Abstract
Cancer genome sequencing is being employed at an increasing rate to identify actionable driver mutations that can inform therapeutic intervention strategies. A comparison of two of the most prominent cancer genome sequencing databases from different institutes (CCLE and COSMIC) revealed marked discrepancies in the detection of missense mutations in identical cell lines (57.38% conformity). The main reason for this discrepancy is inadequate sequencing of GC-rich areas of the exome. We have therefore mapped over 400 regions of consistent inadequate sequencing (cold-spots) in known cancer-causing genes and kinases, in 368 of which neither institute finds mutations. We demonstrate, using a newly identified PAK4 mutation as proof of principle, that specific targeting and sequencing of these GC-rich cold-spot regions can lead to the identification of novel driver mutations in known tumor suppressors and oncogenes. We highlight that cross-referencing between genomic databases is required to comprehensively assess genomic alterations in commonly used cell lines and that there are still significant opportunities to identify novel drivers of tumorigenesis in poorly sequenced areas of the exome. Finally we assess other reasons for the observed discrepancy, such as variations in dbSNP filtering and the acquisition/loss of mutations, to give explanations as to why there is discrepancy in pharmacogenomic studies given recent concerns with poor reproducibility of data.
Keywords: Cancer Genomics, Next Generation Sequencing, Driver Mutation, GC Content
Introduction
Personalised therapeutic approaches that target genetically activated drivers have significantly improved patient outcome in a number of common and rare cancers. The development of personalised therapeutics relies on affordable, efficient, and accurate cancer genomic sequencing to identify genetic aberrations present in a given tumor, from which actionable mutations can then be obtained (1). To aid novel driver and targeted therapy discovery, the Sanger and Broad Institutes have developed extensive catalogues of mutations found in a large cohort of cell lines. These resources, which are readily accessible to most biomedical researchers via database portals, have greatly facilitated the process of driver gene discovery. Through an initial evaluation of genetic dependencies in NSCLC cell lines we observed inconsistencies in the mutational profiles as reported by the Sanger Institute’s COSMIC database and the Broad Institute’s Cancer Cell Line Encyclopaedia (CCLE) (2-4). We therefore investigated the extent and causes of these discrepancies in order to identify opportunities to improve the discovery of driver mutations in oncogenes and tumor suppressors (TSs).
Materials and Methods
18 Cell Line Comparison between COSMIC and CCLE data
Commercially available cell lines previously sequenced by COSMIC were identified from the Greenman et al. paper (5). Eighteen of these cell lines were also sequenced by CCLE using the Hybrid Capture method using the SureSelect Target Enrichment System (Agilent Technologies) and sequencing on Illumina instruments (76bp paired read ends). Mutational data was downloaded from CCLE website on 14th May 2013 (CCLE_hybrid_capture1650_hg19_NoCommonSNPs_NoNeutralVariants_CDS_2012.05.07.m af). COSMIC data was downloaded for each cell line from their respective webpages on 14th May 2013. Common genes reported as sequenced by both institutes were used to compare both datasets. Script A (supplementary data) was written in Groovy programming language to compare the genetic location of missense non-truncating mutations recorded by each institute and compare the lists to find conformity. Sequencing bam files for the CCLE hybrid capture sequencing (COSMIC data unavailable) was viewed using the Integrative Genomics Viewer (IGV: Broad Institute) (6) to categorize the mutations only reported in COSMIC. GC content of the missed mutations was calculated with Ensembl Rest API (version 70) reference genome and capturing the sequence 100bp either side of the mutation.
568 cell line comparison
COSMIC cell line names were compared with the list of cell lines sequenced by CCLE to find 568 mutually sequenced cell lines. CCLE data was downloaded in the filtered MAF file as described above. COSMIC data was downloaded as a complete file from the COSMIC FTP site on 12th November 2013 (CosmicCellLineProject_v67_241013.tsv.gv). The comparison of the sequencing of 1630 mutually sequenced genes by the two data sets was performed using Script B (supplementary data). Mutations were matched by genomic location. Given the variability of gene transcripts from which amino acid changes are calculated, the amino acid change reported was derived from the most common resultant amino acid change and where there was no majority change the CCLE change was reported followed by COSMIC when comparing COSMIC and CRUK MI data only. CCLE data that was unfiltered (data for common polymorphisms, putative neutral variants and mutations located outside of the CDS not filtered out) and containing all variants with an allelic fraction >10% was obtained from the CCLE website on 22nd November 2013 (CCLE_hybrid_capture1650_hg19_allVariants_2012.05.07.maf.gz). The COSMIC only mutations were cross-referenced against the unfiltered CCLE list to identify further mutation matches. Cancer Census genes were identified from the COSMIC Cancer Census webpage (http://cancer.sanger.ac.uk/cancergenome/projects/census/) (7).
Whole Exome Sequencing of 4 cell lines
Cell lines were obtained from ATCC and DNA extracted within 3 passages of delivery from ATCC corresponding to less than one month from time of receipt. ATCC authenticates cell lines through short tandem repeat profiling, morphology analysis, cytochrome C oxidase I (COI) testing, and karyotyping. On arrival from ATCC the total passage number for each cell line was; H2009 = 23, H2087 = 21, H2122 = 21, H1437= 46. Cells are maintained in RPMI Medium 1640 (Invitrogen) with additional 10% FCS (Lonza Group) and 4mM GlutaMAX™ (Invitrogen). Cells are split 1:10 at 80% confluency. DNA extraction is performed using DNeasy Blood and Tissue kit (Qiagen). Whole exome sequencing was performed using Agilent Sure Select XT Target Enrichment System for Illumina Pair-end Multiplex Sequencing, enriching with the SureSelect XT Human All Exon V4 library and performing 2 × 100 bp paired-end sequencing on the Illumina HiSeq 2500 with TruSeq SBS v3 chemistry (read density: Supp. Table 5). Average read density for each sample was calculated using the Lander/Waterman equation as detailed in the Illumina Estimating Coverage Technical Note (http://res.illumina.com/documents/products/technotes/technote_coverage_calculation.pdf). Variant calling was made using the Genome Analysis Toolkit (GATK: Broad Institute) (8). Comparison of conformity with the COSMIC and CCLE mutation calls was made using Script B with data filtered and unfiltered for mutations with dbSNP ids.
Cold-spot analysis
Bam files from hybrid capture used to create the CCLE database are not available for download so 10 independent CCLE whole exome bam files (performed on Illumina HiSeq 2000) were downloaded on 9th Jan 2014 via the Cancer Genomics Hub (bam files and metadata with experimental info available from https://browser.cghub.ucsc.edu). These files were analysed for 986 kinase and Cancer Census genes (Supp. Table 6), among which 969 genes are protein-coding genes as annotated in ENSEMBL human gene database version 70. The lung cancer sequencing files used were: CCLE-NCI-H2286-DNA-08, CCLE-NCI-H1944-DNA-08, CCLE-COR-L95-DNA-08, CCLE-NCI-H1373-DNA-08, CCLE-NCI-H1184-DNA-08, CCLE-HLF-a-DNA-08, CCLE-JL-1-DNA-08, CCLE-HCC-78-DNA-08, CCLE-DV-90-DNA-08, CCLE-DMS153-DNA-08. The reads in the bam files were mapped onto the reference genome hg19. From each bam file the read coverage at each base of the protein-coding exonic regions of the 969 selected genes was obtained using samtools mpileup (9). Sequencing read cold-spots were defined as protein-coding exonic regions spanning 100 nucleotide bps or more and with the averaged read coverage <= 4 at each base. Read cold-spots were identified in the sequencing data and the GC content calculated using the bases corresponding to the read cold-spot. Multiple transcripts of the same gene were removed if the genetic location of the identified cold-spot was identical or the start or end genomic location was the same between same gene transcripts (retaining the transcript with the longest read cold-spot). Top 20 cold-spots are defined as gene transcripts (that were sequenced by CCLE and COSMIC) with the largest cold-spot regions. The average GC content for all coding exons was calculated using the longest transcript (Ensemble Version 70) for each of the 969 genes screened for cold-spots. Circos plots were constructed using the Circos software (from http://www.circos.ca) (10).
Verification of PAK4 mutation
Amplification PCR of region of interest performed using Phusion High Fidelity PCR Master Mix with H.F. Buffer (New England Biolabs) [12.5μl] with Betaine 5M (Sigma) [5μl], 250ng DNA, forward and reverse primers (Eurofin MWG Operon) [1.25 μl each] and water to make reaction volume of 25 μl. PCR was carried out on S1000 Thermal Cycler (Biorad) with the following PCR steps for a total of 40 cycles; 1) 98.0° 30 seconds 2) 98.0° 10 seconds 3) 62.0° 30 seconds 4) 72.0° for 150 seconds. PCR product purification was carried out with Illustra ExoProstar Enzymatic PCR and Sequencing Clean-up (GE Healthcare). Sequencing was carried out using an ABI13130 16 capillary system (Life Technologies) and sequencing data analysed using 4Peaks software (MekenTosj).
PAK4 Transient Overexpression
Wild-type PAK4 plasmid (Addgene 23713) was obtained from Addgene (deposited by Hahn and Root) (11). The plasmid was cloned into a Flag-tagged destination vector. STOP codon and the E119Q mutation was introduced by site-directed mutagenesis (QuickChange II kit, Agilent Technologies). Plasmid was transfected into HEK293T cells in a 12 well format using Attractene according to the manufacturers protocol. Cells were lysed on ice after 48 hours using Triton X-100 Cell Lysis Buffer supplemented with protease inhibitor tablet (Roche). Lysates were resolved on SDS-PAGE gels followed by western blotting. Primary antibodies used were: Flag M2 and alpha tubulin (Sigma); pERK1/2 (T202/Y204) and pJNK (T183/Y185) (Cell Signalling). Mouse or rabbit HRP-conjugated antibodies were used as secondary (Cell Signaling). All western blots are representative of three independent experiments.
Results and Discussion
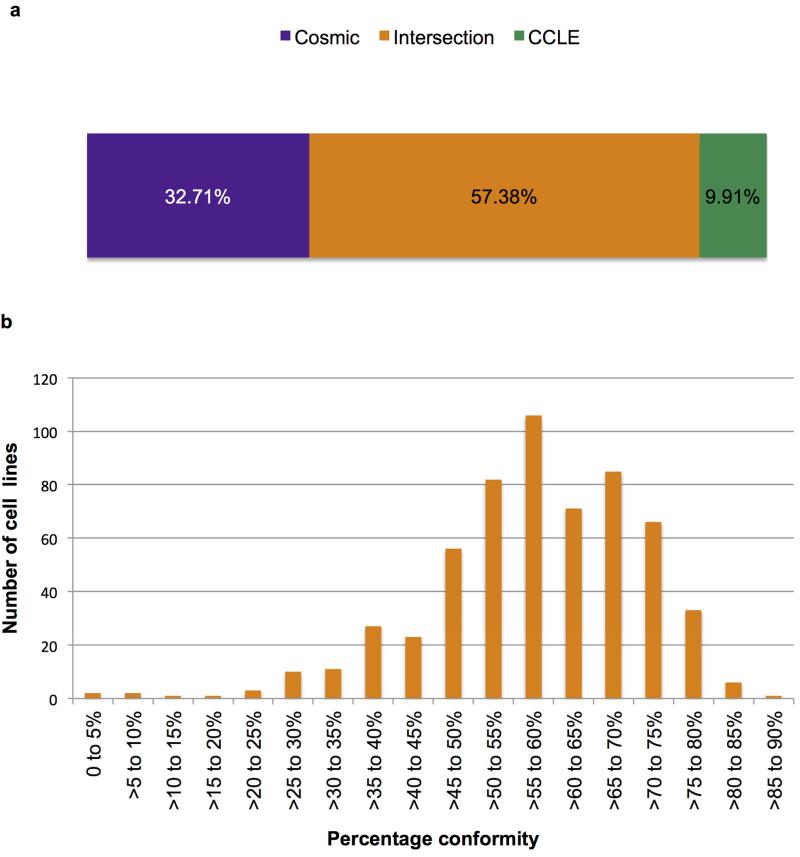
We compared missense mutations found in 568 cancer cell lines sequenced by CCLE and COSMIC (v67) across 1,630 mutually sequenced genes (3). A total of 45,377 mutations were reported, of which 26,038 were consistent between institutes (57.38%). 4,496 (9.91%) and 14,843 (32.71%) mutations were found solely by CCLE or COSMIC respectively (Figure 1). The ISHIKAWAHERAKLIO02ER cell line, sequenced by both institutes using their standard protocols, showed a total of 263 mutations (52 in COSMIC and 213 in CCLE) but no matches, suggesting different cell lines may have been sequenced. Cross referencing to Cancer Census genes (7) found that 4,058 mutations reported in one, but not both, of the databases were in known cancer causing genes (Supp. Table 1). These included mutations in EGFR, TP53, BRAF, MAP2K1 and PIK3CA (Table 1), highlighting the difficulties faced when using NGS to identify driver mutations even in well-known cancer causing genes. Our data reveal a marked discrepancy in mutation reporting between the two most prominent resources and that cross-referencing between the databases is imperative.
Figure 1.
Marked discrepancy is seen in mutation calling between CCLE and COSMIC. a) Overall percentage conformity of 46,409 mutations detected by COSMIC and/or CCLE. The intersection between datasets (mutations found by both institutes) accounted for 57.38%. Cosmic-only mutations comprised 32.71% of the dataset and CCLE only mutations 9.91%. b) The percentage agreement between mutations reported in the 568 cell lines sequenced by both institutes.
Table 1.
Mutations in well-known oncogenes and tumor suppressor genes that were detected by only one institute (COSMIC or CCLE). Mutations in bold occurred multiple times (number of occurrences in parentheses). Supplemental tables 1a and 1b list the mutations, stratified according to the reporting institute.
| BRAF | P74A, S76P, V120I, E296K, I326T, I326V(5), R506G, S727G |
| EGFR | Q71L, R98Q, E282K, S306, V323, K327E, Q408R, L469W, G614S, V654M, G659R, P672R, R677C, T678M, G682V, Q701R, A702D, V738D, A750E, A755D, L815F, L861Q, R973Q, A1076T, T1085N, A1118T, D1127N |
| FGFR2 | R6C, C9S, G89V, E163K, P187S, A315T, Y328S, T341M, A355S, G364E, K401R, L451I, P559H, I643T, C809Y, P814T |
| HRAS | A11T,G12D, L171P |
| IDH2 | Q95R, H358R, S408R(2) |
| JAK2 | V80M, Y96H, T108A, V563I, V617F, L905P, N1129S |
| KRAS | G12D(2), Q61H, I171M, M188V |
| MAP2K1(MEK1) | Q56P(3), V85I, A158T, R160K, K185T, V211A |
| NRAS | Q61K(6), Q61R (2) |
| PIK3CA | K111E, C420R, E542K, E545K, F666L, R770Q |
| STK11 | I46T, Y49D, G56V, K62N, K78N, L105S, D196R, S216F(2), G242W, M392I |
| TP53 | V31I, P47R, D48N, D49H, W53L, A74P, A74S, Y103F, R110L(2), R110P, F113C, F113V, K120N, V122L, C124R, Y126D, M133K, C135R, C176W, E180G, R181C(2), I195T, R213L, V216L(2), V218L, Y220C, N239S, S241F, C242F, M246V, R249S, R273H(14), R283C, R290C, P309S, D324A, R337L, F341C, A347P, G360V, G389W |
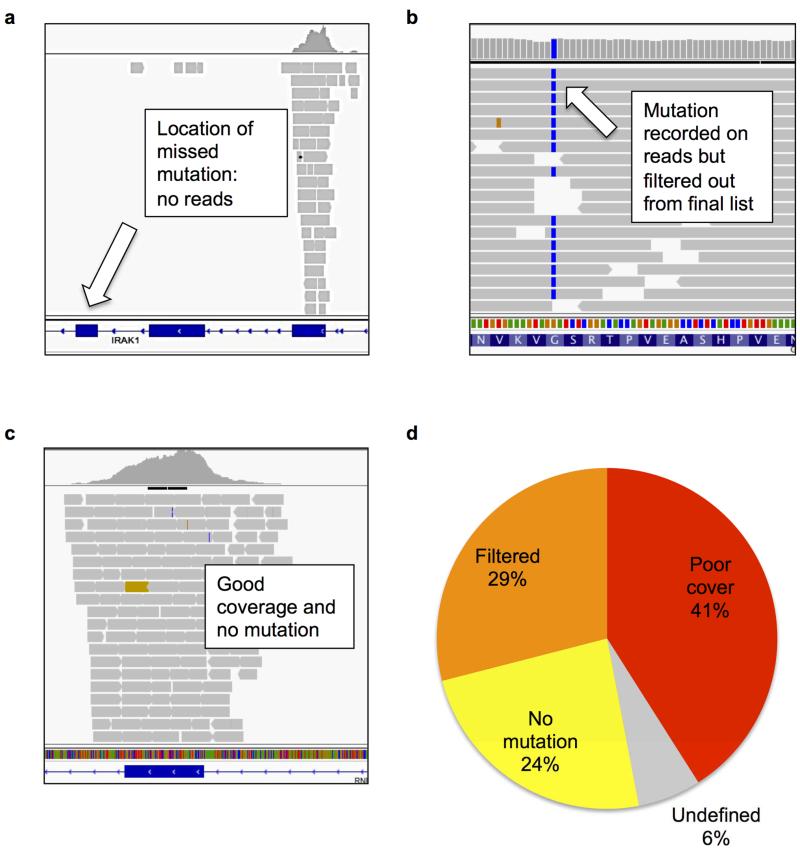
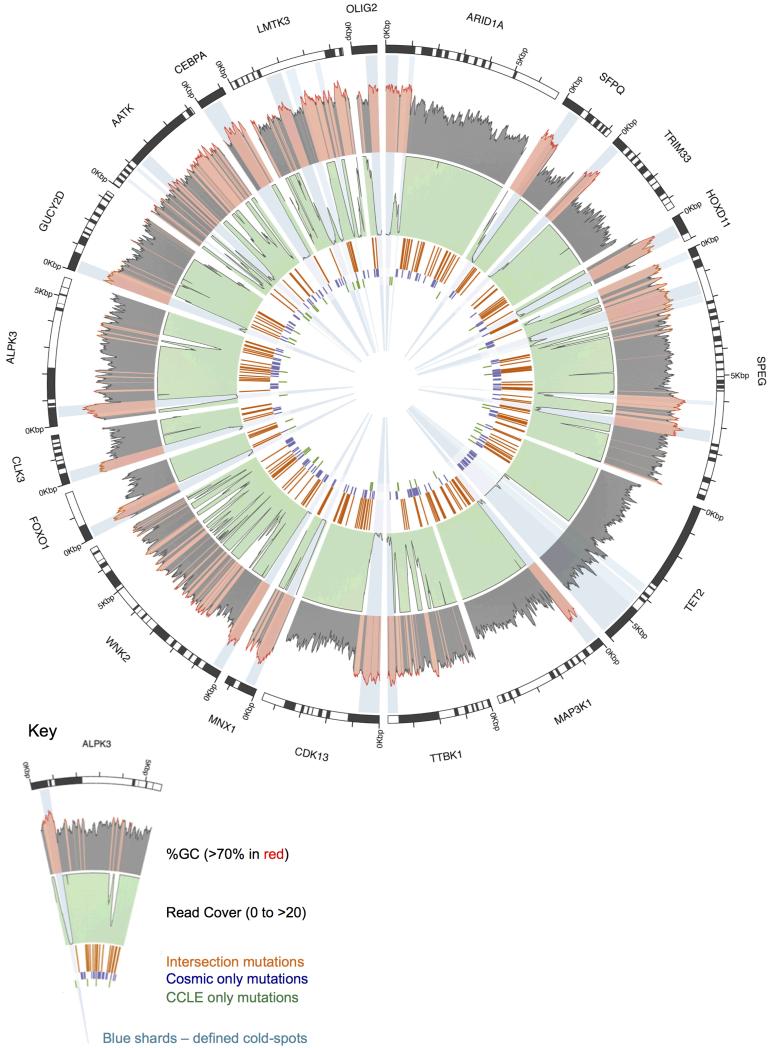
We had previously performed a pilot comparison of mutational profiles in 18 cancer cell lines sequenced by the Broad Institute’s CCLE using Hybrid Capture sequencing (3), and an earlier release of Sanger Institute’s COSMIC database (5,12). Similar to our larger scale comparison we observed low consensus between missense mutation detection in mutually sequenced genes (mean 41.33%; Supp. Figure 1). Analysing the raw read data (6) from CCLE suggested that the most common source of discrepancy was poor sequencing read coverage (41%; Figure 2). We therefore analysed 10 randomly selected CCLE whole exome sequencing files to identify regions of poor coverage (‘cold-spots’). We discovered over 400 cold-spots (100bp or larger) in Cancer Census and kinase genes that we have mapped as a resource for the research community (Figure 3 / Supp. Table 2) (10). These cold-spots are rich in GC nucleotides (63.49% compared to 51.74% average GC-content of all exons in target genes) indicating that high GC-content is a major cause of inadequate sequencing coverage. Importantly, we found for CCLE and COSMIC data combined, a 18-fold reduction in mutation density at these loci relative to the remaining exonic regions in the dataset. Extrapolating these data suggests that an additional 1871 mutations would have been detected in Cancer Census and kinase genes across the 568 cell lines (corresponding to a mean of over 3 new mutations in Cancer Census or kinase genes per cell line) had the read coverage in the cold-spots been adequate. The TET2 cold-spot (Figure 3) is one of the largest of such loci identified, and is not associated with high GC-content. Mutations were reported for this locus in COSMIC, suggesting a sequencing issue specific to the CCLE protocol. This demonstrates that factors other than inadequate sequencing of GC-content, such as library preparation, reagents and amplification efficiency can also affect mutation detection at certain loci.
Figure 2.
In the original 18 cell line comparison, mutations detected by COSMIC but not CCLE were categorised into: poor coverage with 5 or less reads (Panel a); good read coverage (over 20 reads) and mutation detected on reads but annotated as a dbSNP, neutral variant, outside coding region in all transcripts, or detected on less than 10% of reads, and removed (Panel b); and good coverage, no mutation (Panel c). (Panel d) reveals that the most common cause for mutations being missed by CCLE was poor read coverage (41%). Images of read coverage were taken using the Integrative Genomics Viewer.
Figure 3.
The 20 largest cold-spots detected in cancer census or kinase genes transcripts (of those that were sequenced by both COSMIC and CCLE hybrid capture) using CCLE whole exome sequencing data. All but one of these cold-spots is located in a high GC-content area and results in no mutations being detected by either institute. The TET2 cold-spot is not located in high-GC content areas and contains mutations detected by COSMIC, indicating that this cold-spot was not present in the COSMIC data. The outer shaded grey plot shows the GC content at each base (calculated as 50bp either side) with GC content over 70% shaded in red. The middle light green plot shows sequencing read coverage with white troughs representing poor read coverage. The inner 3 rings record the position of mutations found by both institutes (orange), COSMIC only (violet) and CCLE (green). Light blue shards show cold-spots over 100bp in length with the top 20 shaded darker. Data were plotted using a combination of Circos and custom scripts.
We performed whole exome sequencing on 4 of the sequenced lung cancer cell lines (H2009, H1437, H2122, H2087) using an Illumina HiSeq 2500 (achieving over 98% uniquely mapped reads) and a GATK pipeline for mutation detection (8). Our own sequencing identified 27 novel mutations in these 4 cell lines that were undocumented by COSMIC or CCLE (Supp. Table 3). Two thirds of these were located in areas of poor read coverage as defined by the CCLE hybrid capture sequencing (less than 4 reads) but reasonable coverage in our data (mean read depth = 63). The average GC-content 100 bp either side of these newly identified mutations was significantly higher than those where all three institutes were in agreement (60.85% vs. 47.13%: P = < 10−4). These findings suggest that the new mutations were previously missed due to being located in GC-rich cold-spots. Whilst the contribution of factors such as different library preparation and reagents may play a role, our data indicate that NGS efficiency of high GC-rich regions is improving, but earlier datasets are more likely to have missed mutations in GC-rich regions. The majority of The Cancer Genome Atlas and International Cancer Genome Consortium data is of a similar age to CCLE and COSMIC, and therefore subject to similar limitations. Our own more recent sequencing faired better in these regions but still had many GC-rich cold-spots in cancer associated genes. This is a significant problem, particularly in cancers including lung cancers, which have a mutational signature predominantly favouring GC-rich trinucleotides (13).
One of the novel mutations identified by our group was in PAK4 (E119Q) in H2009. This mutation lies in a GC-rich (> 76%) area of poor read coverage in CCLE (2 reads; neither reporting the mutation). By contrast, the locus was covered by 39 reads in our data, of which 51% identified the mutation (Supp. Figure 2). Given the importance of the PAK kinases in cancer proliferation and survival pathways (2,14), we further characterised this mutation. Overexpression of the PAK4 E119Q mutant in 293T cells showed enhanced activation of the ERK pathway compared to the wild type kinase, suggesting this is a gain-of-function mutation (Supp. Figure 3). These data indicate that additional cancer driver mutations in GC-rich regions will be consistently missed by next generation cancer genomic sequencing studies, and highlight the potential of developing sequencing platforms to target cold-spot regions for novel cancer gene discovery.
Difference in computational protocols represent another important cause of discrepancy, and includes differences in dbSNP filtering as well as the threshold allelic fraction required to call a mutation. We investigated the effects of dbSNP filtering by comparing the COSMIC only mutations with unfiltered data from CCLE (the equivalent COSMIC data were unavailable). Conformity increased to 67.85% although 10,091 COSMIC only mutations remained unmatched to CCLE (Supp. Figure 4). Therefore one third of mutations detected only by COSMIC were present on CCLE sequencing reads but discarded since they were thought to be germline variants. This observation recapitulated the original 18-cell line comparison and our own sequencing also confirmed this with a similar percentage of mutations unreported as a consequence of dbSNP filtering (Supp. Figure 5).
By comparing the COSMIC and CCLE data with the 4 cell lines that we sequenced, we found that 86.34% of the mutations reported by only one database were actually present in our data suggesting a minority (approximately 15-20% based on our two comparisons) of the discrepancy between cell lines is due to acquisition / loss of mutations (Supp. Table 4). Although a relatively minor factor in our comparisons, the effect of gaining a mutation in a cell line has the potential to greatly affect pharmacogenomic studies. This is highlighted by eight cell lines in the larger comparison that contained activating codon 61 NRAS mutations that were reported in only one of the databases (7 reported by COSMIC alone; 1 by CCLE alone). Analysis of the sequencing data covering the 7 NRAS mutations not detected by CCLE confirmed good read coverage (mean 220 reads) without evidence of mutation in all 7 cases, suggesting loss or gain of the mutation by cell passaging. Passage number is not generally reported in online databases but would greatly assist researchers characterising the role of specific mutations, by indicating whether a mutation has been lost or acquired during passaging.
Whilst the retrospective nature of our study is unable to control for many sequencing variables such as reagents, polymerases and platform parameters we have identified important factors for the discrepancies between the two main cancer genomics databases. These are important findings in the context of a recent study that identified inconsistencies in large pharmacogenomics studies (15). Comparing only 64 genes, this study found some acceptable discrepancies in mutational profiles of cells reported by CCLE and COSMIC but concluded that they were due to differences in the sequencing platforms and variant filtering. Our analysis of a larger panel of genes shows that there is marked discrepancy in sequencing results caused by inadequate sequencing and acquisition of new mutations in addition to variances in dbSNP calling. The authors also concluded that mutational profile was not a major cause of discrepancy in pharmacogenomics data based on the finding that mutational status was not significantly associated with drug response. However our data show that mutations of cancer causing genes in sequencing read cold-spots will be frequently undetected, and therefore greatly weaken any analysis attempting to correlate mutation status with drug response. These unsequenced regions of the exome will undoubtedly contain driver mutations, thus mapping cold-spot regions will facilitate novel therapeutic target discovery.
Supplementary Material
Acknowledgments
This research was fully supported by Cancer Research UK. We thank members of the Signalling Networks in Cancer Group and RNA Biology Groups for helpful discussions and the Core Facility for their advice and support. We thank Dr. William Newman and Dr. Ged Brady for helpful comments and suggestions
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Tsao A, et al. The BATTLE Trial: Personalizing Therapy for Lung Cancer. Cancer Discovery. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fawdar S, Trotter EW, Li Y, Stephenson NL, Hanke F, Marusiak AA, et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc Natl Acad Sci U S A. 2013;110:12426–31. doi: 10.1073/pnas.1305207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z, Hu W, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 2006;103:15404–9. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–93. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.