Abstract
Reprogramming of epigenetic states in gametes and embryos is essential for correct development in plants and mammals1. In plants, the germ line arises from somatic tissues of the flower necessitating erasure of chromatin modifications accumulated at specific loci during development or in response to external stimuli. If this occurs inefficiently it can lead to epigenetic states being inherited from one generation to the next2-4. However, in most cases accumulated epigenetic modifications are efficiently erased before the next generation. An important example of epigenetic reprogramming in plants is the resetting of expression of the Arabidopsis thaliana floral repressor FLC locus. FLC is epigenetically silenced by prolonged cold in a process called vernalization. However, the locus is reactivated prior to completion of seed development to ensure a vernalization requirement every generation. In contrast to our detailed understanding of the Polycomb-mediated epigenetic silencing induced by vernalization, little is known about the mechanism involved in the re-activation of FLC. Here we show that a hypomorphic mutation in the jumonji domain protein ELF6 impaired the reactivation of FLC in reproductive tissues, leading to inheritance of a partially vernalized state. ELF6 has H3K27me3 demethylase activity and the mutation reduced this enzymatic activity in planta. Consistent with this, H3K27me3 levels at the FLC locus stayed higher and FLC expression remained lower, than in the wild type in the following generation. Our data reveal an ancient role for H3K27 demethylation in the reprogramming of epigenetic states in plant and mammalian embryos5-7.
Many Arabidopsis thaliana accessions overwinter before flowering through FRIGIDA-mediated high-level expression of a floral repressor called FLC8,9. Prolonged cold during the weeks of winter antagonizes this activation and progressively epigenetically silences FLC. This enables other floral promotion signals such as day length to induce flowering in spring. The epigenetic silencing of FLC involves Polycomb-mediated chromatin regulation10-12 and is maintained until embryogenesis, when FLC expression is reset to ensure a vernalization requirement every generation13,14. Resetting of FLC expression occurs in the early globular embryo13,14; then, FLC expression increases throughout embryo development until it reaches maximum levels when the seed is completely formed14. However, the molecular mechanisms underlying FLC resetting are unknown, and the testing of several factors known to be required for the up-regulation of FLC in vegetative tissues showed they were dispensable for FLC expression in the embryo14. One exception is the yeast SWR1 homolog PIE114, although it is not clear if this a resetting-specific defect because pie1 mutations strongly reduce FLC expression across the plant independently of vernalization status.
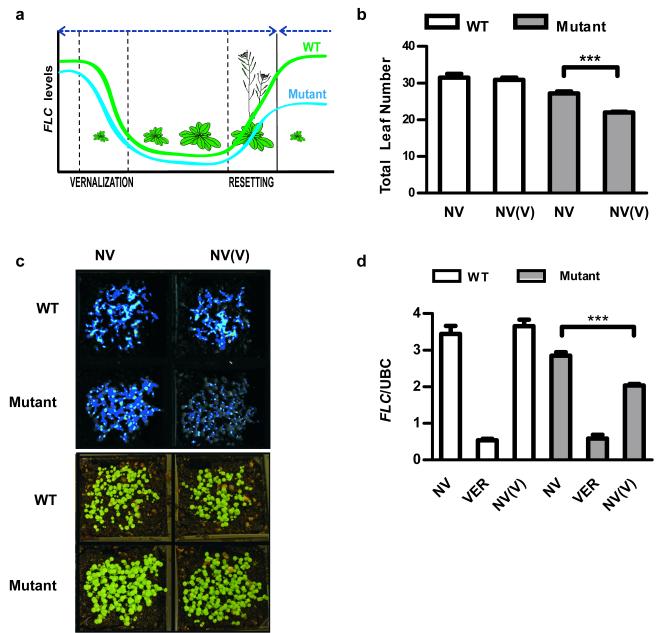
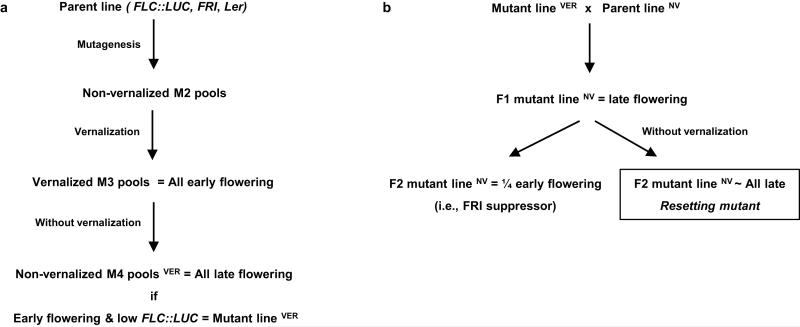
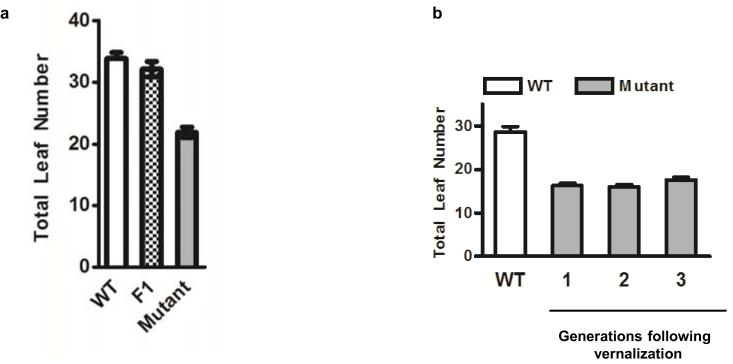
In order to dissect this resetting mechanism we isolated mutants defective in the reactivation of FLC after vernalization (Extended Data Fig. 1a). The parental line was an Arabidopsis Landsberg erecta (Ler) plant carrying an FLC::luciferase (FLC::LUC) translational fusion and an active FRIGIDA (FRI) transgene15. We searched for plants where FLC expression pre-vernalization was silenced by vernalization but unlike wild type was not fully restored in the following generation (Fig. 1a), leading to inheritance of the vernalized state. The frequency of these mutations was low (only 2 mutants identified from progeny of 6,000 mutagenized parent lines), contrasting with the more common class of mutations, which were early flowering before vernalization due to reduced FLC expression (Extended Data Fig. 1b). The first resetting mutant isolated was found to be recessive (Extended Data Fig. 2a) and was slightly earlier flowering without vernalization than the wild type (Fig. 1b). In the generation following vernalization the mutant was even earlier flowering and had significantly reduced FLC expression (Fig. 1c,d), albeit ~four fold higher than fully vernalized seedlings (Fig. 1d). The resetting mutant therefore causes transgenerational inheritance of a partially vernalized state. The early flowering phenotype was stable for at least 3 generations following vernalization (Extended Data Fig. 2b) but was not enhanced by a second vernalization treatment in the later generations. There were no other strong developmental phenotypes.
Figure 1. Isolation and characterization of the resetting mutant.
a, Logic of the genetic screen. Parental wild-type is Ler-FRI FLC::luciferase. b-d, The resetting mutant is early flowering (b) with fewer total leaves and maintains low FLC expression shown by FLC luciferase imaging of 8 day-old seedlings (c) or Q-RT-PCR analysis normalised to UBC (d). NV is non-vernalized, VER is vernalized, NV(V) is non-vernalized following vernalization in the previous generation. Means + s.e.m., n = 20 (b) and n = 3 (d), ***P < 0.001.
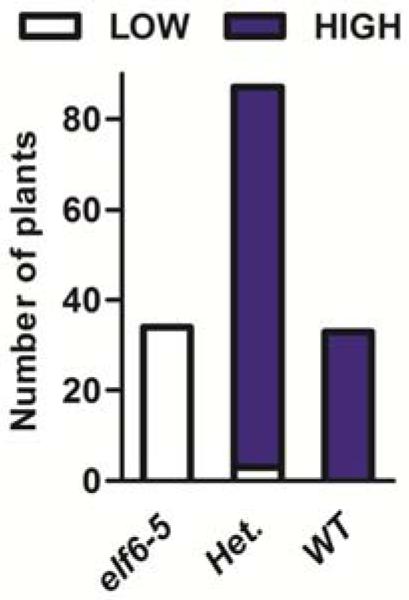
The mutant phenotype was strongly affected by segregation of modifiers in a traditional Ler X Columbia (Col) cross, normally used for genetic mapping, and the mutation could only be narrowed down to a ~500 kb region on chromosome 5. We therefore sequenced the whole genome of the mutant plant and analysed linkage of candidate SNPs in an F2 population generated from a cross between the mutant and the isogenic progenitor line. This strategy identified a single nucleotide polymorphism in ELF6 (At5g04240) that co-segregated with the resetting phenotype (Extended Data Fig. 3). To confirm that the resetting phenotype was indeed caused by the elf6-5 mutant allele, we complemented the mutation using ELF6 gene under its own regulatory sequences. Vernalized T2 transgenic lines carrying the wild-type ELF6 transgene showed wild-type FLC expression levels in the siliques (Fig. 2a,b). Thus, we concluded that the single nucleotide mutation in ELF6 caused the mutant phenotype.
Figure 2. Mapping of the resetting mutant.
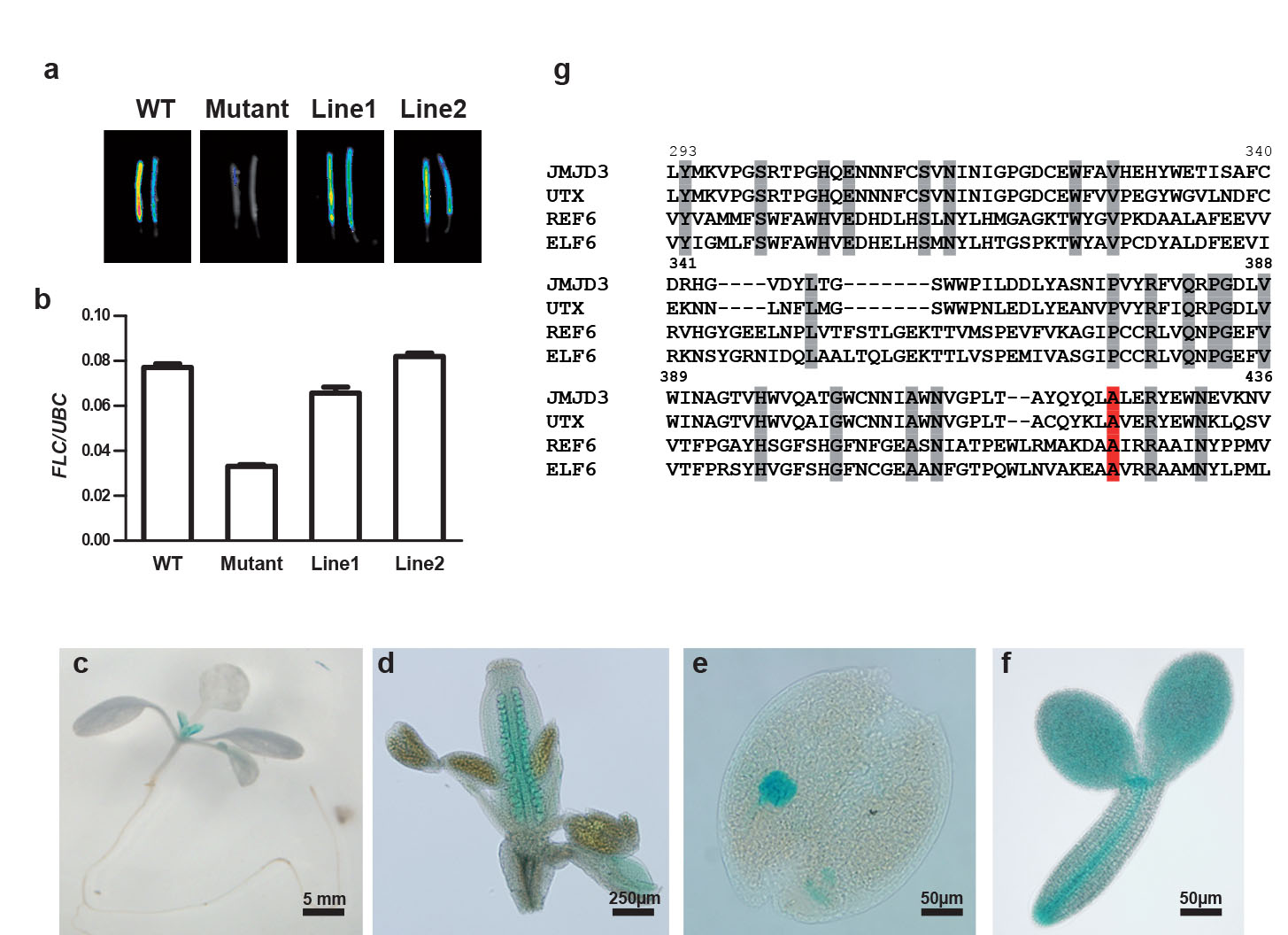
a, b ELF6 genomic construct complements the resetting mutant. FLC luciferase imaging (a) and FLC Q-RT-PCR data (b) of mature siliques from vernalized WT, elf6-5 and representative T2 elf6-5 [pELF6::ELF6] lines. c-f, ELF6::GUS expression profile in 7 day-old seedling (c), ovules (d), globular embryo (e) and mature embryo (f). g, The ELF6 residue mutated in elf6-5 is conserved (red A). Sequence alignment of the JmJC domain of Arabidopsis ELF6 and REF6, and human JMJD3 and UTX proteins. Highly conserved residues are shadowed in grey. Numbering refers to ELF6 amino acid position.
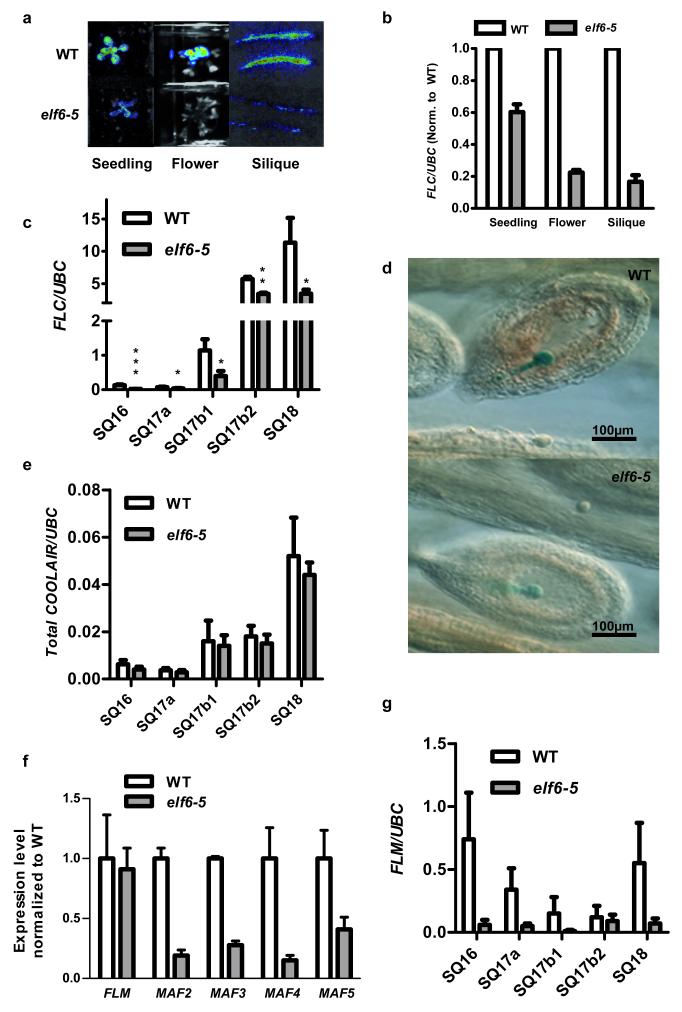
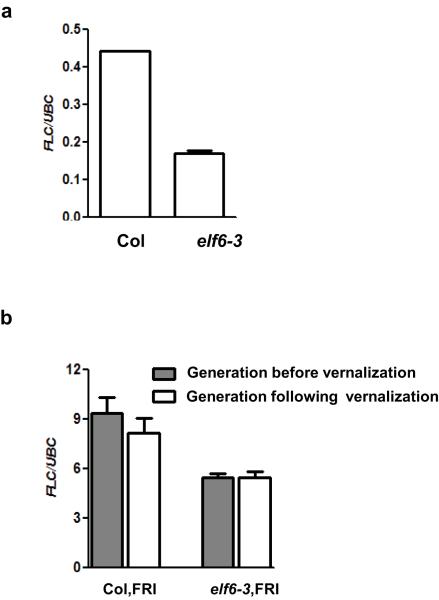
ELF6 is a jumonji C (JmJC) domain protein closely related to the histone H3 lysine 27 (H3K27me3) demethylase REF616 and it is expressed at low levels in seedlings but at high levels in flowers and embryos (Fig 2c-f). The elf6-5 mutation changes an alanine to valine (amino acid 424) at the carboxy terminal end of the JmJC domain (Fig. 2g). This amino acid is conserved in REF6 and the human H3K27me3 demethylases UTX (also known as KDM6A) and JMD3 (also known as KDM6B) (Fig. 2g). This high degree of conservation suggests that this residue may be crucial to the function of the protein. A null ELF6 T-DNA insertion allele is early flowering due to increased expression of FT17, an integrator gene that promotes floral transition. In addition, we found reduced FLC expression in an elf6-3 KO allele compared to wild-type Col (Extended Data Fig. 4a), confirming that ELF6 regulates FLC expression. The early flowering phenotype and low FLC expression in elf6-3 precluded seeing the resetting phenotype (Extended Data Fig. 4b). Although different genetic backgrounds of the two alleles may complicate interpretation these data suggest that the elf6-5 alanine to valine substitution confers a hypomorphic phenotype affecting an activity particularly important for resetting FLC expression during reproductive development.
Consistent with a role in regulating FLC resetting, elf6-5 had a much larger effect on FLC expression in flowers and siliques as compared to seedlings (Figs. 3a,b). To define more precisely when the elf6-5 mutant disrupted FLC expression we measured FLC mRNA levels at different stages of silique development18, a proxy for FLC expression in the embryo13,14. Low FLC mRNA levels could be detected in young siliques from the vernalized parental line (SQ16 and SQ17a) (Fig. 3c), which increased as the silique matured (SQ17b1 and SQ17b2) to a maximum as the silique started to desiccate and the embryo fully developed (SQ18). In the vernalized resetting mutant, FLC mRNA was detected in young developing siliques but it was not up regulated to wild-type levels at the later stages (Fig. 3c). Comparison of sibs differing only by a FLC::GUS reporter19 showed FLC::GUS expression was lower in early globular embryo of elf6-5 compared to wild type (Fig. 3d). This suggests ELF6 increases FLC expression as the embryo develops. There may be no clear mechanistic separation between reprogramming of the epigenetic state and setting of expression level.
Figure 3. Characterization of the elf6-5 resetting mutant.
a, FLC luciferase imaging (b) and FLC Q-RT-PCR data of tissues from WT and elf6-5 the generation following vernalization. Means +/− s.e.m., n = 6. c, Q-RT-PCR data of vernalized WT and elf6-5 siliques18: just after fertilization with petals still attached (SQ16); small without petals (SQ17a); first (SQ17b1) and last (SQ17b2) mature green siliques; yellow siliques (SQ18). Means + s.e.m., n = 4, *P < 0.05, **P < 0.01, ***P < 0.001. d, FLC::GUS expression in vernalized WT and elf6-5 early globular embryos. e, Q-RT-PCR shows COOLAIR levels are not affected in elf6-5 siliques. Means + s.e.m., n = 5. f, Q-RT-PCR showing that MAF2-5 genes are misregulated in elf6-5 seedlings a generation following vernalization. Means + s.e.m., n = 3. g, FLM has reduced expression in elf6-5 vernalized siliques. Means + s.e.m., n = 3.
The FLC locus has a complex transcriptional circuitry including a set of antisense transcripts called COOLAIR that are induced during vernalization but are also expressed in the warm20. We wondered if the resetting mutant also affected COOLAIR expression. Surprisingly, no change between mutant and wild type was found and total COOLAIR transcripts were up regulated normally in the mutant in developing siliques (Fig. 3e). Therefore, in contrast to mutations where both FLC sense and total COOLAIR expression levels are changed co-ordinately, for example fri, the elf6-5 mutation uncouples FLC sense and antisense regulation.
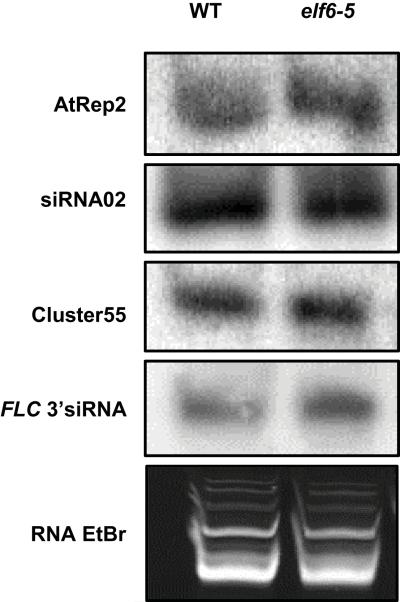
Many other loci are epigenetically modified during Arabidopsis gamete formation and embryo development21,22. We tested if elf6-5 would influence transposon expression by analyzing specific siRNA produced during seed development22; including an siRNA homologous to the flanking 3′ region of the FLC locus that accumulates preferentially in siliques23. Using sensitive northern blot analyses we could not detect any difference in the amount of these siRNAs in elf6-5 when compared to parental line using vernalized siliques (Extended Data Fig. 5). We then asked whether elf6-5 influenced expression of other FLC-family members24. MAF2, MAF3, MAF4 and MAF5 expression were down regulated in elf6-5 seedlings and expressed below detection levels in siliques (Fig. 3f); whereas FLM/MAF1 expression was unchanged in seedlings but strongly down regulated in elf6-5 siliques (Fig. 3f,g). Vernalization response in winter-annual Arabidopsis accessions (containing an active FRI allele) depends predominantly on FLC activity24, but in the rapid-cycling Col (fri) genotype all MAF genes appear to be direct targets of the Polycomb machinery24,25 indicating that the ELF6-regulatory mechanism elaborated for FLC may have more general functions in the Arabidopsis genome.
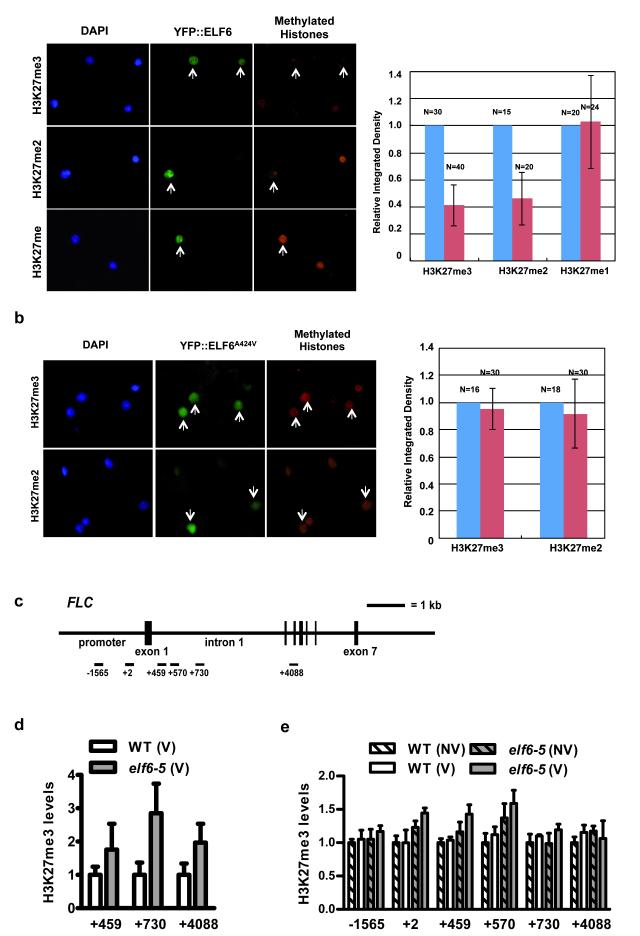
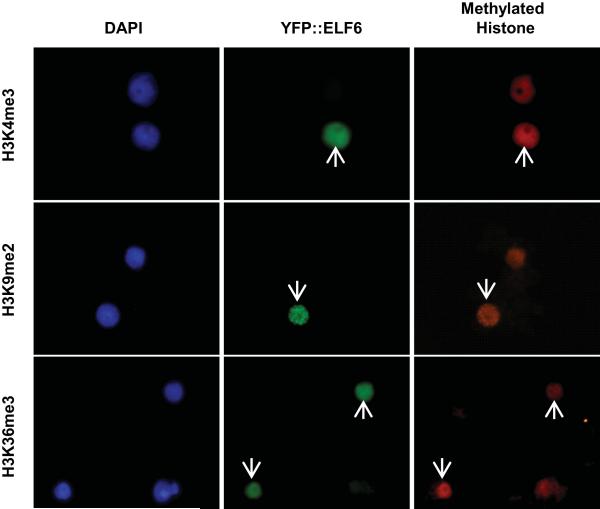
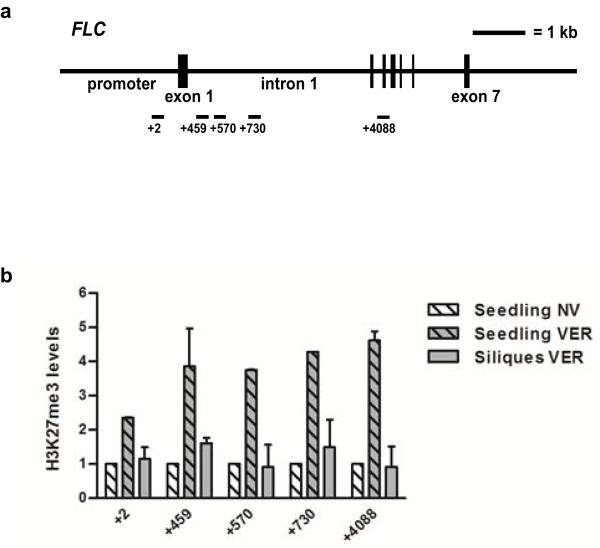
Since ELF6 is closely related to the H3K27me3 demethylase REF6 we tested both wild-type and mutant ELF6 enzymatic activity using an in vivo histone demethylation assay16. We found that transient expression of Arabidopsis ELF6 resulted in reduced H3K27me2 and H3K27me3 levels in tobacco leaves (Fig. 4a); no changes were found in H3K27me1, H3K4me3, H3K9me2 and H3K36me3 (Fig. 4a and Extended Data Fig. 6). These data show that Arabidopsis ELF6 has histone demethylase activity specific for H3K27me2 and H3K27me3. The elf6-5 mutation is an alanine to valine substitution at the carboxy terminal end of the JmJC domain. The mutation of this highly conserved residue (Fig. 2g) thus reduced the H3K27 demethylase activity in our assay (Fig. 4b). To test if this reduced activity influenced H3K27me3 levels in vivo at FLC locus, we performed chromatin immunoprecipitation experiments (ChIP). In wild-type Ler-FRI plants, H3K27me3 levels increase about 2-4 fold in vernalized seedlings, but were reduced to almost non-vernalized levels in vernalized siliques (Extended Data Fig. 7). When the resetting mutant was analysed, we found that H3K27me3 levels were higher at FLC in elf6-5 compared to the parental line on vernalized young siliques (Fig. 4d). ChIP analysis on seedlings the generation following vernalization also showed increased levels of H3K27me3 over different FLC regions in elf6-5 (Fig. 4e). These experiments were performed using whole seedlings or siliques, therefore data should be interpreted cautiously because they comprise a mixture of tissues. The vernalization-independent increase in H3K27me3 in elf6-5 and the phenotype of the elf6 loss-of-function mutant makes it likely that ELF6 has broader functions than just resetting H3K27 methylation after vernalization. Nevertheless, all these data are consistent with reduced FLC expression during embryo development in elf6-5 involving perturbed H3K27me3 dynamics that affects FLC resetting and results in inheritance of a partial vernalized state.
Figure 4. ELF6 shows H3K27 histone demethylase activity.
a, Overexpression of a YFP::ELF6 fusion protein reduces H3K27me3, H3K27me2 but not H3K27me1. b, Overexpression of YFP::ELF6A424V has no effect on H3K27 methylation. Histone methylation was visualized by immunostaining (red; right panels). Transfected nuclei (arrowed) were visualized by YFP signal (green; middle panels), and stained with DAPI (blue; left panels). Graphs quantify methylation levels of transfected (red) versus non-transfected (blue) nuclei. Means ± s.d. c, FLC regions analysed in ChIP. d, H3K27me3 levels in elf6-5 and WT siliques (stage SQ16-SQ17a) from vernalized plants. Means + s.e.m., n = 3. H3K27me3 in WT (V) plants is significantly lower than elf6-5 (V) in FLC region +4088 at *P < 0.05. e, H3K27me3 levels in progeny derived from parents that had (V) or had not (NV) been vernalized. Means + s.e.m., n = 3. H3K27me3 in WT (V) plants is significantly lower than elf6-5 (V) in FLC region +459 at *P < 0.05.
This effective impairment in reducing H3K27me3 levels at FLC leads to transgenerational inheritance of a partially vernalized state (Fig. 1a-d). The consequences of this in nature would be to misalign the developmental program of the plant with respect to the environmental conditions. The sensitivity of FLC resetting to the reduced function elf6-5 allele may indicate the requirement for H3K27me3 demethylase activity is highest at this post-vernalization stage of development, potentially explaining the differences in phenotype between elf6-5 and the null allele elf6-3. Functional redundancy between the three close homologues REF6, ELF6 and AtJMJ13(AT5G46910) may also vary through development16. It will be interesting to see if histone variants known to change in expression during embryogenesis are also involved in FLC resetting26. The large proportion of the chromatin in most eukaryotic genomes decorated with H3K27me3 probably accounts for why its erasure is a tightly controlled event during development and germ cell formation27. Further characterization of the FLC resetting process should provide greater insight into the molecular mechanism underlying genome reprogramming in eukaryotic organisms.
Methods
Plant material and growth conditions
All genotypes except elf6-317 were in a Landsberg erecta (Ler) background; an active FRIGIDA JU223 allele and a genomic FLC::LUC construct were introduced by transformation to generate a vernalization responsive line where in vivo FLC expression could be monitored by luciferase imaging15. Genetic analyses and flowering time experiments were performed with plant sown on soil and grown in controlled environment chambers in long day conditions (16 h light at 22 °C, 8 h darkness at 20 °C). To vernalize, seeds were pre-grown for 7 days at long day warm growing conditions before being transferred to cold (8 h light and 16 h darkness at 5 °C) for 6 weeks, and then returned to warm conditions.
Flowering time
was scored as the total leaf number, including rosette and cauline leaves, before the first flower opened. Statistical evaluations were performed by Student’s t-test.
Reporter gene analysis
Parental Ler-FRI, FLC::LUC line was described previously15 and luciferase imaging was detected using a Nightowl imaging system (Berthold). Siliques valves were opened longitudinally to detect FLC::LUC signal from developing embryos. A complementing pELF6::ELF6::GUS::ELF6-3′UTR construction in elf6-1 mutant background was used to monitor ELF6 expression. FLC::GUS19 was introgressed into the resetting mutant and β-glucuronidase activity was detected in Arabidopsis embryos. Siliques were longitudinally cut, fixed for 2 h at 20°C in 90% acetone and washed three times with 50 mM phosphate buffer (pH 7.0) before incubation at 37°C in reaction buffer for 24h (0.19 mM 5-bromo,4-chloro,3-indolyl-D-glucuronide, 10 mM EDTA, 0.1% Triton X-100, 0.1 mM KFeCN, 50 mM phosphate buffer pH 7.2). Embryos were observed after clearing in Hoyer’s medium using a microscope under bright-field Nomarski optics.
RNA expression analysis
For seedlings RNA analysis seeds were sown on GM media plates, stratified for 2 days and grown in long day conditions for 11 days. RNA extraction from seedlings, DNAse I treatment, cDNA synthesis, real-time quantitative PCR analyses and primers for FLC, total COOLAIR antisense and UBC control gene were described in Crevillen et al (2013)28. Primers for MAF genes detection were described before29,30. For the study of FLC expression in reproductive tissues, each biological replicate was obtained extracting RNA31 from the main inflorescence or 3-5 siliques from 5 different plants. Expression data represents the average a several biological replicates as stated in the figure legends. Statistical evaluations were performed by Student’s t-test.
Illumina sequencing
To identify the resetting mutation we performed genomic DNA deep sequencing from both the parental, Ler-derived plant and the resetting mutant line (EBI accession number PRJEB6498). Total genomic DNA was isolated from 2 g of inflorescences following a standard procedure. Briefly, inflorescences were grinded in liquid nitrogen, samples were homogenized in Extraction Buffer (50mM Tris pH 8; 200 mM NaCl; 2 mM EDTA; 0.5% SDS; 100 mg/ml Proteinase K), and incubated at 37 °C for 30 min. Proteins were removed by phenol/chloroform extraction, and DNA was precipitated using 2.5 volumes ethanol and in presence of 3M sodium acetate (pH 5).
Illumina libraries were prepared using genomic DNA. The Illumina GAIIx sequencing platform was used to generate 76 base, paired-end reads for each sample. The alignment program Maq32 v0.7.1 was used, first of all, to map reads from the parental line against the Col-0 (TAIR6) reference sequence. For this parental line, 10,867,014 reads from a total of 14,455,072 were mapped (with 10,265,540 of these in pairs). 40,573 high confidence SNPs identified with the companion maq.pl Perl script were then used, in combination with a list of Ler SNPs (http://signal.salk.edu/atg1001/data/), to edit the original Col-0 reference sequence for use in the subsequent alignment with reads obtained from the mutant line. 58,556,730 reads from a total of 65,673,308 were aligned against this modified reference sequence, with 55,308,855 of these in pairs, resulting in average 34X coverage. A local instance of the GBrowse genome browser 33 was created and loaded with the modified pseudochromosome sequences and TAIR6 coordinate-based gene model annotations. The mutant SNPs, relative to the Col-0/Ler/parental-adapted reference sequence, were then loaded into the GBrowse MySQL database and made available for visual inspection as an added feature track. By programmatically interrogating the database with a Perl script using Bio::DB::GFF methods, a genome-wide total of 417 EMS candidates (G/A -> C/T in annotated coding sequence and inferred to induce either non-synonymous codon or donor/acceptor splice site mutations) were then identified for further study.
Genetic complementation
A genomic fragment including the ELF6 locus was amplified from wild-type Ler genomic DNA using the primers 5′-ATGCCAATCCCAGAAAGTTG-3′ and 5′-AGGAGTCGTTGTCACGCTTA-3′, and cloned into pGEM-T vector (Promega). A 7.35 Kb SacI – KpnI fragment including full genomic ELF6 and regulatory regions (1.1kb from promoter and 860 bp downstream of stop codon) was cloned into pCAMBIA1300 binary vector (www.cambia.org). Resetting mutant plants were transformed by floral dipping and hygromycin resistant T1 plants isolated. For complementation analyses parental (Ler-FRI, FLC::LUC), mutant and independent T2 lines were vernalized and FLC luciferase activity measured in siliques: 8 out of 10 transgenic T2 lines analysed complemented the resetting phenotype. As a control experiment all T2 lines were also sown without antibiotic selection and plants not carrying the transgene were analysed: all lines without the transgenic ELF6 construct failed to complement the resetting phenotype.
Sequence alignment
The amino acid sequence alignment of the JmJC domain of Arabidopsis ELF6 (Q6BDA0), Arabidopsis REF6 (Q9STM3), human JMJD3 (AAH09994) and human UTX (AAT86073) proteins was performed using the web-based software Multalin tool34 (http://multalin.toulouse.inra.fr/multalin/).
Small RNA analysis
Total RNA extraction and northern blot analysis were performed as described before23 using seeds specific probes22.
In vivo histone demethylation assay
Wild-type ELF6 or mutant elf6-5 full genomic coding sequences were cloned into pEG104 vector35. The demethylation assay was carried out as previously described16. Briefly, N. benthamiana leaves were infiltrated with A. tumefaciens EHA105 strains carrying a functional wild-type 35S::YFP::ELF6 or mutant 35S::YFP::ELF6A424V. Transfected nuclei were isolated after 48 h. Immunolabeling of fixated nuclei was performed using histone methylation-specific antibodies: H3K4me3 Millipore 07-473, 1:100; H3K9me2 Millipore 07-441, 1:200; H3K27me3 Millipore 07-449, 1:100; H3K27me2 Millipore 07-452, 1:200; H3K27me1 Millipore 07-448, 1:200; H3K36me3 Abcam 9050, 1:100. The modified histones were revealed by Alexa Fluor 555-conjugated goat anti-rabbit (Invitrogen, 1:200). Transfected cells were revealed by monitoring the YFP signal. After staining, the slides were mounted in VECTASHIELD mounting medium with DAPI (Vector Laboratory) and then photographed with an OLYMPUS BX51 fluorescence microscope. Histone methylation levels were quantified by comparing staining density of a number of transfected 35S::YFP::ELF6 nuclei versus non-transfected nuclei in the same field. Image density was determined using ImageJ software. A negative result in our assay usually corresponds to 80% or less than total wild-type histone demethylase activity.
Chromatin Immunoprecipitation experiments
(ChIP) were performed using 11 days old seedlings using the protocol described before12. For siliques minor modifications were performed. About 0.5 g of tissue was grinded in liquid mitogen, and then the powder was incubated for 10 min at room temperature in extraction buffer + 1% formaldehyde to fix the tissue. We used anti- trimethyl-histone H3 lysine 27 (Millipore 07-449) and anti-H3 core antibody (Abcam 1791). All ChIP experiments were quantified by Q-PCR and analysed with the primers described before36. ChIP data is represented as the ratio H3K27me3/totalH3 normalized to non-vernalized wild-type levels. ChIP seedling data was also normalized to STM H3K27me3 levels36.
Statistical analysis
Statistical evaluations by Student’s t-test and graphical representation of the data were performed using the Prism software package (GraphPad). Means and s.e.m. are derived from independent biological samples.
Extended Data
Extended Data Figure 1. Screening for mutants impaired in the epigenetic reprogramming of FLC.
a, We started with a population of EMS-mutagenized Arabidopsis Landsberg erecta (Ler) plants carrying an FLC::luciferase (FLC::LUC) translational fusion and an active FRIGIDA transgene. The mutants we screened for were early flowering (from low FLC expression) in the generation following vernalization, but which did not flower early (and whose FLC expression was near normal) without vernalization. b, To discriminate early flowering from resetting mutants early flowering M2 plants were backcrossed to the parental line and the F2 phenotype evaluated without vernalization. Those showing no early flowering segregants were considered to be resetting mutants. In the figure, we use superscript characters to note if the plant was vernalized in the previous generation.
Extended Data Figure 2. Characterization of the first resetting mutant.
a, The first resetting mutation was found to be recessive. F1 plants were generated from a cross between the mutant the generation following vernalization and the parental wild-type line. Flowering time was assayed as total leaf number under non-vernalized long day conditions. Means + s.e.m., n = 8. b, The earlier flowering time of the mutant the generation following vernalization was stable at least three generations without vernalization. Means + s.e.m., n = 10.
Extended Data Figure 3. The elf6-5 SNP is linked to resetting of FLC expression.
Histogram showing the relationship between FLC::LUC levels in reproductive organs of vernalized plants and the elf6-5 SNP (n=154).
Extended Data Figure 4. FLC expression levels in the null elf6-3 T-DNA insertion allele.
a, elf6-3 shows reduced FLC expression compared to Col wild-type. b, the null elf6-3 allele suppresses the high FLC expression induced by FRI before vernalization. This pre-vernalization phenotype of the null allele precludes observing the role of ELF6 during FLC resetting after vernalization. All graphs show 10 day-old non-vernalized seedlings. Means + s.e.m., n = 3.
Extended Data Figure 5. siRNA production in elf6-5.
The production of specific siRNA associated with the epigenetic reactivation of transposable elements is not affected in elf6-5. Total RNA was extracted from vernalized mature siliques and detection of siRNAs was performed as described in the Methods.
Extended Data Figure 6. ELF6 has no H3K4/K9/K36me demethylase activity in a N. benthamiana transient assay.
Overexpression of a YFP-ELF6 fusion protein, using wild-type ELF6 sequence has no effect on H3K4me3/K9me2/K36me3 methylation. Histone methylation was visualized by immunostaining with rabbit polyclonal modification-specific antibodies followed by Alexa fluor 555-conjugated goat anti-rabbit (red; right panels). Transfected nuclei were visualized by the YFP signal (green; middle panels). Nuclei were stained with DAPI (blue; left panels). Arrows indicate transfected nuclei.
Extended Data Figure 7. H3K27me3 accumulation at the FLC locus.
a, Schematic representation of the FLC locus and regions analysed in the chromatin immunoprecipitation. b, H3K27me3 levels at FLC in Ler-FRI seedlings grown without vernalization (Seedling NV), 7 days after vernalization (Seedling VER) and siliques from vernalized seedlings (Siliques VER). Means + s.d., n = 2.
Acknowledgments
We thank Dean lab members and A. Surani for useful discussions. The Dean lab is supported by the UK Biotechnology and Biological Sciences Research Council grants BB/G009562/1 and BB/C517633/1, and a European Research Council Advanced Investigator grant 233039 ENVGENE. The Cao lab is supported by National Basic Research Program of China grants 2013CB967300 and 2011CB915400, and the National Natural Science Foundation of China grant 31271363. C.D. holds stock in Mendel Biotechnology.
Footnotes
The authors declare no competing financial interests.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 2011;14:195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–9. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–73. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansour AA, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 6.Canovas S, Cibelli JB, Ross PJ. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2400–5. doi: 10.1073/pnas.1119112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W, et al. Jmjd3 Inhibits Reprogramming by Upregulating Expression of INK4a/Arf and Targeting PHF20 for Ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johanson U. Molecular Analysis of FRIGIDA, a Major Determinant of Natural Variation in Arabidopsis Flowering Time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 9.Sheldon CC, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–58. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–35. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 11.Song J, Angel A, Howard M, Dean C. Vernalization - a cold-induced epigenetic switch. J. Cell Sci. 2012;125:3723–31. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 12.De Lucia F, Crevillen P, Jones AME, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16831–6. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheldon CC, et al. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2214–9. doi: 10.1073/pnas.0711453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J, et al. Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. 2009;57:918–31. doi: 10.1111/j.1365-313X.2008.03776.x. [DOI] [PubMed] [Google Scholar]
- 15.Mylne J, Greb T, Lister C, Dean C. Epigenetic regulation in the control of flowering. Cold Spring Harb. Symp. Quant. Biol. 2004;69:457–64. doi: 10.1101/sqb.2004.69.457. [DOI] [PubMed] [Google Scholar]
- 16.Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011;43:715–9. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 17.Noh B, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–13. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roeder AHK, Yanofsky MF. Fruit development in Arabidopsis. Arabidopsis Book. 2006;4:e0075. doi: 10.1199/tab.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–7. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 20.Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–53. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–72. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosher R. a, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 23.Swiezewski S, et al. Small RNA-mediated chromatin silencing directed to the 30 region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3633–8. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandre CM, Hennig L. FLC or not FLC: the other side of vernalization. J. Exp. Bot. 2008;59:1127–35. doi: 10.1093/jxb/ern070. [DOI] [PubMed] [Google Scholar]
- 25.Kim D-H, Sung S. Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell. 2013;25:454–69. doi: 10.1105/tpc.112.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingouff M, et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 2010;20:2137–43. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Crevillén P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–8. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–33. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang D, Gu X, He Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2009;21:1733–1746. doi: 10.1105/tpc.109.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oñate-Sánchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–29. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 36.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–8. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]