Abstract
We report the design of a chemically defined platform engineered for the culture of human pluripotent stem cells (hPSCs) that supports the long-term maintenance of self-renewing hPSC populations in a more uniform manner than standard culture systems. Microcontact printing (µCP) of alkanethiol self-assembled monolayers (SAMs) was used to spatially direct hPSC adherence. This technique not only establishes control over hPSC colony size and shape but also preserves genetic stability and provides unprecedented uniformity in the pluripotency of hPSC populations that is quantitatively assessed in the present study.
Keywords: human pluripotent stem cell, self assembled, monolayers, microcontact printing
1. Introduction
Human pluripotent stem cells (hPSCs) possess the extraordinary ability to self-renew indefinitely and to differentiate into any mature cell type found in the human body [1]. These stem cell lines may be broadly categorized into two sub-groups, human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs). The former are pluripotent stem cells derived from the inner cell mass of blastocyst stage embryos [2] while hiPSCs are somatic cells that have been genetically reprogrammed into a hESC-like state [3, 4]. Both hPSC types can have profound impact as models for understanding human disease mechanisms as well as in providing therapies in regenerative medicine. Unfortunately, progress toward deploying stem cell derived materials in clinically relevant protocols has been slowed by a number of technological barriers associated with the culturing of hPSCs. For example, to date, most stem cell culturing protocols require xenogenic substrate materials that are both undefined and expose the hPSCs to contaminants [5–7]. In addition, chemically defined conditions that universally optimize hPSC growth and differentiation remain elusive. Despite recent success toward designing chemically defined alternatives [8–13], hPSCs are still prone to spontaneous differentiation, resulting in colony heterogeneity as well as inefficient and variable differentiation. Thus development of improved culture systems is essential and could lead to standardized hPSC culturing practices.
The primary contention of the present study is that a rationally designed hPSC culturing platform should achieve the following objectives: i) promote stable, long-term growth of hPSCs; ii) avoid xenogenic contamination; iii) promote uniformity to improve differentiation; iv) offer scalability at both laboratory and industrial levels. While viable solutions that address items i) and ii) are emerging, heterogeneities persist as a serious hurdle to the reliable growth of self-renewing hPSCs in vitro [14]. Heterogeneity in conventional hPSC culture results in a gradient of expression of genes regulating the pluripotent state [15].
One attractive approach to reduce stem cell heterogeneity is to regulate colony size and shape [8, 16]. In the present article we report a stem cell culturing platform based upon the well-established technique of microcontact printing (µCP) [17, 18] for patterning self-assembled monolayers (SAMs) [19] that supports long-term maintenance of self-renewing hPSC populations in a more uniform manner than standard culture systems. The surface chemistry at these SAM interfaces is used to spatially direct hPSC adherence, which establishes uniform colony size and shape while preserving genetic stability and pluripotency. Moreover, this culturing system achieves robust control over colony uniformity across multiple hPSC lines, which will have broad technological implications for standardizing how stem cell lines are maintained, expanded, and differentiated in culture.
2. Experimental
2.1. Materials
16-mercaptohexadecanoic acid, triethylene glycol mono-11-mercaptoundecyl ether, octadecanethiol, and anhydrous ethanol were purchased from Sigma Aldrich. 1-propanol was obtained from Fisher Scientific. All reagents and solvents unless otherwise stated were used without further purification.
2.2. Preparation of gold coated substrates
Briefly, 34 mm diameter glass cover-slides (Fisher) were cleaned in a Piranha solution (70% H2SO4, 30% H2O2) at 100°C for twenty minutes followed by thorough rinsing in deionized (DI) water and drying under a stream of nitrogen. Immediately following this cleaning procedure, a 10 nm layer of gold with a 5 nm titanium adhesion layer was deposited onto the cleaned substrate using a CHA E-beam evaporator.
2.3. Preparation of Stamps for µCP
Photolithographically prepared silicon masters were used as templates to cast polydimethylsiloxane (PDMS) stamps for µCP. Masters were fabricated using conventional photolithography techniques. Four-inch Si(100) test wafers (Tech Gopher Inc.) were first cleaned in a Piranha solution at 100°C for twenty minutes. The wafers were then thoroughly rinsed in deionized (DI) water and spun dry at 2000 rpm for 10 min in a wafer drier. Next, a film of the negative photoresist SU-8–50 (MicroChem, Bedford, MA) with an approximate thickness of 50 µm was applied to the surface of a cleaned wafer via spin coating. The SU-8 film was then soft baked on a hotplate using annealing parameters provided by the photoresist supplier. Subsequently, the films were exposed to UV irradiation through a photomask containing the desired pattern with a Karl Suss contact aligner. Photomasks were designed in Adobe Illustrator and printed onto transparencies at a resolution of 24,000 dpi with a high-resolution printer (CAD Art Sciences). Next, the non-exposed photoresist was selectively removed with SU-8 developer solution to reveal the patterned features. Upon pattern development, the final designs were rinsed with isopropyl alcohol, gently dried with N2, and hard baked for 20 minutes at 150°C per instructions from the photoresist supplier. A Veeco Dektak 150 profiler was used to confirm the step heights of the patterned SU-8 features. Prior to replica molding of PDMS stamps, the masters were exposed to Tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane vapor under vacuum for 1 hour, rendering the SU-8 features hydrophobic and inhibiting PDMS delamination.
PDMS stamps were cast by pouring a mixture of Sylgard 184 (Dow Corning, Midland, MI), prepared in the standard 10:1 prepolymer-to-crosslinker ratio, onto the desired master. Samples were held under vacuum overnight at room temperature and subsequently cured at 60°C for one hour. Upon curing, PDMS stamps were then removed from the master and sectioned into roughly 34 mm diameter circular stamps for patterning SAM inserts to standard 6 well tissue culture plates.
2.4. µCP of SAMs
Gold coated cover-slides were first cleaned in a Piranha solution (70% H2SO4, 30% H2O2) for 20 minutes. Prior to stamping, each substrate was rinsed sequentially with DI water and ethanol twice and then dried under a stream of N2. In parallel, a PDMS stamp was exposed to oxygen plasma for 20 minutes. The plasma treated stamp was inked with a 2 mM solution of 16-mercaptohexadecanoic acid in ethanol, dried with N2 after 2 minutes, and placed into conformal contact on top of a freshly cleaned gold coated substrate for 1 minute. Samples were then immersed in a 2 mM ethanol solution of triethylene glycol mono-11-mercaptoundecyl ether for 2.5 hours. During SAM formation, samples were covered under aluminum foil to prevent exposure from ambient light. The substrates were subsequently removed from the alkanethiol solution, rinsed with anhydrous ethanol, dried under a stream of N2, and glued into six-well tissue culture plates. Samples were covered under aluminum foil in a sterile cell culture hood for 1 hr while the epoxy cured. Prior to cell culture the µCP SAM substrates were treated overnight with a 3 ml of solution of extracellular matrix (ECM) proteins in phosphate buffered saline (PBS, Invitrogen), containing 0.6 µg/ml of laminin (Sigma Aldrich), 0.6 µg/ml of collagen (Sigma Aldrich), and 0.09 µg/ml of nidogen I (R&D Systems).
2.5. HPSC Culture and Passaging
Human embryonic stem cells (hESCs) lines, H9 (WiCell) or HSF1 (UCSF), or human induced pluripotent stem cells (hiPSCs)[3] were cultured on gelatin coated plates with mitomycin C-treated mouse embryonic fibroblasts (MEFs) derived from embryonic day 13.5 embryos from CF1 mice. Cells were grown in hPSC medium consisting of Dulbecco’s Modified Eagle Medium-F12 (Invitrogen) supplemented with 20% Knockout Serum Replacement (Invitrogen), 1 mM L-Glutamine, 2 mM 2-Mercaptoethanol, 1mM Non-essential Amino Acids, and 4 ng/ml basic Fibroblast Growth Factor (bFGF, Invitrogen). To generate conditioned medium (CM), MEFs were cultured with hPSC medium for 24 hours. Cell culture on µCP SAM substrates first involved dissociating cells from the desired hPSC line into single cells using 0.05% trypsin-EDTA (Invitrogen) and filtering with a 40 µm nylon mesh strainer (BD Falcon) to remove feeders and aggregates. After the addition of trypsin inhibitor (Invitrogen), the cells were centrifuged at 1,000 rpm and resuspended in either CM or mTeSR defined medium formulation (StemCell Technologies) with 10 µm HA-1077 dihydrochloride (Sigma).[20] The ECM protein solution was then aspirated from the patterned SAM inserts and each well rinsed three times with PBS. A dilution of 500,000 cells in 4 mL was then seeded onto each µCP SAM substrate. It is important to allow the cells to attach undisturbed for 1 day and to remove only half of the medium and replace with fresh medium every other day.
For studies assessing pluripotency maintenance, H9 cells were passaged on the third day after seeding by once again dissociating with trypsin and re-seeding onto freshly prepared patterned SAMs. Cells were typically counted and split in a 1:1.5 or 1:2 ratio for each passage.
2.6. HPSC characterization
A variety of standard techniques were utilized to assess both self-renewal and pluripotency of the hPSC populations cultured on the presently described µCP/SAM platform. Experimental details of the methods used to characterize the patterned hPSC population can be found in the supplementary data.
3. Results and discussion
Earlier human embryonic stem cell studies which utilized µCP involved only a single cell culture passage. The heterogeneities reported within the patterned colonies did not demonstrate any particular improvement in lineage specific differentiation compared to controls [21, 22]. The approach adopted in the present study is significantly different in that we use alkanethiol SAMs to obtain multifunctional, chemically defined surfaces that are tailored to support the uniform growth of self-renewing hPSC colonies. With this µCP/SAM-based platform, we are able to achieve serial passaging of cells and thus demonstrate the continuous culture of hPSCs. Another significant feature of our approach is that colony formation proceeds with unprecedented consistency. The present µCP/SAM platform is capable of supporting hPSC populations in which pluripotency markers displayed by stem cells such as alkaline phosphatase (AP) are uniformly distributed within individual colonies (Fig. 1). This is in sharp contrast to staining patterns observed from standard cultures where variable AP staining and differentiation are seen routinely (Supplementary data, Fig. S1).
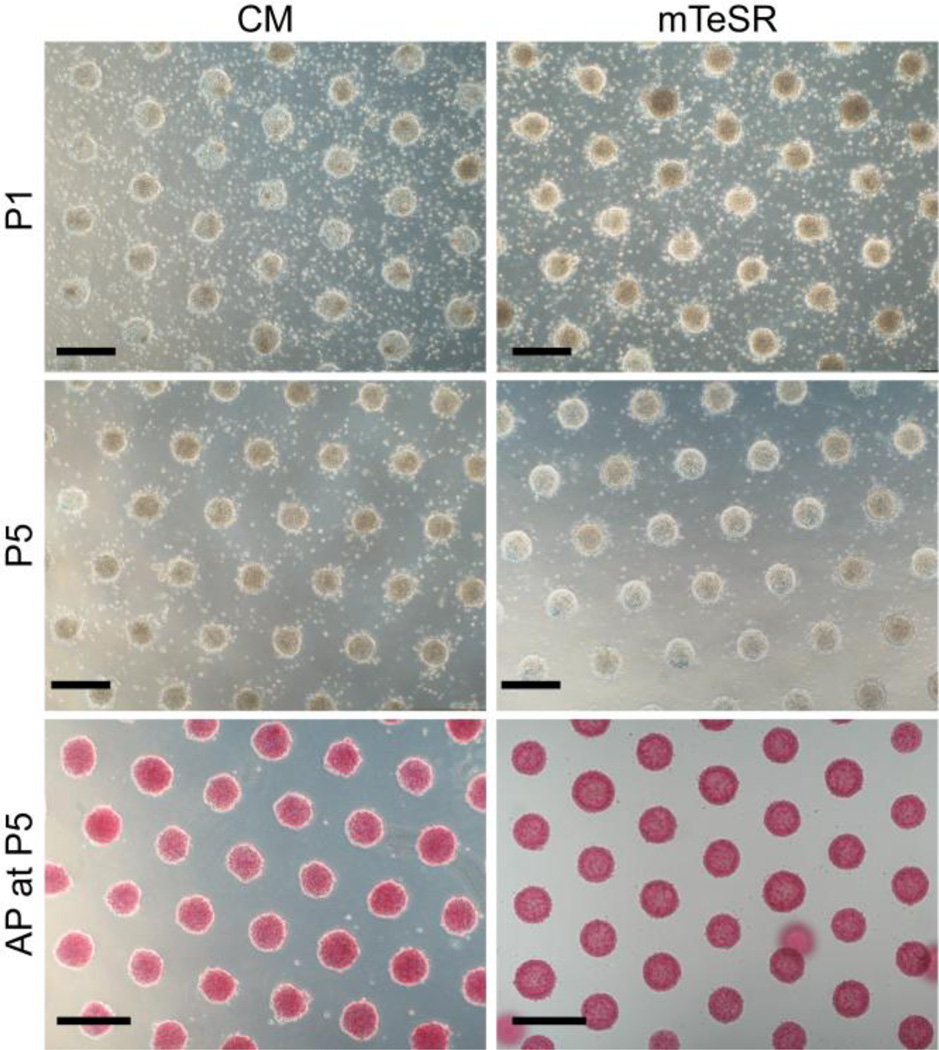
Fig. 1.
Bright-field images of a patterned array of hPSC colonies cultured in conditioned medium (CM, left) and mTeSR (right) after one passage (P1, top) and after five passages (P5, middle). Images were taken at day 2 of culture prior to feeding the cells. The colonies stain positive for alkaline phosphatase (AP) in both CM and mTeSR at P5 (bottom), indicating that the hPSCs have remained pluripotent. The patterned hPSC colonies displayed in this figure are comprised of hESCs from line H9 (WiCell). Scale bars = 400 µm.
To regioselectively direct colony formation, the SAM interfaces are designed such that a SAM that supports stem cell growth is stamped onto a second SAM that resists cell attachment (Supplementary data, Scheme 1). Single cell suspensions of hPSCs are preferentially guided to growth positions at the µCP patterned cell culture interface by selective wetting. The present study utilizes carboxylic acid (−COOH) presenting SAMs to confine stem cell growth within the regions patterned by µCP. The unpatterned background region of the surface consists of SAMs terminated by either methyl (−CH3) or polyethylene glycol (−PEG) moieties. Interestingly, the selection of either −CH3 or −PEG for the background SAM can dictate colony behavior and pattern fidelity during culture (Supplementary data, Fig. S2).
Orner et al have previously identified alkanethiol SAMs terminated by COOH functionalities as being able to support the short-term growth of human embryonic stem cells (hESCs) [23]. Our preliminary research has found that under standard conditions the number of cells that attach and thrive on pristine −COOH SAMs is insufficient for continuing the culture of hESC populations after a single passage (Supplementary data, Fig. S3). To address this limitation, the patterned −COOH SAMs are chemically augmented with extracellular matrix (ECM) proteins. −COOH SAMs are well suited to act as the foundation for the present cell culture platform because they readily adsorb ECM proteins that promote cell adhesion [24]. The selection of purified ECM components was guided by the work of Xu et al [25]. Physical adsorption of laminin onto the patterned −COOH containing regions of the SAM leads to the formation of uniformly sized, well-defined, compact hESC colonies that conform to the µCP pattern (Supplementary data, Fig. S3). Moreover, the laminin treated SAMs support hPSC growth under a variety of culture conditions, including traditional MEF conditioned medium (CM) and the commercial xeno-free preparations mTeSR and StemPro.
An additional refinement to the surface chemistry at the SAM cell culture interface involved combining several ECM proteins for use in the patterned areas. Because the ECM of natural stem cell niches is an amalgam of multiple proteins and factors that together provide structural support and anchorage points for developing cells, we hypothesized that the addition of multiple ECM components would be advantageous for continuous passaging. For the present study a cocktail of ECM factors consisting of laminin, nidogen I, and collagen IV was formulated based on a recent report from Evseenko et al [26]. Nidogen acts as a crosslinker between laminins that helps to organize the basement membrane component of the ECM. When this ECM formulation is applied to the µCP SAMs, robust hPSC colony formation can be achieved with protein concentrations that are an order of magnitude lower than what was needed for SAMs treated only with laminin.
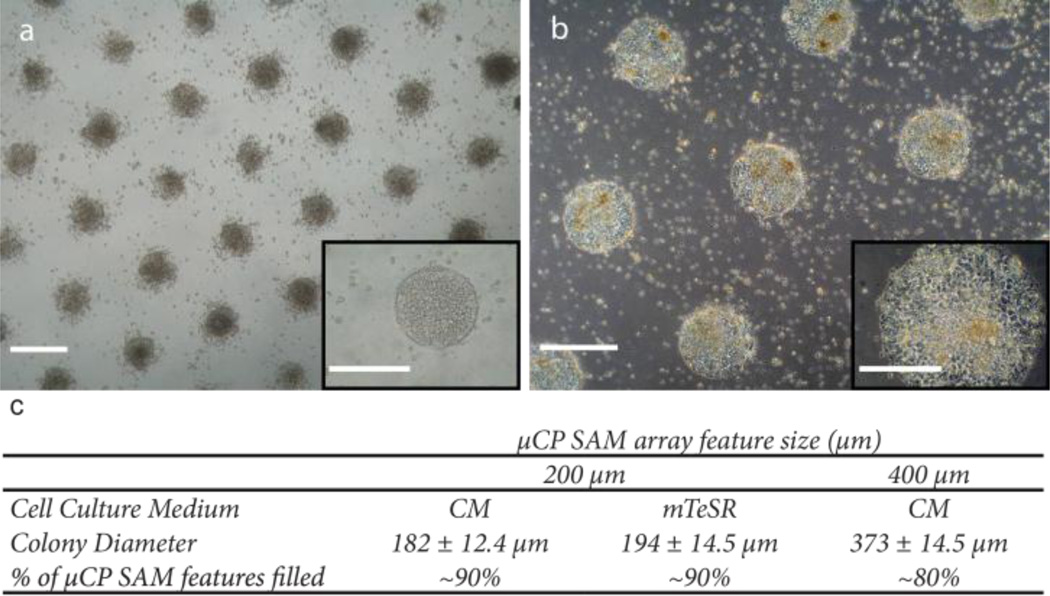
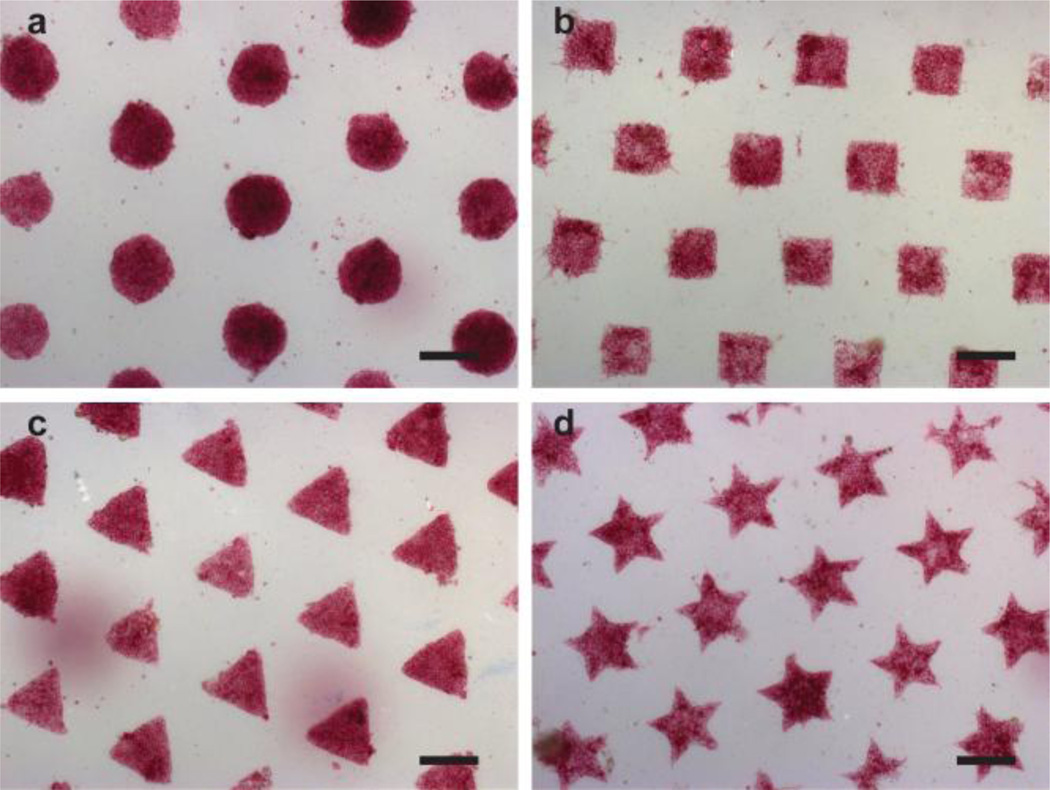
In a typical µCP experiment, we obtain arrays of uniformly distributed hPSC colonies that span the entire area of standard six-well tissue culture plates. For example, for configurations consisting of 200 µm circular features, greater than 90% of the SAM pattern is filled by hPSC colonies that average 194 ± 14.5 µm in diameter for cells cultured in mTeSR and 182 ± 12.4 µm for cells cultured in CM (Fig. 2). Similarly, for SAM arrays consisting of 400 µm features, typically 80% of the pattern is filled with colonies averaging 373 ± 14.2 µm in diameter. Moreover, colony shape can be precisely controlled using the µCP SAMs by simply changing the configuration of the polydimethylsiloxane (PDMS) stamp used for µCP (Fig. 3). This level of precision cannot be obtained with standard hPSC culture conditions (Supplementary data, Fig. S1).
Fig. 2.
Multiple sizes of uniform hPSC colonies can be formed on µCP SAMs. The patterned substrate spatially defines colony growth within circular features that are either 200 µm (a) or 400 µm (b) diameter. Scale bars = 400 µm. Inset images show patterned colonies are compact with well-defined borders that conform to each micro-patterned SAM spot (Insets, scale bars = 200 µm). c) The uniformity of colony size is consistently maintained on the µCP SAMs. hPSC cell line: H9 hESC (WiCell).
Fig. 3.
Representative bright field images depicting arrays of hPSC colonies where the shape of the patterned features used to direct cell growth was varied. Specific configurations examined include circles (a), squares (b), triangles (c), and stars (d). Scale bars = 200 µm. hPSC cell line: H9 hESC (WiCell).
To establish that our platform is compatible with the continuous culture of pluripotent stem cells, we tracked arrays of uniformly sized hPSC colonies over multiple passages. Prior to beginning this stability study, cell lines used in these experiments were subjected to standard assays to determine the karyotype and pluripotent status of the bulk hPSC population (Supplementary data, Fig. S4). Evaluation of the “passage zero” (P0) population confirmed the undifferentiated status of the hPSCs and provided a baseline for examining pluripotency and genetic stability after multiple passages on the µCP SAMs.
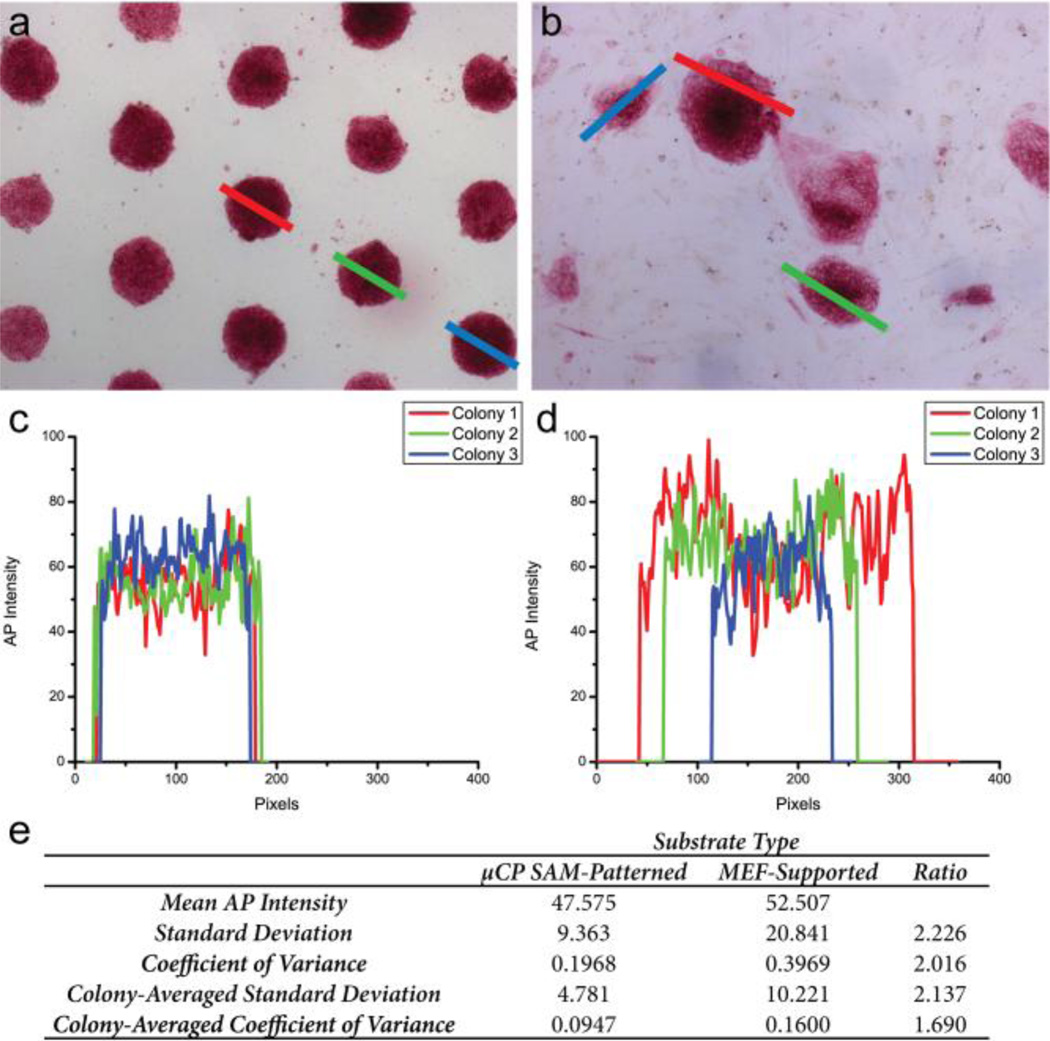
Fig. 1 shows that after five serial passages, hPSCs cultured on the chemically defined substrates consistently form round compact colonies that stain positive for the pluripotency marker alkaline phosphotase (AP). These results hold true when culturing the hPSCs in either traditional CM or the commercial mTeSR defined medium formulation. The uniformity of AP expression in µCP-patterned and MEF-supported hPSC populations was characterized further using an image analysis protocol. Briefly, AP expression was correlated to the intensity of the red signal measured from bright field images of stained hPSC colonies (Supplementary data, Fig. S5). Analysis of the spatial variability of AP staining by this approach shows a two-fold improvement in the coefficient of variance for µCP patterned colonies compared to colonies grown on MEFs, indicating that the patterned population is far more homogeneous (Fig. 4). Additional discussion of the image analysis is provided in the supplementary data.
Fig. 4.
Representative images of µCP-patterned colonies (a) and MEF-supported colonies (b). The lines drawn across the indicated colonies represent the path of the intensity profiles measured for individual colonies. Line colors correspond to the plots of AP intensity displayed in c,d. (e) Comparison of AP-staining intensities from µCP SAM-patterned and MEF-supported populations. Results expressed as calculated standard deviations and average coefficient of variance, defined as the ratio of the standard deviation to the mean. hPSC cell line: H9 hESC (WiCell).
Interestingly, hPSCs comprising the patterned colonies appear more confluent than unpatterned conventional cultures, which are more loosely associated and display more of a migrating border (Fig. 4a,b). Recently, Li et al demonstrated that hPSCs are better able to form colonies when individual stem cells are in close proximity with one another, which promotes the up-regulation of the tight junction protein E-Cadherin leading to decreased apoptosis within colonies and improved clonal efficiency [27]. Moreover, work recently reported by Chen et al suggests that hPSCs utilize E-cadherin to sense the nanotopology of the local cell culture environment and that this mechano-sensory regulation can influence stem cell fate decisions such as self-renewal and clonal expansion [28]. Thus, it is likely that the improved uniformity displayed with the present µCP platform may be attributed to hPSCs within the patterned colonies being able to establish and maintain more intercellular connections via the clustering of tight junctions. In addition, when the shape of the micropatterned features is altered, hPSC colonies continue to display less variability in AP expression than MEF supported cells (Table 1). This observation implies that colony shape has a minimal effect on the behavior of hPSCs cultured under self-renewing conditions.
Table 1.
Uniformity of AP stained hPSC colonies supported on µCP SAMs
| Colony Shape | Mean AP Intensity | Standard Deviation | Coefficient of Variance |
|---|---|---|---|
| Circles | 47.575 | 9.36 | 0.1968 |
| Squares | 54.006 | 11.78 | 0.2181 |
| Triangles | 82.408 | 13.99 | 0.1698 |
| Stars | 60.115 | 10.90 | 0.1814 |
| Unpatterned hPSCs on MEFs | 52.507 | 20.84 | 0.3969 |
hPSC cell line used in analysis: H9 hESC (WiCell)
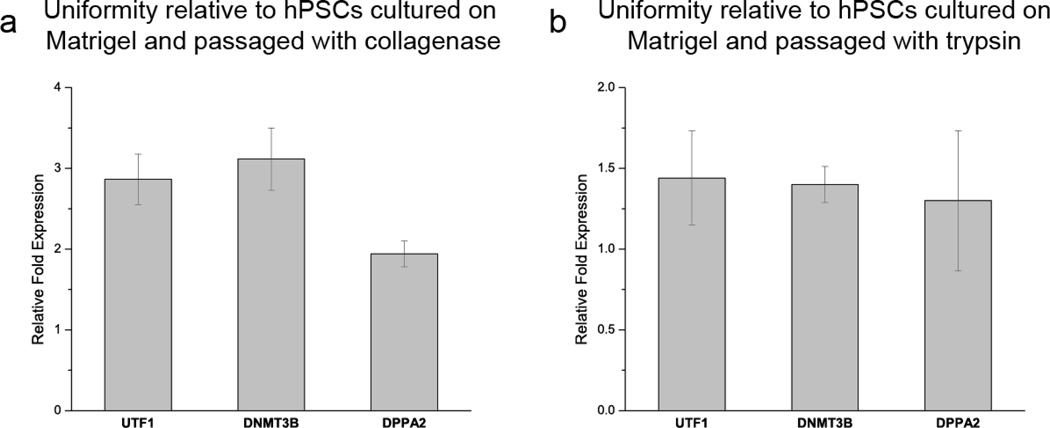
Quantitative polymerase chain reaction (qPCR) studies provide additional support to the observation of improved uniformity within hPSC populations cultured on µCP SAMs (Fig. 5). hPSCs are heterogenous in marker expression and have decreased self renewal potential as expression of stem cell surface markers and pluripotency genes (e.g. UTF1, DNMT3B, and DPPA2) wane. Recent work has shown that evaluation of expression levels of these pluripotency genes can be used to monitor variability within hPSC populations [29]. As much as a 3-fold increase in expression of these markers was observed in µCP-patterned hPSCs relative to control cells cultured on Matrigel, further demonstrating enhanced uniformity on µCP substrates over standard culture conditions.
Fig. 5.
qPCR analysis of the expression of the pluripotency markers UTF1, DNMT3B, and DPPA2 indicates that there is less variability in micropatterned hPSC populations compared to unpatterned Matrigel controls passaged with either (a) collagenase or (b) trypsin. Relative fold expression was calculated by normalizing the expression level for each pluripotency marker to the housekeeping gene GAPDH through a threshold cycle (Ct) value comparison.
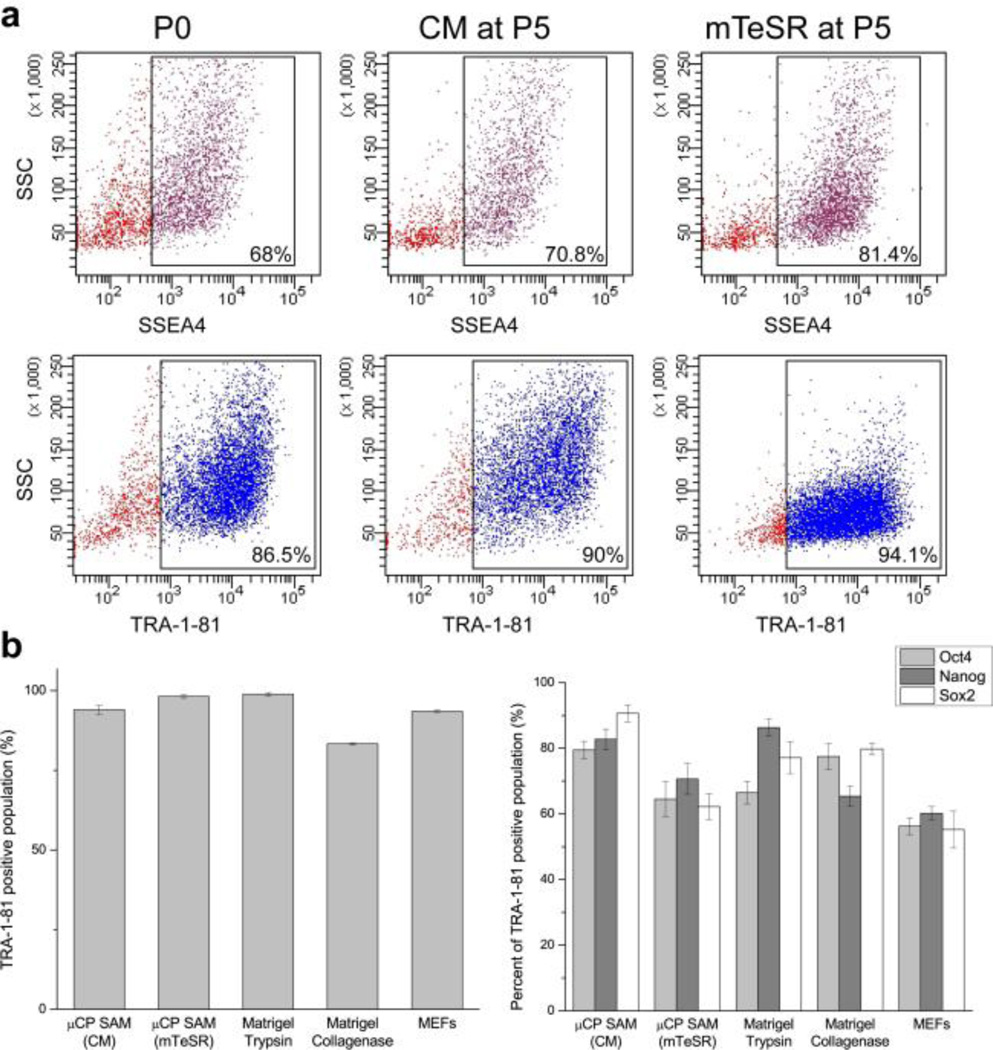
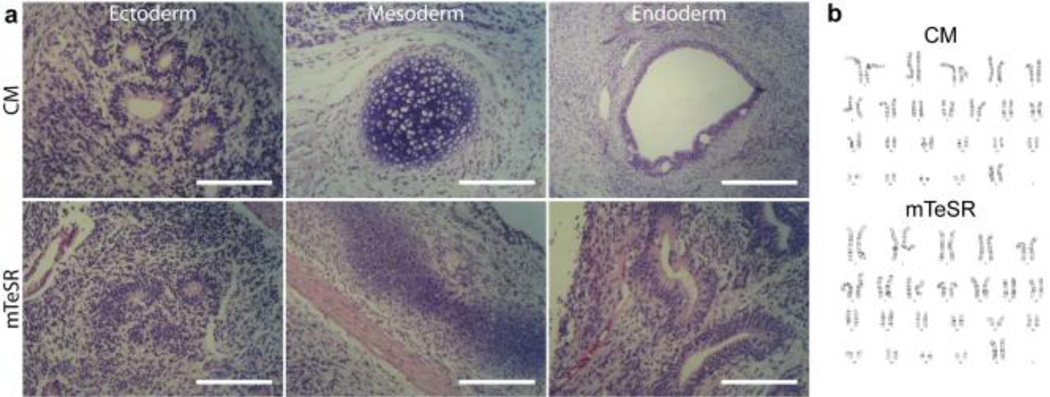
While the positive AP test suggests that the cells have not begun to differentiate, the pluripotent status of the stem cells was more rigorously confirmed using qPCR and fluorescent activated cell sorting (FACS) based assays for examining standard pluripotency markers (Figs. 5 and 6). These data indicate that the µCP patterned hPSCs after five passages (P5) express pluripotency markers at a level that meets and in some cases exceeds the expression displayed by the original P0 population supported on either MEF or Matrigel controls. This observation is particularly evident in the analysis of Oct4, Nanog, and Sox2 expression within the TRA-1–81 positive population from each cell culture substrate (Fig. 6b). For example there is as much as a 1.5-fold improvement in the number of TRA-1–81 positive hPSCs co-expressing these pluripotency markers on µCP SAMs compared to cells grown on MEF feeder layers under identical culture conditions. P5 hPSCs were also able to form teratomas when injected into the testes of immunocompromised mice, which conclusively establishes that the cells are able to produce derivatives from each embryonic germ layer (Fig. 7a). Moreover, the hPSCs remained genetically stable with no evidence of trisomies or other karyotypic abnormalities (Fig. 7b).
Fig. 6.
(a) FACS analysis indicates that expression of pluripotent stem cell surface markers SSEA-4 (a, upper) and TRA-1–81 (a, lower) for hPSCs (line H9, WiCell) cultured on the chemically defined SAM platform in either conditioned medium (CM) or mTeSR after five passages (P5) is similar to the original P0 population. (b) Additional FACS analysis of the TRA-1–81 positive population indicates that the patterned hPSCs cultured in CM and mTeSR continue to express the pluripotency markers Oct4, Nanog, and Sox2 similar to MEF and Matrigel cultured controls.
Fig. 7.
(a) H&E stained sections from teratomas formed from P5 hPSCs cultured in either CM or mTeSR demonstrate potential for generating derivatives from all three embryonic germ layers. (b) P5 hESCs have a normal karyotype for both the CM and mTeSR conditions. hPSC cell line: H9 hESC (WiCell). Scale bars = 200 µm.
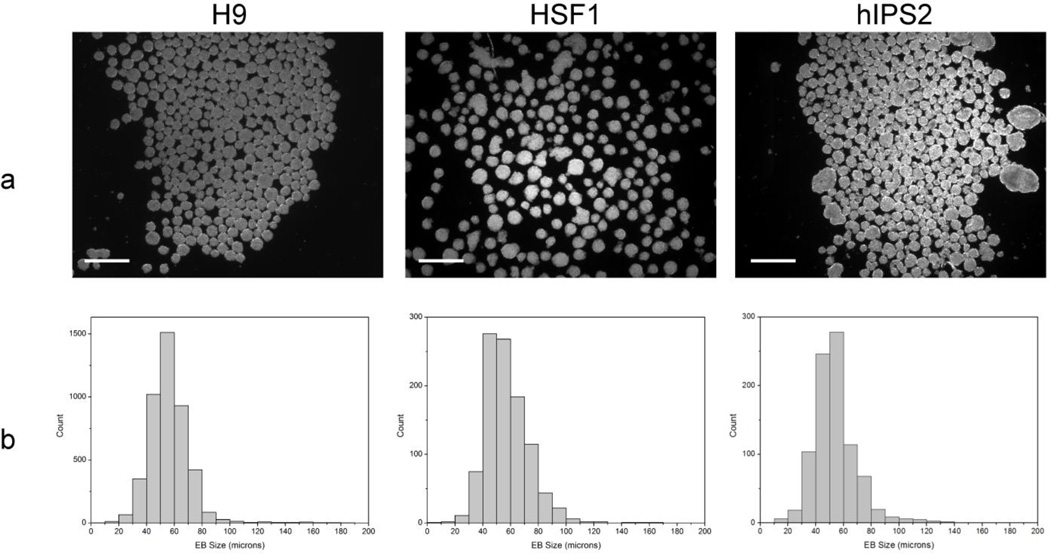
In addition to confirming pluripotency and genetic stability, the differentiation potential of the hPSCs cultured on the chemically defined platform was examined by embryoid body (EB) formation. An interesting consequence of the the patterned surface chemistry of the µCP SAMs is that we are able to tailor the cell culture interface to obtain size selected EBs that express differentiation markers characteristic of endoderm, mesoderm, and ectoderm (Fig. 8). Histograms comparing EB diameters from three independently cultured hPSC lines immediately after removal from the µCP SAMs show that the recovered EBs fall within a size distribution that is centered at ~40% of the original pattern size. Moreover, 74% of the EBs obtained are within 15 µm of the mean diameter, which represents superior size control over standard EB formation techniques such as suspension culture and hanging drop.[30, 31] Directly harvesting the patterned colonies in this manner permits the standardization of lineage specific differentiation studies as multiple size ranges can be achieved using the presently described µCP SAMs (Fig. 2).
Fig. 8.
(a) Colonies patterned on the µCP SAM platform can be harvested to form uniformly sized EBs. EBs were formed from three independently cultured hPSC lines (H9, HSF1, and hIPS2) after 3 days of culture. Phase contrast images of the EBs were obtained immediately after removal from the patterned substrate to quantify the population size distributions (Scale bars = 250 µm). (b) Histograms illustrating the size distribution of EB populations obtained from each cell line.
4. Conclusions
The present study demonstrates a completely synthetic route to stably maintain colonies of self-renewing hPSCs in culture. Although the development of this platform is still at its inception, it clearly offers a number of attractive features for the routine culturing of hPSCs. A significant advantage offered by the µCP approach is the far greater uniformity of hPSC growth as compared to currently available stem cell culture strategies. This unique capability is likely to overcome the heterogeneity observed in the expression of pluripotency factors found in conventional cultures (Figs. 4–5), and may thus provide new insights to achieving improved lineage specification. In addition, the use of patterning and spatially selective surface chemistry enables researchers, for the first time, to consider fundamental questions regarding the role that colony size and shape play in regulating basic stem cell fate decisions such as self-renewal and differentiation. It should be recognized that the platform does not directly evaluate lineage specification. However, by providing a uniform and reproducible population of pluripotent hPSCs, this platform will be particularly useful in studies directed at understanding the inefficiencies in conventional stem cell differentiation techniques. Moreover, this robust stem cell culturing method is cell line independent and can easily be configured for larger scale operations. To achieve the latter, the µCP SAM platform would need to be integrated with an automated microfluidic system that can facilitate the expansion and harvesting of hPSC populations to levels appropriate for stem cell based therapies. In summary, this cell culture platform represents a valuable enabling technology to the stem cell research community that provides a new level of control and standardization to the culture of hPSCs.
Supplementary Material
Highlights.
-
-
hPSC culturing platform based on patterned self assembled monolayers
-
-
Surface chemistry directed hPSC growth, establishing uniform colony size and shape
-
-
Self-renewal and genetic stability of hPSC colonies preserved on patterned µCP SAMs
-
-
Improved uniformity of self-renewing hPSC populations
-
-
First demonstration of serial passaging from µCP patterned SAM interfaces
Acknowledgements
This work has been supported by the National Institutes of Health through Grant P01GM081621. WR acknowledges California Institute of Regenerative Medicine (CIRM) for training grant (T1-00005). SJJ acknowledges the NIH Medical Scientist Training Program and the National Science Foundation IGERT program. We wish to thank our colleagues Dr. Denis Evseenko, Prof. Amander Clark, Prof. Gay Crooks, and Prof. Jerome Zack for their helpful comments and technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://.
References
- 1.Yu J, Thomson JA. Genes & Development. 2008;22:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Hou R, Booth CJ, Yang S-H, Snyder M. Proceedings of the National Academy of Sciences. 2006;103:5688–5693. doi: 10.1073/pnas.0601383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallon BS, Park K-Y, Chen KG, Hamilton RS, McKay RDG. The International Journal of Biochemistry & Cell Biology. 2006;38:1063–1075. doi: 10.1016/j.biocel.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin MJ, Muotri A, Gage F, Varki A. Nature Medicine. 2005;11:228–232. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 8.Saha K, Mei Y, Reisterer CM, Pyzocha NK, Yang J, Muffat J, Davies MC, Alexander MR, Langer R, Anderson DG, Jaenisch R. Proceedings of the National Academy of Sciences. 2011;108:18714–18719. doi: 10.1073/pnas.1114854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. Nat Biotech. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 10.Rodin S, Domogatskaya A, Strom S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. Nat Biotech. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 11.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O'Shea KS, Lahann J, Smith GD. Nat Biotech. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derda R, Li L, Orner BP, Lewis RL, Thomson JA, Kiessling LL. ACS Chemical Biology. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- 13.Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho S-W, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart MH, Bendall SC, Levadoux-Martin M, Bhatia M. Nat Meth. 2010;7:917–922. doi: 10.1038/nmeth.1519. [DOI] [PubMed] [Google Scholar]
- 15.Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. PLoS ONE. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, Kumacheva E, Zandstra PW. EMBO J. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 18.Rogers JA, Nuzzo RG. Materials Today. 2005;8:50–56. [Google Scholar]
- 19.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chemical Reviews. 2005;105:1103–1170. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 20.Damoiseaux R, Sherman SP, Alva JA, Peterson C, Pyle AD. Stem Cells. 2009;27:533–542. doi: 10.1634/stemcells.2008-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 22.Peerani R, Bauwens C, Kumacheva E, Zandstra PW. In: Patterning Mouse and Human Embryonic Stem Cells Using Micro-contact Printing Stem Cells in Regenerative Medicine. Audet J, Stanford WL, editors. Humana Press; 2009. pp. 21–33. [DOI] [PubMed] [Google Scholar]
- 23.Orner BP, Derda R, Lewis RL, Thomson JA, Kiessling LL. Journal of the American Chemical Society. 2004;126:10808–10809. doi: 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- 24.Faucheux N, Schweiss R, Lutzow K, Werner C, Groth T. Biomaterials. 2004;25:2721–2730. doi: 10.1016/j.biomaterials.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Nat Biotech. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 26.Evseenko D, Schenke-Layland K, Dravid G, Zhu Y, Hao Q-L, Scholes J, Wang XC, MacLellan WR, Crooks GM. Stem Cells and Development. 2009;18:919–928. doi: 10.1089/scd.2008.0293. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Wang BH, Wang S, Moalim-Nour L, Mohib K, Lohnes D, Wang L. Biophysical Journal. 2010;98:2442–2451. doi: 10.1016/j.bpj.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Villa-Diaz LG, Sun Y, Weng S, Kim JK, Lam RHW, Han L, Fan R, Krebsbach PH, Fu J. ACS Nano. 2012;6:4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laslett A, Grimmond S, Gardiner B, Stamp L, Lin A, Hawes S, Wormald S, Nikolic-Paterson D, Haylock D, Pera M. BMC Developmental Biology. 2007;7:12. doi: 10.1186/1471-213X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang SM, Kyba M, Perlingeiro R, Daley GQ, Zandstra PW. Biotechnology and Bioengineering. 2002;78:442–453. doi: 10.1002/bit.10220. [DOI] [PubMed] [Google Scholar]
- 31.Kurosawa H. Journal of Bioscience and Bioengineering. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.