Abstract
Long suspected, recently recognized, and increasingly studied, non protein-coding RNAs (ncRNAs) are emerging as key drivers of biological control and pathology. Since their discovery in 1993, microRNAs (miRNAs) have been the subject of intense research focus and investigations have revealed striking findings, establishing that these molecules can exert multiples levels of biological control in numerous tissues. More recently, long ncRNAs (lncRNAs), the lesser-studied siblings of miRNA, have been suggested to have a similar robust role in developmental and adult tissue regulation. Mesenchymal stem cells (MSCs) are an important source of multipotent cells for normal and therapeutic tissue repair. Much is known about the critical role of miRNAs in biogenesis and differentiation of MSCs however; recent studies have suggested lncRNAs may play an equally important role in the regulation of these cells. Here we highlight the role of lncRNAs in the regulation of mesenchymal stem cell lineages including adipocytes, chondrocytes, myoblasts and osteoblasts. In addition, the potential for these noncoding RNAs to be used as biomarkers for disease or therapeutic targets is also discussed.
Keywords: long noncoding RNA, mesenchymal stem cells, osteogenesis, myogenesis, biomarkers, adipogenesis, chondrogenesis
Noncoding RNAs Emerge from Genomic Dark Matter to Become Regulatory Superstars
Non-protein-coding RNAs (ncRNAs), not including ribosomal and transfer RNAs (which have well-established roles in transcription), were formerly defined as “transcriptional noise” and dismissed as predominately non-functional. These vast quantities of “non-functional” RNA were dramatically described as genomic “dark matter”, an allusion to the invisible elements that substantially contribute to the mass of the universe. Like these elements, ncRNAs comprise the majority of transcriptional output in terms of complexity; 75% of the human genome is transcribed, but only 2% encodes proteins (Djebali et al. 2012). As we delve into understanding the roles and responsibilities of ncRNAs, a classification system is emerging. The two major classes are defined by their length; small ncRNAs are under 200 nt, and species 200 nt and longer are classified as long noncoding RNAs (lncRNAs). Small ncRNAs are further subdivided based on function, and cellular location; subclasses include microRNA (miRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA). LncRNAs comprise a heterogeneous mixture with a wide variety of sizes, from just over 200 nt to as long as several kb. Note that the lncRNA classification typically does not include ribosomal RNAs—these abundant, well-characterized transcripts make up their own class of ncRNA.
miRNAs Are ~21 nt ncRNAs That Suppress Translation
miRNAs have been shown to have pivotal regulatory roles in biological control during development, tissue remodeling, and physiological responsiveness; they are now considered principle mediators of gene regulation in mammalian cells. In addition, compelling evidence points to functional linkages of miRNAs with specific cells, tissues and biological processes, as well as with both germline and acquired genetic modifications. Initially identified in plants and lower eukaryotic organisms, mature miRNAs are short (~21–23 nt), single-stranded, ncRNAs. Genes encoding miRNAs may serve as template for a single miRNA, but more commonly encode clusters of miRNAs (Bartel 2004), or a protein and a miRNA in an intronic sequence. They are transcribed by RNA polymerase II and unprocessed transcripts include 5’ cap structures and poly(A) tails (Kim 2005). Mature miRNAs are created via a two-step trimming process that is completed in the cytoplasm. They associate with an Argonaute protein inside a miRNA-induced silencing complex (miRISC) to suppress translation by base-pairing to complementary sequences in the 3’ untranslated regions of target messenger RNAs (mRNAs) (Carthew and Sontheimer 2009). The emerging consensus is that, with perfect sequence homology between target mRNA and nucleotides 2–7 of the miRNA (the “seed region”), mRNA are targeted for degradation. With imperfect homology, mRNA are targeted for translational suppression (van den Berg et al. 2008). Because of this lax requirement for target-sequence homology, a single miRNA may influence the expression of hundreds of genes. The converse is also true; most genes have many potential miRNA-binding sites. Consequently, the effects of miRNA can be additive, and as biomarkers, miRNA expression profiles or signatures, rather than the expression or absence of individual miRNAs, typically provide biologically and clinically relevant information. This complexity and redundancy makes it challenging to understand precisely how the many players integrate their regulatory activities, but provides multiple possible options for miRNA-mediated intervention.
lncRNAs
LncRNA is the most recently recognized class of ncRNA, yet the most diverse. The latest annotation of the human genome (Version 20, GRCh38) identifies 24,489 lncRNA transcripts from 14,470 lncRNA genes (http://www.gencodegenes.org/stats.html). Generally, lncRNAs are expressed at lower levels than protein-coding transcripts, and their expression is more restricted in terms of tissue specificity. As researchers make use of next-generation sequencing in more cell types and tissues, the number of identified lncRNAs will likely increase. For example, a recent analysis of the transcriptome of human erythroid cells identified 100 novel unannotated lncRNA transcripts with highly erythroid-specific expression (Alvarez-Dominguez et al. 2014).
Like mRNAs, lncRNAs are primarily transcribed by RNA polymerase II; are alternatively spliced and have multiple isoforms; contain RNA processing signals, such as poly(A) tails and 5’ caps; and undergo processing to remove intronic sequence. The non-polyadenylated lncRNAs conversely, are generally transcribed by RNA polymerase III. Typically, however, lncRNAs are shorter than mRNAs and show a bias for having a single intron (Derrien et al. 2012). LncRNAs are encoded throughout mammalian genomes; and they are often categorized based on their proximity in the genome to protein-coding genes (Batista and Chang 2013) (see Box 1). Interestingly, lncRNA genes are less conserved than those for protein-coding mRNAs, however, their promoter regions and splice sites are highly conserved. This may reflect the fact that for protein interactions, secondary structure is arguably more important than primary nucleotide sequence, yet lncRNA expression at specific times and places is crucial, therefore promoter sequences are relatively highly conserved.
Mechanisms of lncRNA control
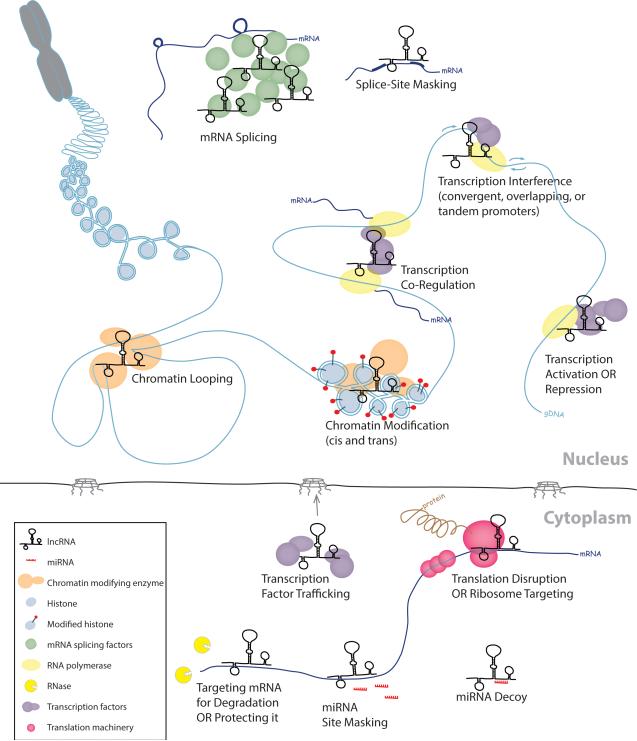
Unlike noncoding miRNAs, that are found primarily in the cytoplasm and regulate expression almost exclusively through mRNA interactions, lncRNAs are largely localized in the nucleus and chromatin, though not exclusively. They function through a surprisingly diverse set of interactions with numerous components of the gene regulatory machinery (see Figure 1). A single lncRNA may have several modular components; each interacting with different molecules (DNA, RNA and/or protein) and a single lncRNA can have multiple functions. Mechanisms include base-pairing interactions with mRNA to mask mRNA splice sites in the nucleus or miRNA binding sites in the cytoplasm (Fatica and Bozzoni 2014). LncRNAs can also base pair with miRNAs, effectively depleting them by acting as a sponge or decoy. Base pairing between lncRNA and cytoplasmic mRNA may mark the mRNA for degradation by RNases or conversely protect them; likewise with translation, base-paired lncRNAs may disrupt translation of bound mRNA or may recruit ribosomes to enhance it. LncRNAs also form associations with transcription factors to affect gene expression. Cytoplasmic lncRNAs can transport transcription factors from the cytoplasm to the nucleus. Nuclear lncRNAs can recruit specific transcription factors or complexes to genomic sites located near or far from their own genomic location for transcription activation of mRNA in cis and trans. They can also act as a scaffold that tethers transcription complexes to bridge chromosomal distances and direct co-regulated transcription. Conversely, lncRNAs can also interfere with transcription of nearby mRNA genes with convergent, overlapping, or tandem promoters, simply by being transcribed themselves. Chromatin modification mediated by lncRNA interactions may be responsible for either suppression or enhancement of target-mRNA transcription. Similarly, interactions between chromatin-binding proteins and lncRNAs can cause chromatin looping. Finally a class of lncRNAs, known as enhancer RNAs (eRNAs), activates gene expression using mechanisms that may include looping to bring transcriptional enhancers closer to promoters. Numerous reviews cover the mechanisms of lncRNAs in much greater detail than can be afforded here. Please refer to the following for more information on the role lncRNAs in the regulation of miRNAs (Rinn 2014), interaction with chromatin (Rinn 2014), nuclear structure (Bergmann and Spector 2014), histone modification (Joh et al. 2014) and transcriptional regulation (Vance and Ponting 2014).
The first well-characterized lncRNAs were found to be key players in X-chromosome inactivation (Borsani et al. 1991) and imprinting (Bartolomei et al. 1991). These initial discoveries set high expectations that these molecules were likely to have important roles in biology. The increasing number of publications on lncRNA involvement in numerous biological processes confirms that they represent an entirely new level of control. LncRNAs have been demonstrated to be essential regulators of numerous development pathways, including maintenance of stem cell pluripotency (Ghosal et al. 2013), regulation of apoptosis (Rossi and Antonangeli 2014) and lineage commitment during hematopoiesis (Paralkar et al. 2014), erythropoiesis (Alvarez-Dominguez et al. 2014) and keratinocyte differentiation (Kretz et al. 2012, Orom et al. 2010). LncRNAs are involved in numerous diseases associated with aberrant cellular control including neurological, autoimmune, cardiovascular conditions and cancer (Batista and Chang 2013). With the increasing evidence of lncRNA involvement in normal and aberrant cell differentiation, examination of the literature of the role of lncRNAs in lineage commitment of mesenchymal stem cells (MSC) is of interest.
Mesenchymal Stem Cells
Mesenchymal stem cells (MSCs) are a source of adult stem/progenitor cells; and are an abundantly researched cell model and an extremely useful medical therapeutic to regenerate or repair a wide variety of mature tissues. The field of MSC research has progressed at a staggering rate, however new discoveries, specifically the involvement of lncRNA in regulating their differentiation, suggest that new avenues of research could unlock the full potential of these cells to prevent disease or repair damaged tissue. MSCs are an abundant non-hematopoetic cell population found in bone marrow and most connective tissues. Following their initial discovery almost 40 years ago (Friedenstein et al. 1976), it was quickly established that MSCs were multipotent, with the ability to differentiate into osteoblasts, chondrocytes, adipocytes, and myoblasts. However, one vexing property of MSCs is that, even though most newly isolated cultures appear to be homogeneous—with a spindle shaped, fibroblastic appearance, the ability to self-renew and sustain a rapid rate of proliferation—they are heterogeneous in terms of differentiation potential (Mets and Verdonk 1981). This is evident when MSCs are plated at low densities and grown as in vitro cultures; they rapidly segregate into clonal populations that have distinct properties and form microenvironmental niches (Prockop 2009). Given their propensity to undergo stochastic commitment, a great deal of research has focused on controlling MSC differentiation to specific lineages by modulating cell signaling pathways or phenotypic transcription factors that regulate lineage specification. The detailed understanding of transcriptional regulators involved in the commitment of MSC to defined lineages has lead to a novel ancillary discovery, these cells can transdifferentiate or switch phenotypes, from an adipocyte to an osteoblast (Ullah et al. 2014). The relative plasticity of these mesenchymal-lineage cells is achieved by modulation of levels of phenotypic transcription factors such as C/EBP-α (adipocytes), Runx2 (osteoblasts), Sox9 (chondrocytes) and MyoD (myoblasts). Although the regulation of these factors has been extensively studied, new evidence suggests that lncRNAs may play a significant role in control of these phenotypic regulators, and they may help establish lineage specification through independent mechanisms.
ncRNA regulation of MSC-derived lineages
Numerous studies have identified, catalogued and functionally analyzed miRNAs involved in the lineage commitment of MSC to myocytes, adipocytes, chondrocytes and osteoblasts and several exemplary reviews have described the role of miRNAs in these processes: myogenesis (Crippa et al. 2012, Goljanek-Whysall et al. 2012, Luo et al. 2013, Sharma et al. 2014, Sokol 2012, Wang 2013), adipogenesis (Alexander et al. 2011, Hilton et al. 2013, Peng et al. 2014, Romao et al. 2011, Trajkovski and Lodish 2013), chondrogenesis (Dong et al. 2012, Hong and Reddi 2012, Le et al. 2013, Shang et al. 2013) and osteogenesis (Dong et al. 2012, Lian et al. 2012, Taipaleenmaki et al. 2012, van Wijnen et al. 2013), and therefore will not be reviewed here.
Over the last few years, the focus has turned to how lncRNAs may play a role in the differentiation of stem cell populations. Here we will focus on what is known to date about lncRNAs expressed in the MSC derived lineages (Table 1). Additionally, we will examine the role of lncRNAs in MSC-associated diseases, including obesity, osteoarthritis, muscular dystrophy and osteosarcoma.
TABLE 1.
LncRNAs expressed in Mesenchymal Stem Cell Derived Lineages.
| Gene ID | Gene Name | Type of IncRNA | MSC lineage(s) Associated With | Reference |
|---|---|---|---|---|
| AK038898 | Brown fat lncRNA (Blnc1)* | linc | adipogenesis | (Zhao et al. 2014) |
| DANCR | Differentiation antagonizing non-protein coding RNA | linc | osteogenesis | (Zhu et al. 2013) |
| H19 | H19, imprinted maternally expressed transcript | linc | chondrogenesis, osteosarcoma | (Steck et al. 2012, Ulaner et al. 2003, Ulaner et al. 2003) |
| HOTTIP | HOXA distal transcript transcript antisense RNA | antisense | chondrogenesis | (Kim et al. 2013) |
| LINC00901 | Long intergenic non-protein coding RNA 901 | antisense | osteosarcoma | (Pasic et al. 2010) |
| lincMD1 | Long intergenic non-protein coding RNA, muscle differentiation 1 | linc | myogenesis | (Cesana et al. 2011, Legnini et al. 2014) |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 | linc | myogenesis | (Watts et al. 2013) |
| MEG3 | Maternally expressed 3 | linc | myogenesis | (Zhou et al. 2010) |
| MSX1-as* | Msh homeobox 1 antisense | antisense | osteogenesis | (Berdal et al. 2002) |
| PU.1 AS* | PU.1 antisense* | antisense | adipogenesis | (Pang et al. 2013) |
| RP11-162L10.1* | Cartilage injury-related lncRNA (lncRNA-CIR)* | pseudogene | chondrogenesis | (Liu et al. 2014) |
| SIRT1-AS* | Sirtuin 1 antisense | antisense | myogenesis | (Wang et al. 2014) |
| slincRAD* | Super-long intergenic non-coding RNA* | linc | adipogenesis | (Yi et al. 2013) |
| SRA | Steroid receptor RNA activator | noncoding isoform | adipogenesis myogenesis | (Caretti et al. 2006, Hube et al. 2011, Xu et al. 2010) |
| TUG1 | Taurine up-regulated 1 | linc | osteosarcoma | (Zhang et al. 2013) |
| TUSC7 | tumor suppressor candidate 7 | antisense | osteosarcoma | (Pasic et al. 2010) |
| YAM1 | YY1-associated myogenesis RNA 1 | antisense | myogenesis | (Lu et al. 2013) |
Note: these are not official HUGO approved names.
Myogenesis and Muscular Dystrophy
Investigations into lncRNA expression and function during muscle development and myogenesis have provided the most insight into the proposed functions of lncRNAs in MSC-derived lineages. Several important lncRNAs have been identified in muscle that have a critical role during myogenesis. The noncoding transcript of steroid receptor activator (SRA) was one of the first lncRNA studied in myogenesis (Caretti et al. 2006). Although SRA has roles in other lineages, this lncRNA interacts with RNA helicases p68 and p72 to promote MyoD recruitment and transcriptional complex activation resulting in myogenic differentiation. Interestingly, lncRNA SRA is regulated in part by the alternatively spliced SRA protein (SRAP), and the ratio of the two is critical for myogenic differentiation (Hube et al. 2011).
Maternally expressed gene 3 (MEG3) is an imprinted gene that encodes for a lncRNA that is expressed in numerous tissues including muscle (Fleming-Waddell et al. 2009). The mouse homolog of MEG3 (also known as Gtl2) was discovered by a gene trap insertion strategy, when mice inheriting a disrupted paternal copy of Gtl2 demonstrated a dwarfism phenotype (Schuster-Gossler et al. 1998). When Gtl2 was deleted in germline cells perinatal lethality and severe skeletal muscle defects were observed, indicating an important role in embryonic development (Zhou et al. 2010). The role of MEG3/Gtl2 in muscle development is complex, as this lncRNA is associated with a large cluster of miRNAs thought to be involved in diverse biological mechanisms including Wnt signaling (Snyder et al. 2013) and angiogenesis (Gordon et al. 2010) as well as a polycomb repressor protein complex (PRC2) cofactor that directs PRC2 to target genes marked for repression (Zhao et al. 2010).
Like other mesenchymal lineages, one of the best-studied lncRNAs in muscle is H19. H19 is transcribed from the IGF2 locus and is highly expressed in the developing embryo and in adult muscle (Gabory et al. 2010, Onyango and Feinberg 2011). MyoD has been demonstrated to induce H19 expression and H19 can in turn repress Igf2 expression in trans during muscle differentiation (Borensztein et al. 2013). Expression of H19 increases with muscle differentiation and knockout of H19 decreases differentiation, a phenotype that can be rescued by the addition of exogenous H19 (Dey et al. 2014). In addition, H19 is a miR-675 host gene, and these miRNAs function by targeting the Smad transcription factors that are required for the BMP pathway. H19 also acts as a molecular sponge for the let-7 miR family, and its deletion can cause precocious muscle differentiation (Kallen et al. 2013). Let-7 miRs can target Hmga2 and Igfbp2 both of which are critical for myoblast proliferation (Li et al. 2012).
Another lncRNA that acts as a molecular sponge, or competing endogenous RNA (ceRNA) is LINCMD1 (Cesana et al. 2011). LINCMD1 expression is induced during myoblast differentiation and acts as a miR-133 and miR-135 sponge to control the expression of the transcription factors MAML1 and MEF2C, which activate muscle-specific gene expression. LINCMD1 is the host transcript for miR-133, although their biogenesis is mutually exclusive. This alternative synthesis is due to a feedforward regulatory loop with HuR (Legnini et al. 2014). HuR binds LINCMD1 and represses Drosha cleavage, which is required for miRNA processing. HuR is itself repressed by miR-133 and the sponging activity of LINCMD1 creates a feedback loop between HuR, miR-133 and LINCMD1.
The lncRNA YAM-1 affects myogenesis via transcriptional activation (Lu et al. 2013). The transcription factor YY-1 silences several genes in myoblasts and YAM-1 was identified as an YY-1 regulated lncRNA. YAM-1 expression is downregulated during myogenesis and its expression inhibits myoblast differentiation via regulation of miR-715, which in turn targets Wnt7b.
Two eRNAs upsteam of MyoD (CERNA and DRRRNA) regulate gene transcription with CERNA functioning in cis to activate MyoD and DRRRNA operating in trans to activate MyoG transcription (Mousavi et al. 2013). Both of these eRNAs are involved in the recruitment of RNA polymerase II.
Malat1 is a ubiquitously expressed lncRNA whose expression was found to increase significantly during the myogenic differentiation of C2C12 cells (Watts et al. 2013). Knockdown of Malat1 by siRNA was found to suppress myoblast proliferation by arresting cell growth at G0/G1. In addition, myostatin stimulation was found to inhibit expression of Malat1. While Malat1 maybe be involved in myogenesis, its expression does not appear to be required as Malat 1 knockout mice are viable and fertile with no visible phenotype, although closer examination of the skeletal muscle may reveal abnormalities not previously detected (Nakagawa et al. 2012). Similar to MEG3, Malat1 has also been linked to epigenetic regulation and mRNA-splicing in muscle cells (Neguembor et al. 2014).
The expression of protein coding genes and lncRNAs appears to be linked in myogenesis. Expression of the protein-coding gene Sirt1 and antisense lncRNA Sirt1-AS both decrease during myogenic differentiation of C2C12 cells and the expression of Sirt1-AS appears to regulate Sirt1 expression (Wang et al. 2014). Overexpression of Sirt1-AS lncRNA was shown to increase the levels of Sirt1 protein and counteract downregulation of Sirt1 by miR-34a.
Duchenne muscular dystrophy is characterized by severe muscle wasting beginning in early childhood. Duchenne muscular dystrophy is caused by mutations in the protein coding dystrophin gene (DMD), however not as much is known about the regulation of dystrophin and interest has turned to determining if lncRNAs play a role. Using tiling arrays, 14 intronic lncRNAs were identified in both sense and antisense orientation to the DMD gene (Bovolenta et al. 2012). Additionally, a decrease in LINCMD1 expression has been associated with muscular dystrophy and myoblasts from these patients have a reduced ability to undergo terminal differentiation (Cesana et al. 2011). In addition, ectopic expression of LINCMD1 was found to rescue myogenic potential of these cells and reinforces LINCMD1 as a relevant lncRNA to myogenesis.
Adipogenesis
The prevalence of obesity has led to a surge of interest in understanding the detailed mechanisms underlying adipocyte development and transdifferentiation between other mesenchyme-derived lineages. Understanding the role lncRNAs may offer future therapeutic targets to combat obesity.
One of the first studies examining lncRNA function in adipogenesis identified a noncoding transcript of the SRA1 protein, that that associates with PPARγ, the master transcriptional regulator of adipogenesis. This transcript was found to be essential for preadipocyte differentiation (Xu et al. 2010).
In a study examining the transcriptome of mouse fat tissue, 175 poly-A tailed lncRNAs were identified as being expressed in primary and in vitro-cultured brown and white mature mouse adipocytes (Sun et al. 2013). Many of these adipose-enriched genes were bound by key transcription factors involved in adipogenesis (PPARγ and CEBPα) at promoter regions. Several of these lncRNAs were strongly induced during differentiation and siRNA-mediated silencing experiments demonstrated 10 specific lncRNAs could independently impair differentiation from preadipocytes. In the pre-adipocyte 3T3-L1 cell line, the non-polyadenylated lncRNA transcriptome was examined during adipocyte differentiation (Yi et al. 2013). Similar to the polyA-tailed transcriptome in primary cells (Sun et al 2010), several lncRNAs appear to show differentiation stage-specific differential expression. In transition between pre-adipocyte to adipocytes, 179 and 61 transcriptionally active regions were up- and downregulated respectively and 698 and 106 regions up- and downregulated (respectively) in adipocytes to compared to mature cells. In addition, studies have examined the relative expression of lncRNA between brown adipose tissue and skeletal muscle. Zhang and colleagues identified 704 up-regulated and 896 down-regulated lncRNAs between these two tissues (Zhang et al. 2014a).
Although the majority of lncRNA studies to date have focused on solely on differential expression, there is some information on the mechanism by which these molecules may regulate molecular processes. A recent study has suggest that the expression of a lncRNA antisense transcript of the PU.1 gene (PU.1-as) promotes adipogenesis by preventing PU.1 mRNA translation by forming a mRNA/AS lncRNA duplex (Pang et al. 2013). This is important as PU.1 is a transcription factor required for normal hematopoiesis and its overexpression has been show to inhibit differentiation of 3T3-L1 preadipocytes (Wang and Tong 2008).
Results from theses studies would strongly suggest that lncRNAs may play a role in differentiation and lineage commitment of progenitor cells and that lncRNAs are involved in the regulation of adipogenesis.
Chondrogenesis and Osteoarthritis
The numerous types of cartilage in the body would suggest that chondrogenesis is a complex process with numerous regulators. Calcified cartilage is very distinct from joint-forming articular cartilage however in many cases they originate from similar cell types. Very little is known is known about lncRNA expression during chondrogenesis; however, there has been some investigation into the expression of lncRNA in osteoarthitic articular cartilage. H19 is one of the most abundant and conserved non-coding transcripts in mammalian development and is downstream of the Igf2 gene (Nordin et al. 2014). Sequences within H19 are the source of miR-675 and H19 has been shown to be involved in the sequestration of let-7 family miRNAs (Keniry et al. 2012). Stuhlmuller and colleagues initially identified H19 in synovial fluid of rheumatoid arthritis patients (Stuhlmuller et al. 2003). Subsequently, H19 was found to be significantly elevated in osteoarthritic cartilage (Steck et al. 2012). Several well-known lncRNAs (PTENP1, HOTAIR, TUG1, HOTTIP, GAS5) were found to have increased expression in osteoarthritic chondrocytes compared to normal tissue, while others were downregulated (SNHG4, Emx2os, DISC2) when lncRNA profiles were investigated by qPCR array (Kim et al. 2013). Interestingly in this study, it was noted that HOTTIP expression was changed 10-fold between in the cartilage of arthritic patients. Concordantly, the levels of the HOTTIP-target gene, Hoxa13 were decreased in osteoarthritic chondrocytes, suggesting a role for this regulatory loop in osteoarthritis.
In other comprehensive studies using microarray analysis, Liu and others found 152 lncRNAs to be differentially expressed (> 8-fold) between normal and osteoarthritic cartilage (Liu et al. 2014a). In this study the authors found that lncRNA-CIR was upregulated 22-fold in osteoarthritic cartilage and IL-1 and TNF-α, two critical mediators of inflammation leading to osteoarthritis, could stimulate the expression of lncRNA-CIR. LncRNA-CIR is a vimentin pseudogene and its knockdown has been demonstrated to increase expression of cartilage associated genes (collagens I/II and aggrecan) while its overexpression causes an increase in several arthritis-associated genes including MMP13 and ADAMTS5. Although there has been significant progress in lncRNA as markers of arthritic articular cartilage, there has not, as-of-yet been a characterization of lncRNAs in calcified or bone-related cartilages. As several of the mechanisms identified in articular cartilage have correlates in calcified tissues, a full lncRNA transcriptome would greatly improve our understanding of cartilage formation and homeostasis. In addition the analysis of the role of lncRNA in cartilage progenitors would help to identify molecular differences in the formation of articular and calcified cartilage as well as the potential role in chronic or acute disease states. This information would be beneficial for models of tissue regeneration and therapeutic intervention during trauma as well as genetic conditions.
Osteogenesis and Osteosarcoma
Prior to the popularization of high-throughput sequencing technologies, several groups had identified a noncoding transcript that was an antisense counterpart to the Msx1 gene. Given the important role of Msx1 in dental and craniofacial development, Msx1-as was shown to be and important osteogenic regulator, negatively affecting Msx1 transcript levels resulting in dose-dependent reduction of Msx1 protein levels (Babajko et al. 2009, Berdal et al. 2002). Moreover, the cellular location of Msx1 protein is modulated by Msx1-as and Msx1-as can abolish the inhibition of Dlx5 by Msx1.
With the flood of interest in lncRNAs over the past several years, a few high throughput studies have examined lncRNA profiles during osteogenesis. A small study using a lncRNA array identified 116 lncRNAs differentially expressed in C3H10T1/2 MSCs undergoing BMP-2 induced early osteogenic differentiation (Zuo et al. 2013). Examination of neighboring genes found 24 lncRNAs had the same expression pattern (up- or down-regulated) as their neighboring gene suggesting cooperative expression and regulation of osteogenesis, but little else is known about the function or regulation of these lncRNAs.
Examination of lncRNA-ANCR (anti-differentiation ncRNA; DANCR) levels in differentiating hFOB1.19 cells demonstrated that during osteoblast differentiation, expression of lncRNA-ANCR was decreased (Zhu and Xu 2013). In addition, the exogenous overexpression of ANCR reduced osteogenic differentiation, through a mechanism suggested to involve EZH2 and Runx2. ANCR was previously identified as critical in the maintenance of undifferentiated cell state within the epidermis (Kretz et al. 2012) and may be functioning in MSCs and/or osteoblasts in a similar manner, as a molecular switch regulating cell commitment.
Although currently no other lncRNAs have been identified in osteoblastic cells, it is conceivable that lncRNAs identified in similar cellular or developmental contexts may have a role in osteogenesis. Numerous lncRNAs have been found to be associated with EZH2 (Benetatos et al. 2013, Guil et al. 2012). EZH2 is a member of the polycomb-repressor complex (PRC2), which functions by methylating histone residues (specifically H3K27) resulting in gene repression (Tan et al. 2014). EZH2 has been shown to be involved in the regulation of mesenchymal lineage specification and epigenetic switching between adipo- and osteogenesis (Hemming et al. 2014). This would suggest that lncRNA involved in EZH2-related mechanisms would conceivably be involved in the commitment of MSCs to the osteogenic lineage. Similarly, the Wnt signaling pathway, which is critical to osteogenesis and MSC differentiation, has been linked to numerous lncRNAs. lncRNA-LALR1 accelerated mouse hepatocyte proliferation and cell cycle progression through activation of Wnt/β-catenin signaling (Xu et al. 2013). Activated β-catenin has been shown to repress the expression of the lncRNA HOTAIR (Zhang et al. 2014b). Interestingly, in a model of aortic valve calcification, knockdown of HOTAIR in human aortic interstitial cells increased expression of the bone-related genes alkaline phosphatase (ALPL) and bone morphogenic protein 2 (BMP2) (Carrion et al. 2014). Members of the let7 miR family have been shown to be a positive regulator of bone formation (Wei et al. 2014) As mentioned above, the lncRNA H19 can act as a molecular sponge to regulate the amount of unbound let7 making H19, suggesting that this gene may play a significant role during osteogenesis.
Osteosarcoma is an aggressive cancer that arises from mesenchymal cells that exhibit osteoblast-like differentiation. Frequently, osteosarcomas are associated with copy number alterations in tumor suppressors or oncogenes giving rise to malignancy. Two noncoding RNAs, tumor suppressor candidate 7 (TUSC7; LOC285194) and LINC00901 (BC040587), along with the tumor suppressor limbic system-associated membrane protein (LSAMP) are in the most frequently altered region associated with osteosarcoma (Pasic et al. 2010). In normal osteoblasts, knockdown of TUSC7 expression by siRNA was found to promote proliferation through regulation of apoptotic and cell-cycle-related genes. TUSC7 has since been identified as a p53-regulated tumor suppressor and its repression of cell growth is related to the repression of miR-211 (Liu et al. 2013). Interestingly, miR-211 is an endogenous negative regulator of Runx2 and inhibits osteogenesis to promotes adipogenesis of MSCs (Huang et al. 2010).
One of the best-characterized lncRNAs that is associated with osteosarcoma is H19. H19 and the adjacent protein-coding gene IGF2 are imprinted in most normal tissues, and this imprinting is often lost in tumors (Ulaner et al. 2003a, Ulaner et al. 2003b). This loss of imprinting is associated with differential methylation of a CTCF-binding site upstream of H19. H19 knockout mice have an overgrowth phenotype that is associated with its control over IGF2 expression (Ripoche et al. 1997). TUG1 is another lncRNA that is highly expressed in osteosarcoma (Zhang et al. 2013). Knockdown of TUG1 expression by siRNA significantly impairs cell proliferation and promoted apoptosis and is an ideal target for therapeutic intervention. Over 25,000 additional lncRNAs were identified by microarray to be associated with osteosarcoma, with over 1200 of these differentially regulated compared to noncancerous tissue (Li et al. 2013). It would be of great interest to determine the expression and function of these osteosarcoma-associated lncRNAs in normal osteogenesis.
Potential for circulating lncRNA biomarkers
Interestingly, the turnover rate of miRNAs has been shown to be highly variable. Most miRNAs persist for long periods of time. This longevity, and the widespread presence of circulating miRNAs in blood, is being exploited to develop miRNA biomarkers for conditions such as cancer, diabetes, and cardiovascular conditions (Bronze-da-Rocha 2014, Farr et al. 2013, Hayes et al. 2014). There are also a few examples, however, of miRNAs that respond rapidly to physiological conditions. Both long- and short-lived miRNAs may function partly as signaling molecules, communicating with protein and or nucleic acid partners located within the same cells cell or in distant cells, respectively. Those that exhibit a very rapid response to external stimuli may represent a new way for cells to respond to changes in their environment.
Recent findings of circulating lncRNAs suggest that they also have potential as disease biomarkers. miRNA are released into bodily fluids, either as part of a protein complex or encased in extracellular vesicles, such as apoptotic bodies, ectosomes and exosomes (Grasedieck et al. 2013). These extracellular vesicles can also transport relatively long nucleic acids such as mRNA, and pre- and pri-miRNAs, so it is logical to propose that they may be involved in long distance transport of lncRNAs. Indeed, there are a handful of published reports documenting circulating lncRNAs in the context of disease, including major depressive disorder and gastric cancer (Arita et al. 2013, Liu et al. 2014b) and PCA3 is a lncRNA that is currently being used as a biomarker for prostate cancer. The concept of “liquid biopsies” to query bodily fluid for indicators of disease is very appealing. Because lncRNAs, like miRNAs, often help control transcription, they have enormous potential as very early biomarkers of disease. miRNAs are typically most useful as biomarkers in combinations of two or more individual species. It will be interesting to evaluate whether lncRNAs, either alone or as another component of a miRNA biomarker signature, will offer predictive or prognostic value.
LncRNAs as therapeutic targets
Recently is has become evident that only 7% of disease-associated single nucleotide polymorphisms (SNPs) are located in protein-coding regions, with the remaining 93% in noncoding regions (Hindorff et al. 2009), making both miRNA and lncRNAs ideal therapeutic targets. To date, no clinical trials are underway targeting lncRNAs (http://clinicaltrials.gov/), however, with the recent advances in biological therapeutics, targeting RNA, and specifically lncRNAs, is a possibility. There are numerous items to consider when targeting lncRNAs. The first is cellular location. The majority of lncRNAs are located in the nucleus and efficient delivery to the nucleus will be a requirement in many instances. Also like proteins, lncRNAs have higher order structure with secondary structure showing hairpins, duplexes, internal loops and junctions. LncRNAs also interact with various other molecules including proteins, small regulatory RNA elements, RNA and DNA. Disruption of these interactions may be an ideal target in certain circumstances (Li and Chen 2013).
To date, most research is focused upon targeting lncRNAs to reduce the intracellular transcript levels, or to attenuate activity and function. Prospective strategies for targeting lncRNAs include: small interfering RNA (siRNA), antisense oligonucleotides (ASO), ribozymes, aptamers, small molecules, post-transcriptional processing pathways and miRNAs that target lncRNAs (Li and Chen 2013). Research in these areas with lncRNAs is still in its infancy and the use of several of these strategies in combination may prove to be the most effective for targeting lncRNAs. As proof-of-concept, it has been demonstrated that ASO are able to knockdown the nuclear lncRNA MALAT1 in muscle for a prolonged period of time (Wheeler et al. 2012).
In instances where expression of a lncRNA is desired, overexpression or delivery of synthetic lncRNA may be an ideal solution. The delivery of proteins as biological therapeutics has been in use for a long time, however, delivery of large RNA molecules is not commonplace. Numerous studies are currently underway investigating the delivery and use of mRNA as biological therapies. The lessons learned from those studies, including targeted organ and subcellular delivery via nanoparticles as well research into how to increase RNA stability, will aid our understanding of using lncRNAs as therapeutics in a similar manner in the future.
Another manner in which therapeutics based on lncRNAs may be used, is by modifying their expression in stem cell populations. One very interesting study performed a knockdown of H19 in embryonic stem cells (ESCs) and demonstrated the possibility of using modified stem cells as a cell-based therapy for conditions such as muscular atrophy (Kwak et al. 2012).
Could lncRNAs Bridge Nuclear Structure and Function?
We have only just begun to understand the functional roles lncRNAs play in normal development and disease. As an understanding of the fundamental properties of lncRNAs becomes more extensively established and their utility as biomarkers is evaluated, it will be important to evaluate how this new class of regulatory molecules fits into the larger framework of gene expression control. We know from work completed so far that both miRNA and lncRNA are involved in epigenetic control of expression. miRNAs, in addition to negative regulation of translation, have also been shown to be responsible for promoting epigenetic silencing of specific genes via DNA methylation or histone modifications (Dai et al. 2014). Conversely, epigenetic silencing of miRNA genes has also been amply demonstrated.
LncRNAs effect epigenetic modifications in both cis and trans using combinations of modular functional components that include RNA-, DNA-, and protein-binding domains. They also defy these linear descriptions of position by inducing chromosomal looping to affect multiple genomic loci at once, creating a scaffold for chromatin-modifying enzymes. These properties make it interesting to speculate that lncRNAs may be a component of a mechanism that integrates structure, function, organization, and activity within the cell. The information amassed so far provides details about how individual lncRNAs interact with associated proteins, genomic DNA, and other RNA molecules. The diverse nature and impact of these reactions suggest that lncRNAs may be candidates for an overarching role in integrating regulatory activities, possibly mediating the organization of regulatory machinery, with the potential to have both dynamic and persistent effects.
Summary
It is evident from the relatively small amount of evidence generated that lncRNAs have a significant and crucial role in the maintenance, commitment and differentiation of MSCs to mature lineages. Understanding the mechanism of lncRNA action in differentiation is important not only to normal tissue development but also to disease states. The use of high throughput and next-generation sequencing technologies has allowed not only for the novel identification of lncRNA but provides a rapid screen to monitor tissue-specific changes in these transcripts. The unique and complex molecular mechanisms associated with lncRNA function will require careful analysis and characterization of in vitro and in vivo and their cellular interacting partners, be it DNA, RNA or protein. This level of analysis is daunting however, in order to fully understand how lncRNAs can be translated into novel therapies that either target these molecules or take advantage of their function, there needs to be detailed understanding of their structure, their regulation in normal and pathological conditions.
Box 1. lncRNA Categories.
Sense: lncRNA gene overlaps with a protein-coding gene sense strand and is often considered transcript variants of the coding gene
Antisense: overlaps with antisense strand of protein-coding gene
Bidirectional: non-overlapping on the opposite strand of a protein-coding gene, but within 1000 bases of the transcription start
Intronic: overlaps an intron of a protein-coding gene on the sense or antisense strand
Intergenic: Located between protein-coding genes, at least 1 kb from the nearest
Acknowledgements
This work was supported by National Institutes of Health (R01 AR039588 to G.S.S. and J.B.L., P01AR048818 and R37 DE012528 to J.B.L.).
REFERENCES
- Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin Ther Targets. 2011;15:623–636. doi: 10.1517/14728222.2011.561317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- Babajko S, Petit S, Fernandes I, Meary F, LeBihan J, Pibouin L, Berdal A. Msx1 expression regulation by its own antisense RNA: consequence on tooth development and bone regeneration. Cells Tissues Organs. 2009;189:115–121. doi: 10.1159/000151748. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E. Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer. 2013;133:267–274. doi: 10.1002/ijc.27859. [DOI] [PubMed] [Google Scholar]
- Berdal A, Lezot F, Pibouin L, Hotton D, Ghoul-Mazgar S, Teillaud C, Robert B, MacDougall M, Blin C. Msx1 homeogene antisense mRNA in mouse dental and bone cells. Connect Tissue Res. 2002;43:148–152. doi: 10.1080/03008200290000970. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M, Monnier P, Court F, Louault Y, Ripoche MA, Tiret L, Yao Z, Tapscott SJ, Forne T, Montarras D, Dandolo L. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development. 2013;140:1231–1239. doi: 10.1242/dev.084665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Bovolenta M, Erriquez D, Valli E, Brioschi S, Scotton C, Neri M, Falzarano MS, Gherardi S, Fabris M, Rimessi P, Gualandi F, Perini G, Ferlini A. The DMD locus harbours multiple long non-coding RNAs which orchestrate and control transcription of muscle dystrophin mRNA isoforms. PLoS One. 2012;7:e45328. doi: 10.1371/journal.pone.0045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. Biomed Res Int. 2014;2014:985408. doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Carrion K, Dyo J, Patel V, Sasik R, Mohamed SA, Hardiman G, Nigam V. The long non-coding HOTAIR is modulated by cyclic stretch and WNT/beta-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PLoS One. 2014;9:e96577. doi: 10.1371/journal.pone.0096577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa S, Cassano M, Sampaolesi M. Role of miRNAs in muscle stem cell biology: proliferation, differentiation and death. Curr Pharm Des. 2012;18:1718–1729. doi: 10.2174/138161212799859620. [DOI] [PubMed] [Google Scholar]
- Dai E, Yu X, Zhang Y, Meng F, Wang S, Liu X, Liu D, Wang J, Li X, Jiang W. EpimiR: a database of curated mutual regulation between miRNAs and epigenetic modifications. Database (Oxford) 2014;2014:bau023. doi: 10.1093/database/bau023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418:587–591. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- Farr RJ, Joglekar MV, Taylor CJ, Hardikar AA. Circulating non-coding RNAs as biomarkers of beta cell death in diabetes. Pediatr Endocrinol Rev. 2013;11:14–20. [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fleming-Waddell JN, Olbricht GR, Taxis TM, White JD, Vuocolo T, Craig BA, Tellam RL, Neary MK, Cockett NE, Bidwell CA. Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PLoS One. 2009;4:e7399. doi: 10.1371/journal.pone.0007399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedenstein AJ, Gorskaja JF, Kulagina NN. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol. 1976;4:267–274. [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goljanek-Whysall K, Sweetman D, Munsterberg AE. microRNAs in skeletal muscle differentiation and disease. Clin Sci (Lond) 2012;123:611–625. doi: 10.1042/CS20110634. [DOI] [PubMed] [Google Scholar]
- Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasedieck S, Sorrentino A, Langer C, Buske C, Dohner H, Mertens D, Kuchenbauer F. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121:4977–4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, Gronthos S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32:802–815. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- Hilton C, Neville MJ, Karpe F. MicroRNAs in adipose tissue: their role in adipogenesis and obesity. Int J Obes (Lond) 2013;37:325–332. doi: 10.1038/ijo.2012.59. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012;18:445–453. doi: 10.1089/ten.TEB.2012.0116. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh RI, Palmieri CM, Hill IT, Motamedi M. Regulation of histone methylation by noncoding RNAs. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song J, Han J, Kim Y, Chun CH, Jin EJ. Two non-coding RNAs, MicroRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-alpha1. Cell Signal. 2013;25:2878–2887. doi: 10.1016/j.cellsig.2013.08.034. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z, Khavari PA. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M, Hong S, Yu SL, Sim BW, Seo JS, Kang J. Parthenogenetic embryonic stem cells with H19 siRNA-mediated knockdown as a potential resource for cell therapy. Int J Mol Med. 2012;29:257–262. doi: 10.3892/ijmm.2011.838. [DOI] [PubMed] [Google Scholar]
- Le LT, Swingler TE, Clark IM. Review: the role of microRNAs in osteoarthritis and chondrogenesis. Arthritis Rheum. 2013;65:1963–1974. doi: 10.1002/art.37990. [DOI] [PubMed] [Google Scholar]
- Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200–206. doi: 10.1016/j.bbrc.2013.02.083. [DOI] [PubMed] [Google Scholar]
- Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, Shavlakadze T, Eash JK, Scaramozza A, Goddeeris MM, Kirsch DG, Campbell KP, Brack AS, Glass DJ. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23:1176–1188. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, Zhou C, Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014a;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, Wang Y, Zhang K. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One. 2014b;9:e93388. doi: 10.1371/journal.pone.0093388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Sun K, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013;32:2575–2588. doi: 10.1038/emboj.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics. 2013;40:107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Mets T, Verdonk G. In vitro aging of human bone marrow derived stromal cells. Mech Ageing Dev. 1981;16:81–89. doi: 10.1016/0047-6374(81)90035-x. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet Muscle. 2014;4:8. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin M, Bergman D, Halje M, Engstrom W, Ward A. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014;47:189–199. doi: 10.1111/cpr.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci U S A. 2011;108:16759–16764. doi: 10.1073/pnas.1110904108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:325–331. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang WJ, Lin LG, Xiong Y, Wei N, Wang Y, Shen QW, Yang GS. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J Cell Biochem. 2013;114:2500–2512. doi: 10.1002/jcb.24595. [DOI] [PubMed] [Google Scholar]
- Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, Dunagin M, Pimkin M, Gore M, Sun D, Konuthula N, Raj A, An X, Mohandas N, Bodine DM, Hardison RC, Weiss MJ. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yu S, Li H, Xiang H, Peng J, Jiang S. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell Signal. 2014;26:1888–1896. doi: 10.1016/j.cellsig.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Prockop DJ. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther. 2009;17:939–946. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL. lncRNAs: Linking RNA to Chromatin. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med (Maywood) 2011;236:997–1004. doi: 10.1258/ebm.2011.011101. [DOI] [PubMed] [Google Scholar]
- Rossi MN, Antonangeli F. LncRNAs: New Players in Apoptosis Control. Int J Cell Biol. 2014;2014:473857. doi: 10.1155/2014/473857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Shang J, Liu H, Zhou Y. Roles of microRNAs in prenatal chondrogenesis, postnatal chondrogenesis and cartilage-related diseases. J Cell Mol Med. 2013;17:1515–1524. doi: 10.1111/jcmm.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Juvvuna PK, Kukreti H, McFarlane C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front Physiol. 2014;5:239. doi: 10.3389/fphys.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140:31–42. doi: 10.1242/dev.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS. The role of microRNAs in muscle development. Curr Top Dev Biol. 2012;99:59–78. doi: 10.1016/B978-0-12-387038-4.00003-3. [DOI] [PubMed] [Google Scholar]
- Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90:1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- Stuhlmuller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, Ulbrich N, Erdmann VA, Burmester GR, Kinne RW. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol. 2003;163:901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmaki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–371. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- Tan JZ, Yan Y, Wang XX, Jiang Y, Xu HE. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol Sin. 2014;35:161–174. doi: 10.1038/aps.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, Gorlick R, Meyers P, Healey J, Ladanyi M, Hoffman AR. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet. 2003a;12:535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- Ulaner GA, Yang Y, Hu JF, Li T, Vu TH, Hoffman AR. CTCF binding at the insulin-like growth factor-II (IGF2)/H19 imprinting control region is insufficient to regulate IGF2/H19 expression in human tissues. Endocrinology. 2003b;144:4420–4426. doi: 10.1210/en.2003-0681. [DOI] [PubMed] [Google Scholar]
- Ullah M, Sittinger M, Ringe J. Transdifferentiation of adipogenically differentiated cells into osteogenically or chondrogenically differentiated cells: phenotype switching via dedifferentiation. Int J Biochem Cell Biol. 2014;46:124–137. doi: 10.1016/j.biocel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta. 2008;1779:668–677. doi: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ, Taipaleenmaki H, Hesse E, Riester S, Kakar S. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Tong Q. Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol. 2008;295:C213–220. doi: 10.1152/ajpcell.00422.2007. [DOI] [PubMed] [Google Scholar]
- Wang XH. MicroRNA in myogenesis and muscle atrophy. Curr Opin Clin Nutr Metab Care. 2013;16:258–266. doi: 10.1097/MCO.0b013e32835f81b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pang WJ, Wei N, Xiong Y, Wu WJ, Zhao CZ, Shen QW, Yang GS. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. 2014;539:117–124. doi: 10.1016/j.gene.2014.01.037. [DOI] [PubMed] [Google Scholar]
- Watts R, Johnsen VL, Shearer J, Hittel DS. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013;304:C995–1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- Wei J, Li H, Wang S, Li T, Fan J, Liang X, Li J, Han Q, Zhu L, Fan L, Zhao RC. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014;23:1452–1463. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, Chen XW, Cawthorn WP, MacDougald OA, Koenig RJ. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- Yi F, Yang F, Liu X, Chen H, Ji T, Jiang L, Wang X, Yang Z, Zhang LH, Ding X, Liang Z, Du Q. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol. 2013;10:991–1001. doi: 10.4161/rna.24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cui X, Shen Y, Pang L, Zhang A, Fu Z, Chen J, Guo X, Gan W, Ji C. Distinct expression profiles of LncRNAs between brown adipose tissue and skeletal muscle. Biochem Biophys Res Commun. 2014a;443:1028–1034. doi: 10.1016/j.bbrc.2013.12.092. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014b;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, Rice KA, Zhang L, Zhang X, Gordon FE, Lidov HG, Bronson RT, Klibanski A. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]