Highlights
-
•
Lipophagy is a transcriptionally regulated process.
-
•
The lysosome as a sensor of lipophagy induction.
-
•
Nuclear receptors link lipophagy to lipid catabolism.
Keywords: Autophagy, lipophagy, lysosome, transcription factors, nuclear receptors, TP53, FOXOs, TFEB, mTORC1
Abstract
Autophagy is a catabolic pathway that has a fundamental role in the adaptation to fasting and primarily relies on the activity of the endolysosomal system, to which the autophagosome targets substrates for degradation. Recent studies have revealed that the lysosomal–autophagic pathway plays an important part in the early steps of lipid degradation. In this review, we discuss the transcriptional mechanisms underlying co-regulation between lysosome, autophagy, and other steps of lipid catabolism, including the activity of nutrient-sensitive transcription factors (TFs) and of members of the nuclear receptor family. In addition, we discuss how the lysosome acts as a metabolic sensor and orchestrates the transcriptional response to fasting.
Overview of lipid catabolism pathway during fasting
Nutrient deprivation is one of the harshest conditions that an organism may face. To sustain cellular metabolism, lipids, proteins, and carbohydrates must be degraded to give rise to fatty acids (FA), amino acids (AA), and pyruvate, respectively (see Glossary). These substrates can in turn generate ATP to sustain energy [1]. Specifically, lipids such as triglycerides (TGs) can be broken down through various mechanisms including lipolysis and autophagy during times of nutrient deprivation.
TGs are the predominant form of fat in the human body and are composed of three chains of FA linked to one molecule of glycerol. TGs are normally stored in specialized organelles named lipid droplets (LDs), which also contain esterified cholesterol. LDs originate from the membrane of the endoplasmic reticulum and are delimited by a monolayer of phospholipids and coated with structural proteins such as perilipins, caveolins, and several members of the Rab family of small GTPases. LDs are the main cytosolic component of adipocytes and are found in virtually every cell type, such as hepatocytes, neurons, glia, macrophages, dendritic cells, and lymphoblasts 2, 3. LDs were traditionally thought to be hydrolyzed by three lipases: LD-associated adipose triglyceride lipase (ATGL); the cytosolic hormone-sensitive lipase (HSL); and monoacylglycerol lipase (MGL), which are all regulated by nutrients and hormonal levels [4]. However, in most non-adipose tissues these lipases exhibit low expression levels, raising the question as to whether other lipases are required for efficient lipolysis.
Nutrient deprivation triggers the breakdown of TGs, which are contained in LDs, into glycerol and FA. In adipose tissue, these intermediates are secreted into the bloodstream, whereas in non-adipose tissues they are oxidized in mitochondria and peroxisomes via the ß-oxidation pathway, which splits long carbon chains of FA into molecules of acetyl-CoA, an important substrate for the tricarboxylic acid (TCA) cycle 2, 4. Several proteins facilitate the cellular import of FA, including the plasma membrane FA translocase CD36. Once in the cytosol, FA are esterified to CoA and then conjugated to carnitine by carnitine palmitoyl transferase I and II (CPTI and CPTII) and carnitine acyltransferase enzymes. These modifications enable FA to move across the inner mitochondrial membrane. The import of FA into mitochondria is considered a rate-limiting step of the ß-oxidation pathway. Once inside the mitochondria a molecule of FA is degraded through cyclic reactions that shorten their chain length by two carbons per cycle, generating FADH2 and NADH molecules. This process is mainly mediated by acyl-CoA synthetase, long- and medium-chain acyl-CoA dehydrogenases, and 3-ketoacyl-CoA thiolase. ß-oxidation of very long FA chains may also occur within peroxisomes.
The expression and activity of key metabolic enzymes and transport proteins involved in lipid catabolism are tightly regulated by transcriptional and post-transcriptional mechanisms, which are mediated by endocrine, paracrine, and autocrine signals. The transcriptional regulation of lipid catabolism in liver and skeletal muscle is mainly controlled by the activities of the nuclear receptor peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARα) and of its co-activator PGC1α, which are significantly induced during fasting [5]. PPARα regulates intracellular lipid metabolism through direct transcriptional control of many genes involved in peroxisomal and mitochondrial β-oxidation pathways, FA uptake, and TG catabolism including CD36, CPT-1, and HMG-CoA synthase, which converts acetyl-CoA units into ketone bodies [84]. Emerging evidence suggests a link between these lipid catabolism pathways and nutrient deprivation, which is reviewed in further detail below.
Lipophagy: a vesicular way to degrade fat
In recent years, the intracellular process known as macroautophagy (hereafter autophagy) has emerged as a critical regulator of energy metabolism during fasting [6] (Figure 1). Autophagy is an evolutionarily conserved catabolic process involved in the first steps of the degradation of intracellular components, such as proteins and glycogen, which yield AA and glucose, respectively (Figure 1) [7]. Consequently, autophagy sustains energy production when extracellular nutrients are not available. A brief overview of the autophagic pathway is described in Box 1.
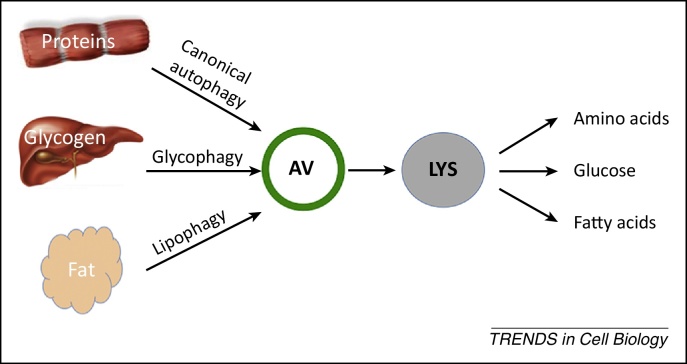
Figure 1.
Autophagy mediates substrate catabolism during fasting. During nutrient deprivation proteins, glycogen and fat are sequestered by autophagosomes and targeted to lysosomes where they are degraded by resident hydrolases and transformed into amino acids, fatty acids, and glucose, which are then released into the cytoplasm to support cellular energetic demands. Abbreviations: AV, autophagosome; LYS, lysosome.
Box 1. Overview of the autophagy pathway.
The autophagy pathway relies on the activities of two organelles: the autophagosome (AV) and the lysosome. AVs are involved in intracellular cargo sequestration and delivery to the lysosomes, initiating the degradation steps. AVs are double-lipid-bilayer membrane vesicles that originate from several membrane sources including the endoplasmic reticulum (ER), mitochondria, Golgi, and plasma membrane [75]. The biogenesis of the isolation membrane occurs mainly at the ER–mitochondria contact sites [76]. During membrane expansion, substrates are sequestered through receptor-mediated mechanisms before membrane sealing. Mature AVs fuse with lysosomes, forming autophagolysosomes, hybrid vesicles in which the degradation of the autophagic cargo occurs. Lysosomes are single-lipid-bilayer membrane organelles with an acidic lumen containing soluble hydrolytic enzymes, which are the executors of the catabolic processes. They include members of protein families such as sulfatases, glycosidases, peptidases, phosphatases, lipases, and nucleases that allow the lysosome to hydrolyze a vast repertoire of biological substrates, including proteins, glycosaminoglycans, lipids, glycogen, and nucleic acids [77]. Fasting leads to an increase in the number and size of AVs and lysosomes, suggesting that the cell has evolved a mechanism to co-regulate biogenesis and function of these two organelles to respond to fasting [27].
In the past few years great progress has been made to understand the molecular mechanisms governing autophagy [78]. Briefly, a core set of essential autophagy proteins (ATG) and several accessory proteins, such as soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors (SNAREs), vacuolar protein sorting-associated protein (VPSs), and RABs, are required for AV biogenesis. Their physiological activities are connected to the energy status of the cell by nutrient-sensitive proteins. For example, the mechanistic target of rapamycin complex 1 (mTORC1) kinase and the AMP-activated protein kinase (AMPK), which sense the cellular levels of nutrients, inhibit or activate autophagy, respectively, through phosphorylation of the mammalian homolog of yeast adipose triglyceride (ATG)1 [Unc-51 like autophagy activating kinase 1 (ULK1)] kinase 79, 80, 81. ULK1, in turn, controls the activity of the protein complexes composed of ATG13, FIP200, and ATG101 and of the Beclin1/class III phosphatidylinositide 3-kinase (PI3K) VPS34, which produces the phosphatidylinositol 3-phosphate (PI3P) necessary for AV membrane formation [82]. Another example is the fasting-induced activation of the c-Jun N-terminal kinase (JNK)/mitogen-activated protein (MAP) kinase, which phosphorylates B cell leukemia factor 2 (Bcl-2) and triggers its dissociation from Beclin1, which activates autophagy [83].
Autophagy has recently been shown to be involved in lipid catabolism and has been termed lipophagy to refer to the specific degradation of lipids by autophagy [8] (Figure 1). Indeed, in cells lacking an essential autophagy gene, LDs accumulate in the cytoplasm due to defective catabolism [8], suggesting that it takes a functional autophagic pathway to break them down. Studies performed in cultured hepatocytes showed that autophagosomes sequester portions of LDs and target them to lysosomes for degradation. This process may also occur in vivo, because it was observed that cells of patients affected by liver steatosis contain lipid-filled lysosomes [9]. In liver, lipophagy participates in constitutive lipid degradation. However, its maximal contribution appears to be during fasting, when the liver imports an increased amount of lipids derived from peripheral lipolysis [8]. Mice deficient in several key autophagy genes such as ATG5, ATG7, ATG14 and VPS34 show elevated TG levels in the liver 8, 10, 11, 12. In addition, several studies have demonstrated that lipophagy also occurs in other tissues and cell types, including fibroblasts, neurons, stellate cells, and macrophages 13, 14.
Furthermore, it has been suggested that intracellular accumulation of LDs may promote autophagy, because LDs provide lipid precursors for the nascent autophagosome membrane. Mechanistically, the LD membrane-associated patatin-like phospholipase domain-containing proteins (PNPLAs) mobilize TGs to form diacylglycerol (DAG), which is used to build phospholipids necessary for autophagosomal membrane formation [15].
In mammals, lysosomes contain a single lysosomal acid lipase (LAL) [16], which is mainly active at an acidic pH and mediates lysosomal degradation of TGs, endocytosed lipoproteins, and membrane lipids. LAL deficiency causes Wolman disease (known as cholesteryl ester storage disease in its milder form), a severe lysosomal storage disorder (LSD) characterized by the storage of lipids in various organs, including liver and brain 17, 18. During lipophagy, LAL mediates the hydrolysis of cholesterol esters, which are delivered from LDs to lysosomes via autophagy 19, 20. In addition, Caenorhabditis elegans strains lacking the ce-lysosomal lipases LIPL-1 and LIPL-3 (for which only one lipase exists in mammals) accumulate LDs as a result of their defective degradation, further suggesting that lysosomal lipases have an important role in fat degradation [21].
The way in which lipophagy is regulated and connected to other lipid catabolic pathways like the intracellular uptake of lipid from the bloodstream and ß-oxidation in mitochondria is still unclear. These processes must be co-regulated to optimize lipid usage during fasting and prevent the lipotoxicity associated with the accumulation of lipids in the cytosol.
In this review, we discuss the evidence suggesting that lipophagy is a transcriptionally regulated process and propose that a gene network links lipophagy to other pathways of lipid catabolism. In addition, we hypothesize that regulation of lipophagy during fasting occurs through a signaling pathway triggered by the lysosome. This pathway requires the coordinated activities of the mechanistic target of rapamycin complex 1 (mTORC1), of nutrient-sensitive transcription factors (TFs), and of members of the nuclear receptor family.
A transcriptional hypothesis for the regulation of lipophagy
Until recently, autophagy regulation was thought to occur exclusively in the cytosol. This dogma was set forth after enucleated cells showed accumulation of LC3 puncta in response to an autophagic stimulus [22]. However, recent reports have challenged this view and have provided evidence for transcriptional and epigenetic control of autophagy. The list of TFs implicated in the regulation of autophagy has continued to grow [23]. Furthermore, microRNAs and epigenetic mechanisms have added an additional layer of complexity to autophagy regulation [24].
Transcriptional regulation coordinates complex metabolic processes, such as the multistep pathways that occur within cellular organelles [25]. In the nematode C. elegans the transcriptional regulation of autophagy occurs concomitantly with transcriptional induction of lipolysis [26], suggesting that the two processes can be co-regulated by the activity of TFs. In mammals, transcription factor EB (TFEB), tumor suppressor protein 53 (p53), and members of the forkhead box protein class O (FOXOs) are emerging as master transcriptional regulators of autophagy [23]. Importantly, these TFs are regulated by nutrients and growth factor levels and, in turn, regulate the expression levels of genes involved in energy metabolism. This regulation occurs either directly or via cooperation with members of the nuclear receptor and co-receptor families, which are master regulators of energy metabolism and lipid catabolism. These observations suggest that these TFs regulate lipophagy and coordinate it with other pathways of lipid catabolism.
TFEB links lipophagy to β-oxidation via nuclear receptors
Among the TFs involved in autophagy, TFEB appears to exert the most global control over autophagy [27]. Indeed, it regulates multiple steps of autophagy, such as autophagosome biogenesis, substrate targeting, and lysosome degradation, by managing expression levels of several autophagy and lysosomal genes [28].
TFEB belongs to the microphthalmia-associated transcription factor (MITF) subfamily of basic helix-loop-helix (bHLH) TFs [29]. The gene network controlled by TFEB was named coordinated lysosomal expression and regulation (CLEAR) [30] and includes several genes with a known role in autophagy and lysosome biogenesis 27, 28. LAL is among TFEB targets, suggesting that TFEB activation may promote lysosomal degradation of lipids [28]. Functional studies indicate that TFEB induces de novo biogenesis of autophagosomes and lysosomes, enhances their fusion, and increases the degradation of substrates. In addition, TFEB enhances other lysosomal functions such as lysosomal exocytosis 31, 32.
Under normal feeding conditions TFEB is retained in the cytosol, whereas during fasting it translocates to the nucleus and becomes active [27]. mTORC1, a key regulator of TFEB, phosphorylates TFEB at conserved serine residues, preventing its nuclear translocation. Nutrient deprivation inhibits mTORC1, thus promoting TFEB nuclear translocation and transcriptional activation of the CLEAR gene network 33, 34, 35.
The transcriptional changes induced by TFEB overexpression in the liver are similar to those observed after fasting, suggesting that TFEB mediates the fasting response [36]. In particular, TFEB regulates the expression of genes involved in several steps of lipid degradation, such as the intracellular import of FA across the plasma membrane [e.g., CD36 and fatty acid binding proteins (FABPs)] and the β-oxidation of FA in mitochondria [e.g., Cpt1, carnitine acetyltransferase (Crat), acyl-CoA dehydrogenase, long chain (Acadl), acyl-CoA dehydrogenase, C-2 To C-3 short chain (Acads)] and in peroxisomes (CYP4a) [36]. Furthermore, TFEB controls the expression of PGC1α and PPARα genes, which are known master regulators of lipid catabolism. Indeed, during fasting nuclear TFEB binds to the promoters of PGC1α [36] and PPARα (Settembre, C. and Ballabio, A., unpublished) and increases their expression levels. Conversely, mice lacking TFEB in the liver show a defective fasting response probably because they lack PGC1α upregulation. Genetic interaction studies have revealed that TFEB activity depends on the presence of PGC1α and PPARα [36], hence suggesting that these nuclear receptors are involved in the regulation of autophagy and lysosomal biogenesis. Consistently, overexpression of PGC1α in muscle increases the number of autophagosomes and lysosomes [37]. Furthermore, TFEB overexpression in the liver of mice lacking the essential autophagy gene ATG7 fails to induce lipid catabolism and results in the accumulation of LDs in the cytosol of hepatocytes [36], indicating that TFEB requires a functional autophagic pathway to have an effect on lipid catabolism. This finding clearly indicates that lipid catabolism requires functional autophagy.
TFEB controls lipophagy in vivo
The phenotypic consequences of TFEB overexpression in mice are striking. Mice overexpressing TFEB in the liver are leaner and show enhanced whole-body energy metabolism and FA catabolism. Cholesterol levels were not significantly altered in TFEB-overexpressing mice and consequently the role of TFEB in the regulation of cholesterol metabolism was not investigated. Mice overexpressing TFEB in liver are protected from diet-induced obesity and metabolic syndrome. Conversely, mice lacking TFEB in the liver display accumulation of LDs in the cytoplasm of hepatocytes, which is consistent with defective lipophagy, and defective peripheral fat mobilization after fasting [36]. Peripheral organs are also affected probably due to enhanced transcription of liver-derived hormones regulating lipid catabolism, such as fibroblast growth factor (FGF)21. FGF21 is directly induced by PPARα in the liver in response to fasting. In addition, it promotes weight loss, lowers blood levels of glucose and triglycerides, increases insulin sensitivity, and stimulates lipolysis in white adipose tissue [38]. FGF21 has been recently linked to autophagy [39]. Inhibiting autophagy in skeletal muscle was proposed to ameliorate whole-body energy metabolism through FGF21 upregulation. As a consequence of defective autophagy, mitochondrial dysfunction activates the TF ATF4, which in turn enhances FGF21 levels [39]. Although these data are difficult to reconcile with those demonstrating that autophagy maintains a positive role in lipid metabolism, they suggest that transcriptional crosstalk exists between autophagy, lipid metabolism, and nuclear receptors.
In addition, recent studies performed in C. elegans suggest TFEB may have a role in longevity. Indeed, a C. elegans mutant strain lacking the TFEB orthologous gene (HLH30) showed that HLH30 mutant animals fail to degrade LDs during starvation and have decreased lifespan 21, 40. In the worm HLH30 promotes lysosomal biogenesis and autophagy and mediates lysosomal lipolysis through the upregulation of the lysosomal lipases LIPL-1 and LIPL-3. When nutrients are available, transcription of LIPL-1 and LIPL-3 is repressed by the transcriptional repressor MLX-3. During fasting MAX-like protein X (MLX-3) is rapidly degraded, and HLH30 is activated [21]. Consistent with lipid metabolism and autophagy as determinants of C. elegans longevity [41], HLH30 overexpression extends the lifespan of C. elegans [40]. Collectively, these data suggest that TFEB coordinates lipophagy with other lipid degradation pathways in response to starvation (Figure 2).
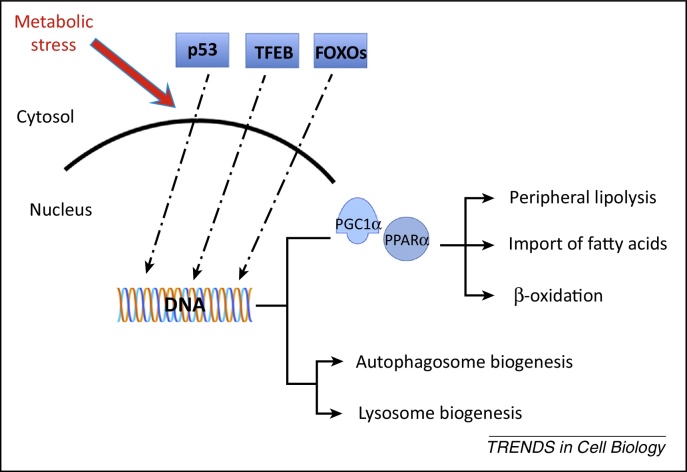
Figure 2.
Proposed model of coordinated transcriptional regulation of lipid catabolism via forkhead box protein class O (FOXO), transcription factor EB (TFEB), p53, and nuclear receptors. The nuclear translocation of TFEB, p53, and FOXOs is induced under conditions of metabolic stress such as nutrient depletion and growth factor deprivation. These transcription factors (TFs) regulate directly the expression of autophagy genes and the nuclear receptor and co-receptor peroxisome proliferator-activated receptor (PPAR)α and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α), respectively, which control lipid catabolism.
Unconventional role of p53 in autophagy and lipid metabolism
p53 is a tumor suppressor protein that is activated in response to several types of cellular stress. Activated p53 can induce cell cycle arrest, DNA repair, senescence, and apoptosis [42]. In the past few years, it has become evident that activated p53 induces lipid catabolism [43]. Indeed, p53 directly controls the expression levels of genes involved in FA β-oxidation (CYP4F) and transport (CPT1A, CROT) [44]. In addition, p53 regulates LIPIN1 protein, which exerts a profound effect on lipid metabolism. LIPIN1 is a phosphatidic acid phosphatase that converts phosphatidic acid to diacylglycerol. In the nucleus, LIPIN1 functions as a transcriptional co-activator and induces PPARα expression [45]. In addition, it represses the transcriptional activity of sterol regulatory element-binding proteins (SREBPs), the master TFs controlling TG and cholesterol biosynthesis, thus inhibiting lipid biosynthesis [46]. During glucose deprivation, p53 also promotes PGC1A1α activity [47], suggesting that in the absence of glucose p53 may induce a metabolic shift toward lipid usage.
p53 is a well-defined transcriptional regulator of autophagy. Originally, it was found to induce autophagy via the lysosomal protein DNA-damage regulated autophagy modulator 1 (DRAM1) [48]. More recently, global genomic profiling has revealed that p53 directly regulates the expression of several autophagy genes such as ATG7, ATG4, ULK1 and UVRAG [49]. Furthermore, p53 controls the transcriptional levels of tuberous sclerosis complex and phosphatase and tensin homolog (PTEN) [50], which regulate autophagy indirectly via mTORC1.
These observations suggest that p53 may be involved in lipophagy, by simultaneously inducing autophagy and FA oxidation. Human Chang liver cells challenged with oleic acid (OA), a monounsaturated FA, display increased levels of LC3II, lysosomal-associated membrane protein 1 (LAMP1), Beclin1, and ATG5 proteins and accumulation of OA in the lysosomes, suggesting an induction of lipophagy. Consequent to OA supplementation, pharmacological inhibition of p53 with pifithrin-α inhibits the activation of lipophagy and increases markers of cellular toxicity [51]. Although preliminary, these data support an essential role of p53 in lipophagy (Figure 2).
FOXOs link autophagy to energy metabolism
FOXO proteins are a family of TFs that integrate information from multiple upstream signals and enable tissue homeostasis during stress [52]. FOXOs activate protein catabolism by inducing two major protein degradation systems, the ubiquitin/proteasome and the autophagic/lysosomal pathways 53, 54.
FOXO3-induced autophagy has been studied in the context of muscle wasting following starvation or denervation and, more recently, during nutrient depletion in hematopoietic stem cells [55]. Upon nuclear translocation, FOXO3 binds the promoters of several autophagy genes 54, 56, including the main mediator of FOXO3a pro-autophagic activity BNIP3, a member of the Bcl2/adenovirus E1B 19 kd-interacting protein (BNIP) family. BNIP3, through its BH3 domain, interacts with Bcl-2, promoting its dissociation from Beclin1 [57]. In addition, BNIP3 regulates mitochondria clearance via autophagy (mitophagy) through its LC3-interacting region (LIR) [58]. FOXO3a can also induce autophagy indirectly via the upregulation of glutamine synthetase, which inhibits mTORC1 activity by increasing cellular levels of glutamine [59]. In addition to FOXO3, the regulation of autophagy by FOXO1 has been linked to cancer progression [60]. The mechanism by which FOXO1 induces autophagy is less-well-defined and it appears to involve transcriptional independent mechanisms. For example, in the cytosol acetylated FOXO1 binds and activates the autophagy protein ATG7 [60].
FOXO proteins have a crucial role in regulating energy metabolism. FOXO activity depends on the insulin/phosphatidylinositide 3-kinases (PI3K)/AKT pathway. Phosphorylation of FOXOs by AKT, a serine/threonine kinase AKT, leads to their cytosolic retention. Fasting inhibits the insulin/PI3K/AKT pathway and induces nuclear translocation and activation of FOXO proteins [52]. FOXOs regulate glucose homeostasis by promoting gluconeogenesis and contribute to the ability of tissues to transition from glucose to lipid metabolism, when a fuel-source is needed during fasting 52, 61. Indeed, activated FOXOs interact with nuclear receptors, such as PGC1α and PPARα, thus supporting induction of gluconeogenenesis, FA oxidation, and ketone body production. Notably, FOXOs can directly induce PGC1α expression [62]. Hence, FOXO factors concomitantly activate autophagy and promote lipid usage. FOXO1/3/4 liver-specific knockout mice develop hepatic steatosis and hypertriglyceridemia and display decreased autophagic flux. Importantly, this liver phenotype can be reverted if autophagy is activated by overexpression of the autophagy gene ATG14 [12]. These data strongly support the notion that lipophagy is an integral part of FOXO-mediated induction of lipid metabolism. In adipocytes, FOXO1 controls the expression of LAL during starvation [63], suggesting that FOXO1 can also enhance the capacity of the lysosome to degrade TGs (Figure 2).
The lysosome as a sensor for lipophagy induction
Lipophagy is activated during starvation, suggesting that cells have developed a mechanism to regulate this selective form of autophagy in response to nutrient availability, or lack thereof. The nutrient-sensing machinery that triggers lipophagy in response to starvation has not been characterized yet. Here, we discuss the evidence suggesting a role for the lysosome as a nutrient-sensing organelle able to trigger lipophagy.
For years, the lysosome has held the reputation of the cell's housekeeper, specifically for the degradation of cellular debris. However, recent data showed that the cell monitors lysosomal content to gather information on its energy status. Specifically, the cell can sense the luminal levels of lysosomal AA and conveys this information to the cytosolic side [64]. A key player in this nutrient-sensing machinery is the mTORC1 kinase, which plays a central part in regulating the equilibrium between anabolic and catabolic processes [65]. mTORC1 associates with the lysosomal surface to become activated. This association is mediated by a growing list of components, including the lysosomal vacuolar adenosine triphosphatase (v-ATPase), the Ragulator complex, and the RAS-related GTP-binding protein (RAG) family of small GTPases (RagA/B and RagC/D). AA in the lumen of the lysosome send signals to Ragulator, which exerts guanine nucleotide exchange factor (GEF) activity on Rags. In their GTP-loaded status, Rags recruit mTORC1 to the lysosomal surface where it interacts with the GTP-binding protein Rheb and becomes active [66]. The proton pump channel v-ATPase transfers the signal from inside the lysosome to the surface of the lysosomal membrane [64]. These findings suggest that the cell influences its metabolic fate by analyzing the products that arise from lysosomal catabolism. When AA concentration decreases inside the lysosome, mTORC1 is inhibited [64]. Active mTORC1 inhibits PPARα by promoting its interaction with nuclear receptor co-repressor 1 (nCoR1) [67] and inhibits TFEB via direct phosphorylation. Hence, as a consequence of mTORC1 inhibition, FA β-oxidation and lipophagy are activated. This regulatory mechanism, which begins at the lumen of the lysosome, may ensure the generation of energy from the oxidation of lipids when AA levels become insufficient (Figure 3).
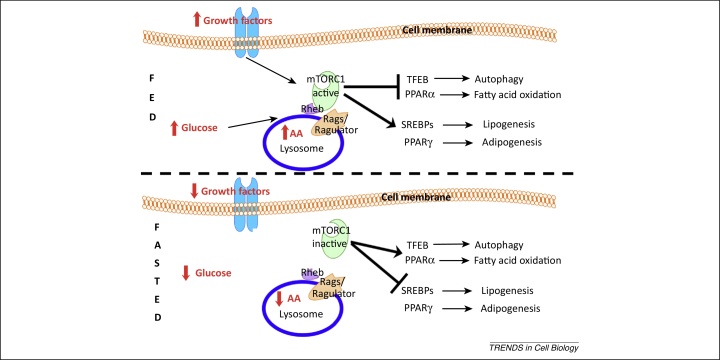
Figure 3.
The lysosome as a regulator of lipid metabolism. In the fed state (upper panel), the presence of lysosomal amino acids (AA), glucose, and growth factors induces activation of mechanistic target of rapamycin complex 1 (mTORC1) on the lysosomal membrane. Active mTORC1 transcriptionally induces lipogenesis and adipogenesis by activating SREBPs and peroxisome proliferator-activated receptor (PPAR)γ transcription factors, respectively. Concomitantly, mTORC1 blocks autophagy and fatty acid oxidation via transcription factor EB (TFEB) and PPARα inhibition, respectively. In the fasted state (lower panel), lower levels of lysosomal AA, glucose, and growth factors induce mTORC1 detachment from the lysosomal membrane and its consequent inhibition. mTORC1 inhibition in turn activates TFEB and PPARα transcription factors, which leads to the transcriptional induction of lipophagy and β-oxidation, respectively. Concomitantly, SREBPs and PPARγ are no longer activated by mTORC1, therefore lipogenesis and adipogenesis are inhibited. Abbreviations: Rheb, Ras homolog enriched in brain; Rags, Ras-related small GTP-binding protein; SREBPs, sterol regulatory element-binding proteins.
Thus, the lysosome is emerging as a central regulator of cellular metabolism, controlling intracellular signaling via mTORC1 and TFEB regulation of several downstream effectors (Figure 2). The physiopathological contribution of this lysosome-to-nucleus signaling mechanism [35] is still unknown; however, one may imagine that perturbing this regulatory machinery would contribute to the onset of common pathological conditions that characterize the processes of aging and obesity. Indeed, in aged and obese mice, mTORC1 seems to be hyperactivated and less sensitive to nutrient fluctuations 67, 68. Consistently, the expression levels of genes involved in autophagy and in lipid degradation processes are downregulated 10, 69, probably due to sustained inhibition of PPARα and TFEB. Intriguingly, lipidomic studies clearly show that aged and obese mice have an altered composition of lysosomal membrane and an accumulation of lipids in the lysosomal lumen [70], which could lead to abnormal lysosomal degradative capacity and disability to fuse with other cellular structures 71, 72. The accumulation of lipofuscin-like material in lysosomes represents a clear hallmark of aging and correlates to cellular dysfunction. The behavior of ‘aged’ lysosomes is similar to that associated with LSDs. In LSD cells cholesterol accumulates in the lysosomal membranes and decreases the capacity of the lysosome to fuse with other membranes by altering soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors (SNAREs), which are key components of the cellular membrane fusion machinery [71]. It will be important to evaluate whether SNARE function is also impaired in cells from aged or obese individuals. Such lysosomal abnormalities may contribute to altered mTORC1 function. Hyperactive mTORC1 can in turn inhibit lysosome and autophagosome biogenesis, further promoting intracellular storage of unwanted material and lipid accumulation. This inevitably leads to a vicious cycle that alters cellular metabolism, predisposing cells to metabolic dysfunction.
Concluding remarks
During nutrient starvation lipophagy, an autophagic process delivers lipids to the lysosome for their degradation. Lysosomal degradation of lipids requires LAL, an enzyme where deficiency causes massive intracellular accumulation of lipids and Wolman disease. The mechanisms by which lipophagy is regulated are currently unknown. The increase in the mRNA level of LAL, as well as of several lysosomal and autophagic genes, observed during starvation suggests that lipophagy is a transcriptionally regulated process. Such regulation may influence the composition of newly formed lysosomes and autophagosomes in a way that allows them to adapt to the nature of the substrates that need to be targeted and degraded. This observation may be important for the identification of proteins that are selective regulators of lipophagy, such as putative lipophagy receptors.
Increasing evidence suggests that the nutrient-sensitive TFs: TFEB, p53, and members of the FOXO family, regulate lipophagy during fasting and that the nuclear receptor and co-receptor family members: PPARα and PGC1α, link lipophagy to other pathways involved in lipid catabolism. The emerging view is that the cell coordinates lipid catabolism by transcriptionally regulating lipophagy and lipid degradation pathways that occur in different organelles.
Moreover, the signals that trigger lipophagy may start from within the lysosome. The lysosome is emerging as a sensor of cellular metabolic cues. The capacity of mTORC1 to associate to the lysosomal membrane, along with the recent observation that members of the extracellular signal-regulated kinase (ERK) pathway localize to autophagosomes [73], are both indications that the lysosomal/autophagy pathway is a critical regulator of cellular metabolism. This observation raises new important biological questions on the role of the lysosome in regulating energy metabolism and suggests that genetic and environmental factors affecting lysosomal homeostasis can influence whole-body metabolism. This concept may have a profound impact on the development of novel therapeutic strategies for a variety of human diseases, ranging from genetic disorders such as LSDs to the more common metabolic processes associated with aging and obesity. Modulation of autophagy has already been shown to ameliorate metabolic syndromes in diet- and genetic-induced preclinical models of obesity [10]. The discovery of TFEB and the modulation of lysosome-mediated cellular clearance by means of TFEB overexpression are examples of how regulation of lysosomal function may have important implications for the development of novel therapeutic strategies. This approach has already shown promising results in the treatment of preclinical models of LSDs 31, 32 and neurodegenerative disease [74]. However, more studies are needed to evaluate its effectiveness and potential side effects. Finally, drugs holding the ability to enhance lysosomal function may represent new therapies for the treatment of a variety of disease conditions.
Acknowledgments
The authors thank Rajat Singh, Babak Razani and Graciana Diez Roux for their critical reading of the manuscript. The authors’ work is supported by the Italian Telethon Foundation grant numbers TGM11CB6 (A.B.) and TCP12008 (C.S.), the Italian Ministry of Research (FIRB) (C.S.), Marie Curie Integration Grant PCIG13-GA-2013-618805 (C.S.), the Beyond Batten Disease Foundation (A.B.), European Research Council Advanced Investigator grant no. 250154 (CLEAR) (A.B.), and US National Institutes of Health (R01-NS078072) (A.B. and C.S.).
Glossary
- Autophagosomes
double-membrane-surrounded organelles that form in the endoplasmic reticulum (ER) in response to cellular stress (e.g. nutrient depletion). Their main role is to target substrates to the lysosomes for degradation.
- ß-oxidation
generates energy from degradation of fatty acids. In a single ß-oxidation cycle the fatty acid chain is shortened by two carbons to generate a molecule of acetyl-CoA. The acetyl-CoA can enter the tricarboxylic acid (TCA) cycle to generate ATP.
- Fatty acids
carboxylic acids (-COOH) with a long tail composed of chains of carbon atoms. Depending on the chain length, fatty acids can be classified as short (<6 carbon atoms), medium (6–12), long (12–21) or very long (>22).
- Gluconeogenesis
the metabolic reaction triggering the biosynthesis of glucose from pyruvate, lactate, glycerol and the amino acids alanine and glutamine.
- Ketone bodies
produced by the liver during fasting and from fatty acid catabolism after prolonged exercise. They are used peripherally as an energy source when glucose is not available. The three types of ketone bodies are acetone, acetoacetic acid and beta-hydroxybutyric acid.
- Lipases
hydrolytic enzymes that catalyze the degradation of fats by hydrolyzing their ester bonds such as those between glycerol and fatty acid chains.
- Lipid droplets
intracellular organelles that store neutral lipids, such as triacylglycerols and sterol esters (including cholesterol).
- Lipogenesis
the generation of fatty acids from acetyl-CoA. It is activated under fed conditions by insulin signaling and by mechanistic target of rapamycin complex 1 (mTORC1) kinase.
- Lysosomes
cellular organelles, containing several classes of hydrolytic enzymes, which are deputed to the degradation of a large repertoire of cellular substrates.
- TCA cycle
(also Krebs cycle or citric acid cycle) is a series of enzyme-catalyzed chemical reactions occurring in mitochondria that generate energy (ATP) through the oxidation of acetate derived from carbohydrates, fats and proteins.
- Triglycerides
fat molecules composed of a sugar alcohol (glycerol) linked to three molecules of fatty acids through ester bonds between the hydroxide groups of the glycerol and the carboxyl group of the fatty acid.
Contributor Information
Carmine Settembre, Email: Settembre@tigem.it.
Andrea Ballabio, Email: Ballabio@tigem.it.
References
- 1.Wang T. The comparative physiology of food deprivation: from feast to famine. Annu. Rev. Physiol. 2006;68:223–251. doi: 10.1146/annurev.physiol.68.040104.105739. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y. Lipid droplets at a glance. J. Cell Sci. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reue K. A thematic review series: lipid droplet storage and metabolism: from yeast to man. J. Lipid Res. 2011;52:1865–1868. doi: 10.1194/jlr.E020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg A.S. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finck B.N., Kelly D.P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R., Cuervo A.M. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabinowitz J.D., White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iancu T.C. What's in a name? -“Lipolysosome”: ultrastructural features of a lipid-containing organelle. Ultrastruct. Pathol. 2013;37:293–303. doi: 10.3109/01913123.2013.799625. [DOI] [PubMed] [Google Scholar]
- 10.Yang L. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaber N. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong X. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J. Biol. Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu K., Czaja M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaushik S. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupont N. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lübke T. Proteomics of the lysosome. Biochim. Biophys. Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson R.A. Mutations at the lysosomal acid cholesteryl ester hydrolase gene locus in Wolman disease. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2718–2722. doi: 10.1073/pnas.91.7.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolman M. Wolman disease and its treatment. Clin. Pediatr. (Phila.) 1995;34:207–212. doi: 10.1177/000992289503400406. [DOI] [PubMed] [Google Scholar]
- 19.Razani B. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15:534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouimet M. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Rourke E.J., Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morselli E. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrocola F. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Füllgrabe J. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 25.Desvergne B. Transcriptional regulation of metabolism. Physiol. Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 26.Lapierre L.R. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Settembre C. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmieri M. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 29.Widlund H.R., Fisher D.E. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 30.Sardiello M. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 31.Medina D.L. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spampanato C. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martina J.A. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roczniak-Ferguson A. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Settembre C. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takikita S. Fiber type conversion by PGC-1α activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PLoS ONE. 2010;5:e15239. doi: 10.1371/journal.pone.0015239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potthoff M.J. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K.H. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 40.Lapierre L.R. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapierre L.R., Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 2012;23:637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vousden K.H., Lane D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein I., Rotter V. Regulation of lipid metabolism by p53 – fighting two villains with one sword. Trends Endocrinol. Metab. 2012;23:567–575. doi: 10.1016/j.tem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Assaily W. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 45.Finck B.N. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Peterson T.R. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen N. PGC-1α, a key modulator of p53, promotes cell survival upon metabolic stress. Mol. Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 48.Crighton D. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 49.Kenzelmann Broz D. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stambolic V. Regulation of PTEN transcription by p53. Mol. Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 51.Park E-J. Role of p53 in the cellular response following oleic acid accumulation in Chang liver cells. Toxicol. Lett. 2014;224:114–120. doi: 10.1016/j.toxlet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 52.Eijkelenboom A., Burgering B.M.T. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 53.Sandri M. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mammucari C. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Warr M.R. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Van der Vos K.E. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat. Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 61.Barthel A. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 62.Daitoku H. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 63.Lettieri Barbato D. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013;4:e861. doi: 10.1038/cddis.2013.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zoncu R. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bar-Peled L., Sabatini D.M. SnapShot: mTORC1 signaling at the lysosomal surface. Cell. 2012;151:1390–1390.e1. doi: 10.1016/j.cell.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 67.Sengupta S. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 68.Menon S. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci. Signal. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettinelli P. Enhancement in liver SREBP-1c/PPAR-α ratio and steatosis in obese patients: correlations with insulin resistance and n-3 long-chain polyunsaturated fatty acid depletion. Biochim. Biophys. Acta. 2009;1792:1080–1086. doi: 10.1016/j.bbadis.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 70.Rodriguez-Navarro J.A. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E705–E714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fraldi A. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Koga H. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Lopez N. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Decressac M. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamb C.A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 76.Hamasaki M. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 77.Settembre C. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mizushima N. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 79.Nazio F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 80.Kim J. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Egan D.F. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russell R.C. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei Y. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lefebvre P. Sorting out the roles of PPAR in energy metabolism and vascular homeostasis. J. Clin. Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]