Abstract
Background
Brain abnormalities of subcortical and limbic nuclei are common in schizophrenia and variation in these structures is considered a putative endophenotype for the disorder. Multiplex-Multigenerational families afflicted by schizophrenia provide an opportunity to investigate the impact of shared genetic ancestry, but have not been previously examined to study structural brain abnormalities. Here we estimate the heritability of subcortical and hippocampal brain volumes in such families and the heritability of sub-regions using advanced shape analysis.
Methods
439 participants from two sites completed 3-Tesla structural magnetic resonance imaging. They included 190 European-Americans from 32 Multiplex- Multigenerational families with schizophrenia and 249 healthy comparison subjects. Subcortical and hippocampal volume and shape were measured in 14 brain structures. Heritability was estimated for volume and shape.
Results
Volume and shape were heritable in families. Estimates of heritability in subcortical and limbic volumes ranged from 0.45 in the right hippocampus up to 0.84 in the left putamen. The shape of these structures was heritable (range: 0.40–0.49) and specific sub-regional shape estimates of heritability tended to exceed heritability estimates of volume alone.
Conclusions
These results demonstrate that volume and shape of subcortical and limbic brain structures are potential endophenotypic markers in schizophrenia. The specificity obtained using shape analysis may improve selection of imaging phenotypes that better reflect the underlying neurobiology. Our findings can aid in the identification of specific genetic targets that affect brain structure and function in schizophrenia.
Keywords: heritability, schizophrenia, hippocampus, neuroimaging-genetics, endophenotypes, structural MRI
Introduction
The recent identification of genetic variants that influence brain structure (1–3) is an important step in elucidating the biological mechanisms underlying neuropsychiatric disorders. Structural brain abnormalities are common in schizophrenia (4–6) and variation in regional brain volume is considered a putative endophenotype for the disorder (7–9). Reduced gray matter volume associated with schizophrenia is present before illness onset and is heritable (8, 10). Consistent with the endophenotype concept (11), unaffected first-degree relatives of patients also exhibit reduced regional brain volume compared to healthy controls, but of a lesser extent than that observed in patients (12–16). There is evidence that quantitative brain measurements, such as volume and cortical thickness, are heritable in healthy (17–22) and neuropsychiatric samples (8, 10, 23–25). However, little is known about specific genetic targets underlying structural brain variation in schizophrenia. The gap may relate to the heterogeneity of the disorder (26) or the morphometric abnormalities (4, 7, 27). As heritability differs across brain structures (22) it is possible that particular regions, or sub-regions, will show greater heritability (28). Indeed, neuroimaging studies in healthy individuals (17, 29–31) and in large extended pedigrees (22), show a substantial range of heritability estimates across brain structures (22); a pattern that also extends to subcortical brain regions and hippocampus (17, 32). These findings suggest that some brain structures and measures are more heritable than others and may serve as better endophenotypes. The few studies supporting heritability of brain volume in schizophrenia employed twin pairs (33) or mostly nuclear families (24). No previous study has examined heritability of brain structures in large extended families affected with schizophrenia. Here we evaluate the influence of shared genetic ancestry on brain structure within large, multiplex-multigenerational families afflicted by schizophrenia.
In this report, we focus on subcortical and limbic brain structures, which are heritable in healthy (17, 29, 31, 34) and clinical populations (35, 36). These regions show consistent volumetric reductions in patients with schizophrenia (4, 37–43) and to some extent in family members (13, 44–47). More recently, morphometric changes in schizophrenia have been further scrutinized using shape analysis (13, 39, 43, 44, 48, 49), which allows for the estimation of disease-related regional deformation. This complex approach is a reliable (48, 50–53) and sensitive measure (54) of subtle, localized morphological changes in brain structure in schizophrenia (27, 39, 43, 49), and to a lesser extent in family members (13, 44). Such localized alterations may be related to distinctive dimensions of psychopathology and may be determined by specific genetic risk factors that are unique to subsets of patients with schizophrenia, or particular families (13, 28, 55). Thus, the specificity obtained using shape analysis may improve the selection of imaging phenotypes that are closer to schizophrenia pathophysiology and that may be affected by risk gene variants.
Here, we estimate heritability of subcortical and limbic brain regions in multiplex-multigenerational families and healthy comparison subjects. We focus on estimating heritability of 1) volume of subcortical and limbic brain regions, including the amygdala, caudate, hippocampus, accumbens, pallidum, putamen and thalamus, and 2) the local deformation patterns of these brain structures.
Methods & Materials
Participants
The sample consisted of 439 participants from two sites (223 from University of Pennsylvania, 216 from University of Pittsburgh), including 190 European-Americans from 32 multiplex-multigenerational families with schizophrenia and 249 healthy volunteers (Table 1). This cohort is a sub-sample of a previously characterized cohort (56, 57) with the addition of new family members. Patients had an extended multigenerational family and a consensus best-estimate DSM-IV diagnosis of schizophrenia or schizoaffective disorder. An example pedigree is shown in Figure 1. Participants were older than 15 years of age at initial contact and provided signed informed consent. The Institutional Review Boards of University of Pennsylvania and the University of Pittsburgh approved the study. For minors <18 years old, assent was obtained from the child, and consent from a parent. These data were collected as part of a larger project examining genetic mechanisms of schizophrenia. In order to reduce genetic heterogeneity the sample was restricted to Caucasian individuals.
Table 1.
Sample characteristics for the sample as a whole and at each study site.
| Group | Site | N | Sex (M/F) |
Age (years) |
GAFa | SANSb | SAPSc |
|---|---|---|---|---|---|---|---|
| Schizophrenia (SZ) |
All | 33 | 23/10 | 52 (11)* | 50 (16)*‡ | 34 (21)*‡ | 31 (25)*‡ |
| Penn | 20 | 14/6 | 54 (9) | 46 (17) | 41 (18) | 36 (27) | |
| Pitt | 13 | 9/4 | 49 (12) | 56 (11) | 18 (17) | 18 (15) | |
| Family Members (FAM) |
All | 153 | 74/79 | 43 (18) | 82 (14)* | 9 (13)* | 1 (4)* |
| Penn | 75 | 41/34 | 41 (19) | 78 (14) | 12 (14) | 2 (5) | |
| Pitt | 78 | 33/45 | 45 (17) | 88 (12) | 3 (9) | 1 (3) | |
| Healthy Comparison (HC) |
All | 246 | 115/131 | 39 (16) | 90 (11) | 3 (7) | 0 (1) |
| Penn | 125 | 57/68 | 40 (16) | 87 (8) | 6 (9) | 0 (1) | |
| Pitt | 121 | 58/63 | 39 (16) | 93 (13) | 1 (1) | 0 (1) | |
N=sample size; M=males; F=females; GAF=Global Assessment of Functioning; SANS=Scale for the Assessment of Negative Symptoms; SAPS=Scale for the Assessment of Positive Symptoms; HC=Healthy comparison subjects; SZ=Patients with schizophrenia; FAM=Multiplex family members. Mean and standard deviation shown.
significantly different than HC;
different than SZ;
; permutation tests, 100,000 permutations, p<0.01.
Tests based on sample of 222, HC; 30, SZ; 129 FAM.
Tests based on sample of 222, HC; 29, SZ; 127 FAM.
Tests based on sample of 222, HC; 29, SZ; 127 FAM.
Figure 1.
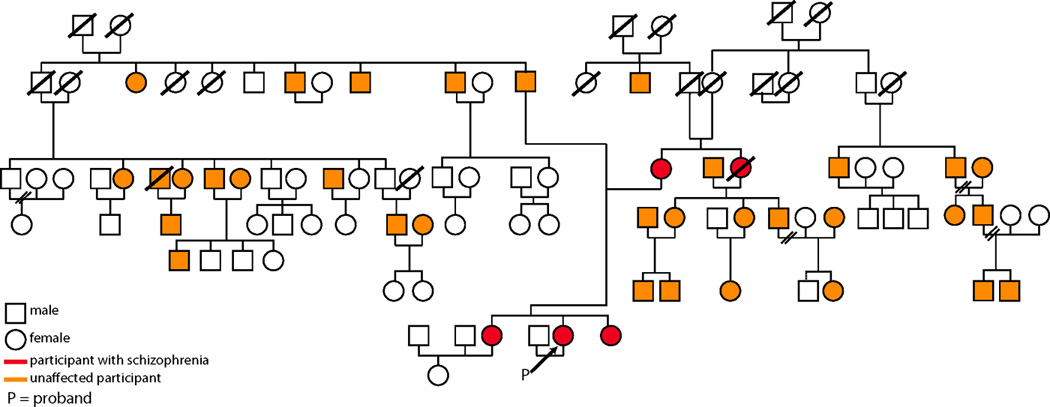
An example pedigree of a multiplex-multigenerational family with schizophrenia. This pedigree consists of 99 identified family members. Thirty-eight of which were enrolled in this study, and fourteen were eligible and completed structural magnetic resonance imaging.
Schizophrenia patients were competent to provide informed consent, capable to participate, and not exhibiting acute positive symptomatology that required medication adjustment or hospitalization. Twenty-three patients were medicated with second-generation antipsychotics, two with first-generation antipsychotics, and four with a combination of first and second-generation antipsychotics. One individual was not medicated and medication information was not available for one other patient. Family members were excluded if they had mental retardation (IQ<70), a CNS disorder that could potentially impact brain function, or were not proficient in English. Global functioning was measured using the Global Assessment of Functioning (GAF; (58)) with higher scores indicating better functioning. The Scale for the Assessment of Negative Symptoms (SANS; (59)) and the Scale for the Assessment of Positive Symptoms (SAPS; (60)) were used to rate the presence and severity of negative and positive symptoms. Twenty-one families (one-hundred thirty-five individuals) provided to the sample at least one patient with schizophrenia and at least one family member, two families (three individuals) provided only patients, and nine families (fifty-two individuals) provided only family members. Overall, the multiplex sample included 33 patients with schizophrenia and 156 family members. There is a higher prevalence of mood (~26% vs. ~10%; 61) and substance related disorders (~15% vs. ~6%) as compared to the general population (62).
The healthy comparison group included 249 psychiatrically, medically, and neurologically healthy European–Americans with no axis I or axis II cluster A disorders and no history of psychosis or mood disorder in their first-degree relatives. Healthy comparison subjects were recruited from the same communities as patients and families and also underwent urine drug testing to rule out current substance use. There were no related individuals in the comparison group, thus this randomly sampled group was included in analyses to estimate normal shape of subcortical brain structures and improve the accuracy of the statistical model used to estimate familial variance. No comparisons of heritability between multiplex families and the comparison group were performed due to the lack of family inclusion within the controls.
Demographic and clinical information for those that passed imaging quality control analysis (see below) is provided in Table 1. Permutation tests (100,000 permutations) were used to assess pairwise group differences in age, GAF, SANS, and SAPS. Permutation tests were used in place of t-tests because the data were not normally distributed based on quantile-quantile plots and the Shapiro-Wilk test for normality.
Image acquisition
A 5-minute magnetization-prepared, rapid acquisition gradient echo T1-weighted (MPRAGE) image (repetition time 1680 msec, echo time 4.67 msec, field of view 180 × 240 mm, matrix 192 × 256, flip angle =15 degrees, effective voxel resolution of 0.94 × 0.94 × 1 mm) was acquired as part of a larger imaging protocol. Data were acquired with Siemens Tim Trio 3T (Erlangen, Germany) systems at both sites. Radio frequency transmission used a quadrature body coil and reception used an 8-channel head coil. Every effort was made to minimize potential differences between sites by using identical scanners, head coils, and acquisition protocols. In addition, all data were checked for quality assurance and site was accounted for in the analyses. The results of a pilot study demonstrated good comparability between the two imaging sites in both image quality and functional activation. Image SNR varied more between subjects than between sites as in a previous study (63).
Image analysis
Subcortical Volumetric Analysis
Structural images were segmented and vertex meshes were created within FSL v4.1.7 (FMRIB’s Software Library, www.fmrib.ox.ac.uk/fsl; (64–66) using the FIRST subcortical segmentation procedure (53). FIRST segments 15 regions, including the brainstem, bilateral nucleus accumbens, amygdala, caudate, hippocampus, pallidum, putamen, and thalamus. The brainstem was not considered for analysis. The segmented subcortical regions of interest (ROIs) were corrected for intracranial volume (ICV) using FSL’s SIENAX procedure. In addition, outlier detection (> 2.5 S.D.) was performed for uncorrected brain volumes, ICV-corrected brain volumes, and laterality (LAT = 2*(L-R)/(L+R)) in each FIRST region. These parameters were selected to flag observations that may have had poor subcortical segmentation or ICV estimation. An expert analyst (SNV) visually inspected flagged regions for final determination of inclusion or exclusion. Seven subjects (3 HC; 1 SZ; 3 FAM) with poor imaging data quality failed inspection on >5 ROIs and were excluded from analysis. The final sample size for each ROI is detailed in Supplemental Table S1. False Discovery Rate (FDR) correction was used to control for multiple comparisons in all analyses of the 14 subcortical volumes.
To ensure comparability across the two acquisition sites data from three human phantoms was acquired at the inception of this study. The same structural (MPRAGE) scan was collected at both sites and these data were analyzed according to the description in the Methods section. Overall, we find moderate to high intraclass correlations across brain structure (Table 3) indicating high reliability of measurement and analysis within this study.
Table 3.
Intraclass correlations coefficients (ICC) and 95% confidence intervals (CI) for each subcortical region in three human phantoms. The same structural scan was collected at each site for each participant.
| Region | Hemisphere | ICC | Lower CI | Higher CI |
|---|---|---|---|---|
| Accumbens | L | 0.86 | −0.10 | 1.00 |
| R | 0.60 | −0.60 | 0.99 | |
| Amygdala | L | 0.52 | −0.67 | 0.98 |
| R | 0.60 | −0.60 | 0.99 | |
| Caudate | L | 0.99 | 0.84 | 1.00 |
| R | 0.99 | 0.93 | 1.00 | |
| Hippocampus | L | 0.90 | 0.09 | 1.00 |
| R | 0.83 | −0.20 | 1.00 | |
| Pallidum | L | 0.97 | 0.63 | 1.00 |
| R | 0.95 | 0.38 | 1.00 | |
| Putamen | L | 0.91 | 0.15 | 1.00 |
| R | 0.94 | 0.38 | 1.00 | |
| Thalamus | L | 0.96 | 0.54 | 1.00 |
| R | 0.89 | 0.02 | 1.00 |
Subcortical Shape Analysis
Changes in local regional shape of all structures were measured using FSL’s vertex analysis utility in FSLv5.0.0. Briefly, during FIRST segmentation a mesh was created for each subject that is a net of points (vertices) in 3-dimensions that describe the shape of each ROI. For each ROI, averaging the location of each vertex across all subjects was performed to generate a mean shape mesh. To quantitatively measure the overall shape difference in each ROI, a distance (in mm) from the average shape mesh was calculated at each vertex using only data from healthy individuals. Distance was calculated in the direction perpendicular to the surface of the average shape mesh, which indicated whether a vertex was inward (e.g. smaller) or outward (e.g. larger) relative to the average. Measured distances for each subject were projected onto a mean shape image template for analysis. All vertex analyses were conducted in standard space (Montreal Neurological Institute) space to control for between-subject differences in ICV.
Heritability Analysis
Heritability estimates were generated for brain volumes and shapes similar to previous studies (22, 67). Briefly, standard maximum likelihood variance component methods (68–71) were implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR; Department of Genetics, Texas Biomedical Research Institute). Covariance among family members was modeled as a function of additive genetic effects, with this variance component structured by a kinship matrix, and heritability estimated as the ratio of the additive genetic variance to the total phenotypic variance. Likelihood ratio tests were used to compare models in which the additive genetic component was estimated versus constrained to zero. Thus, each individual’s volume (or shape) was modeled as a function of measured covariates, specifically age, sex, and site, additive genetic effects estimated from correlations among family members, and individual-specific residual environmental factors. For the shape analysis, heritability was estimated at each voxel and corrected p-values were generated using FDR. Finally, all statistical images were projected onto the group average template using nearest neighbor interpolation in R statistical software (72) allowing for visualization in KWMeshvisu software (73).
Results
Volumetric Analysis
Significant heritability (h2) of subcortical and limbic volumes among families was present (Figure 2). Heritability was significant in 12 of the 14 volumes (Figure 2, inset), including the bilateral accumbens, caudate, hippocampus, pallidum, putamen, and thalamus. Estimates of heritability did not reach significance in bilateral amygdala. We also estimated heritability of ICV. The heritability of normalized ICV is 0.36; where as the heritability of raw ICV was 0.68. These data indicate that ICV is heritable and is not accounted through normalization. However, all measures reported are corrected for ICV and thus the estimates of heritability of these regions should not be unduly influenced by ICV.
Figure 2.
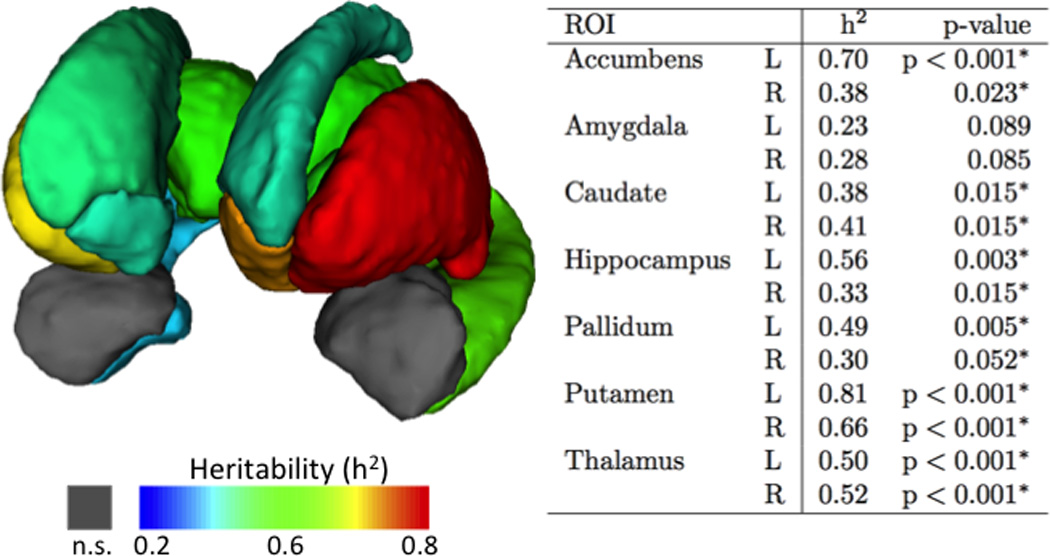
Three-dimensional reconstruction of subcortical brain structures. Most, but not all, subcortical volumes were heritable in multiplex-multigenerational families. Individual regions are color-coded according to heritability estimates presented in the accompanying table. Maps show are FDR corrected thresholded at p<.05
A comparison of subcortical and limbic volumes between patients, family members and healthy comparison subjects are provided in Table 4. There were significant group differences for bilateral accumbens, caudate, hippocampus, putamen and thalamus, but not for amygdala or pallidum. In general, patients with schizophrenia had smaller volumes as compared to family members and healthy comparison subjects. As a whole, multiplex family members were similar to healthy comparison subjects; however, when comparisons were limited to 1st degree family members an intermediate pattern emerged in several regions (putamen, caudate and hippocampus—See Table 4). Overall, in subcortical and limbic regions there is significant genetic contribution of volume among multiplex-multigenerational families with schizophrenia. Importantly, many of these regions with high familial heritability have been frequently found to be abnormal in schizophrenia, including the bilateral caudate, hippocampus, and thalamus (4).
Table 4.
A comparison of subcortical and hippocampal volumes across diagnositic groups. Number of individuals included for each regional analysis can be found in Supplemental Table S1.
| Region | Hemisphere | SZ | FAM | HC |
F-test p-value |
Comparisons (p-value) |
|---|---|---|---|---|---|---|
| Accumbens | L | 537(28) | 631(13) | 641(10) | 1.4 × 10−5 |
SZ<FAM (0.006) SZ<HC (0.001) FAM=HC (0.80) |
| R | 437(26) | 550(12) | 544(9) | 2.1 × 10−7 |
SZ<FAM (<0.001) SZ<HC (<0.001) FAM=HC (0.93) |
|
| Amygdala | L | 1554(44) | 1578(20) | 1588(16) | 0.89 | SZ<FAM (0.87) SZ<HC (0.74) FAM=HC (0.92) |
| R | 1552(55) | 1587(26) | 1549(20) | 0.54 | SZ<FAM (0.83) SZ<HC (0.99) FAM=HC (0.47) |
|
| Caudate | L | 4316(82) | 4658(37) | 4709(29) | 2.4 × 10−10 |
SZ<FAM (<0.001) SZ<HC (<0.001) FAM=HC (0.52) |
| R | 4378(84) | 4678(39) | 4754(30) | 7.5 × 10−11 |
SZ<FAM (0.003) SZ<HC (<0.001) FAM=HC (0.27)* |
|
| Hippocampus | L | 4272(97) | 4618(43) | 4658(34) | 1.2 × 10−5 |
SZ<FAM (0.003) SZ<HC (<0.001) FAM=HC (0.74) |
| R | 4297(97) | 4649(45) | 4757(35) | 2.7 × 10−6 |
SZ<FAM (0.003) SZ<HC (<0.001) FAM=HC (0.14)# |
|
| Pallidum | L | 2244(43) | 2288(19) | 2263(15) | 0.52 | SZ<FAM (0.62) SZ<HC 0.90) FAM=HC (0.58) |
| R | 2245(43) | 2299(19) | 2284(15) | 0.51 | SZ<FAM (0.48) SZ<HC (0.66) FAM=HC (0.82) |
|
| Putamen | L | 5898(113) | 6314(52) | 6335(41) | 2.1 × 10−7 |
SZ<FAM (0.002) SZ<HC (<0.001) FAM=HC (0.94) |
| R | 6028(99) | 6232(46) | 6352(36) | 2.1 × 10−7 | SZ<FAM (0.15) SZ<HC (<0.001) FAM=HC (0.09)* |
|
| Thalamus | L | 9961(133) | 10641(60) | 10659(47) | 5.6 × 10−12 |
SZ<FAM (<0.001) SZ<HC (<0.001) FAM=HC (0.97) |
| R | 9731(126) | 10391(58) | 10410(45) | 1.0 × 10−12 |
SZ<FAM (<0.001) SZ<HC (<0.001) FAM=HC (0.96) |
1st degree family members significantly smaller than HC; #1st degree family members nomially smaller than HC (p=0.06).
Vertex Analysis
Shape analyses were performed on the subcortical and limbic surfaces to identify specific loci that may be heritable. Sub-regions of bilateral accumbens, amygdala, caudate, putamen, and thalamus, and portions of left hippocampus and left pallidum were found to be heritable (Figure 2). Specific sub-regional estimates of heritability tended to exceed heritability estimates of volume alone. For example, heritability of a sub-region of right ventral amygdala was 0.76 (as seen on Figure 3b) as compared to 0.28 (Figure 2—table insert) for the entirety of right amygdala volume. Cumulatively, significant heritability was found across the surface of each structure. These effects ranged from 3% of right amygdala up to 97% of right thalamus (Table 2). These data indicate that distinct local shape patterns of subcortical brain structures are heritable in multiplex-multigenerational families.
Figure 3.
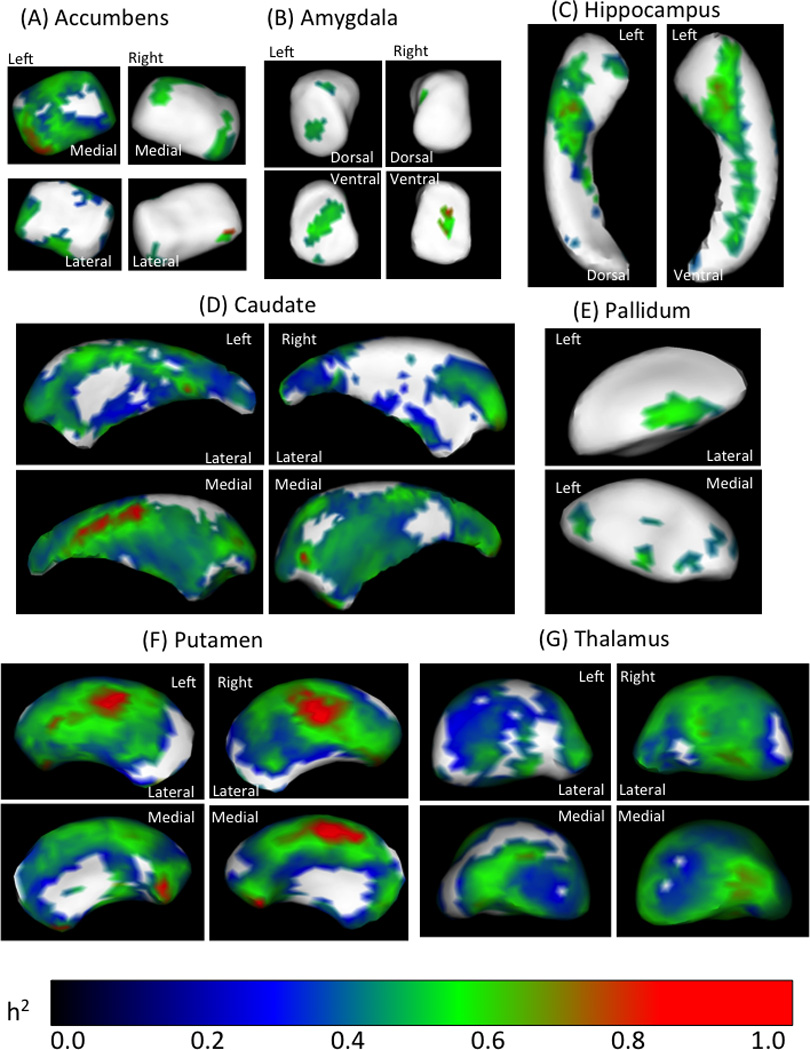
Estimates of heritability in subcortical shape. Varying extents of the bilateral accumbens (a), amygdala (b), caudate (d), putamen (f) and thalamus (g), and portions of left hippocampus (c) and left pallidum (e) were found to be significantly heritable. Many of these subfields have high heritability estimates (h2 >0.8). Notably, subcortical volumes that were not heritable (e.g. amygdala) do have focal subfields that are heritable based on shape analysis. Maps show are FDR corrected thresholded at p<.05
Table 2.
Prevalence rates of Axis I and II disorders in multiplex multigenerational family members. Axis I and II diagnoses were grouped into the follow subcategories: Mood Disorders, Substance Abuse, Other and None. Mood disorders were the most prevalent followed by substance related disorders. Substance Related Disorders included abuse of alcohol, cannabis and opioids. Other disorders included Attention-Deficit/Hyperactivity disorder, Bereavement, Intermittent Explosive Disorder, Brief Psychotic Disorder, Delusional Disorder, and Paranoia-delusional Disorder.
| Family Members | Current | Percent Current | Past | Percent Past |
|---|---|---|---|---|
| Mood Disorders | 40 | 26.14% | 9 | 5.88% |
| Substance Related Disorders |
24 | 15.68% | 8 | 5.23% |
| Other | 12 | 7.84% | 0 | 0% |
| None | 77 | 50.98% | 0 | 0% |
Discussion
The current study estimated heritability in both volume and shape of subcortical and limbic brain structures in multiplex-multigenerational families affected with schizophrenia. Heritability estimates for most brain volumes were moderate to high. The largest heritability estimates of brain volume were observed in bilateral putamen and left nucleus accumbens. Estimates of heritability of shape were moderate, but substantial extents of surface shape of most subcortical and limbic regions were found to be significantly heritable, with high heritability estimates in focal subfields of each region. Indeed, specific sub-regional estimates of heritability actually exceed heritability estimates of volume alone in some instances. Overall, these data confirm previous reports that subcortical regional volumes are heritable. Our results show, for the first time, heritability of localized sub-fields within these subcortical structures. These data add to recent findings using healthy populations that subcortical brain volume as well as shape may aid in the selection of imaging endophenotypes associated with genetic variants underlying structural brain abnormalities in schizophrenia.
Our volumetric findings confirm and extend previous work. Patients with schizophrenia had smaller volumes than family members or healthy comparison subjects. In comparison to other studies in healthy individuals (29, 30, 74) we report similar heritability, albeit slightly lower, in subcortical brain volumes using a cohort of affected patients with schizophrenia and their relatives. Specifically, our heritability estimates in most brain regions, including the hippocampus (23, 34), caudate (17, 30), putamen (29, 31, 34), pallidum (29, 31) and thalamus (29, 31), but see (34), align with previous findings. Our pattern of heritability estimates in the accumbens (higher h2 in left than right accumbens) is consistent with one previous report (29). We also report no significant heritability in amygdala volume, which is at odds with a prior finding of significant heritability in this region (29). These discrepancies may be due to differences in the technical approach, such as image acquisition parameters, bias in regional partitioning (17, 32) or processing software (75), sample composition (e.g. age, family size) or the ascertainment strategy employed.
The few studies estimating heritability of brain volume in schizophrenia were encouraging, yet most employed twin pairs (33) or were limited to nuclear families (24). Thus, our findings in large extended pedigrees of families affected with schizophrenia corroborate previous work and further solidify subcortical volumes as meaningful endophenotypes. Our heritability estimates of brain volume in multiplex families with schizophrenia are lower in comparison with den Braber et al. (2013), the only study to estimate heritability of subcortical and limbic structures in a healthy sample. However, our use of multiplex multigenerational families is likely to represent a more homogeneous and targeted group of instrumental genes and pathways (76), as compared to studies of unrelated individuals (76). As previously discussed (56), it is unlikely that heritability estimates in extended pedigrees are inflated by shared environment, if that were the case each rung on the family tree would require a fixed proportional decrease in shared environment for it to mimic the genetic heritability estimated here, which is unlikely.
Significant heritability of sub-regional shape deformations in subcortical and limbic brain structures establishes for the first time the shape of these structures as promising endophenotypic markers of schizophrenia. Despite the absence of heritability in some subcortical volumes (e.g. amygdala), we found significant heritability subregionally. Notably, measurement of sub-regional variation with shape analysis provides a more sensitive method to detect subtle abnormalities compared to traditional volumetric methods (13, 54, 77). Local shape may imply volumetric change within that specific area, but also suggest differences deeper in a given structure. Systematic investigation of subcortical shape may provide insight into the underlying genetics of microscopic cytoarchitecture; for example changes in shape may indicate neurodevelopmental changes in parenchymal volume or physiological compensation due to variations in activity (55). Moreover, shape deformations may provide significant information about neurodevelopmental abnormalities seen in schizophrenia and family members (13, 44). Previous work suggests that physical tension during neurodevelopment may lead to specific, localized structural abnormalities (78, 79). Additionally, the onset of schizophrenia typically occurs during a critical period of dynamic, progressive gray matter reductions (80). This may lead to localized changes within gray matter structures and possibly affect cognition. Furthermore, it is possible that sub-regions with high heritability may help differentiate families with high burden of illness from those with a lower burden. Recent epidemiological work suggests that schizophrenia is associated with a large number of individually rare mutations that likely differ among families (81–83). If indeed unique genes are responsible for illness among families, the phenotypes (e.g. brain volume or shape) may differ enough such that pooling results across families would preclude identification of ‘common’ abnormalities. For example, a previous study of patients and unaffected family members reported that family explained approximately 10% of the variance in hippocampus volume (23). Thus, it is possible that particular families contribute more than others to these markers and on-going work in these multiplex-multigenerational families is aimed at identifying endophenotypes that result from rare alleles with large effect, which is another specific advantage of using large extended pedigrees (82, 83). In general, it appears that localized shape of subcortical structures in large families may be useful as endophenotypic markers of illness in frank schizophrenia. While we have focused on subcortical and limbic structures, future work is required to assess heritability of cortical structures, including cortical thinning (22), which may represent another informative endophenotype in schizophrenia.
It is likely that the multiplex multigenerational aspects (e.g. mixture of relationships— within and across multiple generations and the unequal numbers of observations per family) of this study add to the specificity of calculated heritability (84). It may result in preferential selection of patients and families with less genetic loading for pathological endophenotypic values, which may lower the value, but not reduce the significance of our heritability estimates. Moreover, the inclusion of affected individuals likely impacts our heritability results given known changes in brain structure in patients with schizophrenia (4). It is possible that lower heritability, as compared to den Braber et al., (2013), reflects disease specific differences in the variation of brain volumes in and across families affected with schizophrenia. In addition, the effect of environmental biases like antipsychotic medication introduces non-genetic variation and reduces our estimate of heritability (4). Our sample is a specifically selected subpopulation that likely has lower genetic variability than the population at large and this restricted range may affect estimates of heritability. Yet, we find significant heritability in many, but not all brain regions, which likely speaks to the heightened liability that schizophrenia has on regional brain volume in multiplex families. Other factors, such as, sample size, age, degree of relationship, or even measurement type (1.5T vs. 3T data acquisition) may affect comparison across studies. Yet, given the similar patterns observed in our data and in den Braber et al., (2013) we believe that our estimates of heritability are appropriate and useful. Thus, the heritability estimates presented may provide more specific, relevant targets for genomic studies of brain development in schizophrenia.
Our use of multiplex family members provides a unique perspective on subcortical volume and shape, but these data may not translate to simplex families because multiplex families may have higher incidences of other axis I or axis II disorders. While we did not consider other psychiatric diagnoses, future studies are aimed at using a dimensional approach to assess phenotypic heritability. Our sample does have higher prevalence of mood and substance related disorders as compared to the general population, but we did not observe any effect on volumetric measures. However, the numbers here are small, particularly if family is considered as an additional factor, making it difficult to interpret the influence of Axis I and II disorders on brain volume in multiplex families. Furthermore, the older age and inclusion of only Caucasian individuals makes broad generalizations complex. In comparison to twin studies, which typically report higher heritability, the use of extended pedigree in the estimation of heritability is more susceptible to uncontrolled age-related influence. However, we have attempted to mitigate this by statistically correcting for age. One advantage over twin studies is that heritability estimates in extended pedigrees are less likely to be unduly inflated by shared environment (76). Importantly, relatively few genes are known to be associated with both brain structure and are relevant in schizophrenia (85, 86). This scarcity may reflect the genetic heterogeneity of schizophrenia and suggests that different alleles in different families may be responsible for morphological abnormalities. Our use of an automated segmentation tool takes advantage of recent developments in the neuroimaging community, however this technique is not without its limitations. Regions that showed the lowest ICC were also the least heritable. This is in agreement with a recent manuscript that assessed heritability of these same structures in healthy individuals (17). Thus, there may be higher measurement error in some regions as compared to others, as has been previously suggested (32, 75). Nonetheless, automated segmentation in most subcortical and limbic structures appears robust and repeatable. Finally, our approach can also reduce heterogeneity by allowing for the analysis of subgroups or subfamilies, and follow-up studies in large pedigrees should consider family-specific effects. In the current study, we are unable to compare volume or family heritability within specific regions to a comparable group of healthy subjects due to a lack of related individuals within the healthy sample. Future endeavors can incorporate large pedigrees of healthy comparison subjects to determine if there are subtypes of families with schizophrenia that show especially large abnormalities in brain structure.
In conclusion, we find that subcortical brain volume and shape are heritable in multiplex multigenerational families with schizophrenia. Such localized features may be related to distinct dimensions of psychopathology or may be determined by specific genetic risk factors unique to patients with schizophrenia, or to particular families. The specificity obtained using shape analysis of brain structures may improve the selection of imaging phenotypes that better reflect the underlying neurobiology and aid in the identification of specific genetic targets that affect brain structure and function in schizophrenia.
Supplementary Material
Table 5.
Heritabiilty of Subcortical Shape in Multiplex-Multigenerational Families with Schizophrenia.
| Region | Hemisphere | Heritability |
% surface heritable |
|---|---|---|---|
| Accumbens | L | 0.48(0.27–0.75) | 61 |
| R | 0.44(0.31–0.75) | 12 | |
| Amygdala | L | 0.48(0.38–0.59) | 11 |
| R | 0.62(0.52–0.76) | 3 | |
| Caudate | L | 0.45(0.21–1.00) | 78 |
| R | 0.43(0.20–0.80) | 61 | |
| Hippocampus | L | 0.49(0.28–0.72) | 28 |
| R | 0.42(0.22–0.82) | 64 | |
| Pallidum | L | 0.46(0.35–0.66) | 10 |
| R | n.s. | n.s. | |
| Putamen | L | 0.47(0.20–1.00) | 83 |
| R | 0.49(0.20–1.00) | 82 | |
| Thalamus | L | 0.41(0.21–0.69) | 76 |
| R | 0.48(0.21–0.70) | 97 |
All results are False Discovery Rate (FDR) corrected and thresholded at p<0.05. Heritability = mean(minimum-maximum) heritability in the thresholded surfaces after controlling for age and site; % Surface = the percent of the subcortical surface that has significant heritability; ROI = Region Of Interest; n.s.= not significant.
Acknowledgements
We would like to thank J. Eric Schmitt MD, PhD for his insightful comments on the final draft of this manuscript.
We are grateful to the participants and their families for volunteering for this research.
The authors thank Robert Witalec, B.A. Sue Clifton and Amy Cassidy M.A. for data gathering and processing.
This work was supported by the National Institute of Mental Health (R01MH042191 and T32 MH019112 to [REG], R01MH063480 to [VLN], R01MH061622 to [LAA], and R01MH084856 to [RCG]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
All authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikram MA, Fornage M, Smith AV, Seshadri S, Schmidt R, Debette S, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44:539–544. doi: 10.1038/ng.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shepherd AM, Laurens KR, Matheson SL, Carr VJ, Green MJ. Systematic meta-review and quality assessment of the structural brain alterations in schizophrenia. Neurosci Biobehav Rev. 2012;36:1342–1356. doi: 10.1016/j.neubiorev.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Gur RE, Turetsky BI, Bilker WB, Gur RC. Reduced gray matter volume in schizophrenia. Arch Gen Psychiatry. 1999;56:905. doi: 10.1001/archpsyc.56.10.905. [DOI] [PubMed] [Google Scholar]
- 6.Takayanagi Y, Takahashi T, Orikabe L, Masuda N, Mozue Y, Nakamura K, et al. Volume reduction and altered sulco-gyral pattern of the orbitofrontal cortex in first-episode schizophrenia. Schizophr Res. 2010;121:55–65. doi: 10.1016/j.schres.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Nenadic I, Gaser C, Sauer H. Heterogeneity of Brain Structural Variation and the Structural Imaging Endophenotypes in Schizophrenia. Neuropsychobiology. 2012;66:44–49. doi: 10.1159/000338547. [DOI] [PubMed] [Google Scholar]
- 8.Fusar-Poli P, Howes O, Allen P, Broome M, Valli I, Asselin M, et al. Abnormal prefrontal activation directly related to pre-synaptic striatal dopamine dysfunction in people at clinical high risk for psychosis. Mol Psychiatry. 2009;16:67–75. doi: 10.1038/mp.2009.108. [DOI] [PubMed] [Google Scholar]
- 9.Almasy L, Gur RC, Haack K, Cole SA, Calkins ME, Peralta JM, et al. A genome screen for quantitative trait loci influencing schizophrenia and neurocognitive phenotypes. Am J Psychiatry. 2008;165:1185–1192. doi: 10.1176/appi.ajp.2008.07121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner JA, Calhoun VD, Michael A, van Erp TGM, Ehrlich S, Segall JM, et al. Heritability of multivariate gray matter measures in schizophrenia. Twin Res Hum Genet. 2012;15:324. doi: 10.1017/thg.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 12.Keshavan MS, Prasad KM, Pearlson G. Are brain structural abnormalities useful as endophenotypes in schizophrenia? Int Rev Psychiatry. 2007;19:397–406. doi: 10.1080/09540260701486233. [DOI] [PubMed] [Google Scholar]
- 13.Ho BC, Magnotta V. Hippocampal volume deficits and shape deformities in young biological relatives of schizophrenia probands. Neuroimage. 2010;49:3385–3393. doi: 10.1016/j.neuroimage.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boos HBM, Cahn W, van Haren NEM, Derks EM, Brouwer RM, Schnack HG, et al. Focal and global brain measurements in siblings of patients with schizophrenia. Schizophr Bull. 2012;38:814–825. doi: 10.1093/schbul/sbq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusar-Poli P, Borgwardt S, Crescini A, Deste G, Kempton MJ, Lawrie S, et al. Neuroanatomy of vulnerability to psychosis: a voxel-based meta-analysis. Neurosci Biobehav Rev. 2011;35:1175–1185. doi: 10.1016/j.neubiorev.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Karnik-Henry MS, Wang L, Barch DM, Harms MP, Campanella C, Csernansky JG. Medial temporal lobe structure and cognition in individuals with schizophrenia and in their non-psychotic siblings. Schizophr Res. 2012;138:128–135. doi: 10.1016/j.schres.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Braber A, Bohlken MM, Brouwer RM, van’t Ent D, Kanai R, Kahn RS, et al. Heritability of subcortical brain measures: A perspective for future genome-wide association studies. Neuroimage. 2013;83:98–102. doi: 10.1016/j.neuroimage.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Baaré WFC, Pol HEH, Boomsma DI, Posthuma D, de Geus EJC, Schnack HG, et al. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- 19.Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, et al. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25:805–816. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- 20.Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glahn DC, Paus T, Thompson PM. Imaging genomics: mapping the influence of genetics on brain structure and function. Hum Brain Mapp. 2007;28:461–463. doi: 10.1002/hbm.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honea RA, Meyer-Lindenberg A, Hobbs KB, Pezawas L, Mattay VS, Egan MF, et al. Is gray matter volume an intermediate phenotype for schizophrenia? A voxel-based morphometry study of patients with schizophrenia and their healthy siblings. Biol Psychiatry. 2008;63:465–474. doi: 10.1016/j.biopsych.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldman AL, Pezawas L, Mattay VS, Fischl B, Verchinski BA, Zoltick B, et al. Heritability of brain morphology related to schizophrenia: a large-scale automated magnetic resonance imaging segmentation study. Biol Psychiatry. 2008;63:475–483. doi: 10.1016/j.biopsych.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 25.van der Schot AC, Vonk R, Brans RGH, van Haren NEM, Koolschijn P, Nuboer V, et al. Influence of genes and environment on brain volumes in twin pairs concordant and discordant for bipolar disorder. Arch Gen Psychiatry. 2009;66:142. doi: 10.1001/archgenpsychiatry.2008.541. [DOI] [PubMed] [Google Scholar]
- 26.Tandon R, Gaebel W, Barch DM, Bustillo J, Gur RE, Heckers S, et al. Definition and description of schizophrenia in the DSM-5. Schizophr Res. 2013 doi: 10.1016/j.schres.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 27.Nugent TF, III, Herman DH, Ordonez A, Greenstein D, Hayashi KM, Lenane M, et al. Dynamic mapping of hippocampal development in childhood onset schizophrenia. Schizophr Res. 2007;90:62–70. doi: 10.1016/j.schres.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SL, Wang L, Alpert KI, Greenstein D, Clasen L, Lalonde F, et al. Hippocampal shape abnormalities of patients with childhood-onset schizophrenia and their unaffected siblings. J Am Acad Child Adolesc Psychiatry. 2013;52:527–536. doi: 10.1016/j.jaac.2013.02.003. e522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace GL, Eric Schmitt J, Lenroot R, Viding E, Ordaz S, Rosenthal MA, et al. A pediatric twin study of brain morphometry. J Child Psychol Psychiatry. 2006;47:987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- 31.Yoon U, Perusse D, Lee J-M, Evans AC. Genetic and environmental influences on structural variability of the brain in pediatric twin: Deformation based morphometry. Neurosci Lett. 2011;493:8–13. doi: 10.1016/j.neulet.2011.01.070. [DOI] [PubMed] [Google Scholar]
- 32.Blokland GAM, de Zubicaray GI, McMahon KL, Wright MJ. Genetic and environmental influences on neuroimaging phenotypes: A meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 2012;15:351–371. doi: 10.1017/thg.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brans RGH, van Haren NEM, van Baal GCM, Schnack HG, Kahn RS, Pol HEH. Heritability of changes in brain volume over time in twin pairs discordant for schizophrenia. Arch Gen Psychiatry. 2008;65:1259. doi: 10.1001/archpsyc.65.11.1259. [DOI] [PubMed] [Google Scholar]
- 34.Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- 35.Kaymaz N, Van Os J. Heritability of structural brain traits: an endophenotype approach to deconstruct schizophrenia. Int Rev Neurobiol. 2009;89:85–130. doi: 10.1016/S0074-7742(09)89005-3. [DOI] [PubMed] [Google Scholar]
- 36.Schneider B, Prvulovic D. Novel biomarkers in major depression. Curr Opin Psychiatry. 2013;26:47–53. doi: 10.1097/YCO.0b013e32835a5947. [DOI] [PubMed] [Google Scholar]
- 37.Lawrie SM, Abukmeil SS. Brain abnormality in schizophrenia. A systematic and quantitative review of volumetric magnetic resonance imaging studies. Br J Psychiatry. 1998;172:110–120. doi: 10.1192/bjp.172.2.110. [DOI] [PubMed] [Google Scholar]
- 38.Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- 39.Mamah D, Harms MP, Barch D, Styner M, Lieberman JA, Wang L. Hippocampal shape and volume changes with antipsychotics in early stage psychotic illness. Front Psychiatry. 2012;3 doi: 10.3389/fpsyt.2012.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckers S, Konradi C. Behavioral Neurobiology of Schizophrenia and its Treatment. Springer; 2010. Hippocampal pathology in schizophrenia; pp. 529–553. [DOI] [PubMed] [Google Scholar]
- 41.Keshavan MS, Haas GL, Kahn CE, Aguilar E, Dick EL, Schooler NR, et al. Superior temporal gyrus and the course of early schizophrenia: progressive, static, or reversible? J Psychiatr Res. 1998;32:161–167. doi: 10.1016/s0022-3956(97)00038-1. [DOI] [PubMed] [Google Scholar]
- 42.Ebdrup BH, Glenthøj B, Rasmussen H, Aggernaes B, Langkilde AR, Paulson OB, et al. Hippocampal and caudate volume reductions in antipsychotic-naive first-episode schizophrenia. J Psychiatry Neurosci. 2010;35:95. doi: 10.1503/jpn.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClure RK, Styner M, Maltbie E, Lieberman JA, Gouttard S, Gerig G, et al. Localized differences in caudate and hippocampal shape are associated with schizophrenia but not antipsychotic type. Psychiatry Res. 2013 doi: 10.1016/j.pscychresns.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tepest R, Wang L, Miller MI, Falkai P, Csernansky JG. Hippocampal deformities in the unaffected siblings of schizophrenia subjects. Biol Psychiatry. 2003;54:1234–1240. doi: 10.1016/s0006-3223(03)00702-9. [DOI] [PubMed] [Google Scholar]
- 45.van Erp TGM, Saleh PA, Rosso IM, Huttunen M, Lönnqvist J, Pirkola T, et al. Contributions of genetic risk and fetal hypoxia to hippocampal volume in patients with schizophrenia or schizoaffective disorder, their unaffected siblings, and healthy unrelated volunteers. Am J Psychiatry. 2002;159:1514–1520. doi: 10.1176/appi.ajp.159.9.1514. [DOI] [PubMed] [Google Scholar]
- 46.Staal WG, Pol HEH, Schnack H, van der Schot AC, Kahn RS. Partial volume decrease of the thalamus in relatives of patients with schizophrenia. Am J Psychiatry. 1998;155:1784–1786. doi: 10.1176/ajp.155.12.1784. [DOI] [PubMed] [Google Scholar]
- 47.Harms MP, Wang L, Mamah D, Barch DM, Thompson PA, Csernansky JG. Thalamic shape abnormalities in individuals with schizophrenia and their nonpsychotic siblings. J Neurosci. 2007;27:13835–13842. doi: 10.1523/JNEUROSCI.2571-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46:177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JM, Kim SH, Jang DP, Ha TH, Kim JJ, Kim IY, et al. Deformable model with surface registration for hippocampal shape deformity analysis in schizophrenia. Neuroimage. 2004;22:831–840. doi: 10.1016/j.neuroimage.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 51.Ashburner J, Csernansk JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003;2:79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- 52.McHugh TL, Saykin AJ, Wishart HA, Flashman LA, Cleavinger HB, Rabin LA, et al. Hippocampal volume and shape analysis in an older adult population. Clin Neuropsychol. 2007;21:130–145. doi: 10.1080/13854040601064534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, et al. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamah D, Harms MP, Wang L, Barch D, Thompson P, Kim J, et al. Basal ganglia shape abnormalities in the unaffected siblings of schizophrenia patients. Biol Psychiatry. 2008;64:111–120. doi: 10.1016/j.biopsych.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gur RE, Nimgaonkar VL, Almasy L, Calkins ME, Ragland JD, Pogue-Geile MF, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–819. doi: 10.1176/ajp.2007.164.5.813. [DOI] [PubMed] [Google Scholar]
- 57.Roalf DR, Gur RC, Almasy L, Richard J, Gallagher RS, Prasad K, et al. Neurocognitive performance stability in a muliplex multigenerational study of schizophrenia. Schizophr Bull. 2013;39:529–537. doi: 10.1093/schbul/sbs078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endicott J, Spitzer RL, Fleiss JL, Cohen J. The global assessment scale. A procedure for measuring overall severity of psychiatric disturbance. Arch Gen Psychiatry. 1976;33:766–771. doi: 10.1001/archpsyc.1976.01770060086012. [DOI] [PubMed] [Google Scholar]
- 59.Andreasen NC. The Scale for the Assessment of Negative Symptoms (SANS) Iowa City, Iowa: Unversity of Iowa; 1984. [Google Scholar]
- 60.Andreasen NC. The Scale for the Assessment of Positive Symptoms (SAPS) Iowa City, Iowa: Unversity of Iowa; 1984. [Google Scholar]
- 61.NIH. The Numbers Count: Mental Disorders in America. 2013 http://www.nimh.nih.gov/health/publications/the-numbers-count-mental-disorders-in-america/index.shtml.
- 62.WHO. Global Health Observatory. 2013 http://www.who.int/gho/substance_abuse/burden/alcohol_prevalence/en/
- 63.Roalf DR, Ruparel K, Gur RE, Bilker W, Gerraty R, Elliott MA, et al. Neuroimaging predictors of cognitive performance across a standardized neurocognitive battery. Neuropsychology. doi: 10.1037/neu0000011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 65.Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009;45:S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 66.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 67.Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64:1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lange K, Westlake J, Spence M. Extensions to pedigree analysis III. Variance components by the scoring method. Ann Hum Genet. 1976;39:485–491. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 69.Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet. 1982;46:373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 70.Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535. [PMC free article] [PubMed] [Google Scholar]
- 71.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Granert O. Rniftilib: Rniftilib -R Interface to NIFTICLIB (v2.0.0) 2013 [Google Scholar]
- 73.Oguz I, Gerig G, Barre S, Styner M. KWMeshVisu: a mesh visualization tool for shape analysis. Insight Journal, MICCAI Open-Source Workshop. 2006 [Google Scholar]
- 74.Stein JL, Hibar DP, Madsen SK, Khamis M, McMahon KL, de Zubicaray GI, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N= 1198) using genome-wide search. Mol Psychiatry. 2011;16:927–937. doi: 10.1038/mp.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morey RA, Selgrade ES, Wagner HR, Huettel SA, Wang L, McCarthy G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp. 2010;31:1751–1762. doi: 10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borecki IB, Province MA. Genetic and genomic discovery using family studies. Circulation. 2008;118:1057–1063. doi: 10.1161/CIRCULATIONAHA.107.714592. [DOI] [PubMed] [Google Scholar]
- 77.Csernansky JG, Schindler MK, Splinter NR, Wang L, Gado M, Selemon LD, et al. Abnormalities of thalamic volume and shape in schizophrenia. Am J Psychiatry. 2004;161:896–902. doi: 10.1176/appi.ajp.161.5.896. [DOI] [PubMed] [Google Scholar]
- 78.Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 79.Van Essen DC, Drury HA, Joshi S, Miller MI. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. Proc Natl Acad Sci. 1998;95:788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClellan JM, Susser E, King M-C. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 82.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 83.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wray N, Visscher P. Estimating trait heritability. Nature Education. 2008;1:29. [Google Scholar]
- 85.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Scheltinga AFT, Bakker SC, Kahn RS, Kas MJH. Fibroblast Growth Factors in Neurodevelopment and Psychopathology. Neuroscientist. 2013;19:479–494. doi: 10.1177/1073858412472399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


