Summary
Protein interactions from painful states form a coherent network that can be used to inform further study. We identify proteins key to painful subnetworks.
Keywords: Networks, Protein–protein interactions, Gene expression, Neuropathic pain, Inflammatory pain, Text mining
Abstract
Understanding the molecular mechanisms associated with disease is a central goal of modern medical research. As such, many thousands of experiments have been published that detail individual molecular events that contribute to a disease. Here we use a semi-automated text mining approach to accurately and exhaustively curate the primary literature for chronic pain states. In so doing, we create a comprehensive network of 1,002 contextualized protein–protein interactions (PPIs) specifically associated with pain. The PPIs form a highly interconnected and coherent structure, and the resulting network provides an alternative to those derived from connecting genes associated with pain using interactions that have not been shown to occur in a painful state. We exploit the contextual data associated with our interactions to analyse subnetworks specific to inflammatory and neuropathic pain, and to various anatomical regions. Here, we identify potential targets for further study and several drug-repurposing opportunities. Finally, the network provides a framework for the interpretation of new data within the field of pain.
1. Introduction
Acute pain has evolved as a key physiological alert system for avoiding noxious stimuli and protecting damaged regions of the body by discouraging physical contact and movement [50]. This form of pain is crucial; in its absence, for example, in individuals with congenital insensitivity to pain, we are more prone to damaging or nonprotective behaviors that can hinder our quality of life [44], [46]. Conversely, persistent or chronic pain can be similarly debilitating, with those affected typically suffering psychological disturbance and significant activity restrictions [16]. The incidence of chronic pain is widespread across the global population, with estimates in the adult general population of 12.7% to 29.9% in developed and 14.5% to 33.9% in developing nations [11]. Pharmacological therapeutics such as opioids, and nonsteroidal anti-inflammatory drugs (NSAIDs) such as cyclooxygenase-2 (COX-2) inhibitors, are often prescribed as the standard treatment regimens for patients with chronic pain [3], [27]. However, although these and a huge range of other treatment options are available, their efficacy often proves at best modest, and their use is limited by unwanted side effects [13], [49]. There is therefore an urgent need to better understand the molecular systems that mediate chronic pain and to use this knowledge to develop improved therapeutics.
Pain researchers have published hundreds of thousands of articles, many of which detail knowledge of the molecular interactions involved in pain. However, digesting and using this knowledge is impractical without the use of text mining. In our previous work [24], we used state-of-the-art computational methods to retrieve molecular interactions associated with pain from the primary literature: the whole of Medline and open-access articles in PubMed Central (PMC). These data are catalogued at wiki-pain.org, which contains 93,271 molecular interactions derived from 765,692 pain-related articles. Each interaction is annotated with detailed contextual information such as anatomy, associated point mutations, and disease relevance. However, as fully automated text-mining results can be error prone [43], we implemented a novel strategy to curate mentions of protein–protein interactions (PPIs) grouped from multiple publications to create the first pain-specific dataset of interactions. Through ongoing curation, this dataset now contains more than 1000 unique contextualized PPIs [24].
Here we explore the relevance and accuracy of our pain related PPIs using network and functional enrichment analyses and gene bias assessment methods. To emphasize the quality and effectiveness of this approach of sourcing interaction data, we provide comparisons with pain-related interaction networks derived from gene expression data, a manually curated list of pain genes, and the known targets of pain drugs. Our results demonstrate that a semi-automated text-mined interaction network allows us to interpret the sum knowledge of the biomedical domain of pain in an integrated manner, providing a more complete portrait than is possible from other common means of inferring disease networks. The network has immediate utility to researchers in the field as a framework for the interpretation of new findings and high-throughput ‘omic’ datasets. Importantly, this approach has broad applicability to other diseases or syndromes to which a combination of text mining and network biology might be applied.
2. Methods
2.1. Data availability
The data generated in this study are available in Supplementary Tables 1 to 24.
2.2. Curation procedure for PPIs
In a previous study detailing our text-mining methodology, we curated more than 1500 unique PPIs involving mouse, rat, and/or human proteins, ranked by their overall relevance to pain [24]. Raw interactions were extracted from text automatically and displayed on wiki-pain.org to be verified by an expert. For a PPI to be considered accurate, the proteins, including underlying species and associated Entrez Gene IDs, and interactions had to have been extracted accurately in at least 1 instance when all mentions of that interaction were grouped together. We continued curating interactions in this study following these guidelines. We focused on those interactions that had a text mining confidence score above a threshold (28%) that was empirically determined to be a good indicator of true-positive interactions [24]. We grouped orthologous proteins from rats, mice, and humans (using NCBI Homologene IDs) and simplified the interactions to positive regulation, negative regulation, regulation, or binding to remove superfluous data.
PPIs for the neuropathic and inflammatory pain tasks were curated in the same way as with general pain-associated interactions, with the addition of 1 more condition: each interaction had to have a specific association with the relevant pain disorder, for example, “activiation of c-Jun in DRGs induces VIP and NPY upregulation and contributes to the pathogenesis of neuropathic pain” [48]. Those interactions that were selected for curation had neuropathic or inflammatory pain relevance of greater than 90% [24] and, again, a text mining confidence threshold of 28%. The involvement was noted as being either part of the mechanism of that disorder or having an inhibitory effect on it. We note that interactions were curated from literature published over decades, and so any changes in the formal definition of these indications used may not have been accounted for.
2.3. Network analysis
Networks were analyzed using iGraph for R [9] and visualised using Cytoscape 3.0 [45].
Enrichment analysis of proteins was performed using Fisher’s exact test to determine proteins that had a statistically significant number of interactions in the subgraph under study compared to the relevant main graph. This follows similar implementations of Fisher’s exact test as described in Poirel et al. [40] and Wuchty [51]. Enrichment was determined by calculating the number of interactions that each protein features in the subgraph (a) and those that it does not (c), as well as that it features in a comparison, main graph (b) and those that it does not (d). The probability that a protein is enriched is then determined using the hypergeometric distribution, such that
iRefIndex (version 06062013) was used as a source of generic PPIs to construct a comparison main graph representative of the human interactome. iRefIndex is a large generic molecular interaction database containing interactions that have been sourced from numerous manually curated databases [42]. Using only human proteins from this database, the network contains 14,818 nodes and 167,413 edges, with an average degree of 22.6.
2.4. Gene functional enrichment
To determine enriched GO terms, we used the DAVID functional annotation tool to assign genes with their affiliate terms and to order them by enrichment [21].
2.5. Pain category assignment
To determine which drugs are used to treat pain-associated indications, we manually assigned pain categories to all pharmacologically treatable indications. Indications were assigned to 1 of the 4 categories: (1) “Pain specific” are indications that are specifically associated with pain (eg, neuropathic pain and headaches); (2) “typically painful” are indications that are typically painful, where pain is consistently presented as a symptom of the disorder (eg, endometriosis and arthritis); (3) “Can be painful” are indications that can be painful but can also manifest in a pain-free state (eg, certain cancers and diabetes); and (4) “typically nonpainful” are indications that are typically not associated with pain (eg, alopecia and wrinkling skin), including mental illnesses (eg, schizophrenia and depression). Protein targets of drugs were sourced from an in-house database and were then assigned a pain category using the most pain-related indication.
2.6. Anatomical categorization
To build pain networks specific to the brain, spinal chord, peripheral nervous system (PNS), and immune system, all interactions that had at least 1 mapping to an anatomical term derived from wiki-pain.org data were used. Anatomical terms were then mapped into 1 or more of the 4 anatomical regions or other (see Supplementary file 24 for mappings). Each network was then built for the 4 anatomical regions according to interactions that had an associated anatomical term.
2.7. Microarray analysis
We performed tibial nerve transection (TNT) surgery [18] on adult female rats (n = 8) alongside sham controls (n = 8). Rats were confirmed for tactile allodynia in response to mechanical pressure and both dorsal root ganglia (DRG) and spinal cord were harvested at 7 days after surgery. Gene expression analysis was performed using the Affymetrix Rat 230 2.0 chip. After quality control (QC), data were robust multi-array average (RMA) normalized, and limma was used to identify differentially expressed genes versus sham, which were considered significant if their false discovery rate (FDR) corrected P value was <.05 and their fold change was >1.5. These experiments were approved and monitored by the local ethics committee. The data from this experiment are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-2260.
3. Results
3.1. Literature-derived pain PPI network
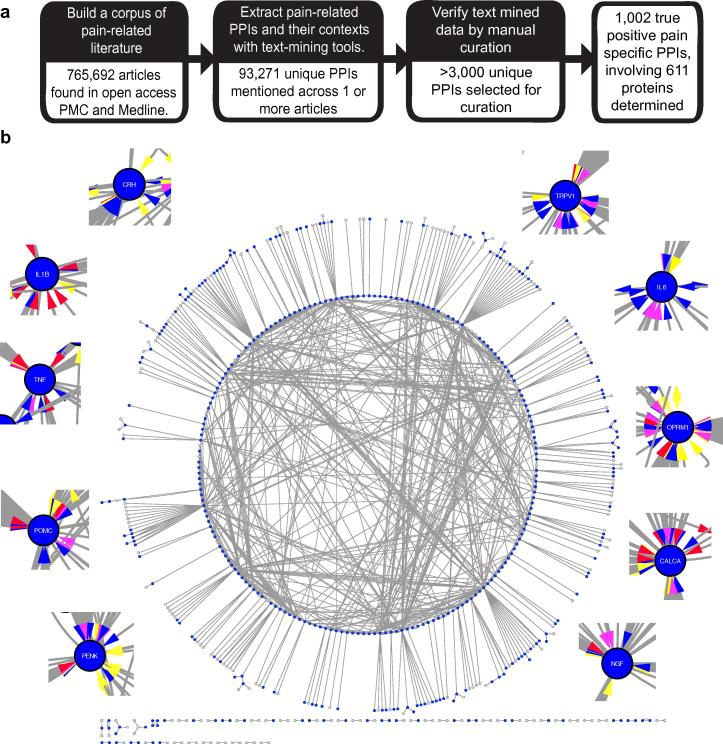
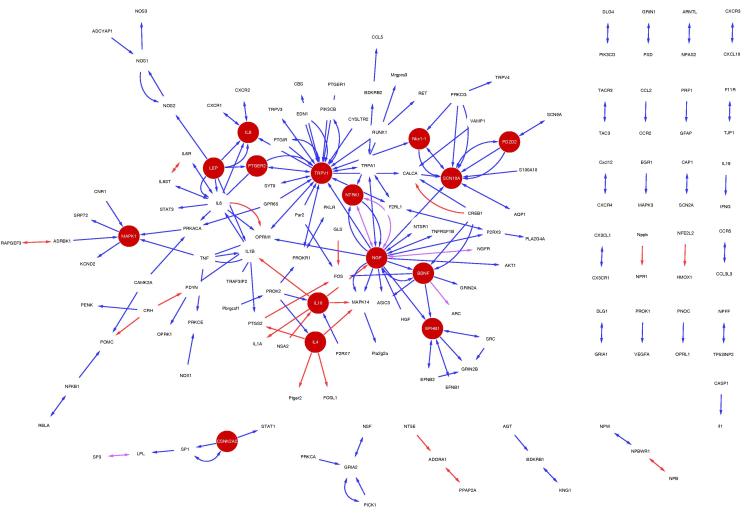
Using a semi-automated text-mining procedure [24], we identified 1002 unique PPIs associated with pain, involving 611 different proteins (Fig. 1a and Supplementary Table 1; see Methods). In total, there are 124 interactions classed as negative regulation, 403 as positive regulation, 180 as regulation (either positive or negative) and 295 as binding. When connected as a network, the PPIs form a highly interconnected and coherent structure, with the largest component containing 481 (79%) of the 611 proteins (Fig. 1b). The network has an average degree of 2.8, a clustering coefficient of 0.07, and a power law fits the node degree distribution with 0.993 correlation indicating that it is scale free, consistent with other molecular interaction networks [1]. The proteins in the network show a statistically significant enrichment for pain associated Gene Ontology (GO) biological processes (eg, response to wounding and inflammatory response), cellular components (eg, neuron projection and postsynaptic membrane), and molecular functions (eg, ion channel activity and neurotrophin binding) (Supplementary Table 2).
Fig. 1.
The pain interaction network. (a) Workflow for creating a pain-specific protein–protein interaction (PPI) network. (b) The PPI network for all pain-associated proteins derived from the curated data. Proteins enriched against iRefIndex (P < .05) are highlighted in blue (3887 total; see Supplementary Table 13). Insets show the top 10 enriched proteins. Colored arrows refer to interaction type: Blue corresponds to positive regulation, red to negative regulation, turquoise to regulation, and yellow to binding (these edges are bi-directional).
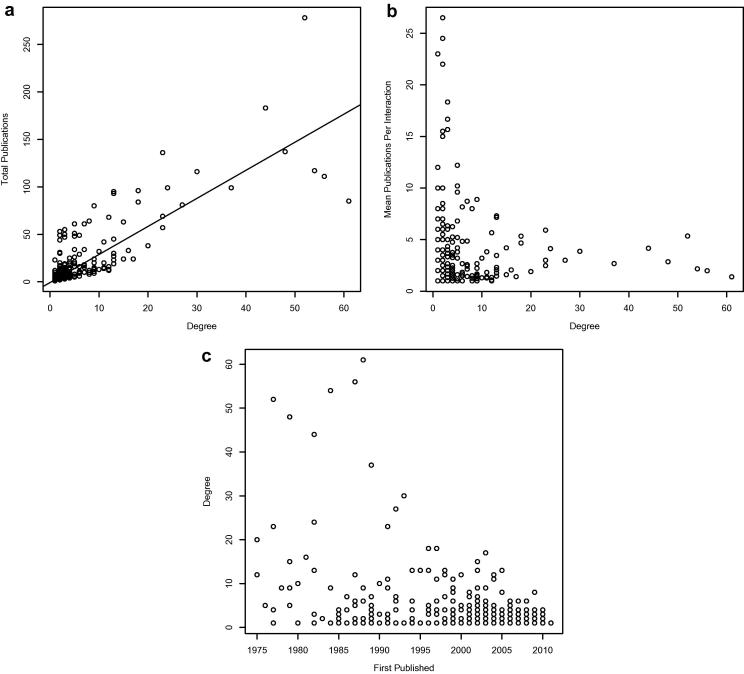
Given that our interaction data are derived from the primary literature, there is potential for ascertainment bias in our network [39]. For example, we will have data only for proteins that have been studied in a pain context, and the most central nodes to our network could be biased by the fact that they have been studied for the longest time. As expected [39], there is a positive linear trend between the degree of a protein and the number of publications describing its interactions (Fig. 2a, Supplementary Figs 1 and 2, and Supplementary Table 3), but there is no significant increase in the average number of documents per interaction observed as degree increases (rho = 0.08, Fig. 2b). However, there is an inverse correlation between the date of a node’s first publication in our dataset and its degree (rho = −0.4; Fig. 2c and Supplementary Table 4).
Fig. 2.
Bias in the pain interaction network. (a) Correlation between the number of publications and degree for nodes in our network showing a linear trend (rho = 0.83). (b) The average number of publications per interaction for a pain protein remains flat (rho = 0.08), suggesting that most interactions are reported individually (see Supplementary Figs. 1 and 2, and Supplementary Table 3). (c) There is an inverse relationship between the date of first publication on a protein’s interactions and the protein’s degree (rho = −0.4) (see Supplementary Table 4).
The first interactions in our network were published as early as 1975, with 25% of interactions published before the year 2000. Those published before 2000 include the 17 highest degree proteins; supporting the assertion that degree correlates with length of study and knowledge of a protein’s perceived importance. We therefore need to be aware of the fact that, within literature-derived networks, the longer and more thoroughly a protein has been studied, the more interactions it is likely reported to have.
3.2. Comparative analyses between alternative pain protein datasets
To investigate the scope and relevance of our text-mined network to pain, we compared it to networks derived from 2 other commonly used sources of disease-associated gene datasets, using generic interaction data from iRefIndex to determine known interactions between the proteins in these datasets (see Methods). It would be preferable to provide comparisons with datasets the interactions of which are derived entirely from pain-specific experiments, but there are no such datasets currently available. First, we generated gene expression data from DRG and spinal cord in the rat TNT model of neuropathic pain (see Methods) to derive a set of pain-associated differentially expressed genes. Second, we used a list of pain-associated proteins from the Pain Genes DB [29] that have been manually curated from the literature. We reasoned that the gene expression data would not be prone to the same biases as literature-associated data (ie, data derived from small-scale experiments), whereas the Pain Genes DB list is curated from the literature but is not dependent on text mining.
From the gene expression experiment, we find 237 genes to be differentially regulated across both DRG and spinal cord tissue; also, there are 399 genes in the Pain Genes DB dataset (Supplementary tables 5 and 6). These are considerably fewer than the 611 proteins in the text-mining derived dataset. Using the generic interactions from iRefIndex to connect proteins in these datasets, it was possible to make only 67 connections between 63 proteins in the gene expression data (Supplementary Fig. 3a and Supplementary Table 7). Therefore, we expanded this dataset to include first-order neighbors with high-betweeness, stipulating that they have interactions with at least 2 of the input genes and so act as bridges to connect the network. As a consequence, 192 (81%) of the differentially expressed genes are included in the network. The resulting networks from Pain Genes DB and gene expression contain, respectively, 272 and 901 nodes, 510 and 12,318 edges, average degrees of 3.75 and 27.34, clustering coefficients of 0.115 and 0.264, and power law correlations of 0.959 and 0.645 (Supplementary Figs. 3b and 4; Supplementary Tables 8 and 9). These networks include relatively few of the proteins from our text-mined network, 125 (20%) and 137 (22%), respectively. We note in particular the high average degree of 27.34 in the gene expression network, whereas the Pain Genes DB and text-mined networks both have similar ratios of 3.75 and 2.8, respectively.
These data indicate that our text-mined network has properties similar to those of a network derived from manual curation. The network derived from gene expression data has a far higher average degree and so presumably contains many more nonspecific/nonrelevant interactions despite the constraints that we placed on introducing new nodes. Indeed, both the gene expression data (without first-order neighbors) and the Pain Genes DB curated data show similar pain-relevant–enriched GO terms to the text-mined proteins (Supplementary Tables 10 and 11). However, when analyzing only those bridge proteins that were added to the gene expression network, there is much lower enrichment of pain-related GO terms in comparison to the original gene expression gene list (Supplementary Table 12), which would suggest that there is considerable noise introduced into this network.
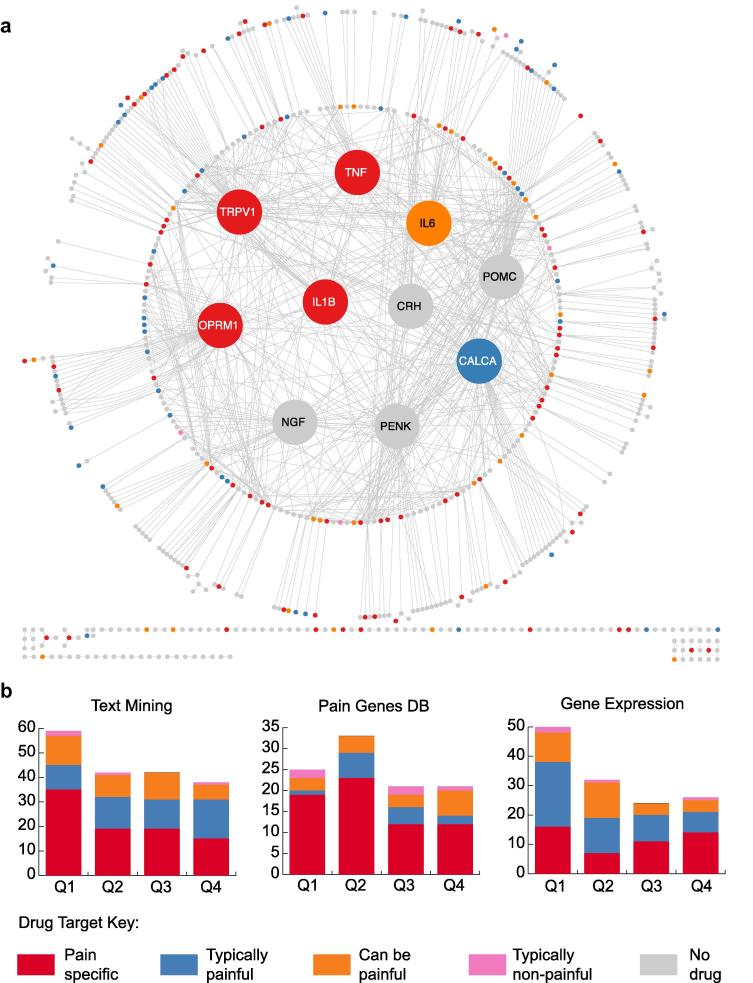
We next cross-referenced nodes in all 3 networks (text-mining derived, gene expression, and Pain Genes DB) with known therapeutic targets of Food and Drug Administration (FDA)–approved drugs (see Methods), taking this as an additional measure of relevance to pain. In the text-mined network, we find 181 targets for existing therapeutics, with 88 targets for anesthetics and pain-specific indications (eg, migraines, neuropathic pain, abdominal pain) and 51 targets for typically painful indications (eg, arthritis, endometriosis) (Fig. 3a and Supplementary Table 13). Examples of pain-specific targets in the text-mined network include OPRM1, the target of analgesics such as morphine [8], and key pro-inflammatory cytokines such as tumor necrosis factor (TNF), which is targeted by numerous drugs for rheumatoid arthritis [35]. The Pain Genes DB and gene expression networks have fewer therapeutic targets than the text-mined network, with 100 and 132, respectively (Supplementary Tables 14 and 15). We also note that the gene expression network contains a much lower proportion of pain-specific targets (36%) in comparison to the Pain Genes DB (66%) and the text-mined network (49%) (Fig. 3b).
Fig. 3.
Drug targets in the pain interaction network. (a) Drug targets are color coded by the contribution of pain to their primary indication (see Methods), as indicated in the key. The 10 most enriched nodes are enlarged and moved into the center for clarity. (b) Drug target profiles of each pain network. Proteins from each dataset are ranked by their enrichment P value and binned into quartiles. Numbers of associated drugs that target proteins in each quartile are then indicated. There is a significant relationship between the enrichment of a node in the text-mined network and the likelihood of it being a drug target for a pain specific indication (χ2 test for trends in proportions, P = .002). However, neither the Pain Genes DB network nor the gene expression data show the same significant trend (P = .05 and P = 0.9, respectively).
There is a significant relationship between the enrichment (see Methods) of a node in the text-mined network and the likelihood of it being a drug target for a painful indication (χ2 test for trends in proportions, P = .002; Fig. 3b), which is not the case for either the Pain Genes DB (P = .05) or gene expression networks (P = .9). We see strong enrichment for targets of drugs currently in development for pain indications, for example, nerve growth factor (NGF; Tanezumab) [5] and genes that have been earmarked as potential therapeutic options, for example, brain derived neurotrophic factor (BDNF) [7], [47]. Moreover, the highly enriched IL6 and SST are currently targets for other indications (diabetes and prostate tumors), and thus their associated drugs may represent promising repurposing opportunities to treat more typically painful and pain-specific indications.
3.3. Insights into the pathology of pain
We next explored the molecular biology of pain apparent from our network. There are a number of proteins in the pain network with a high degree, indicating the importance of these nodes to the structure of the network [25]. As this pain network is a subgraph of the much larger human interactome, we confirmed this connectivity by controlling for proteins that are highly connected in general and thus more likely to appear highly connected in our network. To do this, we developed a method to identify proteins with a significant enrichment of their known interactions within our pain network. We again used iRefIndex as a source of generic interactions to facilitate this [42] (see Methods).
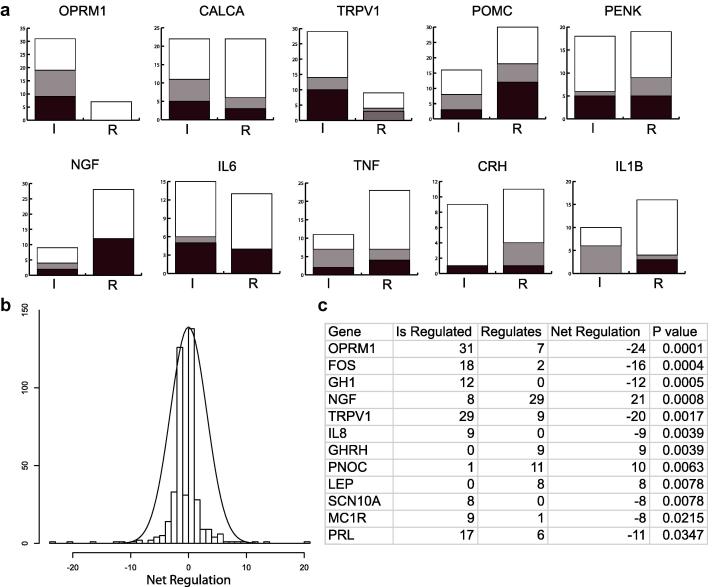
Of the most enriched proteins in the network (Figs. 1b and 4a; Supplementary Table 13), many are key to the pathology of pain, for example OPRM1 [19], TRPV1 [15], [41], and NGF [17], [20]. We find that enriched nodes have multiple regulatory roles, both up and down regulating numerous proteins (Fig. 4a, b, and Supplementary Table 16). There are 8 enriched proteins that are more significantly regulated by others, for example, OPRM1, TRPV1, and FOS, and 4 proteins that more significantly regulate others: NGF, GHRH, PNOC, and LEP (Fig. 4c). This is consistent with the known roles of NGF and nociceptin (PNOC) as mediators of pain signaling [34], [38]. Interestingly, growth hormone (GH), but not growth hormone–releasing hormone (GHRH), has been associated with the chronically painful condition fibromyalgia [30]. Furthermore, recent evidence has suggested a role for leptin (LEP) in the modulation of pain [31], [32]. Our data suggest that both GHRH and leptin might play a more prominent regulatory role in pain than has hitherto been appreciated.
Fig. 4.
Protein regulation in the pain interaction network. (a) The top 10 most enriched genes in the pain network are shown with their regulation profiles broken down by incoming (is regulated, “I”) and outgoing (regulates others, “R”) interactions. Black denotes positive regulation, gray denotes negative regulation, and white denotes other types of interaction. Undirected binding interactions are excluded. (b) The distribution of net regulation for all proteins in the pain network shows a normal distribution with long tails. This indicates that only a few proteins act as master regulators. (c) These master regulators were determined using the exact binomial test (see Supplementary Table 16). The proteins that are significantly more regulated than they are regulators and vice versa are shown; nerve growth factor (NGF) is the most significant net regulator.
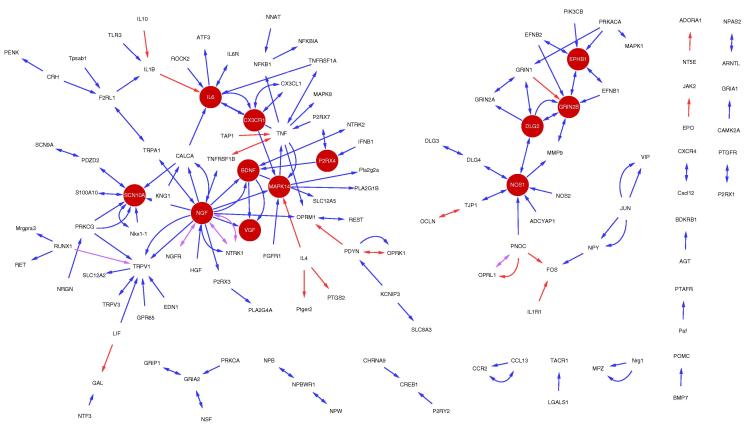
To demonstrate the utility of our contextualized interaction dataset, we chose to investigate inflammatory and neuropathic pain, 2 fundamental aetiologies that manifest chronic pain states [2], [4]. In the construction of these subnetworks, we have also curated the overall effect of the interaction on the outcome of the pain type, that is, an inhibitory or positive effect. There are 144 interactions associated with neuropathic pain in our dataset, with 122 found to be contributory to its pathology, 17 inhibitory, and 5 denoted as both (Fig. 5; Supplementary Table 17). In comparison, 181 interactions are related to inflammatory pain, including 154 contributory interactions, 22 inhibitory interactions, and 5 that have been documented as both (Fig. 6; Supplementary Table 18). In all, 61% of the proteins from inflammatory pain form a coherent core graph, and there are 2 distinct subgraphs in the neuropathic pain network that together account for 73% of the proteins.
Fig. 5.
Protein–protein interactions (PPIs) specific to neuropathic pain. (a) Neuropathic pain specific subnetwork. Blue edges represent those interactions that have been curated as increased in a neuropathic pain state, red edges decreased, and pink edges are those that have been denoted as both. Dark red nodes are those that are enriched against the general pain network (see Supplementary Table 17).
Fig. 6.
Protein–protein interactions (PPIs) specific to inflammatory pain. (a) Inflammatory pain specific subnetwork. Blue edges are those interactions that have been curated as increased in a neuropathic pain state, red edges decreased, and pink edges are those that have been denoted as both. Dark red nodes are those that are enriched against the general pain network (see Supplementary Table 18).
The neuropathic and inflammatory pain networks contain 127 and 157 proteins, respectively, with 80 featuring in both datasets. This large overlap in proteins demonstrates that both forms of pain have underlying core pathology even if the initiation of pain is distinct. This is highlighted by the biological processes that the 80 proteins are highly enriched for, for example, response to wounding (P = 2.88 × 10−14) and sensory perception of pain (P = 6.41 × 10−16) (Supplementary Table 19). However, the proteins unique to the inflammatory and neuropathic pain datasets also reveal the more subtle differences between these disorders. For example, proteins unique to inflammatory pain show a much higher enrichment of inflammatory associated biological processes in comparison to proteins unique to neuropathic pain, such as inflammatory response (P = 5.16 × 10−12 vs P = 3.54 × 10−4) and defense response (P = 1.30 × 10−10 vs P = 6.03 × 10−5) (Supplementary Table 19). The most enriched biological processes unique to the neuropathic pain dataset include regulation of membrane potential (P = 4.41 × 10−5) and regulation of action potential (P = 4.66 × 10−5). Moreover, there are 12 proteins in the intersection between the neuropathic pain dataset and the rat TNT gene expression dataset in comparison to just 3 from the intersection of the inflammatory pain dataset and the gene expression data. An odds ratio test confirms that proteins in the neuropathic pain network are more likely to feature in the gene expression dataset compared to proteins from the inflammatory pain network (odds ratio = 5.36, z = 2.55, P = .01). Given that the gene expression dataset was derived from a neuropathic pain model, this would suggest that our neuropathic pain curated data are indeed more relevant to neuropathic than to inflammatory pain.
To identify the key molecules within the 2 pain types, we repeated our method to reveal enriched proteins against the human interactome and, in addition, against the main pain network. This revealed 116 and 135 proteins for which the majority of their interactions were present in the neuropathic and inflammatory pain subnetworks, respectively, compared to the generic human interactome (Supplementary Tables 20 and 21). There were 12 and 15 proteins enriched in each subnetwork compared to our main pain network. Of these, only NGF, SCN10A (NaV1.8), BDNF, and EPHB1 (ephrin receptor) feature in both the neuropathic and inflammatory pain datasets. The neurotrophic NGF and BDNF [47] as well as ephrin [6] play a key role in neuronal growth and axonal guidance and have been linked to multiple pain aetiologies. There are 9 genes that encode α-subunits of voltage-gated sodium channels, many of which have been linked to multiple types of pain [33]. It is therefore surprising that only NaV1.8 is identified here. This raises an interesting perspective on our data, which does not seek to identify general gene–disease functional associations but rather to uncover which proteins are highly interacting within a diseased state compared to a normal state. Based on this measure, although other ion channels are important to pain, NaV1.8 appears to be the only sodium channel with an interactome that spans multiple pain etiologies.
There are 8 enriched proteins that are specific to the neuropathic pain network and 11 to the inflammatory pain network. Of the proteins specific for neuropathic pain, GRIN2B and NOS1 are already targeted for pain-specific indications. MAPK14 and IL6 are targets for other indications and so might represent drug re-purposing opportunities for neuropathic pain-specific disorders. In addition, DLG2, CX3CR1, P2RX4, and VGF appear to be promising leads for the specific investigation of neuropathic pain [10], [12], [36]. Similarly, IL10, PTGER2, and IL4 are existing targets for inflammatory pain-associated disorders (eg, rheumatoid arthritis), whereas TRPV1 is targeted by analgesics (eg, propofol [14] and capsaicin [26]). LEP, Nkx1-1, PDZD2, NTRK1, IL8, MAPK1, and CSNK2A2 would also appear to be specifically important to inflammatory pain. NTRK1 (TrkA) is the receptor for NGF, which we previously saw to be enriched in both types of pain. That NGF’s receptor is enriched only in the inflammatory pain dataset (although it is present in the neuropathic pain network) emphasizes the need to apply caution when interpreting such data as complete. The curated data, albeit extensive, is not complete, and indeed the body of published work itself does not detail the full pain interactome.
Finally, to illustrate further the possibilities associated with our data, we repeated the same style of analysis but this time create networks for different anatomical regions. From the 1,002 PPIs, we used the anatomy context in wiki-pain.org to determine 607 interactions that could be mapped to 1 or more of the following pain-relevant anatomical associations: brain, spinal cord, peripheral nervous system (PNS), immune system, and other (Supplementary Table 22). We determined 245, 204, 162, and 92 interactions associated with the brain, spinal cord, PNS and immune system, with 211, 190, 152 and 106 proteins in each, respectively (Supplementary Figs. 5–8). We used our enrichment analysis to identify proteins more highly connected in each of the anatomical regions compared to the general pain network (Supplementary Table 23). We find NGF and BDNF to be key to the network in multiple anatomical locations, being enriched in the brain, spinal cord and PNS networks. PENK, OPRL1, and GHRH were specifically enriched in the brain, FOS in the spinal cord, and CALCA, TRPV1, RUNX1, RUNX3, NTRK2, TNFRSF1A, and GDNF were enriched only in the PNS networks. There are also 20 proteins enriched in immune-related anatomical regions, for example, CCL5 and IL8. These data allow us to explore the anatomical interplay that contributes to the development of pain, in particular the interplay between the peripheral and central nervous systems. In addition, this also aids drug development by informing the necessary central or peripheral distribution of a drug candidate.
4. Discussion
We have shown that our large semi-automated text-mining–derived network is relevant to pain and forms a more complete representation of the molecular mechanisms underlying the disease than is possible using other common starting points. We identify several drug repurposing opportunities and use our enrichment method to identify novel mediators of pain. In particular, we show that NaV1.8 is a key ion channel for both neuropathic and inflammatory pain. Furthermore, as we are able to extract specific context with each interaction, we can create and explore networks specific to individual pain indications or anatomical regions. Recent studies have undertaken meta-analyses of gene expression data from pain models [28] or have described resources that enable the network visualization of known pain genes by incorporating PPIs from nondiseased contexts [37]. Our method, using disease-specific interactions identified from the pain-relevant literature, offers a considerable advance in specificity and relevance.
Text mining has long been heralded as the practical solution to efficiently retrieving data denoted in the ever-expanding body of published biomedical literature [43], but poor precision and recall have restricted its wider use in delivering reliable data [52]. Instead, the majority of data derived from free text that is subsequently used in biological analyses is identified and extracted by manual curation, a process that is costly, time consuming, and often unable to offer more exhaustive coverage [23]. As a method of extracting and characterizing key proteins and interactions that are denoted in the literature, our study offers a strong case for a semi-automated approach that uses text mining to rapidly generate the data and manual curation of the results to achieve high precision. Although the protein interaction data that we have retrieved and curated in this study is not complete, the datasets have proved sufficiently broad, accurate, and relevant to make compelling biological findings.
The results in this study represent the most extensive summary of all of the published research conducted on pain-associated proteins. The power of such an approach comes from integrating the data at the network level, which allows novel hypotheses to be drawn in the context of the global picture. Furthermore, the network can be used as a framework to provide context to the interpretation of datasets generated by researchers within the field. This is increasingly recognized as a successful approach to the study of disease biology [22]. It is foreseeable, therefore, that a similar approach to data retrieval and analysis could be applied to a huge range of biomedical disorders under various different contexts, to provide networks and targets for further study.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgements
D.J. is funded by a BBSRC CASE studentship to G.N. and D.L.R. with industrial partner B.S. We dedicate this paper to the memories of our colleagues Dr Phoebe M Roberts and Michael Kennedy. Phoebe contributed to the text mining of the pain data and sadly died on December 8, 2013. Co-author Michael passed away on February 7, 2014.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.pain.2014.06.020.
Appendix A. Supplementary data
References
- 1.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 2.Basbaum A.I., Bautista D.M., Scherrer G., Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breivik H., Collett B., Ventafridda V., Cohen R., Gallacher D. Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur J Pain. 2006;10:287–333. doi: 10.1016/j.ejpain.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Calvo M., Dawes J.M., Bennett D.L. The role of the immune system in the generation of neuropathic pain. Lancet Neurol. 2012;11:629–642. doi: 10.1016/S1474-4422(12)70134-5. [DOI] [PubMed] [Google Scholar]
- 5.Cattaneo A. Tanezumab, a recombinant humanized mAb against nerve growth factor for the treatment of acute and chronic pain. Curr Opin Mol Ther. 2010;12:94–106. [PubMed] [Google Scholar]
- 6.Cibert-Goton V., Yuan G., Battaglia A., Fredriksson S., Henkemeyer M., Sears T., Gavazzi I. Involvement of EphB1 receptors signalling in models of inflammatory and neuropathic pain. PLoS One. 2013;8:e53673. doi: 10.1371/journal.pone.0053673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coull J.A., Beggs S., Boudreau D., Boivin D., Tsuda M., Inoue K., Gravel C., Salter M.W., De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- 8.Crist RC, Berrettini WH. Pharmacogenetics of OPRM1. Pharmacol Biochem Behav 2013, in press. http://www.sciencedirect.com/science/article/pii/S0091305713002645. [DOI] [PMC free article] [PubMed]
- 9.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal 2006; Complex Systems: 1695. http://cran.r-project.org/web/packages/igraph/citation.html.
- 10.D’Haese J.G., Friess H., Ceyhan G.O. Therapeutic potential of the chemokine-receptor duo fractalkine/CX3CR1: an update. Expert Opin Ther Targets. 2012;16:613–618. doi: 10.1517/14728222.2012.682574. [DOI] [PubMed] [Google Scholar]
- 11.Elzahaf R.A., Tashani O.A., Unsworth B.A., Johnson M.I. The prevalence of chronic pain with an analysis of countries with a Human Development Index less than 0.9: a systematic review without meta-analysis. Curr Med Res Opin. 2012;28:1221–1229. doi: 10.1185/03007995.2012.703132. [DOI] [PubMed] [Google Scholar]
- 12.Ferrini F., Trang T., Mattioli T.A., Laffray S., Del’Guidice T., Lorenzo L.E., Castonguay A., Doyon N., Zhang W., Godin A.G., Mohr D., Beggs S., Vandal K., Beaulieu J.M., Cahill C.M., Salter M.W., De Koninck Y. Morphine hyperalgesia gated through microglia-mediated disruption of neuronal Cl(-) homeostasis. Nat Neurosci. 2013;16:183–192. doi: 10.1038/nn.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finnerup N.B., Sindrup S.H., Jensen T.S. The evidence for pharmacological treatment of neuropathic pain. PAIN®. 2010;150:573–581. doi: 10.1016/j.pain.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Fischer M.J., Leffler A., Niedermirtl F., Kistner K., Eberhardt M., Reeh P.W., Nau C. The general anesthetic propofol excites nociceptors by activating TRPV1 and TRPA1 rather than GABAA receptors. J Biol Chem. 2010;285:34781–34792. doi: 10.1074/jbc.M110.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunthorpe M.J., Chizh B.A. Clinical development of TRPV1 antagonists: targeting a pivotal point in the pain pathway. Drug Discov Today. 2009;14:56–67. doi: 10.1016/j.drudis.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Gureje O., Von Korff M., Simon G.E., Gater R. Persistent pain and well-being: a World Health Organization Study in Primary Care. JAMA. 1998;280:147–151. doi: 10.1001/jama.280.2.147. [DOI] [PubMed] [Google Scholar]
- 17.Hefti F.F., Rosenthal A., Walicke P.A., Wyatt S., Vergara G., Shelton D.L., Davies A.M. Novel class of pain drugs based on antagonism of NGF. Trends Pharmacol Sci. 2006;27:85–91. doi: 10.1016/j.tips.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann H.A., De Vry J., Siegling A., Spreyer P., Denzer D. Pharmacological sensitivity and gene expression analysis of the tibial nerve injury model of neuropathic pain. Eur J Pharmacol. 2003;470:17–25. doi: 10.1016/s0014-2999(03)01753-9. [DOI] [PubMed] [Google Scholar]
- 19.Holliday K.L., Nicholl B.I., Macfarlane G.J., Thomson W., Davies K.A., McBeth J. Do genetic predictors of pain sensitivity associate with persistent widespread pain? Mol Pain. 2009;5:56. doi: 10.1186/1744-8069-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes D. Anti-NGF painkillers back on track? Nat Rev Drug Discov. 2012;11:337–338. doi: 10.1038/nrd3732. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Jacunski A., Tatonetti N.P. Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Ther. 2013;94:659–669. doi: 10.1038/clpt.2013.168. [DOI] [PubMed] [Google Scholar]
- 23.Jamieson DG, Gerner M, Sarafraz F, Nenadic G, Robertson DL. Towards semi-automated curation: Using text mining to recreate the HIV-1, human protein interaction database. Database (Oxford) 2012;2012:bas023. [DOI] [PMC free article] [PubMed]
- 24.Jamieson DG, Roberts PM, Robertson DL, Sidders B, Nenadic G. Cataloging the biomedical world of pain through semi-automated curation of molecular interactions. Database (Oxford) 2013;2013:bat033. [DOI] [PMC free article] [PubMed]
- 25.Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 26.Knotkova H., Pappagallo M., Szallasi A. Capsaicin (TRPV1 agonist) therapy for pain relief: farewell or revival? Clin J Pain. 2008;24:142–154. doi: 10.1097/AJP.0b013e318158ed9e. [DOI] [PubMed] [Google Scholar]
- 27.Kroenke K., Krebs E.E., Bair M.J. Pharmacotherapy of chronic pain: a synthesis of recommendations from systematic reviews. Gen Hosp Psychiatry. 2009;31:206–219. doi: 10.1016/j.genhosppsych.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 28.LaCroix-Fralish M.L., Austin J.S., Zheng F.Y., Levitin D.J., Mogil J.S. Patterns of pain: meta-analysis of microarray studies of pain. PAIN®. 2011;152:1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Lacroix-Fralish M.L., Ledoux J.B., Mogil J.S. The Pain Genes Database: an interactive Web browser of pain-related transgenic knockout studies. PAIN®. 2007;131:e1–e4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 30.Leal-Cerro A., Povedano J., Astorga R., Gonzalez M., Silva H., Garcia-Pesquera F., Casanueva F.F., Dieguez C. The growth hormone (GH)-releasing hormone-GH-insulin-like growth factor-1 axis in patients with fibromyalgia syndrome. J Clin Endocrinol Metab. 1999;84:3378–3381. doi: 10.1210/jcem.84.9.5982. [DOI] [PubMed] [Google Scholar]
- 31.Li X., Kang L., Li G., Zeng H., Zhang L., Ling X., Dong H., Liang S., Chen H. Intrathecal leptin inhibits expression of the P2X2/3 receptors and alleviates neuropathic pain induced by chronic constriction sciatic nerve injury. Mol Pain. 2013;9:65. doi: 10.1186/1744-8069-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim G., Wang S., Zhang Y., Tian Y., Mao J. Spinal leptin contributes to the pathogenesis of neuropathic pain in rodents. J Clin Invest. 2009;119:295–304. doi: 10.1172/JCI36785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M., Wood J.N. The roles of sodium channels in nociception: implications for mechanisms of neuropathic pain. Pain Med. 2011;12:S93–S99. doi: 10.1111/j.1526-4637.2011.01158.x. [DOI] [PubMed] [Google Scholar]
- 34.Mika J., Obara I., Przewlocka B. The role of nociceptin and dynorphin in chronic pain: implications of neuro-glial interaction. Neuropeptides. 2011;45:247–261. doi: 10.1016/j.npep.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Miyasaka N. Etanercept for therapy of rheumatoid arthritis. Nihon Rinsho. 2005;63:521–525. [PubMed] [Google Scholar]
- 36.Moss A., Ingram R., Koch S., Theodorou A., Low L., Baccei M., Hathway G.J., Costigan M., Salton S.R., Fitzgerald M. Origins, actions and dynamic expression patterns of the neuropeptide VGF in rat peripheral and central sensory neurones following peripheral nerve injury. Mol Pain. 2008;4:62. doi: 10.1186/1744-8069-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins J.R., Lees J., Antunes-Martins A., Diboun I., McMahon S.B., Bennett D.L., Orengo C. PainNetworks: a Web-based resource for the visualisation of pain-related genes in the context of their network associations. PAIN®. 2013;154:e1–e12. doi: 10.1016/j.pain.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pezet S., McMahon S.B. Neurotrophins: mediators and modulators of pain. Annu Rev Neurosci. 2006;29:507–538. doi: 10.1146/annurev.neuro.29.051605.112929. [DOI] [PubMed] [Google Scholar]
- 39.Pfeiffer T., Hoffmann R. Temporal patterns of genes in scientific publications. Proc Natl Acad Sci U S A. 2007;104:12052–12056. doi: 10.1073/pnas.0701315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirel C.L., Owens C.C., 3rd, Murali T.M. Network-based functional enrichment. BMC Bioinformatics. 2011;12:S14. doi: 10.1186/1471-2105-12-S13-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomonis J.D., Harrison J.E., Mark L., Bristol D.R., Valenzano K.J., Walker K. N-(4-Tertiarybutylphenyl)-4-(3-cholorphyridin-2-yl)tetrahydropyrazine-1(2H)-carbox-amide (BCTC), a novel, orally effective vanilloid receptor 1 antagonist with analgesic properties: II. In vivo characterization in rat models of inflammatory and neuropathic pain. J Pharmacol Exp Ther. 2003;306:387–393. doi: 10.1124/jpet.102.046268. [DOI] [PubMed] [Google Scholar]
- 42.Razick S., Magklaras G., Donaldson I.M. IRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebholz-Schuhmann D., Oellrich A., Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat Rev Genet. 2012;13:829–839. doi: 10.1038/nrg3337. [DOI] [PubMed] [Google Scholar]
- 44.Rotthier A., Baets J., Timmerman V., Janssens K. Mechanisms of disease in hereditary sensory and autonomic neuropathies. Nat Rev Neurol. 2012;8:73–85. doi: 10.1038/nrneurol.2011.227. [DOI] [PubMed] [Google Scholar]
- 45.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson A., Graham M.E., Williams J. Chronic plantar ulcer secondary to congenital indifference to pain. J Wound Care. 2011;20:542. doi: 10.12968/jowc.2011.20.11.540. [DOI] [PubMed] [Google Scholar]
- 47.Siniscalco D., Giordano C., Rossi F., Maione S., de Novellis V. Role of neurotrophins in neuropathic pain. Curr Neuropharmacol. 2011;9:523–529. doi: 10.2174/157015911798376208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son S.J., Lee K.M., Jeon S.M., Park E.S., Park K.M., Cho H.J. Activation of transcription factor c-jun in dorsal root ganglia induces VIP and NPY upregulation and contributes to the pathogenesis of neuropathic pain. Exp Neurol. 2007;204:467–472. doi: 10.1016/j.expneurol.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 49.Turk D.C., Wilson H.D., Cahana A. Treatment of chronic non-cancer pain. Lancet. 2011;377:2226–2235. doi: 10.1016/S0140-6736(11)60402-9. [DOI] [PubMed] [Google Scholar]
- 50.Woolf C.J. What is this thing called pain? J Clin Invest. 2010;120:3742–3744. doi: 10.1172/JCI45178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wuchty S. Controllability in protein interaction networks. Proc Natl Acad Sci U S A. 2014;111:7156–7160. doi: 10.1073/pnas.1311231111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zweigenbaum P., Demner-Fushman D., Yu H., Cohen K.B. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8:358–375. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available in Supplementary Tables 1 to 24.