tree (Fig. 3B) was obtained using parsimony based on the presence/absence of the ten other mutations, five of which were phylogenetically informative. By way of comparison, identity of the corresponding chlorotypes obtained from analysis of the same tree material with the RFLP method is also indicated. Two most-parsimonious trees required a minimum of 11 steps to account for the ten mutations. The single reversion was observed for length mutation 4. The consistency and retention indices were 0·909 and 0·800, respectively. The consensus tree had confidence values ranging from 65 to 85 % of the major branches (Fig. 3B) and showed overall similarity with the tree obtained from RFLP analysis over the whole cpDNA molecule. However, in Fig. 3B, chlorotype B, observed in the Italian population 8 and corresponding to S2, was the closest to the root as it shared mutation 10 with the outgroup species (Q. cerris and Q. trojana). Moreover, two distinct chlorotypes, A and C, instead of a single one (S1), were identified in Q. suber populations from continental Italy (except no. 8), Sardinia and Corsica, and in those from Sicily, respectively. Conversely, chlorotype D was found in all the populations analysed from Algeria, Tunisia, Sardinia, Corsica and Provence. By contrast, the populations from the eastern and western parts of Provence were characterized by RFLP chlorotypes S3 and S4, respectively. Chlorotype E was identified in populations from northern Morocco, Iberia and the south-western part of France characterized by chlorotypes S5 or S6, according to the RLFP method.
Table 2.
Restriction fragment length changes (kb) compared with the fragment size observed in the Q. suber individuals possessing chlorotype A (Reference) in cork oak material and their occurrence in the other chlorotypes of ‘suber’ cpDNA lineage
| Primers |
Enzyme |
Mutation no. |
Reference |
Fragment size mutant |
Chlorotypes |
|
|---|---|---|---|---|---|---|
| trnC-trnD | TaqI | 1 | 1100 | → | 1050 | E |
| 2 | 1100 | → | 1000 | B | ||
| 3 | 540 | → | 555 | D, E | ||
| 4 | 335 | → | 340 | C, D | ||
| trnS-trnR | HinfI | 5 | 215 | → | 240 | E |
| trnT-trnF | HinfI | 6 | 290 | → | 280 | D, E |
| 7 | 120 | → | 125 | B | ||
| trnH-trnK | TaqI | 8 | 70 + 40 | → | 110 | D, E |
| trnF-trnV | TaqI | 9 | 940 + 520 | → | 1460 | D |
| 10 | 320 | → | 310 | B |
Cork oak material was scored for PCR–RFLP cpDNA variation using five combinations of primers/restriction enzymes.
In the 15 Q. suber populations (mixed stands) which included individuals characterized by an ‘ilex’ cpDNA lineage, six distinct chlorotypes were observed. Five of these had been previously identified in Q. ilex as chlorotypes 12, 15, 21, 23 and 24 (Lumaret et al., 2002). The sixth chlorotype, chlorotype 26, is closely related to Q. ilex chlorotype 20 but differs from it by two mutations, which had never been reported for that species. Comparison of chlorotype 1, the reference ‘ilex’ chlorotype (Lumaret et al., 2002), to chlorotype 26, shows that a 6770-bp fragment and a 1700-bp fragment in chlorotype 1 are replaced by a 6750-bp fragment and by two fragments (1410 and 290 bp) in chlorotype 26, respectively, using DraI. In cork oak populations, chlorotype 12 was observed in the southern and eastern parts of Morocco; chlorotype 15 was found in Portugal (site 70); chlorotype 21 was identified in site 50 (Andalusia) and in several sites of the Spanish and French Catalonia region where chlorotype 23 was also observed. Chlorotype 24 was found exclusively in the French part of Catalonia, whereas chlorotype 26 was restricted to site 46 (Fig. 1). For 14 populations where cork oak was mixed with other evergreen oak species, and which included individuals showing an ‘ilex’ cpDNA lineage, chlorotype comparisons among Q. suber, Q. ilex and Q. coccifera (when this species occurs) are presented in Table 3. In five of these populations, the chlorotypes identified in Q. suber were distinct from those observed in the other oak species.
Table 3.
Chlorotype identity (followingLumaret et al., 2002) and sampling size (in parentheses) in 14 mixed Quercus suber, Q. ilex and Q. coccifera populations in which all Q. suber individuals possessed a cpDNA molecule of the ‘ilex’ lineage
| Chlorotype no. |
|||||
|---|---|---|---|---|---|
| Population no. |
Q. suber |
Q. ilex |
Q. coccifera |
||
| 39 | 23 (7) | 21 (11) | 21 (4) | ||
| 40 | 23 (10) | 21 (8) | – | ||
| 24 (2) | |||||
| 41 | 21 (1) | 21 (8) | 21 (5) | ||
| 23 (5) | 23 (1) | ||||
| 42 | 21 (4) | 21 (4) | – | ||
| 43 | 23 (4) | 22 (4) | – | ||
| 44 | 21 (2) | 21 (9) | – | ||
| 23 (8) | |||||
| 45 | 23 (4) | 21 (4) | – | ||
| 46 | 26 (4) | 19 (4) | 21 (3) | ||
| 21 (1) | |||||
| 50 | 21 (5) | 21 (4) | – | ||
| 82 | 12 (7) | 12 (2) | – | ||
| 13 (9) | |||||
| 83 | 12 (5) | 12 (5) | – | ||
| 84 | 12 (5) | 12 (6) | – | ||
| 85 | 12 (3) | 12 (3) | – | ||
| 86 | 12 (4) | 12 (4) | – | ||
In the present study, 76, 12 and 3 populations consisted of cork oak trees showing exclusively a ‘suber’ cpDNA lineage, an ‘ilex’ lineage and a mixture of individuals possessing either cpDNA lineage, respectively (Fig. 1). A single chlorotype was observed in 81 of the 91 cork oak populations analysed and two chlorotypes were identified in ten polymorphic sites. Total genetic diversity (Ht) and the proportion of diversity attributable to differentiation between the populations (Gst) were equal to 0·689 and 96 %, respectively, in the 76 Q. suber populations containing exclusively ‘suber’ cpDNA types. For the same genetic parameters, the values were 0·705 and 84 %, respectively, in the 12 populations showing exclusively the ‘ilex’ lineage, and 0·770 and 93 % in all the populations.
DISCUSSION
General characteristics of cpDNA variation in Q. suber
As expected, a higher proportion of site mutations were identified by using the RFLP than the PCR–RFLP analysis, which revealed mostly small length variation (insertion/deletion) and the two methods may be considered as complementary. Eight, five and nine distinct chlorotypes were identified using cpDNA restriction over the whole molecule, PCR–RFLPs, and both techniques together, respectively. Most cork oak populations possessed chlorotypes of the ‘suber’ lineage, which is shared with Q. cerris and Q. trojana, the two closest relatives of cork oak. In a previous study, Petit et al. (2002) identified closely related cpDNA molecules in Q. suber and Q. cerris. Consistent with other studies based on PCR–RFLP techniques (e.g. Belahbib et al., 2001; Jimenez et al., 2004) by using a RFLP analysis over the whole cpDNA molecule, substantial genetic differentiation (24 % distinct restriction fragments on average) was revealed between the chlorotypes of the ‘suber’ and ‘ilex’ lineage, respectively. As a comparison, in Q. ilex, using the same endonucleases, 9 % distinct restriction fragments were observed between the genetically most distant chlorotype groups which corresponded to subspecies ilex and rotundifolia, respectively (Lumaret et al., 2002). This result justifies the separate phylogenetic analysis of the two lineages and supports previous discrimination of the taxonomic groups (or subgenera) Cerris and Sclerophyllodris, in which Q. suber and Q. ilex are classified, respectively (Nixon, 1993; Tutin et al., 1993; Bellarosa et al., 2005). In addition to the six chlorotypes of the ‘ilex’ lineage, in cork oak the approach used allowed the detection of nine ‘suber’ chlorotypes, a similar value to those reported in most other surveys of Fagaceae (Magni et al., 2005), with the exception of Q. ilex where substantially higher values were reported (Lumaret et al., 2002; Jimenez et al., 2004). In the present study, only four cork oak populations possessing exclusively a ‘suber’ chlorotype were polymorphic and most chlorotype diversity was attributable to differentiation among populations, as reported previously by most range-wide studies of oak species, including Q. ilex (for a review see Magni et al., 2005).
cpDNA phylogeographical pattern within lineage ‘suber’
Consistent chlorotype phylogeographical patterns were observed in Q. suber from the two complementary cpDNA analyses used in the study. Examination of both chlorotype phylogenetic structure and present geographical distribution showed that the genetically closest chlorotypes to those observed in related oak species (especially Q. cerris) were found exclusively in the eastern part of the present Q. suber distribution. In addition, in the two phylograms the main clusters of chlorotypes, which are differentiated successively from the base, were identified in geographical areas distributed from east to west (i.e. continental and insular Italy, the eastern and central parts of North Africa, the western part of North Africa and the Iberian Peninsula), suggesting that migration and genetic differentiation occurred initially in that direction. Moreover, because each cluster is defined by several mutations, this genetic differentiation may be very ancient and may characterize distinct geographical ice-age refugia (i.e. southern Italian Peninsula, North Africa and southern Iberian Peninsula) from which, after the last glaciations, Q. suber may have begun to migrate northward to the southern part of France. The finding that the Q. suber individuals of site 8 (south-eastern Italy) share a specific cpDNA PCR–RFLP fragment with the Q. cerris and Q. trojana individuals analysed from Greece and Macedonia, may constitute additional evidence to support Bellarosa's assessment of an eastern origin for cork oak (Bellarosa, 2003). However, comparisons of cpDNA variants between species rely on homology of the bands scored, which has not been tested in the present study. On the basis of the variation for the intergenic spacer length of rDNA genes, Bellarosa et al. (2003) suggested that the primary centre of origin of Q. suber may be located in south-eastern Europe, whereas the Iberian Peninsula (see above) is more likely to be a secondary centre of origin for this species. As shown in Fig. 2, substantial chlorotype differentiation was observed between the populations sampled in the eastern part of the species range, i.e. in Provence, Corsica, continental and island Italy, eastern and central North Africa, and those of the far western range, i.e. Morocco, Iberia and the south-western part of France. Using nuclear markers (allozymes and ITS), significant genetic differentiation between the eastern and the western cork oak population was observed and was interpreted as the result of northward post-glacial re-colonization from distinct refugia located in the eastern and western parts of the distribution range, respectively (Toumi and Lumaret, 1998; Bellarosa et al., 2003).
As shown in the present study, within the eastern cork oak range, the only chlorotype (S3) observed in Tunisia and Algeria was also identified in Sardinia, Corsica and Provence, suggesting that, in Q. suber, a North African refuge may have contributed successfully to the re-colonization of this part of Europe after the last glaciations, contrary to what was reported for Q. ilex (Lumaret et al., 2002) and for deciduous European oak species (Hewitt, 1999). An additional chlorotype S1, observed predominantly in continental Italy and in Sicily, was identified in a few populations from Sardinia, and from Corsica which also shared the rare chlorotypes S7 with Tunisia. This situation probably reflects the occurrence of rare natural events of long-distance dispersal from several geographical sources located in the closest island or continental areas to those islands. In addition, the possibility of intentional acorn transport by people to extend cork oak areas for economic purposes cannot be ruled out and its impact on the geographical patterns of cork oak genetic variation should not be underestimated. In a recent paper, Lopez-de-Heredia et al. (2005) suggested the possibility of long-distance dispersal events to explain the sharing of a rare chlorotype by cork oak populations located in Minorca and in Sardinia, respectively.
In the western species range, the occurrence of chlorotype S5 in north-western Morocco and in a very large part of Iberia, implies intercontinental migration. Cork oak could have migrated between North Africa and Iberia, in either direction, through the Strait of Gibraltar at the end of the Tertiary period or later during the interglacial episodes of the Quaternary period (Sauvage, 1961). In the phylogram (Fig. 3A), in addition to several specific mutations, chlorotype S8 contains some (b and c) but not all of the mutations characteristic of the western cork oak range (chlorotypes S5 and S6). Therefore, chlorotype S8 can be considered as an evolutionary intermediate between eastern chlorotypes (chlorotypes S3, S4 and S7) and the western chlorotype (S5) from which S6 is probably derived. Chlorotype S8 was observed exclusively in a natural relict area located in southern Portugal (site 72, Fig. 1). The possibility that S5 originated in Morocco and migrated subsequently to Iberia cannot be ruled out. However, to account for the presence of both S5 and S8 in Iberia, this interpretation implies that several events of cork oak migration from North Africa to Iberia occurred in different episodes and assumes that S8 and/or other closely related ancestral chlorotypes became extinct in Morocco. Alternatively, it can be considered that S5 originated in Iberia, and subsequently migrated to the north-western part of Morocco. In Q. ilex, a north-to-south migration pattern was reported previously on the basis of a wide range in cpDNA variation (Lumaret et al., 2002) in approximately the same geographical area.
cpDNA phylogeographical variation within lineage ‘ilex’
This large geographical survey of cpDNA variation in cork oak confirms that populations possessing exclusively an ‘ilex’ chlorotype are mainly confined to the few regions identified previously (see above), which constitute <20 % of the species range. As mapped in Fig 1, ‘ilex’ chlorotypes were observed in a few additional small areas of southern Iberia where Q. suber and Q. ilex constituted mixed populations or were very close geographically. With the exception of chlorotype 26, the ‘ilex’ chlorotypes found in Q. suber, i.e. chlorotype 12 found in Morocco and chlorotype nos 15, 21, 23 and 24 identified in Iberia and French Catalonia, had been observed previously in the same areas in Q. ilex populations (Lumaret et al., 2002). This result indicates the occurrence of multiple events of local hybridization and of subsequent genetic introgression between the two oak species, as reported previously (e.g. Toumi and Lumaret, 1998; Belhabib et al., 2001). However, as shown in Table 3, in mixed stands of Q. suber and Q. ilex the populations do not always possess the same ‘ilex’ chlorotype, suggesting that the two oak species may have undergone distinct colonization dynamics. This situation may in part be due to human-mediated range expansion of Q. suber in areas where Q. ilex, a species not intensively managed, is naturally present.
A striking point raised by this study is the absence of cork oak populations possessing ‘ilex’ chlorotypes in the eastern range of the species. Similarly, Lopez-de-Heredia et al. (2005) did not identify cork oak individuals possessing an ‘ilex’ chlorotype in the few populations they studied in the east. However, using larger sample sizes, Staudt et al. (2004) observed an ‘ilex’ chlorotype in one of the 22 Q. suber individuals they analysed from a population on Port-Cros Island (Provence, France). In a Corsican population, one of the 50 cork oak individuals scored for cpDNA RFLPs was shown to possess an ‘ilex’ chlorotype (J.-Ph. Ripoll and R. Lumaret, unpublished data). In both populations, ‘ilex’ chlorotype 4, predominant in Provencal and Corsican Q. ilex populations (Lumaret et al., 2002), was identified in Q. suber. This result suggests that cytoplasmic introgression of Q. suber by Q. ilex does occur in the eastern range of cork oak, but that it may be restricted to a few isolated individuals. A substantial number of oak trees showing intermediate morphology between both species, possessing predominantly an ‘ilex’ chlorotype (Lumaret et al., 2002) and for many of which the hybrid origin was confirmed on the basis of nuclear interspecific diagnostic markers, have been observed in south-eastern continental Italy (Bellarosa et al., 2005), in Sardinia and in Provence (Lumaret et al., 2002). So, interspecific hybridization is likely to have happened quite frequently in the eastern part of the range of Q. suber. The very rare occurrence of ‘ilex’ chlorotypes in Q. suber individuals of that area may be attributable either to ‘intrinsic’ genetic difficulties of backcrossed hybrids or, more probably, to ‘extrinsic’ ecological factors during establishment of backcrossed (introgressed) hybrids in these areas.
In the present study, several ‘ilex’ chlorotypes (e.g. 23 and 26) were observed either exclusively or predominantly in Q. suber populations. The possibility cannot be ruled out that these ‘ilex’ chlorotypes were not found in Q. ilex as a simple result of low sample sizes, as suggested previously (Belhabib et al., 2001; Jimenez et al., 2004). However, in very intensively sampled regions, e.g. in Spanish Catalonia, and specifically in French Catalonia where chlorotype 23 was found to be widely distributed in Q. suber (Lumaret et al., 2003; present paper) but was extremely rare in Q. ilex (Lumaret et al., 2002), the possibility exists that this chlorotype originated in Q. suber. As a result of predominantly unidirectional cytoplasmic introgression between the two species, several of the chlorotypes which were never observed in Q. ilex, and which originated directly in Q. suber, have a very low probability of being introgressed into the Q. ilex genome. In addition, as shown in the present work, ‘ilex’ chlorotype 26, observed exclusively in Q. suber, differed by several mutations from the closest chlorotype identified in Q. ilex. This suggests that, from the cpDNA variants of ‘ilex’ lineage that were recovered through interspecific introgression, additional successive cpDNA changes may have occurred in Q. suber. Based on this assumption, the occurrence of two distinct cpDNA lineages in this species is predicted.
Acknowledgments
We are grateful to the following people who collected material used in our analyses: H. Brustel, S. Dettori, A. Ducousso, B. Fady, S. Fineschi, L. Gil-Sanchez, J. P. Huvelin, N. Laszlo, S. Leonardi, B. Noïtsakis, N. Ouazzani, E. Sahuquillo-Balbuena, B. Schirone, L. Toumi, B. Valdès and M. C. Varela. We also thank M. Borne, D. Claret, M. Debiais-Thibaud, C. Graulle, R. Jabbour-Zahab, S. Melki, D. Ranc, J. Ph. Ripoll and V. Sarda for technical assistance, H. Bohbot for help in data mapping, E. Douzery for helpful advice on data treatments, and J. Aronson and T. Burg for making useful suggestions on how to improve the manuscript. The research was supported by the European Union Programme CREOAK, QLK5-CT-2002-01594, and by the GIP-ECOFOR Programme ‘Biodiversité et gestion forestière’ No. 2001-23.
LITERATURE CITED
- Belahbib N, Pemonge MH, Ouassou A, Sbay H, Kremer A, Petit RJ. 2001. Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Molecular Ecology 10: 2003–2012. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikli L, Raufaste N, Bonhomme F. 2001.GENETIX, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions CNRS UMR 5000, UMII, Montpellier, France. [Google Scholar]
- Bellarosa R. 2003. Brief synthesis of the current knowledge on cork oak. In: Varela MC, ed. Handbook of the EU concerted action on cork oak, FAIR I CT 95 0202. Lisbon: INIA, 11–22 (ISBN 972-9573670). [Google Scholar]
- Bellarosa R, Simeone MC, Schirone B. 2003. Germplasm conservation of Mediterranean oaks in Italy: distribution and genetic structure of cork oak (Quercus suber L.). In: Bozzano M, Turok J, eds. Mediterranean Oaks Network, Report of the second meeting 2–4 May 2002, Gozo, Malta. Rome, Italy: International Plant Genetic Resources Institute, 5–12. [Google Scholar]
- Bellarosa R, Simeone M, Papini A, Schirone B. 2005. Utility of ITS sequence data for phylogenetic reconstruction of Italian Quercus spp. Molecular Phylogenetics and Evolution 34: 355–370. [DOI] [PubMed] [Google Scholar]
- Boavida LC, Silva JP, Feij JA. 2001. Sexual reproduction in the cork oak (Quercus suber L.). II. Crossing intra- and inter-specific barriers. Sexual Plant Reproduction 14: 143–152. [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. 1995. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology 4: 129–131. [DOI] [PubMed] [Google Scholar]
- Duggleby RG, Kinns H, Rood J. 1981. A computer program for determining the size of DNA restriction fragments. Critical Review of Plant Science 110: 49–55. [DOI] [PubMed] [Google Scholar]
- Dumolin S, Demesure B, Petit RJ. 1995. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Dumolin-Lapègue S, Pemonge MH, Petit RJ. 1997. An enlarged set of consensus primers for the study of organelle DNA in plants. Molecular Ecology 6: 393–397. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1993. PHYLIP Phylogeny, inference package, version 3.57. Department of Genetics, University of Washington, Seattle, WA. [Google Scholar]
- Grivet D, Heinse B, Vendramin GG, Petit RJ. 2001. Genome walking with consensus primers: application to the Large Single Copy region of chloroplast DNA. Molecular Ecology Notes 1: 345–349. [Google Scholar]
- Hewitt GM. 1999. Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society 68: 87–112. [Google Scholar]
- Jimenez P, Lopez de Heredia U, Collada C, Lorenzo Z, Gil L. 2004. High variability of chloroplast DNA in three Mediterranean evergreen oaks indicates complex evolutionary history. Heredity 93: 510–515. [DOI] [PubMed] [Google Scholar]
- Kvacek Z, Walther H. 1989. Paleobotanical studies in Fagaceae of the European Tertiary. Plant Systematics and Evolution 162: 213–229. [Google Scholar]
- Lopez-de-Heredia U, Jimenez P, Diaz-Fernandez P, Gil L. 2005. The Balearic islands: a reservoir of cpDNA genetic variation for evergreen oaks. Journal of Biogeography 32: 939–949. [Google Scholar]
- Lumaret R, Mir C, Michaud H, Raynal V. 2002. Phylogeographical variation of chloroplast DNA in holm oak (Quercus ilex L.). Molecular Ecology 11: 2327–2336. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Mir C, Royer J. 2003. Chêne vert et chêne liège: de vieux frères. Les Nouvelles Feuilles Forestières 73: 6–8. [Google Scholar]
- Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A. 2005. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Molecular Ecology 14: 513–524. [DOI] [PubMed] [Google Scholar]
- Mariac C, Trouslot P, Poteaux C, Bezançon G, Renno JF. 2000. A simple method for extraction of chloroplast DNA from herbaceous and woody plants for RFLP analysis. Biotechniques 28: 110–113. [DOI] [PubMed] [Google Scholar]
- Nixon KC. 1993. Infrageneric classification of Quercus (Fagaceae) and typification of sectional names. Annales des Sciences Forestières 50: 25S–34S. [Google Scholar]
- Nei M. 1987.Molecular evolutionary genetics. New York, NY: Columbia University Press. [Google Scholar]
- Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamarev E. 1989. Paleobotanical evidences of the tertiary history and origin of the Mediterranean sclerophyll dendroflora. Plant Systematics and Evolution 162: 93–107. [Google Scholar]
- Petit RJ, Csaikl U, Bordacs S, Burg K, Coart E, Cottrell J, et al. 2002. Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2,600 populations. Forest Ecology and Management 156: 5–26. [Google Scholar]
- Sauvage Ch. 1961. Recherches géobotaniques sur les Subéraies marocaines. Travaux de l'Institut Scientifique Chérifien, Série Botanique, Rabat 21: 1–462. [Google Scholar]
- Smit A. 1973. A scanning electron microscopical study of the pollen morphology in the genus Quercus Acta Botanica Neerlandesis, Leiden 22: 655–665. [Google Scholar]
- Staudt M, Mir C, Joffre R, Rambal S, Bonin A, Landais D, Lumaret R. 2004. Isoprenoid emissions of Quercus spp. (Q. suber and Q. ilex) in mixed stands contrasting in interspecific genetic introgression. New Phytologist 163: 573–584. [DOI] [PubMed] [Google Scholar]
- Swofford D. 2000.PAUP*: phylogenetic analysis using parsimony (and other methods). Beta version 4.0b10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Taberlet P, Gielly L, Patou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Toumi L, Lumaret R. 1998. Allozyme variation in cork oak (Quercus suber L.): the role of phylogeography, genetic introgression by other Mediterranean oak species and human activities. Theoretical and Applied Genetics 97: 647–656. [Google Scholar]
- Toumi L, Lumaret R. 2001. Allozyme characterisation of four Mediterranean evergreen oak species. Biochemical Systematics and Ecology 29: 799–817. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. 1993.Flora Europaea. London, UK: Cambridge University Press. [Google Scholar]