Summary
This study extends the functional meaning of lung cancer susceptibility loci to gene expression in the lung and suggests that the risk alleles on 15q25 and 6p21 mediate their effect by regulating the expression levels of CHRNA5 and APOM, respectively.
Abstract
Recent studies identified three genetic loci reproducibly associated with lung cancer in populations of European ancestry, namely 15q25, 5p15 and 6p21. The goals of this study are first to confirm whether these loci are associated with lung cancer in a French Canadian population and second to identify disease-associated single nucleotide polymorphisms (SNPs) influencing messenger RNA (mRNA) expression levels of genes in the lung, that is expression quantitative trait loci (eQTLs). SNPs were genotyped in 420 patients undergoing lung cancer surgery and compared with 3151 controls of European ancestry. Genome-wide gene expression levels in non-tumor lung tissues of the same 420 patients were also measured to identify eQTLs. Significant eQTLs were then followed-up in two replication sets (n = 339 and 363). SNPs found in the three susceptibility loci were associated with lung cancer in the French Canadian population. Strong eQTLs were found on chromosome 15q25 with the expression levels of CHRNA5 (P = 2.23 × 10− 22 with rs12907966). The CHRNA5-rs12907966 eQTL was convincingly validated in the two replication sets (P = 3.46 × 10− 16 and 2.01 × 10− 15). On 6p21, a trend was observed for rs3131379 to be associated with the expression of APOM (P = 3.58 × 10− 4) and validated in the replication sets (P = 1.11 × 10− 8 and 6.84 × 10− 4). On 5p15, no significant eQTLs were found. This study confirmed that chromosomes 15q25, 5p15 and 6p21 harbored susceptibility loci for lung cancer in French Canadians. Most importantly, this study suggests that the risk alleles at 15q25 and 6p21 may mediate their effect by regulating the mRNA expression levels of CHRNA5 and APOM in the lung.
Introduction
Lung cancer has an important heritable component (1). Genome-wide association studies have identified three loci reproducibly associated with lung cancer (2–10). The most strongly associated locus in populations of European descent is on chromosome 15q25, which is a region also associated with smoking behavior (11,12), nicotine addiction (3) and chronic obstructive pulmonary disease (13). Six genes are located in this region including three nicotinic acetylcholine receptor subunits (CHRNA5, CHRNA3 and CHRNB4). It is still unknown whether genes located on 15q25 are causally involved in the pathogenesis of lung cancer or an effect mediated by changing smoking behavior. The lack of association between the 15q25 locus and lung cancer among never smokers (9,14–19) is consistent with genetic variants conferring risk of smoking-related lung diseases through their effects on tobacco addiction. In contrast, other evidences argue against this hypothesis showing significant effect of the 15q25 locus on smoking-related diseases after adjustment for smoking exposure (4,20,21). Multiple distinct loci affecting both smoking behavior (11,20) and lung cancer (8) were reported on 15q25. It is also important to note that the 15q25 lung cancer susceptibility locus was not replicated in populations of Asian ancestry (19,22–26). More functional studies are needed to find the causal alleles and genes on 15q25.
The two additional lung cancer risk loci identified by genome-wide association studies map to 6p21 and 5p15. The 6p21 locus is a 600 kb region located in the classical class III subregion of the major histocompatibility complex (27) harboring candidate genes for lung cancer including BAG6 (also known as BAT3), APOM, TNXB and MSH5. The 5p15 susceptibility locus is more specifically associated with lung adenocarcinoma (8,9) and the most strongly associated single nucleotide polymorphisms (SNPs) are located within, or in proximity to, two biologically relevant genes, CLPTM1L and TERT. Further functional studies are needed to identify causal genes and genetic variants in these lung cancer susceptibility loci.
Previous studies suggest that risk alleles on chromosome 15q25 may mediate their effects by modulating the messenger RNA (mRNA) expression levels of the CHRNA5 gene in the brain (28,29) and lung (30) tissues. We therefore hypothesized that the causal genetic variants for lung cancer on chromosomes 15q25, 6p21 and 5p15 act by modulating the expression levels in the lung of transcripts residing in the same chromosomal regions. To test this hypothesis, we first verified whether the three lung cancer susceptibility loci replicate in a French Canadian population. We then performed a large-scale lung eQTL study in the same population in order to identify SNPs regulating the expression levels of genes residing in the 15q25, 6p21 and 5p15 regions. Significant lung eQTLs were then replicated in two additional European populations.
Materials and methods
Study participants
Blood and non-tumor lung specimens were collected from patients undergoing lung cancer surgery and stored at the ‘Institut universitaire de cardiologie et de pneumologie de Québec’ site of the Respiratory Health Network Biobank of the ‘Fonds de recherche du Québec-Santé’ (www.tissuebank.ca). Genotype data and a detailed pathology report were available for 426 patients. Lung volumes, forced expiratory volume in 1 s and forced vital capacity were determined following the American Thoracic Society guidelines (31) preoperatively. Chronic obstructive pulmonary disease was derived from lung function measurements following the recommendations of the Global initiative for chronic Obstructive Lung Disease (32). Table I shows the clinical characteristics of the subjects. Written informed consent was obtained from all subjects. The study was approved by the local ethics committee.
Table I.
Clinical characteristics of French Canadian participants according to histological types of lung cancer
| All subjects (n = 420) | Adenocarcinoma (n = 240) | Squamous (n =108) | P valuesa | |
|---|---|---|---|---|
| Gender (male:female) | 235:185 (56.0% m) | 111:129 (46.3% m) | 85:23 (78.7% m) | 3.2×10–8 |
| Age (years) | 63.4±10.0 | 62.8±9.2 | 67.5±8.5 | 4.6×10–6 |
| Body mass index (kg/m2) | 26.7±5.3 | 26.3±4.8 | 26.4±5.0 | 0.78 |
| Smoking status | 0.04 | |||
| Never smokers | 37 (8.8%) | 22 (9.2%) | 2 (1.9%) | |
| Former smokers | 289 (68.8%) | 165 (68.8%) | 78 (72.2%) | |
| Current smokers | 94 (22.4%) | 53 (22.1%) | 28 (25.9%) |
Values are mean ± standard deviation.
aStatistical comparisons between patients with adenocarcinoma and squamous cell carcinoma were performed using t-tests for continuous variables and chi-square tests for categorical variables.
Genotyping
Genotyping was carried out using the Illumina Human1M-Duo BeadChip. Selected susceptibility loci for lung cancer were checked for Illumina quality score (GenCall), call rate, Hardy–Weinberg equilibrium and minor allele frequency. Among the 426 samples, 420 participants remained in the study after exclusions for (i) genotypic and phenotypic gender mismatch (33), (ii) patients with outlying average identity-by-state estimates, which indicated possible duplicates and cryptic genetic relatedness (33), and (iii) population outliers detected by STRUCTURE (34) with HapMap subjects as internal controls.
Control population
Cases were compared with publicly available controls of 3294 Caucasian individuals genotyped on the HumanHap550 genotyping BeadChip. The latter were taken from the Illumina® iControlDB. Controls consisted of 1260 males aged 20.9 ± 20.8 and 2034 females aged 32.2 ± 21.5. Among them, 3151 subjects of European ancestry passed the same quality controls described previously and were considered for subsequent analyses. Five out of the seven lung cancer susceptibility SNPs selected in this study were directly genotyped in the control group. MaCH (35) (version 1.0.18) was used for imputation of SNPs (rs16969968 and rs402710) not genotyped in the control group. Reference haplotypes were taken from the 1000 genome project, 1000G Interim Phase I Haplotypes (2010–11 data freeze, 2011-06 haplotypes), which are available on the MaCH Web site (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-PhaseI-Interim.html). A two-step imputation procedure was utilized as recommended by the developers including a pre-phasing step using MaCH and an imputation step using minimac.
SNPs selection
Linkage disequilibrium (LD) was calculated for SNPs previously associated with lung cancer in the three susceptibility loci using the genotyping data from the HapMap CEU population. HapMap data covering chromosome 15q25 (78 806–78 909kb), 5p15 (1286–1343kb) and 6p21 (31 620–32 065kb) were downloaded into Haploview 4.1 (36). Only uncorrelated SNPs (r 2 < 0.8) were selected in this study.
Gene expression in the lung
The lung samples explanted at surgery were examined by an experienced pathologist for clinical diagnosis and staging. For each specimen, a non-neoplastic pulmonary parenchymal sample was harvested from a site distant from the tumor, snap-frozen in liquid nitrogen and stored at −80°C until further processing. Total RNA was extracted using the SV96 Total RNA Isolation System (Promega). Expression profiling was performed using an Affymetrix custom array (see GEO platform GPL10379) and testing 51 562 probe sets. Expression values were extracted using the robust multichip average method (37) as implemented in the Affymetrix Power Tools software. The quality of the arrays was judged using standard quality control parameters (38). Following quality controls, 409 patients had both genotypes and gene expression levels available for eQTL analyses. The expression data set is accessible through GEO series accession number GSE23546.
Statistical analyses
Genetic association tests were performed using the basic allelic test as implemented in PLINK (20) (version 1.07) comparing allele frequencies between cases and controls. The allele frequencies of cases were compared with 3151 Caucasian controls. Association tests were first conducted on all cases and controls, and then the same analyses were repeated with cases stratified based on the two major histologic types of cancer, namely adenocarcinoma and squamous cell carcinoma. Heterogeneity across histological groups was assessed by using the Cochran Q test. These genetic association tests were performed to replicate robust lung cancer susceptibility loci. Accordingly, P values <0.05 were considered statistically significant.
Robust multichip average expression values were analyzed using the R statistical software. Expression traits were adjusted for age, sex and smoking status using robust residuals obtained with the rlm function in the R statistical package MASS. Residual values deviating from the median by more than three standard deviations were filtered as outliers. Association tests between adjusted expression traits and SNPs were performed using quantitative association tests implemented in PLINK. Significant eQTLs were those passing Bonferroni correction considering the number of SNPs and genes tested in each region.
Replication sets
Significant eQTLs discovered in the French Canadian population were replicated in lung specimens collected at two other sites, namely University of British Columbia, Vancouver, Canada and University of Groningen, The Netherlands. At Vancouver, subjects provided written informed consent and the study was approved by the ethics committees at the UBC-Providence Health Care Research Institute Ethics Board. At Groningen, the study protocol was consistent with the Research Code of the University Medical Center Groningen and Dutch national ethical and professional guidelines (‘Code of conduct; Dutch federation of Biomedical Scientific Societies’; http://www.federa.org). Whole-genome gene expression profiling and genotyping were carried out with the same arrays described previously. Quality controls and eQTL analyses were conducted as described previously. The final analyses were performed with 339 and 363 lung specimens from UBC and Groningen, respectively. Supplementary Table 1, available at Carcinogenesis Online, shows the clinical characteristics of the replication sets.
Results
Study participants
A total of 420 subjects passed genotyping quality controls (Table I). Subjects were afflicted with various types of cancer, but mostly adenocarcinoma (n = 240) and squamous cell carcinoma (n = 108). The data set included 37 never smokers, 289 former smokers and 94 current smokers. As expected, adenocarcinoma was more prominent among never smokers compared with squamous cell carcinoma. Chronic obstructive pulmonary disease was present in 56.5% of subjects.
SNP selection and genotyping
Supplementary Figure 1, available at Carcinogenesis Online, shows the LD plots for SNPs previously associated with lung cancer on chromosomes 15q25, 5p15 and 6p21. Seven uncorrelated SNPs were selected for replication (Supplementary Table 2, available at Carcinogenesis Online). rs8034191 on 15q25 was also included as it was directly genotyped in both cases and controls. rs402710, missing in the latest release of HapMap (HapMap 3 Release 3), had r 2 values ranging from 0.04 to 0.68 with other lung cancer-associated SNPs on 5p15 in an older version of HapMap (version 2, release 21 January 2007). All SNPs are intronic or intergenic except one coding non-synonymous variant located in exon 5 of the CHRNA5 gene (rs16969968, D398N). The characteristics of the SNPs are shown in Supplementary Table 3, available at Carcinogenesis Online. High genotyping quality scores (GenCall) and call rates were observed. Minor allele frequencies were similar to the HapMap CEU population and all SNPs were in Hardy–Weinberg equilibrium.
Genetic association
The two uncorrelated SNPs on chromosome 15q (rs16969968 and rs8042374, r 2 = 0.15) were associated with lung cancer (Table II). rs16969968 was the most significant SNP in this study. Analyses by histological types revealed that the two chromosome 15q loci were associated with both adenocarcinoma and squamous cell carcinoma (P = 0.0001 and 0.007, respectively). For rs16969968, the minor allele (A) was associated with an increased risk of lung cancer [odds ratio (OR) = 1.38, 95% confidence interval (CI) = 1.19–1.60]. It should be noted that rs16969968 was imputed in the control population. To validate the results, we have tested a second SNP (rs8034191) in LD with rs16969968 (r 2 = 0.9) and genotyped in both cases and controls. As expected, the results for rs8034191 were similar to rs16969968, with minor allele (C) associated with an increased risk of lung cancer (OR = 1.42, 95% CI = 1.22–1.64; Table II). For rs8042374, the minor allele (G) was associated with a decreased risk of lung cancer (OR = 0.78, 95% CI = 0.65–0.93).
Table II.
Association of selected SNPs on 5p15, 15q25 and 6p21 loci with lung cancer risk by cancer type
| Marker (minor allele) | Type | MAF case/controla | P value | OR (95% CI)b | P het |
|---|---|---|---|---|---|
| Chromosome 15q25 | |||||
| rs16969968 (A) | All cancers | 0.43/0.36 | 0.00001 | 1.38 (1.19, 1.60) | |
| Adenocarcinoma | 0.44/0.36 | 0.0001 | 1.45 (1.20, 1.74) | 0.99 | |
| Squamous cell carcinoma | 0.44/0.36 | 0.007 | 1.45 (1.10, 1.91) | ||
| rs8042374 (G) | All cancers | 0.19/0.24 | 0.006 | 0.78 (0.65, 0.93) | |
| Adenocarcinoma | 0.19/0.24 | 0.032 | 0.77 (0.61, 0.98) | 0.26 | |
| Squamous cell carcinoma | 0.16/0.24 | 0.007 | 0.60 (0.42, 0.87) | ||
| rs8034191c (C) | All cancers | 0.43/0.35 | 0.000003 | 1.42 (1.22, 1.64) | |
| Adenocarcinoma | 0.44/0.35 | 0.00006 | 1.46 (1.21, 1.77) | 0.96 | |
| Squamous cell carcinoma | 0.44/0.35 | 0.007 | 1.45 (1.11, 1.91) | ||
| Chromosome 5p15 | |||||
| rs2736100 (T) | All cancers | 0.46/0.50 | 0.056 | 0.87 (0.75, 1.00) | |
| Adenocarcinoma | 0.43/0.50 | 0.004 | 0.76 (0.63, 0.92) | 0.03 | |
| Squamous cell carcinoma | 0.52/0.50 | 0.492 | 0.91 (0.69, 1.19) | ||
| rs4635969 (T) | All cancers | 0.19/0.19 | 0.708 | 0.97 (0.80, 1.16) | |
| Adenocarcinoma | 0.19/0.19 | 0.796 | 0.97 (0.76, 1.23) | 0.72 | |
| Squamous cell carcinoma | 0.18/0.19 | 0.547 | 0.90 (0.63, 1.28) | ||
| rs402710 (T) | All cancers | 0.31/0.33 | 0.339 | 0.93 (0.79, 1.08) | |
| Adenocarcinoma | 0.30/0.33 | 0.129 | 0.85 (0.70, 1.05) | 0.99 | |
| Squamous cell carcinoma | 0.31/0.33 | 0.306 | 0.86 (0.64, 1.15) | ||
| rs401681 (T) | All cancers | 0.42/0.44 | 0.370 | 0.94 (0.81, 1.08) | |
| Adenocarcinoma | 0.41/0.44 | 0.282 | 0.91 (0.75, 1.09) | 0.73 | |
| Squamous cell carcinoma | 0.40/0.44 | 0.248 | 0.87 (0.64, 1.12) | ||
| Chromosome 6p21 | |||||
| rs3131379 (T) | All cancers | 0.07/0.10 | 0.032 | 0.74 (0.57, 0.98) | |
| Adenocarcinoma | 0.08/0.10 | 0.336 | 0.85 (0.61, 1.19) | 0.68 | |
| Squamous cell carcinoma | 0.07/0.10 | 0.266 | 0.75 (0.45, 1.25) | ||
P het, P value for heterogeneity by histology derived from the Cochran Q test. P values <0.05 are shown in bold. aMinor allele frequency.
bEstimated ORs are for the minor alleles.
cIn LD with rs16969968 (r 2 = 0.9), but directly assayed (not imputed) in cases and controls.
None of the four SNPs on chromosome 5p15 were significantly associated with lung cancer of all cell types (Table II). Analyses by histology revealed that rs2736100 was associated with adenocarcinoma (P = 0.004, OR = 0.76, 95% CI = 0.63–0.92). More specifically, the minor allele (T) decreased the risk of adenocarcinoma.
rs3131379 on chromosome 6p21 was associated with lung cancer (P = 0.032, OR = 0.74, 95% CI = 0.57–0.98, Table II). The minor allele (T) was associated with lowered risk. This SNP was not associated with adenocarcinoma or squamous cell carcinoma independently.
eQTL analyses
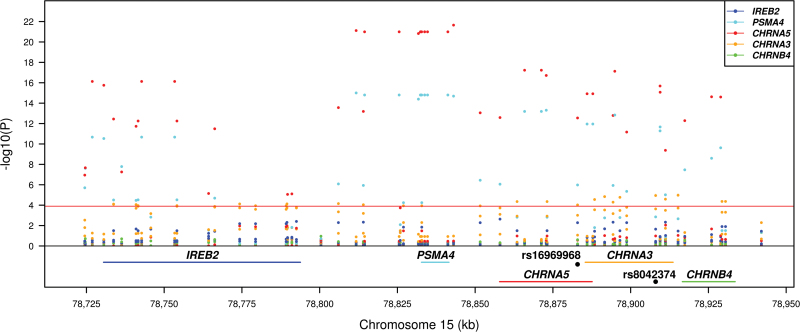
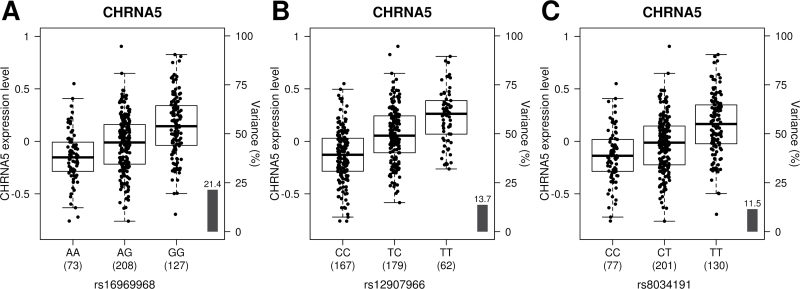
Five genes (IREB2, PSMA4, CHRNA5, CHRNA3, and CHRNB4) found in the 15q25 locus were tested for lung eQTLs. The 223kb region (chromosome 15: 78 720 517–78 943 587; build 37) covering these genes including 10kb downstream of the IREB2 gene and 10kb upstream of the CHRNB4 gene was analyzed. A total of 66 SNPs were genotyped in this region. A Bonferroni P value threshold was set to 1.52 × 10–4 for this region (5 genes × 66 SNPs = 330 tests). Figure 1 shows the association results for the 5 genes and 66 SNPs in the 15q25 region. Strong eQTLs regulating the expression levels of CHRNA5 and PSMA4 were observed in this region. Supplementary Table 4, available at Carcinogenesis Online, shows all gene–SNP combination results. Figure 2 shows the expression levels of CHRNA5 in the lung for the 409 patients according to the genotypes for SNPs rs16969968 (D398N, previously associated with lung cancer; P = 2.83 × 10–13), rs12907966 (the strongest lung eQTL SNP; P = 2.23 × 10–22) and rs8034191 (the strongest SNP associated with lung cancer in this study; P = 2.73 × 10–14). Risk alleles for lung cancer were associated with lower expression of CHRNA5. Although less significant, cis-acting variants were also found to regulate the mRNA levels of CHRNA3 (Figure 1). rs8042374, associated with lung cancer (Table II) was associated with the expression levels of CHRNA3 (P = 1.35 × 10–5). In this case, the minor allele (G) lowered the risk of lung cancer and was associated with higher mRNA CHRNA3 levels.
Fig. 1.
eQTL results for SNPs and genes residing in the 15q25 locus. The positions of the genes are illustrated at the bottom. The red horizontal line represents the Bonferroni threshold (5 genes × 66 SNPs = 330 tests). Positions of genes are from build 37.
Fig. 2.
Gene expression levels of the CHRNA5 gene in the lungs according to genotyping groups for SNPs rs16969968, rs12907966 and rs8034191. The y-axis represents gene expression levels in the lung (n = 409). The x-axis represents the three genotyping groups for SNPs (A) rs16969968, (B) rs12907966 and (C) rs8034191. The number of subjects per genotyping groups is indicated in parenthesis.
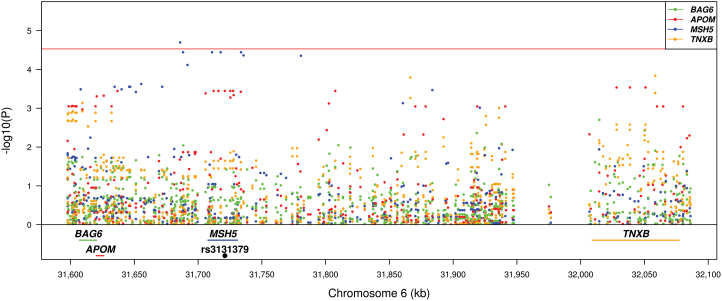
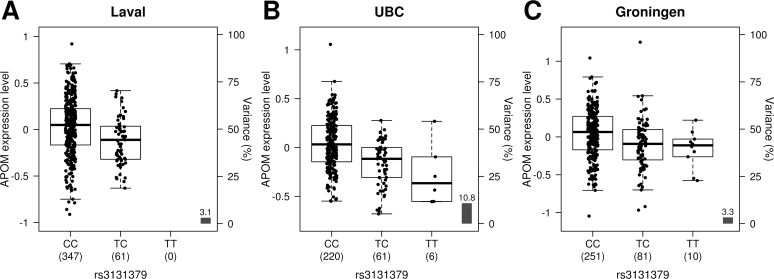
eQTLs analyses were also performed for the 6p21 and 5p15 loci. For the 6p21 locus, a 490kb region (chromosome 6: 31 596 804–32 087 151; build 37) covering 10kb downstream of the BAG6 gene and 10kb upstream of the TNXB gene were analyzed. This region was typed by 422 SNPs and tested for four candidate genes (BAG6, APOM, TNXB and MSH5). A Bonferroni P value threshold was set to 2.96 × 10–5 for this region (4 genes × 422 SNPs = 1688 tests). Figure 3 shows the eQTL results of the 6p21 locus. One SNP (rs405722) was significantly associated with expression levels of MSH5 (P = 2.02 × 10–5). The SNP associated with lung cancer on 6p21, rs3131379 (Table II), was associated with the expression levels of APOM (P = 3.58 × 10–4), but this was not significant after correction for multiple testing. The risk allele for lung cancer (C) was associated with higher mRNA expression of APOM (Figure 4A). rs3131379 and rs405722 were in complete equilibrium (r 2 = 0). We also calculated lung eQTLs for other genes located on 6p21 (Supplementary Figure 2, available at Carcinogenesis Online). New eQTLs were identified, but eQTL-SNPs were not correlated with lung cancer-associated SNPs.
Fig. 3.
eQTL results for SNPs and genes residing in the 6p21 locus. The positions of the genes are illustrated at the bottom. The red horizontal line represents the Bonferroni threshold (4 genes × 422 SNPs = 1688 tests). Positions of genes are from build 37.
Fig. 4.
Gene expression levels of the APOM gene in the lungs according to genotyping groups for the SNP rs3131379 in French Canadian (A; n = 408; P = 3.58×10–4), in UBC (B; n = 287; P = 1.11×10–8) and in Groningen (C; n = 342; P = 6.84×10–4). The y-axis represents gene expression levels in the lung. The x-axis represents the three genotyping groups for SNPs rs3131379 with the number of subjects in parenthesis.
For the 5p15 locus, a 112kb region (chromosome 5: 1 243 286–1 355 002; build 37) covering 10kb downstream of the TERT gene and 10kb upstream of the CLPTM1L gene was analyzed. This region was typed by 46 SNPs and harbored two genes. Supplementary Figure 3, available at Carcinogenesis Online, shows the eQTL results of the 5p15 locus. No SNP was significantly associated with the expression levels of the TERT and CLPTM1L genes following Bonferroni correction. The strongest eQTL was found for SNP rs2853672 and the expression levels of CLPTM1L (P = 0.0015). Supplementary Tables 5 and 6, available at Carcinogenesis Online, show all gene–SNP combination results for the 6p21 and 5p15 loci, respectively.
Replication sets
The most significant eQTLs in the three lung cancer susceptibility loci were followed-up in two replication sets. On 15q25, the association between CHRNA5 and rs16969968 was strongly replicated in the two data sets (Supplementary Figure 4, available at Carcinogenesis Online). Consistent with the French Canadian population, the expression levels of CHRNA5 decrease with the number of risk (A) alleles. The most significant eQTL observed on 15q25 in the French Canadian population (CHRNA5-rs12907966) was also strongly replicated (Supplementary Figure 5, available at Carcinogenesis Online). The association between the strongest SNP associated with lung cancer in this study (rs8034191) and CHRNA5 was also confirmed (Supplementary Figure 6, available at Carcinogenesis Online). In contrast, the eQTL CHRNA3-rs8042374 was not significant in the replication sets (Supplementary Figure 7, available at Carcinogenesis Online). On 6p21, the MSH5-rs405722 eQTL observed in the French Canadian population was validated in the Groningen data set (P = 0.01), but not in the UBC data set (Supplementary Figure 8, available at Carcinogenesis Online). The orientation of effect was similar in the three populations. The SNP associated with lung cancer on 6p21 (rs3131379) and the expression of APOM was also followed-up. Figure 4 shows that this eQTL was strongly validated in the two replication sets. The strongest eQTL on 5p15 in the French Canadian cohort, CLPTM1L-rs2853672, was not supported in the replication sets (Supplementary Figure 9, available at Carcinogenesis Online).
Discussion
This study provides further evidence that genetic variants at 15q25, 5p15, and 6p21 are associated with the risk of lung cancer. ORs of similar magnitude were observed in this French Canadian population compared with previous studies in Caucasians. Most importantly, we have identified cis-acting polymorphisms affecting the expression levels of CHRNA5 in the lung that may explain, at least partly, the molecular mechanism underpinning the risk of lung cancer on chromosome 15q25. We also provided evidence that the SNP associated with lung cancer on 6p21 (rs3131379) acts by modulating the expression levels of APOM in the lung.
The most compelling genetic association with lung cancer was observed on chromosome 15q with a non-synonymous variant (rs16969968, D398N) located in exon 5 of the CHRNA5 gene. The minor allele (here typed as A) was linked to increased risk of lung cancer, which is consistent with previous studies (2–4). In other studies, this allele was also shown to increase the risk of nicotine dependency (39–41). A previous in vitro functional study demonstrated that the nicotinic receptors containing the minor allele (N398) exhibit reduced response to a specific nicotinic agonist (40) suggesting a functional role for this missense mutation. Wang et al. (28) measured gene expression levels by real-time PCR in 104 brain samples from individuals of European descent and showed strong evidence of association between mRNA expression levels of CHRNA5 and polymorphisms located 5ʹ of or within this gene. However, the variants strongly associated with CHRNA5 mRNA expression were not the same as those strongly associated with lung cancer. Accordingly it is not clear from this study whether high or low expression of CHRNA5 in the brain increase the risk of lung cancer. Complex molecular mechanisms may explain these observations. Alternatively, the brain might not be the most relevant tissues to assess molecular mechanisms underlying the risk of lung cancer on chromosome 15q25 locus. In this study, we investigated lung tissue and showed that the risk alleles for lung cancer were associated with lower expression of CHRNA5. The risk allele at rs16969968 (A, coding for N398) was associated with both lung cancer and lower expression of CHRNA5 in the lung. Interestingly, our results confirm those obtained by Falvella et al. (30), who observed decreasing mRNA CHRNA5 levels with increasing dosage of the A allele at rs16969968 in 69 non-tumor lung samples from patients who had adenocarcinoma. Together, these observations suggested that this non-synonymous polymorphism (rs16969968, D398N) located in exon 5 of the CHRNA5 gene may confer susceptibility to lung cancer through regulation of CHRNA5 in the lung. However, this study revealed additional eQTL-SNPs most strongly associated with mRNA expression levels of CHRNA5 in the lung. rs12907966 in the promoter region of CHRNA5 located nearly 15kb from the transcriptional start site of that gene was the most significant SNP associated with CHRNA5 (see Supplementary Figure 5, available at Carcinogenesis Online). Accordingly, our study further refines the causal SNP(s) associated with lung cancer on 15q25. Additional functional studies are warranted.
The SNP associated with lung cancer on 6p21 (rs3131379 and Table II) was more strongly associated with the expression of APOM. Although borderline significant in the French Canadian set (P = 0.0004), the effect of this SNP on APOM mRNA levels was validated in the two replication sets (Figure 4). This provides suggestive evidence that the risk allele on 6p21 acts by modulating the expression levels of APOM in the lung. This gene encoded apolipoprotein M involved in the transport of lipids and its potential role in lung cancer remains to be determined. SNPs flanking the APOM gene were recently associated with lung function and emphysema (42). The 6p21 region contains many genes and more detailed functional evaluation and validation will be required to identify the causal gene/variant at this locus.
Our replication analyses further support that the risk conferred by the 5p15 locus differs across histological subtypes of lung cancer. We observed a significant association at 5p15 (rs2736100) with adenocarcinoma, but not with all lung cancers or with squamous cell carcinoma. The stronger association with adenocarcinoma is consistent with previous studies (8,9). rs2736100 is located in intron 2 of the TERT gene. TERT encodes the enzyme telomerase reverse transcriptase that is potentially implicated in lung tumorigenesis by maintaining telomeres length (43). How this intronic variant affects TERT function and in turn predisposes to lung adenocarcinoma is unknown. Telomere length in peripheral white blood cells was recently associated with lung cancer in a cohort of Asian females (44). The allele G of rs2736100 was associated with longer telomere length, but further studies are needed to validate this finding. In this study as well as in previous large-scale genetic association studies on lung cancer (10,19,45), the G allele was associated with higher risk of lung cancer (Supplementary Table 7, available at Carcinogenesis Online). In this study, no SNP in the 5p15 locus were found to affect the expression of TERT in the lung. A trend was observed for SNPs within the TERT gene to influence the mRNA expression levels of CLPTM1L in the French Canadian cohort (Supplementary Figure 9, available at Carcinogenesis Online), but this was not supported in two replication sets. More functional studies will be required to identify the most likely causal gene in this region.
In conclusion, we have replicated three susceptibility loci for lung cancer in a French Canadian population. Confirmation of these risk loci suggests that functional studies in French Canadian patients are valuable and likely relevant for other Caucasian populations as well. We thus conducted a lung eQTL study to improve our molecular understanding underpinning the association between lung cancer development and the three replicated loci. We strongly confirmed that risk alleles for lung cancer on 15q25 modulate the mRNA expression levels of CHRNA5 in the lung. A disease-associated genetic variant regulating mRNA expression levels of APOM residing at 6p21 was also identified in this study. The role of this gene in lung cancer development is unknown. In contrast, SNPs located on 5p15 were not found to regulate the expression levels of known candidate genes for lung cancer (i.e. CLPTM1L and TERT) in this region. Together, these results are an important step forward to understand the underlying biology of the most consistent susceptibility loci for lung cancer.
Supplementary material
Supplementary Figures 1–9 and Tables 1–7 can be found at http://carcin.oxfordjournals.org/
Funding
Y.B. was a research scholar from the Heart and Stroke Foundation of Canada and recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec-Santé. Chaire de pneumologie de la Fondation JD Bégin de l’Université Laval, the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the Fonds de recherche Québec-Santé, the Cancer Research Society and Read for the Cure and the Canadian Institutes of Health Research (MOP–123369).
Supplementary Material
Acknowledgements
The authors would like to thank Christine Racine and Sabrina Biardel at the Respiratory Health Network Biobank of the Fonds de recherche Québec-Santé for their valuable assistance. We also acknowledge the staff at Calcul Québec for IT support with the high-performance computer clusters.
Conflict of Interest Statement: D.N. is a full-time employee of Merck. D.S.P. received consultancy fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Takeda, TEVA and a grant from Chiesi. D.D.S. has served on advisory boards of Almirall, Nycomed, Talecris, AstraZeneca, Merck Frosst, Novartis and GlaxoSmithKline, received grants from AstraZeneca, GlaxoSmithKline and Wyeth, and received honoraria for speaking engagements from Takeda, AstraZeneca, GlaxoSmithKline and Boehringer Ingelheim. W.T. received a grant from Merck Sharp Dohme, received consultancy fees from Pfizer and received lecture fees from GlaxoSmithKline, Chiesi and Roche Diagnosis. M.Lamontagne is the recipient of a doctoral studentship from the Fonds de recherche Québec-Santé.
Glossary
Abbreviations:
- CI
confidence interval
- eQTL
expression quantitative trait loci
- LD
linkage disequilibrium
- mRNA
messenger RNA
- OR
odds ratio
- SNP
single nucleotide polymorphism.
References
- 1. Matakidou A., et al. (2005). Systematic review of the relationship between family history and lung cancer risk. Br. J. Cancer, 93, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hung R.J., et al. (2008). A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 3. Thorgeirsson T.E., et al. (2008). A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature, 452, 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amos C.I., et al. (2008). Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., et al. (2008). Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKay J.D., et al. (2008). Lung cancer susceptibility locus at 5p15.33. Nat. Genet., 40, 1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu P., et al. (2008). Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl Cancer Inst., 100, 1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Broderick P., et al. (2009). Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res., 69, 6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Landi M.T., et al. (2009). A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Timofeeva M.N., et al. (2012). Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet., 21, 4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J.Z., et al. (2010). Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet., 42, 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tobacco and Genetics Consortium. (2010). Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet., 42, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pillai S.G., et al. (2009). A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet., 5, e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y., et al. (2010). Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis, 31, 234–238. [DOI] [PubMed] [Google Scholar]
- 15. Spitz M.R., et al. (2008). The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J. Natl Cancer Inst., 100, 1552–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsiung C.A., et al. (2010). The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet., 6, e1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y., et al. (2010). Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet. Oncol., 11, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahn M.J., et al. (2012). The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum. Genet., 131, 365–372. [DOI] [PubMed] [Google Scholar]
- 19. Lan Q., et al. (2012). Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet., 44, 1330–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saccone N.L., et al. (2010). Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet., 6, e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chappell S.L., et al. (2011). The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Med. Genet., 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoon K.A., et al. (2010). A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum. Mol. Genet., 19, 4948–4954. [DOI] [PubMed] [Google Scholar]
- 23. Miki D., et al. (2010). Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet., 42, 893–896. [DOI] [PubMed] [Google Scholar]
- 24. Hu Z., et al. (2011). A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet., 43, 792–796. [DOI] [PubMed] [Google Scholar]
- 25. Dong J., et al. (2012). Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat. Genet., 44, 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shiraishi K., et al. (2012). A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat. Genet., 44, 900–903. [DOI] [PubMed] [Google Scholar]
- 27. Horton R., et al. (2004). Gene map of the extended human MHC. Nat. Rev. Genet., 5, 889–899. [DOI] [PubMed] [Google Scholar]
- 28. Wang J.C., et al. (2009). Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum. Mol. Genet., 18, 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J.C., et al. (2009). Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry, 14, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Falvella F.S., et al. (2009). Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin. Cancer Res., 15, 1837–1842. [DOI] [PubMed] [Google Scholar]
- 31. American Thoracic Society. (1995). Standardization of spirometry, 1994 update. Am. J. Respir. Crit. Care. Med., 152, 1107–36. [DOI] [PubMed] [Google Scholar]
- 32. Rabe K.F., et al. (2007). Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med., 176, 532–555. [DOI] [PubMed] [Google Scholar]
- 33. Purcell S., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Falush D., et al. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y., et al. (2009). Genotype imputation. Annu. Rev. Genomics Hum. Genet., 10, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barrett J.C., et al. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 37. Irizarry R.A., et al. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- 38. Heber S., et al. (2006). Quality assessment of Affymetrix GeneChip data. OMICS, 10, 358–368. [DOI] [PubMed] [Google Scholar]
- 39. Berrettini W., et al. (2008). Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry, 13, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bierut L.J., et al. (2008). Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry, 165, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stevens V.L., et al. (2008). Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol. Biomarkers Prev., 17, 3517–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burkart K.M., et al. (2014). APOM and high-density lipoprotein cholesterol are associated with lung function and per cent emphysema. Eur. Respir. J., 43, 1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Janknecht R. (2004). On the road to immortality: hTERT upregulation in cancer cells. FEBS Lett., 564, 9–13. [DOI] [PubMed] [Google Scholar]
- 44. Lan Q., et al. (2013). Longer telomere length in peripheral white blood cells is associated with risk of lung cancer and the rs2736100 (CLPTM1L-TERT) polymorphism in a prospective cohort study among women in China. PLoS One, 8, e59230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Truong T., et al. (2010). Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl Cancer Inst., 102, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.