Abstract
• Background and Aims Isoetes sinensis (Isoeteaceae) is a critically endangered aquatic quillwort in eastern China. Rapid decline of extant population size and local population extinction have occurred in recent years and have raised great concerns among conservationists.
• Methods Amplified fragment length polymorphisms (AFLPs) were used to investigate the genetic variation and population structure of seven extant populations of the species.
• Key Results Eight primer combinations produced a total of 343 unambiguous bands of which 210 (61·2 %) were polymorphic. Isoetes sinensis exhibited a high level of intra-population genetic diversity (HE = 0·118; hs = 0·147; I = 0·192; P = 35·2 %). The genetic variation within each of the populations was not positively correlated with their size, suggesting recent population decline, which is well in accordance with field data of demographic surveys. Moreover, a high degree of genetic differentiation (FST = 0·535; GST = 0·608; θB = 0·607) was detected among populations and no correlation was found between geographical and genetic distance, suggesting that populations were in disequilibrium of migration-drift. Genetic drift played a more important role than gene flow in the current population genetic structure of I. sinensis because migration of I. sinensis is predominantly water-mediated and habitat range was highly influenced by environment changes.
• Conclusions Genetic information obtained in the present study provides useful baseline data for formulating conservation strategies. Conservation management, including both reinforcement for in situ populations and ex situ conservation programmes should be carefully designed to avoid the potential risk of outbreeding depression by admixture of individuals from different regions. However, translocation within the same regional population should be considered as a measure of genetic enhancement to rehabilitate local populations. An ex situ conservation strategy for conserving all extant populations to maximize genomic representation of the species is also recommended.
Keywords: AFLP, genetic diversity, genetic differentiation, population structure, Isoetes sinensis, pteridophyte
INTRODUCTION
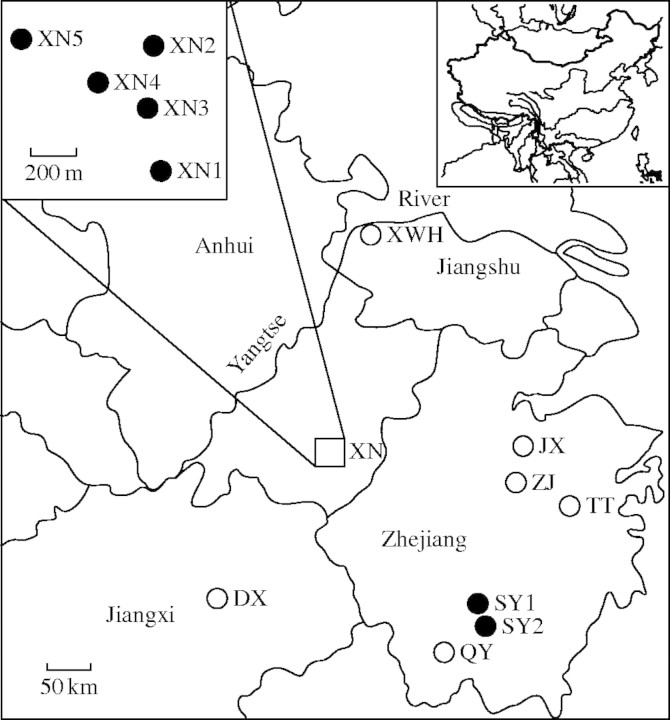
Isoetes sinensis is a member of the Isoeteaceae, a family that consists of a single relict lycopsid genus with approx. 150 species occurring in lake, wetland and terrestrial habitats all over the world (Taylor and Hickey, 1992). Four species, I. hypsophila, I. sinensis, I. taiwanensis and I. yunguiensis, have been identified and documented in China (Wang et al., 2002). All Chinese Isoetes species are highly threatened with extinction due to habitat loss, agricultural land-use, and invasion by exotic species, and are therefore listed as endangered species (Fu and Jin, 1992; Ye and Li, 2003). Isoetes sinensis has four cytotypes, but only allotetraploid (2n = 4x = 44) plants occur in China (He et al., 2002), while hexaploid (2n = 6x = 66) and two aneuploids (2n = 65, 2n = 68) of the species are found in Japan (Takamiya et al., 1994). A recent phylogenetic study suggested that I. sinensis was probably derived from hybridization between two diploid species: I. yunguiensis and I. taiwanensis endemic to China (Taylor et al., 2004). Isoetes sinensis is now a globally threatened taxon and is also listed as an endangered species in Japan (Takamiya 2001; Red List of Threatened Plants of Japan, http://www.biodic.go.jp/english/rdb/red_plants.csv). The species is discretely distributed in eastern China, mostly in limited areas in Anhui, Jiangshu, Jiangxi and Zhejiang provinces (Fu and Jin, 1992). A recent exhaustive survey of herbarium specimen and literature confirmed that the natural range of the species was very limited. Isoetes sinensis was previously documented in Dongxiang (DX) (Chen et al., 1998), Jiangxi province; Jiuxi (JX) (voucher nos HHBG 60934 and ZJFC 0188), Qinyuan (QY) (S. C. Zhu, pers. comm.), Tiantai (TT) (B. Y. Ding, pers. comm.) and Zhuji (ZJ) (voucher nos HZU 9754 and HZU 9810), Zhejiang province; and Xuanwuhu (XWH) (voucher no. NAS 00070205), Jiangshu province (Fig. 1). However, field surveys conducted in the past five years concluded that I. sinensis could be extinct in these six sites (Fig. 1) (Pang et al., 2003; Ye and Li, 2003). In addition, sizes of extant populations have decreased dramatically (Ye and Li, 2003). The rapid decline of the species and extinction of local populations have caused conservation concerns to save this endangered species in China (Pang et al., 2003; Wen et al., 2003; Ye and Li, 2003; Chen et al., 2004) and I. sinensis was recently categorized as a critically endangered species by IUCN (2004).
Fig. 1.
Map of the historical and present distribution of Isoetes sinensis and sampling locations (open circles, historical sites where I. sinensis was previously documented but identified as extinct during field surveys in 2000–2003; closed circles, extant sites where samples were collected for the present study).
Small and isolated populations of endangered species are vulnerable to demographic, environmental and genetic stochasticity, and therefore face a higher risk of local extinction (Lande, 1999). Although the relative importance and consequence of these factors is still controversial (Lande, 1988; Spielman et al., 2004), genetic factors have been demonstrated to have direct and/or indirect impacts on the viability of endangered species (Frankham et al., 2002). Many endangered plant species with small population sizes in isolated or fragmented habitats suffer higher extinction risks due to the loss of genetic variation caused by genetic drift and inbreeding (Barrett and Kohn, 1991). Some critically endangered species may still go to extinction even when their natural habitats are restored because of severely depauperate genetic diversity and the lack of adaptive evolutionary potential in the face of an ever-changing environment (Frankham et al., 2002). It has been widely accepted by conservation geneticists that understanding the level of genetic diversity and population genetic structure is a prerequisite for formulating conservation programmes for endangered plant species. Such knowledge could provide critical information needed to understand the evolutionary history of populations and population genetics underlining potential risk in the long term, and also make available information for the genetic management of endangered species both in situ and ex situ (Frankham et al., 2002).
Of the available techniques in use to investigate population genetic diversity and genetic structure, the amplified fragment length polymorphism (AFLP) technique (Vos et al., 1995) has the combined advantages of RAPD and RFLP, with the promise of generating a large number of markers representative of the target genome. The technique is highly reproducible and cost-efficient, and has been used successfully in population genetic studies of several endangered plants (Travis et al., 1996; Gaudeul et al., 2000; Evans et al., 2001; Tero et al., 2003). However, few such studies have been attempted in pteridophyte species (Keiper and McConchie, 2000).
In this study, AFLP analysis was employed to address the following questions of conservation concern in I. sinensis: (a) What is the level of genetic diversity currently retained in the remnant populations? (b) How is the genetic variation distributed within and among populations? (c) Is population size associated with the level of population genetic variation, and geographic distance associated with genetic distance? These baseline data should be essential to understand the genetic consequences of population decline in remnant I. sinensis growing in isolated and fragile wetland habitats, so that adequate conservation measures could be taken without potential risk of admixture of genetically differentiated populations either for in situ conservation by translocation or for ex situ conservation.
MATERIALS AND METHODS
Study species
Isoetes sinensis Palmer is a small, aquatic and facultative evergreen quillwort with numerous spirally arranged leaves (sporophylls) arising from a globose, three-lobed corm. Pliant and hollow leaves are 2–4 mm wide and up to 30 cm long, depending on water depth in the wetland habitat. Sporangia are embedded in the broadened base of leaves. Megasporangia are usually borne on outer leaves, with a few white, granular and tetrahedric megaspores, while microsporangia are borne on inner leaves, with numerous grey-powdery and bilateral microspores (Fu and Jin, 1992). Both sexual and asexual reproduction has been observed in this species (Takamiya et al., 1994; Chen et al., 2004). Isoetes sinensis occurs in wetland habitats of rice paddy land, shallow ponds, swamps and creeks and appears to be a weak competitor with dominant companion species of Eriocaulon decemflorum Maxim, Polygonum sagittatum Linn., Miscanthus sinensis Anderss, Prunella asiatica Nakai and Hydrangea L. sp.
Field survey and sample collection
First an exhaustive literature search conducted on I. sinensis and visits made to the main Chinese herbaria (AAUB, ANU, ANUB, HHBG, HIB, HZU, NAS, PE and ZJFC) to look for specimen records resulted in a comprehensive compilation of historical and updated information of the geographic distribution and detailed locations of the species. Then, extensive field surveys across the entire range of I. sinensis were conducted during for four years (2000–2003), but this species was only found in two remnant sites (Fig. 1). Two populations (SY1 and SY2), approx. 1 km apart, were found in Songyang county, Zhejiang province (28°16′N, 119°16′E); while five small patches, 162–520 m apart, were observed in Xiuning county, Anhui province (29°30′N, 118°09′E). As this quillwort is a semi- and submerged aquatic the migration of spores or root-runners are restricted; therefore, each patch was presumed to be a population and was designated XN1, XN2, XN3, XN4 or XN5 (Fig. 1).
Population area and population size were determined for each population by directly measuring the area and counting individuals within each population. Then, fresh leaves (sporophylls) were harvested from adult individuals, at least 2 m apart, to avoid sampling the same plant. Ten to 30 individuals were randomly sampled from each population, except population XN2, depending on sampling availability of adult plants in each of the seven populations. In population XN2, a total of 20 individuals were identified, but only five adult plants could be sampled keeping the minimum sampling distance (2 m). As a result, a total of 106 samples were collected from these seven populations at the two remnant sites (Fig. 1). Plant materials were immediately dried in silica gel before DNA isolation.
DNA isolation and AFLP analysis
Total genomic DNA was extracted from 0·3–0·5 g of dried leaves using a modified CTAB protocol outlined in Doyle and Doyle (1987). AFLP analysis was performed essentially as described previously by Vos et al. (1995) with a slight modification. Total genomic DNA (200 ng) was digested using 3 units of EcoRI and MseI endonuclease mixture (Biolab) in a total volume of 25 µL for 2·5 h at 37 °C and digests were confirmed by electrophoresis on 1·5 % agarose gels. Then, 20-μL of ligation solution containing 4 pmol EcoRI adapter, 40 pmol MseI adapter, and 1 unit of T4 DNA ligase were added to the digests in a total volume of 40 µL. The ligation mixture was incubated overnight at 21 °C in a thermocycler. The resulting DNA (template DNA) was then diluted 1:5 in TE buffer. PCR pre-amplification was performed in a 20 µL solution containing 75 mm TRIS–HCl (pH 8·8), 20 mm (NH4)2SO4, 1·5 mm MgCl2, 0·1 ‰ Tween20, 0·20 mm each of dNTPs, 40 ng of primer EcoRI-A, 40 ng of primer MseI-C, 0·5 units Taq polymerase (Fermentas), and 5 µL of template DNA for 20 cycles of the thermal profile: 94 °C for 30 s, 56 °C for 60 s and 72 °C for 60 s. The pre-amplification DNA was diluted 1:25 in TE buffer and used as template DNA for selective amplification. After pre-screening of 48 primer pairs, eight selective primer pairs were chosen for this study (Table 1). An aliquot of 2·5 µL diluted pre-amplification DNA was added to 7·5 µL of selective amplification cocktail (30 ng EcoRI primer, 30 ng MseI primer, 0·20 mm dNTPs, 1× PCR buffer, 0·5 units Taq polymerase, 1·5 mm MgCl2), and amplified with the thermal cycle profile: one cycle with 94 °C for 30 s, 60 °C for 1 min and 72 °C for 1 min, then 12 cycles of 94 °C for 30s, 65 °C (decreasing by 0·7 °C) for 1 min and 72 °C for 1 min, followed by 23 cycles of 94 °C for 30 s, 56 °C for 1 min and 72 °C for 1 min. The amplification products added with 7·5 µL loading buffer (98 % formamide, 10 mm EDTA (pH 8·0), 0·25 % bromophenol blue and 0·25 % xylene cyanol) were denatured at 94 °C for 5 min and electrophoresed on a 6 % denaturing polyacrylamide gel on a 40 × 45 cm EC160 DNA Sequencing System (Thermo). Silver staining was conducted according to protocol of the Silver Sequence™ (Promega) with slight modification.
Table 1.
Polymorphic bands generated by each of eight pair primers in seven extant populations of Isoetes sinensis
| Primer pairs |
Primer sequence (5′–3′) |
Total bands |
Polymorphic bands |
Polymorphism (%) |
|---|---|---|---|---|
| E-AAC/M-CTG | GACTGCGTACCAATTCAACGATGAGTCCTGAGTAACTG | 46 | 36 | 78·3 |
| E-ACT/M-CTC | GACTGCGTACCAATTCACTGATGAGTCCTGAGTAACTC | 25 | 18 | 72·0 |
| E-ATG/M-CGT | GACTGCGTACCAATTCATGGATGAGTCCTGAGTAACGT | 52 | 36 | 69·2 |
| E-ACC/M-CGT | GACTGCGTACCAATTCACCGATGAGTCCTGAGTAACGT | 42 | 29 | 69·0 |
| E-ACG/M-CAT | GACTGCGTACCAATTCACGGATGAGTCCTGAGTAACAT | 38 | 17 | 44·7 |
| E-ACG/M-CTG | GACTGCGTACCAATTCACGGATGAGTCCTGAGTAACTG | 47 | 24 | 51·1 |
| E-AAC/M-CAT | GACTGCGTACCAATTCAACGATGAGTCCTGAGTAACAT | 50 | 25 | 50·0 |
| E-ACT/M-CTG | GACTGCGTACCAATTCACTGATGAGTCCTGAGTAACTG | 43 | 25 | 58·1 |
| Mean | 42·9 | 26·3 | 61·6 | |
| Total | 343 | 210 |
Marker scoring and data analysis
Only intense and unambiguous bands (100–500 bp) were manually scored as present (1) or absent (0). After scoring all bands for all samples, the bands that were monomorphic across all individuals were then discarded for further analyses to reduce the statistical bias (Keiper and McConchie, 2000).
Due to the dominant nature of AFLPs, estimating heterozygosity and population differentiation could be problematic (Zhivotosky, 1999; Tero et al., 2003). Traditional approaches to estimating heterozygosity and population differentiation using dominant markers assume that populations are in Hardy–Weinberg equilibrium, or require a known inbreeding coefficient, or treat multilocus DNA phenotypes as haplotypes (Zhivotovsky, 1999; Holsinger et al., 2002). Earlier studies have suggested that such approaches may lead to severely biased estimates (Lynch and Milligan, 1994; Zhivotovsky, 1999). However, Krauss (2000) has demonstrated that accurate estimates of population genetic parameters can be obtained from dominant markers when a large number of polymorphic loci were used. More recently, Holsinger et al. (2002) introduced a Bayesian approach to estimating genetic diversity and population structure without prior information of the inbreeding coefficient (FIS) and demonstrated that the Bayesian method could provide nearly unbiased estimates of heterozygosity, genetic distance and population differentiation.
In the present study, the following parameters for revealing genetic diversity and population differentiation were estimated using different computer softwares: percentage of polymorphic loci (P at the 0·99 level), Nei's unbiased expected heterozygosity (HE) (Nei, 1978) and Wright' F-statistics (FST) (Weir and Cockerham, 1984) were estimated using the program TFGPA (Miller, 1997a). Shannon and Weaver's index (I) (Shannon and Weaver, 1949) and genetic differentiation (GST) (Nei, 1987) were calculated under the assumption of full selfing (FIS = 1) within populations using the program POPGENE version 1.32 (Yeh et al., 1997). The Bayesian approach described by Holsinger et al. (2002) was also used to determine genetic diversity (hs, analogous to HE) and population differentiation (θB, analogous to FST) using the program HICKORY 0.8 (Holsinger and Lewis, 2003), with five runs of each of three different models (full model, f = 0 model and f-free model) to ensure consistency of the results (burn-in = 50 000, no. of samples = 250000, thinning = 50 in each run). The model chosen was based on the deviance information criterion (DIC) (Spiegelhalter et al., 2002). Models with smaller DIC are preferred, but a difference of >6 DIC units among different models is required to indicate that there is strong favouring of one model over another (Holsinger and Lewis, 2003).
Analysis of molecular variance (AMOVA) (Excoffier et al., 1992) was used to determine hierarchical genetic structure of the populations. The program Amova-prep (Miller, 1997b) was used to construct a Euclidean distance matrix. Then, AMOVA was carried out on the Euclidean distance matrix with the program WINAMOVA, Version 1.55 (Excoffier, 1995), with 1000 random bootstraps of the data set to partition the overall genetic variation into different hierarchical levels of population differentiation.
UPGMA cluster analysis was used to generate a dendrogram of all individual plants based on Nei's (1978) unbiased genetic distance. The analysis was performed using SAHN and TREE programs in NTSYS pc2.0 (Rohlf, 1997) and the resulting dendrogram was tested through 1000 bootstrap replicates. Linear regression was used to assess the relationship between population area (m2) or population size (N) and the amount of genetic variation (HE, hs and I) within populations. The analysis was performed using SPSS version 10.0 (SPSS, 1999). A Mantel test (Mantel, 1967) was used to determine if there was an association between genetic distances (pairwise ΦST) and geographical distances for the five Xiuning populations, and the significance of the test was also determined, using the program TFPGA with 5000 permutations (Miller, 1997a). The amount of gene flow (Nm) among the five Xiuning populations was estimated based on the mean value of pairwise genetic distances using the formula, Nm = (1 – ΦST)/4ΦST.
RESULTS
AFLP polymorphism and genetic diversity within populations
The eight primer combinations generated a total of 343 reliable fragments ranging from 100 bp to 500 bp. Of these, 210 (61·6 %) were polymorphic across 106 individuals of I. sinensis (Table 1). Each individual sampled showed a unique AFLP phenotype. The number of fragments generated by different primer pairs varied from 25 by E-ACT/M-CTC to 52 by E-ATG/M-CGT with an average of 42·9 fragments. The percentage of polymorphic bands of the eight different primer pairs varied from 44·7 % to 78·3 %, with an average value of 61·6 % (Table 1).
The estimates of genetic diversity for each population are summarized in Table 2. The percentage of polymorphic loci ranged from 25·2 % in population XN2 to 53·8 % in population SY1, with a mean value of 35·2 %. The estimates of heterozygosity (HE) based on the Lynch–Milligan procedure were generally lower than Bayesian estimates (hs). There was no significant correlation between estimates of HE and hs (R2 = 0·429, P = 0·337) and between estimates of HE and I (R2 = 0·500, P = 0·253), however, the correlation between hs and I was highly significant (R2 = 0·964, P = 0·000). Population SY1 exhibited the highest heterozygosity in all three parameters (HE = 0·171; hs = 0·200; I = 0·288) (Table 2). The mean values of HE, hs and I across the seven populations were 0·118, 0·147 and 0·192, respectively.
Table 2.
Population area (PA), population size (PS), and within population genetic diversity in seven extant populations of Isoetes sinensis*
| Population |
PA |
PS |
N |
P (%) |
HE |
hs |
I |
|---|---|---|---|---|---|---|---|
| SY1 | 550 | 400 | 30 | 53·8 | 0·171 | 0·200 | 0·288 |
| SY2 | 278 | 1000 | 25 | 34·8 | 0·115 | 0·147 | 0·198 |
| XN1 | 50 | 60 | 11 | 34·3 | 0·117 | 0·141 | 0·186 |
| XN2 | 18 | 20 | 5 | 25·2 | 0·083 | 0·132 | 0·146 |
| XN3 | 70 | 60 | 10 | 38·6 | 0·125 | 0·135 | 0·172 |
| XN4 | 60 | 30 | 15 | 29·0 | 0·104 | 0·148 | 0·206 |
| XN5 | 53 | 50 | 10 | 31·0 | 0·109 | 0·125 | 0·147 |
| Mean | 154 | 231 | 15 | 35·2 | 0·118 | 0·147 | 0·192 |
PA, Population area (m2); PS, actual population size; N, sample size; P(%), percentage of polymorphism; HE, Nei's (1978) unbiased expected heterozygosity; hs, genetic diversity using Bayesian approach; I, Shannon and Weaver's (1949) index.
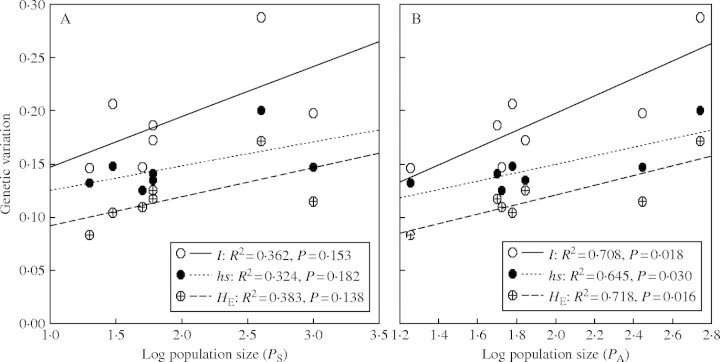
The seven populations surveyed for population size and area varied in size from approx. 20 plants (XN2) to 1000 plants (SY2), and the population areas ranged from 18 m2 (XN2) to 550 m2 (SY1) (Table 2). Larger populations tended to harbour higher levels of genetic variation (P, HE and hs), although no significant correlation was detected (Fig. 2A). However, significant correlations were found between population area and each of the three genetic diversity measures (Fig. 2B).
Fig. 2.
Relationship of (A) population size and (B) population area vs. genetic variation measured as HE, hs and I.
Population genetic differentiation
Using the Bayesian approach, the θB values obtained from the three different models are presented in Table 3. The smallest mean DIC 2858.8 was obtained under the f-free model, suggesting that the f-free model is more suitable than other models for this data set. Therefore, θB = 0·607 was determined to be an unbiased estimate of population genetic differentiation under the f-free model, with an inbreeding coefficient f = 0·504. On the other hand, our estimate of FST, under the assumption of Hardy–Weinberg equilibrium was 0·535, which is smaller than θB obtained from the Bayesian approach, while the value of GST = 0·608 was obtained under the assumption of total selfing, which is almost equal to the θB = 0·607 derived from the f-free model. Thus, the results suggested that natural populations of I. sinensis should neither be completely outcrossed nor randomly mated. The values of θB = 0·607 and GST = 0·608 would be more logical indexes to explain the population genetic differentiation for I. sinensis. Although there is a slight difference among the three measures, all of them indicate a strong genetic structure among I. sinensis populations.
Table 3.
Wright's F-statistics θB calculated under three models of Bayesian approach (95 % confidence intervals are shown in parentheses)
| Model |
f |
θB |
DIC |
|---|---|---|---|
| f = 0 | 0·000 | 0·565 (0·540–0·589) | 2873·4 |
| Full | 0·006 | 0·566 (0·543–0·590) | 2875·2 |
| f-free | 0·504 | 0·607 (0·577–0·635) | 2858·8 |
DIC, Deviance information criterion; f, inbreeding index within populations.
AMOVA further revealed significant genetic differentiation across the sampled distribution of I. sinensis (Table 4). At the population level, 63·5 % of the total molecular variation was attributed to inter-population differentiation, and 36·5 % to individual differentiation within populations. However, when the total variance was partitioned into three hierarchical levels, the largest variance component (46·9 %) was found between the two regions sampled, while only 23·2 % and 29·9 % variance was found among populations within regions and among individuals within populations, respectively.
Table 4.
Analysis of molecular variance (AMOVA) of 210 polymorphic AFLP markers for different hierarchical analyses of Isoetes sinensis populations
| Source of variation |
d.f. |
MSD |
Variance component |
Percentage of total |
P-value* |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Two hierarchical levels: | ||||||||||
| Among populations | 6 | 422·12 | 28·24 | 63·5 | <0·001 | |||||
| Within populations | 99 | 16·22 | 16·22 | 36·5 | <0·001 | |||||
| Three hierarchical levels: | ||||||||||
| Between regions | 1 | 1606·42 | 25·49 | 46·9 | <0·001 | |||||
| Among populations | 5 | 185·26 | 12·60 | 23·2 | <0·001 | |||||
| Within populations | 99 | 16·22 | 16·22 | 29·9 | <0·001 | |||||
Statistic significance is based on 1000 permutations.
The mean pairwise genetic distance among populations (pairwise ΦST) was 0·530 ± 0·192, ranging from 0·093 between population XN2 and XN3 to 0·736 between population SY2 and XN5 (Table 5). All ΦST for each pairwise comparison was significant (P < 0·001), except for ΦST between XN2 and XN3. In addition, the Mantel test revealed that no correlation between genetic distances (ΦST) and geographical distances existed among five local populations in Xiuning region (r = 0·267, P = 0·708), and the amount of gene flow estimated is Nm = 0·378.
Table 5.
Pairwise estimated values of ΦST among Isoetes sinensis populations*
| Populations |
SY1 |
SY2 |
XN1 |
XN2 |
XN3 |
XN4 |
|---|---|---|---|---|---|---|
| SY2 | 0·495*** | |||||
| XN1 | 0·664*** | 0·723*** | ||||
| XN2 | 0·652*** | 0·728*** | 0·265*** | |||
| XN3 | 0·655*** | 0·724*** | 0·221*** | 0·093ns | ||
| XN4 | 0·657*** | 0·720*** | 0·398*** | 0·399*** | 0·396*** | |
| XN5 | 0·668*** | 0·736*** | 0·503*** | 0·564*** | 0·552*** | 0·315*** |
Statistic significance based on 1000 permutations was shown: ns, not significant;
P < 0·001.
Cluster analysis
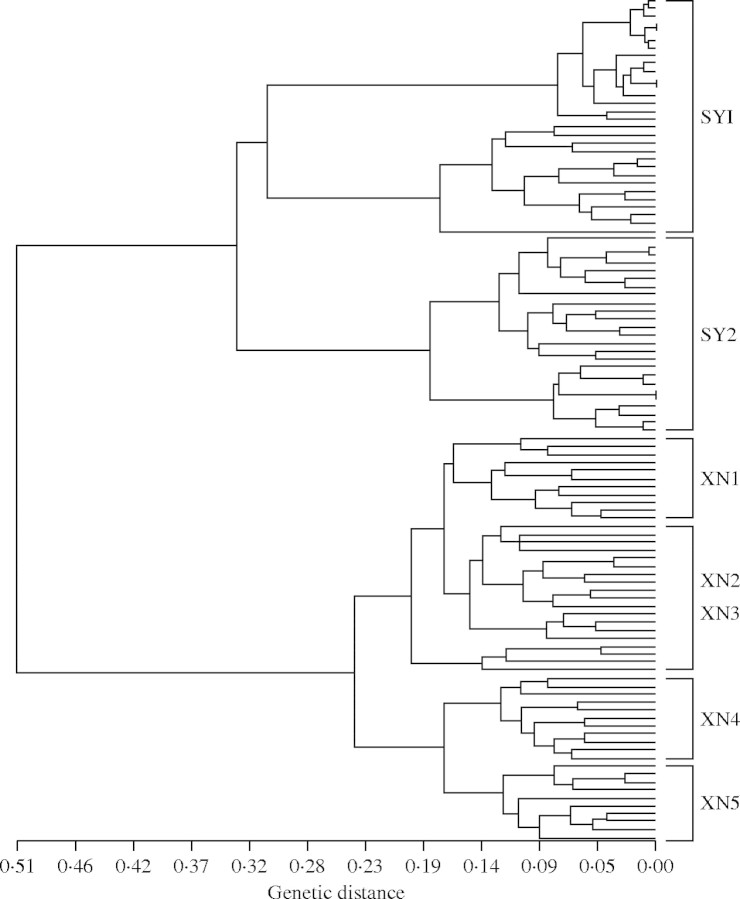
The UPGMA dendrogram (Fig. 3) was supported with high bootstrap values (>90 %), suggesting a strong population structure. The dendrogram of the 106 individual plants based on Nei's genetic distance coefficients revealed two well-defined clusters, which is well in accordance with their geographical distributions, namely the clusters SY and XN (Fig. 3). Furthermore, individuals from each population were mostly clustered into population-specific subclusters, except that individuals from population XN2 and XN3 grouped into a single subcluster and individuals in population XN4 were not clustered together. Individuals of SY1 and SY2 formed two distinct subclusters consistent with their habitats.
Fig. 3.
UPGMA dendrogram of 106 individuals of seven extant Isoetes sinensis populations based on 210 polymorphic AFLP markers.
DISCUSSION
Genetic variation within populations
Most population genetic studies in pteridophyte species have used allozymes to assess the genetic diversity and population structures because the inbreeding coefficient (f) can be directly inferred from co-dominant allozyme markers. However, the advantage is largely compromised when the species is polyploid, such as the tetraploid I. sinensis (Chen et al., 2004), although there are recently developed protocols for analysing tetraploid populations (Thrall and Young, 2000). DNA-based genetic markers have been widely used in population studies of many seed plants and have proven to be a useful and effective tool for the investigation of genetic diversity and population genetic structures in natural populations of pteridophyte species (Schneller et al., 1998; Keiper and McConchie, 2000; Landergott et al., 2001; Pryor et al., 2001; Vitalis et al., 2002; Kingston et al., 2004). In the present study, heterozygosity within populations estimated by the Lynch–Milligan procedure was not consistent with that estimated by the Bayesian procedure recently suggested by Holsinger et al. (2002). For unbiased estimates of heterozygosity, the Bayesian procedure was believed to be more suitable over Lynch–Milligan procedure (Zhivotovsky, 1999), although Krauss (2000) found the Lynch–Milligan and Bayesian procedures could yield equivalent and accurate estimates of average heterozygosity in Persoonia mollis, which is a predominantly outbreeding species and exhibits high levels of AFLP polymorphism. For I. sinensis, the observed relatively low proportion of polymorphisms (P = 35·2 %) (Table 2) may explain the bias. Recently, Tero et al. (2003) detected a high correlation between Lynch–Milligan and Bayesian estimates for Silene tatarica with the percentage of polymorphic loci ranging from 25·9 % to 54·9 %, which is comparable with I. sinensis, but they sampled a larger number of individuals per population (24–30 individuals). This might reduce the statistical bias. In the present study, sampling a larger number of individuals was prohibited due to the status of the critically endangered species with extremely small numbers of adult plants available for sampling in several populations (populations XN1–5). Therefore, a certain degree of statistical bias for estimating average heterozygosity might be expected when using the Lynch–Milligan procedure under the assumption of Hardy–Weinberg equilibrium. On the other hand, estimating population differentiation resulted in nearly identical values; this should provide reasonable confidence that Bayesian estimates of heterozygosity obtained for I. sinensis are adequately accurate.
Estimating the genetic diversity of I. sinensis at the population level revealed relatively high genetic diversity in I. sinensis (P = 35·2 %, HE = 0·118, hs = 0·147 and I = 0·192) in comparison with other available data in pteridophytes. Caplen and Werth (2000) reported very low intra-population diversity for six diploid Isoetes species in North America based on isozymes, with P ranging from 0·0 to 22·9 % and HE ranging from 0·000 to 0·093. Similarly, a low percentage of polymorphic loci (P = 23·7 %) and Nei's heterozygosity (HE) = 0·073 were found in Sticherus flabellatus populations using AFLP (Keiper and McConchie, 2000). In a survey of allozyme data in fern species, the value of P and HE at the population level for 32 diploid homosporous fern species was 31·5 % and 0·110, respectively (Maki and Asada, 1998). The relatively high genetic diversity observed in I. sinensis may be attributed to its allopolyploid origin (He et al., 2002). High genetic diversity was also detected in our previous allozyme analysis (HE = 0·189; P = 43·3 %; Chen et al., 2004). As far as is known, most genetic diversity studies based on AFLP analysis so far conducted were for seed plants, particularly endangered seed plants. For example, the intra-population heterozygosity in a critically endangered Astragalus cremnophylax var. cremnophylax ranged from 0·037 to 0·134 (Travis et al., 1996). However, Evans et al. (2001) found very high genetic diversity within populations in the rare plant Banksia saxicola, with the value of HE ranging from 0·19 to 0·26. Recently, Tero et al., (2003) reported genetic diversity for an endangered perennial plant Silene talarica, ranging from 0·075 to 0·176 (Nei's estimate) and from 0·131 to 0·190 (Bayesian estimate), which were similar to these values detected for I. sinensis (Table 2). Apparently, I. sinensis maintains high levels of genetic diversity in spite of small population sizes. It is likely that the reduction in population size was a relatively recent event in the natural range of I. sinensis. This is consistent with existing documentation that the species was more widely distributed in eastern China previously, especially in Zhejiang province (Fig. 1). Many populations became extinct very recently, such as the DX and TT populations (Fig. 1; field survey data). This recent decline of populations may not yet have had sufficient time to result in a decrease of the intra-population genetic diversity in I. sinensis.
Many empirical studies have confirmed the theoretical prediction that smaller populations harbour lower genetic variation than larger populations due to genetic erosion (Frankham, 1996; Travis et al., 1996; Gaudeul et al., 2000). However, in the present study, no significant correlation was detected between population size and the amount of genetic variation within populations (Fig. 2A). Similar results were also found in other rare and endangered species (Schmidt and Jensen, 2000; Tero et al., 2003). The lack of correlation between population size and genetic variation has been interpreted to be a consequence of recent population decrease or expansion (Tero et al., 2003). Interestingly, the genetic variation was significantly correlated to population area (Fig. 2B), suggesting that within-population migration of propagules was related to the spatial scale of isolated wetland habitat patches. A similar result was found in small natural populations of Calystegia collina (Wolf et al., 2000) in which genetic variation was not correlated to the number of individuals in the population, but significantly correlated to population area. Seemingly, the genetic diversity within each population of I. sinensis has been highly influenced by changes in the size of wetland habitats, possible reflecting fluctuations in annual rainfall and anthropogenic activities such as farmland irrigation management and fertilization methods with downstream effects on water quality. The complex dynamics of wetland habitats could be the greatest threat to I. sinensis. In addition, Wen et al. (2003) found that water quality had deteriorated highly in the wetland habitat of I. sinensis, which contained higher concentration of nitrates, dissolved carbon dioxide and heavy metal ions.
Population structure
Population differentiation in I. sinensis, both at a regional scale and across its distribution range, was evident as measured by a range of different statistics (FST, GST and θB). The values of FST and GST differed slightly when different breeding systems were assumed (FST = 0·535 vs. GST = 0·608). Furthermore, the Bayesian estimate of θB = 0·607 is almost identical to Nei's GST = 0·608 when total selfing (FIS = 1) was assumed for the species. The partitioning of genetic variation among populations has previously been reported in other Isoetes species. For example, the values of FST based on allozyme analysis for two diploid Isoetes species were 0·607 for I. flaccida and 0·947 for I. engelmannii (Caplen and Werth, 2000). Similar results have also been documented in other pteridophyte species (Schneller and Holderegger, 1996; Keiper and McConchie, 2000; Pryor et al., 2001). Using 469 AFLP markers, Keiper and McCondie (2000) reported a high mean FST estimate (0·783) among eight populations of the umbrella fern Sticherus flabellatus. High genetic differentiation was attributed to restricted gene flow, genetic drift and inbreeding (Loveless and Hamrick, 1984). Similar to other aquatic Isoetes species, populations of I. sinensis are isolated from one another by their disjunct habitats, i.e. streams, hills and farmlands (Caplen and Werth, 2000). Dispersal of propagules can be achieved via transportation by water and occasionally by animals. Natural migration between the two populations in the Songyang region appears very limited since the populations are located on the opposing sides of the Anmin Mountain. During the 4-year field survey carried out for this study, no individuals were found in surrounding areas, which suggests current migration by human or animal activities is minimal. The populations in Xiuning region are much closer to each other, but they are also isolated by farmland and hills. The AMOVA analysis further suggested that a large proportion of genetic differentiation was attributed to geographic isolation (Table 4). Additional evidence from the UPGMA dendrogram revealed distinct clustering by geographic locations (Fig. 3), suggesting that gene flow was highly restricted between populations. As an aquatic quillwort, I. sinensis usually grows in damp sites that usually result in small clusters within each population. Therefore, the dispersal of this species is believed to be very limited and highly impacted by water flow regimes even at a microgeographical scale. This is in agreement with Taylor and Hickey (1992) who suggested that low levels of gene flow occur among Isoetes populations.
According to the theory of isolation by distance (Wright, 1943), when populations reach equilibrium between gene flow and genetic drift, a positive correlation should be detected between genetic distance and geographical distance. However, when populations are in disequilibrium, either gene flow or genetic drift can dominantly influence the population structure, and thus no correlation would be detected (Hutchison and Templeton, 1999). When the distance is not a limiting factor for the dispersal and gene flow is unlimited, populations would form a single uniform genetic unit, but when gene flow rate is greatly reduced, population genetic differentiation will increase due to genetic drift (Slatkin, 1977; Hutchison and Templeton, 1999). In the present study, the estimate of gene flow is very small (Nm = 0·378), and no positive correlation between genetic distances (ΦST) and geographical distances was detected among the populations in Xiuning site, suggesting that genetic drift rather than gene flow played an important role in forming the present population structure. On the other hand, no significant genetic differentiation was found between population XN2 and XN3 (ΦST = 0·093) which might indicate adequate gene flow occurred between these two populations. This can be explained by their habitat situation: these two small populations were directly connected by a stream, although they were not the closest populations with respect to geographical distance (Fig. 1). This result confirms the assumption that hydrochory is the most important factor influencing gene flow among I. sinensis populations. A similar observation was also found in Dryopteris cristata, a fern species with high population differentiation and no significant correlation between genetic and geographical distances (Landergott et al., 2001).
Implications for conservation
It has been suggested that genetic variation is important for a species to maintain its evolutionary potential to cope with ever-changing environments. The information obtained in this study provides the first set of population genetic data to address conservation concerns for this critically endangered species. Isoetes sinensis populations appeared to maintain adequate genetic diversity in terms of neutral genetic variation. But in the absence of gene flow among populations and given the small population sizes, genetic drift might lead to a rapid genetic erosion and increase the extinction risk of local populations. An in situ conservation programme is urgently needed to mitigate further reduction of population size and deterioration of remnant habitats. Genetic reinforcement by translocation is an alternative measure to rehabilitate this species. However, considering the high differentiation among populations, the potential for inter-population outbreeding depression should be carefully investigated before such measures are implemented. If population differentiation was caused by different environmental adaptations, a mixture of individuals from genetically distinct populations may result in outbreeding depression (Hufford and Mazer, 2003). In the present study, since there are obvious ecological differences between Xiuning and Songyang regions, translocations between these two regions cannot currently be recommended. Alternatively, if population differentiation is caused by genetic drift, inter-population translocation will improve the performance of populations (Hufford and Mazer, 2003). Inter-population transplantations within regions would increase gene flow among populations and counteract the negative effects of genetic drift and inbreeding.
For ex situ conservation, the same caution should be taken to prevent mixture of individuals from different regions. Ideally, regional populations should be preserved separately in well-designed plots (ponds), while individuals from the same region need to be kept in the same plot (or pond) for genetic enhancement of outcrossing. A field sampling strategy should be developed based on the population genetic data so that sufficient genetic diversity of the extant populations could be maintained for the long-term survival of the species. Given the current situation of rapid decline of populations and natural habitats, it is strongly recommended that all extant populations are extensively sampled to maximize genomic representation of the species before further genetic erosion can occur.
Acknowledgments
This research was partially supported by KIP Pilot Project of Chinese Academy of Sciences (KSCX2-SW-104) and National Natural Sciences Foundation of China (30370153). WZ no. 050820 of Conservation Genetic Laboratory, WBG-CAS. We thank Mr Genyou Li and Mr Bingyang Ding for their information on field surveys; and Dr Siegy Krauss and Dr Tianhua He for their critical discussion and suggestions on the manuscript. We also thank Dr Steve Waldren and another anonymous reviewer for their valuable comments and suggestions.
LITERATURE CITED
- Barrett SCH, Kohn JR. 1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants. New York: Oxford University Press, 3–30. [Google Scholar]
- Caplen CA, Werth CR. 2000. Isozymes of the Isoetes riparia Complex. I. Genetic variation and relatedness of diploid species. Systematic Botany 25: 235–259. [Google Scholar]
- Chen JK, Wang HY, He GQ. 1998. A survey on the habitats of Oryza rufipogon and Isoetes sinensis in Jiangxi Province. Biodiversity Science 6: 260–266. [Google Scholar]
- Chen Y, Ye Q, Li Z, Huang H. 2004. Genetic structure of Xiuning population of Isoetes sinensis, a critically endangered species in China. Biodiversity Science 12: 564–571. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytotchemical Bulletin 19: 11–15. [Google Scholar]
- Evans KM, Ladiges PY, Newbigin E, Ades PK. 2001. Genetic variation in Banksia saxicola (Proteaceae), a rare Australian plant with a markedly disjunct distribution. Plant Systematics and Evolution 227: 105–115. [Google Scholar]
- Excoffier L. 1995.AMOVA 1.55 (analysis of molecular variance). Switzerland: Genetics and Biometry Laboratory, University of Geneva. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variation inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Conservation Biology 10: 1500–1508. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. 2002.Introduction to conservation genetics. Cambridge: Cambridge University Press. [Google Scholar]
- Fu LK, Jin JM. 1992.China plant Red Data Book—rare and endangered plants. Beijing: Science Press, 538–539. [Google Scholar]
- Gaudeul M, Taberlet P, Till-Bottraud I. 2000. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Molecular Ecology 9: 1625–1637. [DOI] [PubMed] [Google Scholar]
- He ZC, Cai Q, Liu HT, Li JQ, Huang H. 2002. Chromosome number of Isoetes sinensis Palmer, a rare and endangered pteridophyte plant. Journal of Wuhan Botanical Research 20: 241–242. [Google Scholar]
- Holsinger KE, Lewis PO. 2003.HICKORY: a package for analysis of population genetic data, Version 8·0. University of Connecticut, Storrs. [Google Scholar]
- Holsinger KE, Lewis PO, Dey DK. 2002. A Bayesian approach to inferring population structure from dominant markers. Molecular Ecology 11: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Hufford KM, Mazer SJ. 2003. Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends in Ecology and Evolution 18: 147–155. [Google Scholar]
- Hutchison DW, Templeton AR. 1999. Correlation of pairwise genetic and geographic distance measures: inferring the relative influence of gene flow and drift on the distribution of genetic variability. Evolution 53: 1898–1914. [DOI] [PubMed] [Google Scholar]
- IUCN. 2004.2004 IUCN Red List of Threatened Species, www.redlist.org, downloaded on 20 March 2005. [Google Scholar]
- Keiper FJ, McConchie R. 2000. An analysis of genetic variation in natural populations of Sticherus flabellatus [R. Br. (St John)] using amplified fragment length polymorphism (AFLP) markers. Molecular Ecology 9: 571–581. [DOI] [PubMed] [Google Scholar]
- Kingston N, Waldren S, Smyth N. 2004. Conservation genetics and ecology of Angiopteris chauliodonta Copel. (Marattiaceae), a critically endangered fern from Pitcairn Island, South Central Pacific Ocean. Biological Conservation 117: 309–319. [Google Scholar]
- Krauss SL. 2000. Accurate gene diversity estimates from amplified length polymorphism (AFLP) markers. Molecular Ecology 9: 1241–1245. [DOI] [PubMed] [Google Scholar]
- Lande R. 1988. Genetics and demography in biological conservation. Science 241: 1455–1460. [DOI] [PubMed] [Google Scholar]
- Lande R. 1999. Extinction risk from anthropogenic, ecological and genetic factors. In: Landweber LA, Dobson AP, eds. Genetics and extinction of species. Princeton, NJ: Princeton University Press, 1–22. [Google Scholar]
- Landergott U, Holderegger R, Kozlowski G, Schneller JJ. 2001. Historical bottlenecks decrease genetic diversity in natural populations of Dryopteris cristata Heredity 87: 344–355. [DOI] [PubMed] [Google Scholar]
- Loveless MD, Hamrick JL. 1984. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15: 65–95. [Google Scholar]
- Lynch M, Milligan BG. 1994. Analysis of population genetic structure with RAPD markers. Molecular Ecology 3: 91–99. [DOI] [PubMed] [Google Scholar]
- Maki M, Asada Y. 1998. High genetic variability revealed by allozymic loci in the narrow endemic fern Polystichum otomasui (Dryopteridaceae). Heredity 80: 604–610. [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Research 27: 209–220 [PubMed] [Google Scholar]
- Miller MP. 1997.Tools for population genetics analysis (TFPGA), Version 1.3. A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author. [Google Scholar]
- Miller MP. 1997.AMOVA-PREP. A program for the preparation of input files for use with Win-AMOVA. Computer software distributed by author. [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1987.Molecular evolution genetics. New York: Columbia University Press. [Google Scholar]
- Pang XA, Liu X, Liu H, Wu C, Wang JY, Yang SX, et al. 2003. The geographic distribution and habitat of the Isoetes plants in China. Biodiversity Science 11: 288–294. [Google Scholar]
- Pryor KV, Young JE, Rumsey FJ, Edwards KJ, Bruford MW, Rogers HJ. 2001. Diversity, genetic structure and evidence of outcrossing in British populations of the rock fern Adiantum capillus-veneris using microsatellites. Molecular Ecology 10: 1881–1894. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. 1997. NTSYS-pc, numerical taxonomy and multivariate analysis system, Version 2.0. Setauket, NY: Exeter Software. [Google Scholar]
- Schmidt K, Jensen K. 2000. Genetic structure and AFLP variation of remnant populations in the rare plant Pedicularis palustris (Schropulariacae) and its relation to population size and reproductive components. American Journal of Botany 87: 678–689. [PubMed] [Google Scholar]
- Schneller JJ, Holderegger R. 1996. Genetic variation in small, isolated fern populations. Journal of Vegetation Science 7: 113–120. [Google Scholar]
- Schneller JJ, Holderegger R, Gugerli F, Eichenberger K, Lutz E. 1998. Patterns of genetic variation detected by RAPDs suggest a single origin with subsequent mutations and long-distance dispersal in the apomictic fern Dryopteris remota (Dryopteridaceae). American Journal of Botany 85: 1038–1042. [PubMed] [Google Scholar]
- Shannon CE, Weaver W. 1949.The mathematical theory of communication. Urbana, IL: University of Illinois Press. [Google Scholar]
- Slatkin M. 1977. Gene flow and genetic drift in a species subject to frequent local extinctions. Theoretical Population Biology 12: 253–262. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. 2002. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society Series B 64: 483–489. [Google Scholar]
- Spielman D, Brook BW, Frankham R. 2004. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences of the USA 42: 15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPSS. 1999.Statistical software package, Version 10.0. Chicago, IL: SPSS Inc. [Google Scholar]
- Takamiya M. 2001.Isoetes sinensis var. sinensis in Korea (Isoetaceae: Pteridophyta). Fern Gazette 16: 169–177. [Google Scholar]
- Takamiya M, Watanabe M, Ono K. 1994. Biosystematic studies on the genus Isoetes (Isoetaceae) in Japan. I. Variation of somatic chromosome numbers. Journal of Plant Research 107: 289–297. [Google Scholar]
- Taylor WC, Hickey RJ. 1992. Habitat, evolution and speciation in Isoetes. Annals of the Missouri Botanical Garden 79: 613–622. [Google Scholar]
- Taylor WC, Lekschas AR, Wang QF, Liu X, Napier, NS, Hoot SB. 2004. Phylogenetic relationships of Isoetes (Isoetaceae) in China as revealed by nucleotide sequences of the nuclear ribosomal ITS region and the second intron of a LEAFY homolog. American Fern Journal 94: 196–205. [Google Scholar]
- Tero N, Aspi J, Siikamäki P, Jäkäläniemi A, Tuomi J. 2003. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica Molecular Ecology 12: 2073–2085. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Young A. 2000. AUTOTET: a program for analysis of autotetraploid genotypic data. Journal of Heredity 91: 348–349. [DOI] [PubMed] [Google Scholar]
- Travis SE, Maschinski J, Keim P. 1996. An analysis of genetic variation in Astragalus cremnophylax var. cremnophylax, a critically endangered plant, using AFLP markers. Molecular Ecology 5: 735–745. [DOI] [PubMed] [Google Scholar]
- Vitalis R, Riba M, Colas B, Grillas P, Olivieri I. 2002. Multilocus genetic structure at contrasted spatial scales of the endangered water fern Marsilea strigosa Willd. (Marsileaceae, Pteridophyta). American Journal of Botany 89: 1142–1155. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QF, Liu X, Taylor WC, He ZR. 2002.Isoetes yunguiensis (Isoetaceae), a new basic dioploid quillwort from China. Novon 12: 587–591. [Google Scholar]
- Wen MZ, Pang XA, Wang QF, Taylor WC. 2003. Relationship between water chemistry and the distribution of the endangered aquatic quillwort Isoetes sinensis Palmer in China. Journal of Freshwater Ecology 18: 361–367. [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wolf AT, Harrison SP, Hamrick JL. 2000. Influence of habitat patchiness on genetic diversity and spatial structure of a serpentine endemic plant. Conservation Biology 14: 454–463. [Google Scholar]
- Wright S. 1943. Isolation by distance. Genetics 28: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Li J. 2003. Distribution status and causation of endangerment Isoetes sinensis Palmer in Zhejiang province. Journal of Wuhan Botanical Research 21: 216–220. [Google Scholar]
- Yeh FC, Yang RC, Boyle T. 1997.POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotchnology Centre, University of Alberta: Edmonton. [Google Scholar]
- Zhivotovsky LA. 1999. Estimating population structure in diploid with multilocus dominant DNA markers. Molecular Ecology 8: 907–913. [DOI] [PubMed] [Google Scholar]