Summary
Our study highlights the clinical significance of metastasis-associated lncRNAs expression in GC cohorts and the functional role of HOTAIR as a metastasis driver in gastric neoplasia using a series of in vitro and in vivo experimental approaches.
Abstract
The prognosis of gastric cancer (GC) patients with peritoneal dissemination remains poor, and a better understanding of the underlying mechanisms is critical for the development of new treatments that will improve survival in these patients. This study aimed to clarify the clinical and biological role of two key metastasis-associated long non-coding RNAs (lncRNAs) in GC. We analyzed the expression levels of two lncRNAs—Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and HOX-Antisense Intergenic RNA (HOTAIR)—by real-time reverse transcription PCR in 300 gastric tissues (150 GC and 150 adjacent normal mucosa), and in seven GC cell lines. Functional characterization for the role of HOTAIR in GC was performed by small interfering RNA (siRNA) knockdown, followed by series of in-vitro and in-vivo experiments. Expression of both lncRNAs was significantly higher in cancerous tissues than in corresponding normal mucosa, and higher expression of these lncRNAs significantly correlated with peritoneal metastasis in GC patients. In addition, elevated HOTAIR expression emerged both as an independent prognostic and risk factor for peritoneal dissemination. SiRNA knockdown of HOTAIR in GC cells significantly inhibited cell proliferation, migration and invasion, but concurrently enhanced the anoikis rate in transfected cells. In an in vivo assay, HOTAIR siRNA-transfected MKN45 cells injected into nude mice inhibited the growth of xenograft tumors and peritoneal metastasis compared with controls. Our data provide novel evidence for the biological and clinical significance of HOTAIR expression as a potential biomarker for identifying patients with peritoneal metastasis, and as a novel therapeutic target in patients with gastric neoplasia.
Introduction
Metastasis of cancer cells is a critical event in tumor progression and determining the prognosis of patients with malignant disease. Recent advances in multimodal therapeutic regimens that combine systemic chemotherapy with radiation therapy and surgery have helped improve the overall prognosis in advanced cancer patients with various type of distant metastasis; nevertheless the manifestation of peritoneal metastasis is commonly considered a terminal condition with an inevitably fatal outcome.
Gastric cancer (GC) is the second most common cause of cancer-related deaths worldwide (1), and peritoneal dissemination represents the most common metastatic pattern in GC (2,3). Furthermore, peritoneal dissemination is the most difficult type of metastasis to treat due to the lack of effective treatment regimen, which results in an extremely poor prognosis of ~3–6 months (4,5). Therefore, a better understanding of the molecular mechanisms underlying peritoneal dissemination is essential for the development of new treatments that might lead to improved survival of GC patients with peritoneal dissemination.
Most studies investigating mechanisms of metastasis have focused on specific protein-encoding genes or transcription factors controlling expression of these genes. However, recent transcriptomic analyses have provided convincing evidence that an overwhelming amount of the transcribed but non-translated non-coding RNAs (6,7) in the human genome, once perceived as ‘junk’, actually plays an important physiologic role in tissue homeostasis, and may be important in various diseases including cancer. Long non-coding RNAs (lncRNAs), which are commonly defined as transcripts >200 nucleotides in length, have emerged as a class of key regulatory RNAs (8). Recent evidence suggests that lncRNAs are differentially expressed in cancer cells and play a major role in the development of cancer progression, including metastasis (9–13).
One of the first lncRNAs identified in lung metastasis was MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1, also known as NEAT2, for Nuclear-Enriched Abundant Transcript 2). MALAT1 is a nuclear lncRNA with a length of more than 8000 nt, and is encoded on chromosome 11q13 (14,15). MALAT1 regulates alternative splicing by modulating the phosphorylation and distribution of pre-messenger RNA splicing factors (SR proteins) (15). Binding of MALAT1 to unmethylated polycomb 2 proteins mediates the translocation of growth-control genes from the repressive environment of polycomb bodies to a gene activating function in interchromatin granules in response to growth signals, which leads to activation of a growth control program (16). Recently, upregulated expression of MALAT1 was observed in various types of human solid carcinomas (17–22), raising the possibility that MALAT1 may be involved in cancer metastasis (14,23–26).
HOTAIR (HOX-Antisense Intergenic RNA), another lncRNA, has also been suggested to play a role in cancer metastasis. HOTAIR is a 2158-bp long lncRNA located within the homeobox C cluster, and regulates the HOXD cluster of genes through trimethylation of histone H3 lysine-27 (H3K27me3) by the polycomb-repressive complex 2 (27,28). Increased expression of HOTAIR has been observed in various solid tumors (29,30), and this feature correlates with enhanced metastasis in breast and colon cancer patients (28,31). In spite of the growing number of studies highlighting their importance in cancer, none of the previous studies has systematically investigated the potential role of metastasis-associated lncRNAs in human GC. In light of these observations, we hypothesized that these two key lncRNAs might be intimately involved in the development of peritoneal metastasis in GC, which is the most representative and serious metastasis pattern in GC patients. We asked whether exploration of these lncRNAs expression might help establish not only their functional role, but possibly reveal their clinical usefulness as potential cancer metastasis biomarkers in GC patients.
Accordingly, we focused on the two key metastasis-related lncRNAs and evaluated their expression statuses in gastric tissues to evaluate their clinical significance as prognostic biomarkers and risk factors for peritoneal metastases in GC. Furthermore, we also performed extensive in vitro and in vivo experiments to decipher the functional role of HOTAIR in GC.
Materials and methods
Patients and sample collection
We analyzed a cohort of 300 gastric tissues, which consisted of 150 pairs of GC tissues and corresponding non-cancerous gastric mucosa. These patients all underwent surgery for GC between January 2000 and November 2009 at the Mie University Hospital, Japan. Further information on patient demographics and clinic-pathological characteristics is provided in the Supplementary Materials and methods, available at Carcinogenesis Online. All tissue specimens were preserved immediately after surgical resection in RNAlater (QIAGEN, Chatsworth, CA) and stored at −80°C until RNA extraction. Written informed consent was obtained from each patient, and the study was approved by the institutional review boards of all the involved institutions.
Total RNA extraction and cDNA synthesis
RNAlater-preserved surgical specimens were homogenized with a Mixer Mill MM 300 homogenizer (QIAGEN). Total RNA from tissues and cell lines was isolated using RNeasy Mini kits (QIAGEN) according to the manufacturer’s instructions. cDNA was synthesized from 5.0 ug total RNA with random hexamer primers and SuperScript® III Reverse Transcriptase (Invitrogen™, Carlsbad, CA).
Real-time quantitative RT–PCR and relative expression levels
Quantitative reverse transcriptase PCR analysis was performed using the StepOne™ Real Time PCR System (Applied Biosystems®, Foster City, CA). The relative abundance of target transcripts was measured using TaqMan probes for MALAT1 (Hs00273907_s1, TaqMan Gene Expression Assays, Applied Biosystems®) and HOTAIR (Hs03296631_m1, Applied Biosystems®), and the expression of these lncRNAs was normalized to the expression level of GAPDH (Hs02758991_g1, Applied Biosystems®); expression levels were evaluated using Applied Biosystems StepOne Software v2.1. All other primer sets and sequences are shown in Supplementary Table S1, available at Carcinogenesis Online. The relative expression of each messenger RNA and lncRNA was determined by the standard curve method. Further information is provided in the Supplementary Materials and methods, available at Carcinogenesis Online.
In situ hybridization
Five-micrometer-thick formalin fixed paraffin embedded tissue sections were hybridized with the HOTAIR probe (LNA-modified and 5′- and 3′-DIG labeled oligonucleotide; Exiqon, Woburn, MA), followed by incubation with anti-DIG-AP Fab fragments conjugated to alkaline phosphatase, and the hybridization signal was detected by applying nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color substrate (Roche Applied Science, Mannheim, Germany). Positive controls (U6 snRNA, LNA-modified and 5′-and 3′-DIG-labeled oligonucleotide; Exiqon) and negative controls (scrambled, LNA-modified and 5′-and 3′-DIG-labeled oligonucleotide; Exiqon) were included in each hybridization experiment.
Cell lines
Human GC cell lines MKN7 (well differentiated tubular adenocarcinoma), MKN45 (poorly differentiated adenocarcinoma), MKN74 (well differentiated tubular adenocarcinoma), NUGC4 (signet-ring cell carcinoma) and AZ521 (tubular adenocarcinoma) were obtained from the Cell Resource Center for Biomedical Research, Tohoku University. AGS (moderately-poorly differentiated adenocarcinoma) and KATOIII (signet-ring cell carcinoma) were obtained from the American Type Culture Collection (ATCC, Rockville, MD). All cell lines were tested and authenticated using a panel of genetic and epigenetic markers every few months. These cell lines were maintained in Iscove's Modified Dulbecco's Medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and antibiotics at 37°C in 5% humidified CO2 atmosphere.
HOTAIR RNA interference studies
HOTAIR-specific small interfering RNA (siRNA) (Silencer Validated siRNA, standard purity) and control siRNA (Silencer Select Negative Control #1 siRNA) were purchased from Ambion® (Austin, TX). Reverse transfection was performed by mixing cell suspensions with the siRNA oligonucleotides, Optimem I (Invitrogen™) and Lipofectamine 2000 (Invitrogen™) before plating. The final siRNA oligonucleotide concentrations for MKN45 and KATOIII were 30 and 50nM, respectively. Cells were transfected for 24 h followed by additional 24 h incubation in culture media before harvesting for analysis.
Cell proliferation, cell cycle analysis and colony formation assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Sigma) was used to determine cellular proliferation. For cell cycle analysis, the DNA content of HOTAIR siRNA and control siRNA-transfected GC cells was evaluated using the Muse™ Cell cycle assay kit (Millipore, Billerica, MA) according to the manufacturer’s instructions using the Muse Cell Analyzer (Millipore). Colony formation assays were performed 10 days after transfection with HOTAIR or control siRNAs, and the number of colonies with >50 cells was counted using GeneTools image analysis software (Syngene, Cambridge, UK). Additional experimental details on these assays are provided in the Supplementary Materials and methods, available at Carcinogenesis Online.
Cell invasion, migration and wound healing assays
The invasion assay was performed as described previously (32). Two days following transfection with HOTAIR siRNA or negative control siRNA, invasion and migration assays were performed with Boyden chambers (BD Biosciences) that had 8 μm pore size membranes with Matrigel (for the invasion assay) or without Matrigel (for the migration assay). For wound healing assays, cell monolayers transfected with HOTAIR siRNA or control siRNA were scratched with a sterile 200 μl pipette tip, and cell migration was observed for up to 48h. All experiments were conducted with replicates in three independent experiments and results were normalized against the cell numbers in 48 h siRNA transfection.
Anoikis assays
Anoikis assays were performed in 6-well Costar Ultra Low Attachment Microplates (Corning, Corning, NY). After a 24 h induction of anoikis, the rate of apoptosis was measured using the Annexin V dead cell assay (Millipore) and the MTT assay (Sigma) following the manufacturer’s instructions. Further information is provided in the Supplementary Materials and methods, available at Carcinogenesis Online.
In vivo studies
Male athymic nude mice were obtained from Harlan Laboratories (Houston, TX) at 5 weeks of age and kept under controlled conditions (12 h light and dark cycles). The animal protocol was approved by the Institutional Animal Care and Use Committee of the Baylor Research Institute. To establish a xenograft tumor model, MKN45 GC cells transfected with HOTAIR siRNA or negative control siRNA were injected subcutaneously into the abdominal flanks of each mouse (3 × 106 cells per mouse). Six to seven mice were used in each group. The mice were monitored for 18 days following injection, and subcutaneous tumors were measured every 2 days. Tumor size was measured using calipers and the volume was calculated using the following formula: (L × W × H)/2, where L represents length, W width, and H height. In the mouse peritoneal metastasis model, MKN45 GC cells (3 × 106 cells/ml/mouse) transfected with HOTAIR siRNA or negative control siRNA were injected intraperitoneally into mice (n = 14). To examine the effect of HOTAIR inhibition on the peritoneal dissemination potential of GC cells, we killed these mice 60 days postinjection and evaluated the number of nodules in the mesentery and peritoneal walls.
Statistical analyses
Statistical analysis was performed using Medcalc version 12.3.0 (Broekstraat 52, 9030; Mariakerke, Belgium). Results are expressed as means ± SE. Differences between groups were estimated by Wilcoxon’s signed rank test, the χ2 test, Mann–Whitney U-test, and one-way analysis of variance, as appropriate. Receiver operating characteristic curves were established to determine the cut-off values for analyzing prognosis by Youden’s index. Overall patient survival was measured from the date of surgery until the date of death resulting from any cause, or last known follow-up for patients that were still alive. Actuarial survival curves were obtained using the Kaplan–Meier method, and comparisons were made using log-rank tests. For assessment of the performance as a prognostic marker for overall survival, the power calculations are based on the detection difference of 0.05 between favorable and unfavorable prognosis groups. The total sample size of 146 (distribution equally between the two group) achieve over 95% power to detect when the proportions surviving in each group are 0.6 and 0.3 at a significance level of 0.05 using a two-sided log-rank test. Cox proportional hazards models were used to estimate hazard ratios for death. Multivariate logistic regression models were used to predict factors influencing peritoneal metastasis. For multivariate testing, all clinicopathological parameters significant in univariate analysis were included. Two-sided P-values <0.05 were considered to be statistically significant.
Results
Overexpression of metastasis-related lncRNAs in gastric cancer
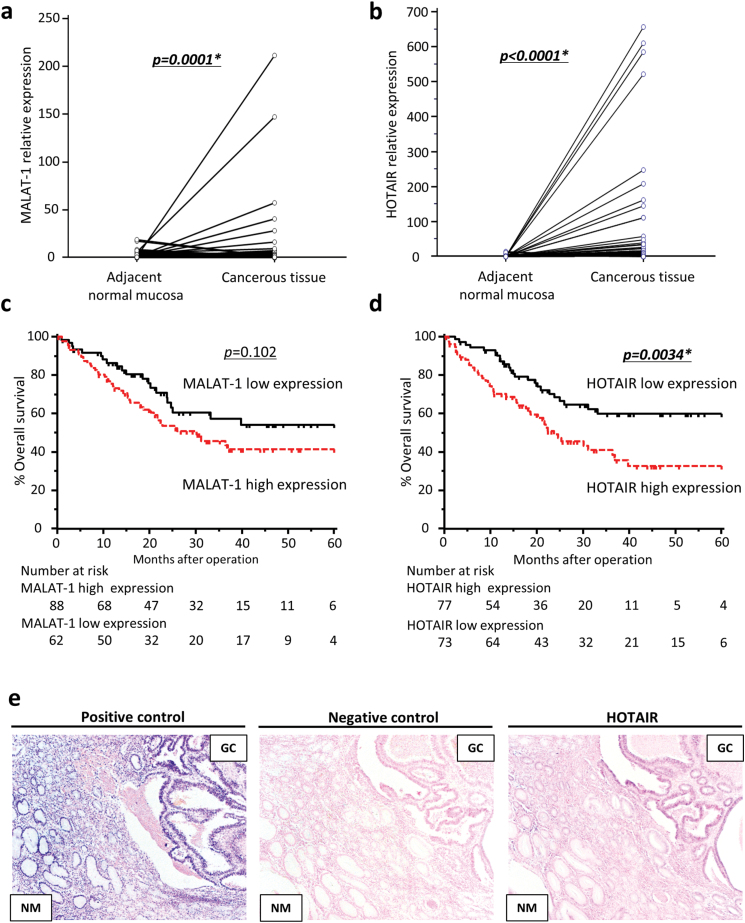
Expression levels of MALAT1 and HOTAIR lncRNAs in 150 GC tissues and paired adjacent normal mucosa were examined by quantitative real-time PCR. The expression levels of both MALAT1 and HOTAIR were significantly higher in neoplastic tissues compared with matching adjacent normal mucosa (MALAT1; P = 0.0001, HOTAIR; P < 0.0001; Figure 1a and b).
Fig. 1.
Metastasis associated lncRNA expression status in GC tissues and adjacent normal mucosa. (a and b) Quantitative real-time PCR was performed on 150 GC and paired normal samples. MALAT1 and HOTAIR expression was significantly higher in cancer samples than in adjacent normal mucosa (P = 0.0001, P < 0.0001, respectively, Wilcoxon rank correlation test). (c and d) Kaplan–Meier survival curves of GC patients according to MALAT1 (c) and HOTAIR (d) expression levels. Patients with high expression of HOTAIR had significantly poorer prognosis compared with those in the low-expression group. (e) In situ hybridization analysis of HOTAIR in GC tissues and adjacent normal mucosa. NM, normal mucosa; positive control, U6 snRNA; negative control, scramble control.
High expression of HOTAIR is associated with peritoneal metastasis and poor outcome in GC patients
Next, we analyzed the expression patterns of both lncRNAs with various clinicopathological factors to determine whether the expression status has any prognostic significance in GC patients (Table I). The expression cut-off thresholds for each lncRNA were determined according to receiver operating characteristic analyses with Youden’s index to determine overall survival of GC patients. Based upon these determinations, we selected a cut-off threshold of 0.985 for MALAT1 and 0.239 for HOTAIR for further analysis. GCs with expression values higher than the cutoff thresholds for both MALAT1 and HOTAIR lncRNAs were assigned to a high-expression group (MALAT1, n = 88; HOTAOR, n = 77), whereas the remainder were assigned to a low-expression group (MALAT1, n = 62; HOTAIR, n = 73).
Table I.
Clinicopathological variables and MALAT1 and HOTAIR expression in gastric cancer patients
| Variable | n | MALAT-1 expression | P value | HOTAIR expression | P value | |||
|---|---|---|---|---|---|---|---|---|
| High (n = 88) | Low (n = 62) | High (n = 77) | Low (n = 73) | |||||
| Gender | Male | 119 | 68 | 51 | 0.459 | 59 | 60 | 0.399 |
| Female | 31 | 20 | 11 | 18 | 13 | |||
| Age (years) | <69 (median) | 71 | 41 | 30 | 0.828 | 35 | 36 | 0.636 |
| ≧69 | 79 | 47 | 32 | 42 | 37 | |||
| Tumor size | ≧5.5cm (median) | 76 | 46 | 30 | 0.639 | 39 | 37 | 0.997 |
| <5.5 cm | 74 | 42 | 32 | 38 | 36 | |||
| Histological type | Intestinal type | 75 | 42 | 33 | 0.507 | 38 | 37 | 0.871 |
| Diffuse type | 75 | 46 | 29 | 39 | 36 | |||
| Pathological T category | pT1/2 | 52 | 30 | 22 | 0.859 | 21 | 31 | 0.051 |
| pT3/4 | 98 | 58 | 40 | 56 | 42 | |||
| Vessel involvement | Absent | 31 | 16 | 15 | 0.371 | 15 | 16 | 0.626 |
| Present | 119 | 72 | 47 | 62 | 57 | |||
| Lymphatic vessel involvement | Absent | 12 | 5 | 7 | 0.212 | 5 | 7 | 0.485 |
| Present | 138 | 83 | 55 | 72 | 66 | |||
| Lymph node metastasis | Absent | 45 | 22 | 23 | 0.111 | 18 | 27 | 0.069 |
| Present | 105 | 66 | 39 | 59 | 46 | |||
| Hepatic metastasis | Absent | 138 | 80 | 58 | 0.557 | 69 | 69 | 0.268 |
| Present | 12 | 8 | 4 | 8 | 4 | |||
| Peritoneal carcinomatosis | Absent | 123 | 67 | 56 | 0.026* | 56 | 67 | 0.002* |
| Present | 27 | 21 | 6 | 21 | 6 | |||
*P < 0.05.
Elevated MALAT1 expression significantly correlated with peritoneal dissemination (Table I), although patients in the high-expression group did not correlate with poor prognoses (Figure 1c). In contrast, HOTAIR over-expression was significantly associated with both peritoneal metastases (Table I) and with significantly poorer prognosis compared with GCs in the low-expression group (Figure 1d). Moreover, by multivariate analysis, high-HOTAIR expression emerged as an independent prognostic and risk factor for peritoneal metastases in GC patients (Tables II and III). To further confirm the pathological expression pattern of HOTAIR in clinical tissue specimens, in situ hybridization staining was performed in primary GC tissues and adjacent normal mucosa. In situ hybridization experiments revealed nuclear and cytoplasmic staining in GC cells, an observation consistent with previous reports in breast cancer (33). Moreover, HOTAIR expression was upregulated in the primary GC cells compared with the corresponding normal mucosa. Furthermore, in situ hybridization data were in agreement with our quantitative reverse transcriptase PCR results for HOTAIR expression in primary GC and adjacent normal mucosa (Figure 1e). Considering these two metastasis-related lncRNAs, only HOTAIR expression served as an independent prognostic and risk factor for metastasis. Therefore, we focused the rest of our study on HOTAIR for assessment of its biological function in gastric neoplasia.
Table II.
Uni- and multivariate analysis for risk of peritoneal metastasis in gastric cancer patients
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | |
| Gender (male) | 0.28 | 0.11–0.69 | 0.006* | 0.29 | 0.10–0.85 | 0.024* |
| Age [≧69 (median)] | 0.96 | 0.42–2.21 | 0.925 | |||
| Tumor size [≧5.5cm (median)] | 3.42 | 1.35–8.67 | 0.01* | 3.54 | 1.19–10.5 | 0.023* |
| T classification (pT3/4) | 5.3 | 1.51–18.6 | 0.009* | 4.12 | 1.03–16.5 | 0.046* |
| Histological type (intestinal type) | 0.52 | 0.22–1.24 | 0.141 | |||
| Lymphatic invasion (present) | NA | NA | 0.995 | |||
| Venous invasion (present) | 8.39 | 1.09–64.5 | 0.041* | 9.8 | 1.10–87.6 | 0.041* |
| Lymph node metastasis (present) | 2.88 | 0.93–8.86 | 0.066 | |||
| HOTAIR expression (high) | 4.19 | 1.58–11.1 | 0.004* | 4.08 | 1.37–12.2 | 0.012* |
| MALAT1 expression (high) | 2.93 | 1.10–7.75 | 0.031* | 2.88 | 0.96–8.63 | 0.06 |
CI, confidence interval; OR, odds ratio.
*P < 0.05.
Table III.
Uni- and multivariate analysis for predictors of survival in gastric cancer patients
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Gender (male) | 0.83 | 0.46–1.47 | 0.521 | |||
| Age [≧69 (median)] | 1.2 | 0.74–1.95 | 0.455 | |||
| Tumor size [≧5.5cm (median)] | 1.60 | 0.98–2.62 | 0.061 | |||
| T classification (pT3/4) | 3.76 | 1.99–7.09 | <0.001* | 2.06 | 0.93–4.55 | 0.074 |
| Histological type (intestinal type) | 0.99 | 0.61–1.60 | 0.956 | |||
| Lymphatic invasion (present) | 2.07 | 0.65–6.58 | 0.219 | |||
| Venous invasion (present) | 3.08 | 1.40–6.76 | 0.005* | 2.07 | 0.93–4.61 | 0.076 |
| Node involvement (present) | 4.02 | 1.98–8.13 | <0.001* | 1.84 | 0.81–4.18 | 0.147 |
| TNM stage (stage III/IV) | 5.6 | 2.65–11.8 | <0.001* | 2.09 | 0.73–5.97 | 0.167 |
| HOTAIR high expression | 2.09 | 1.26–3.45 | 0.004* | 1.77 | 1.06–2.95 | 0.028* |
| MALAT1 high expression | 1.54 | 0.92–2.58 | 0.105 | |||
CI, confidence interval; HR, hazard ratio.
*P < 0.05.
Inhibition of HOTAIR suppresses cell proliferation, tumorigenicity, migration and invasion in gastric cancer cells
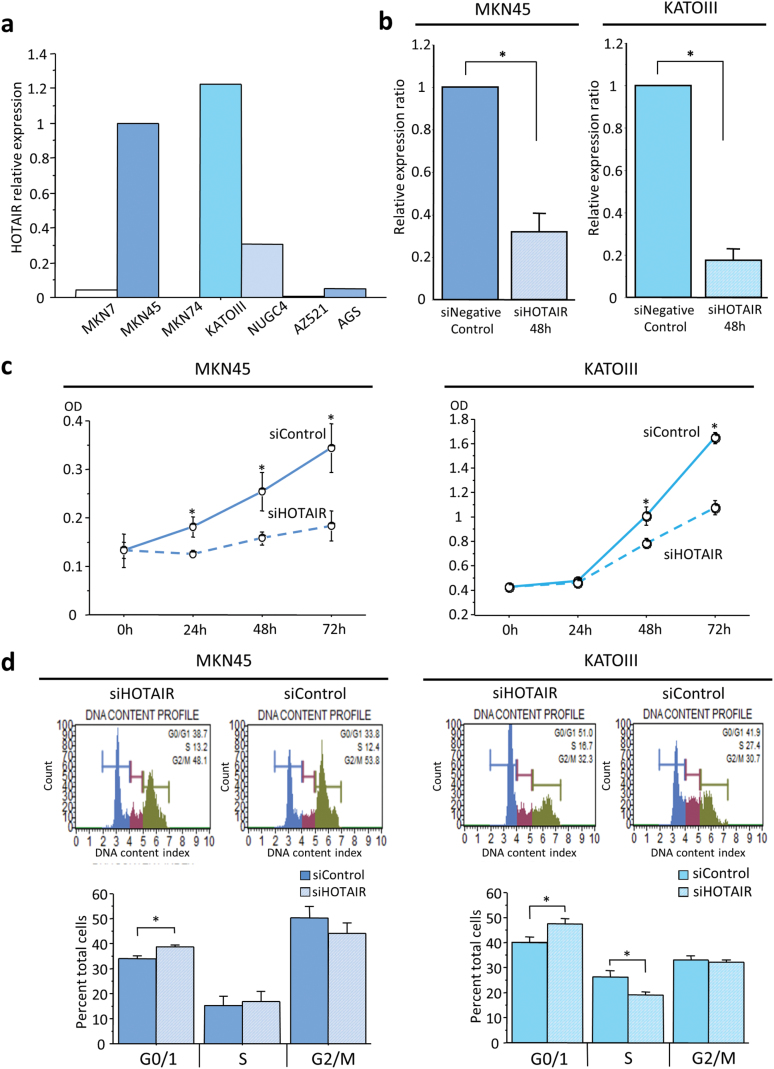
As described above, we found that HOTAIR expression was not only upregulated in cancerous tissue compared with adjacent normal mucosa, its expression also correlated with disease progression in GC patients. In view of these findings, we next determined the functional role of HOTAIR in the pathogenesis of GC. To investigate whether HOTAIR alters the biological characteristics of GC cells, HOTAIR gene silencing was performed using siRNA. HOTAIR expression was assessed by quantitative reverse transcriptase PCR in MKN7, MKN45, MKN74, KATOIII, NUGC4, AZ521 and AGS cells. MKN45 and KATOIII cell lines were subsequently chosen for siRNA transfection experiments since these cells demonstrated the highest levels of HOTAIR expression (Figure 2a). Forty-eight hours after transfection of MKN45 and KATOIII cells with HOTAIR siRNA, its expression was considerably reduced in the HOTAIR siRNA-transfected cells (Figure 2b).
Fig. 2.
Expression status of HOTAIR in GC cell lines and functional analysis using siRNA transfection. (a) HOTAIR expression status in gastric cancer cell lines. (b) Knockdown of HOTAIR expression 48h after transfection in MKN45 and KATOIII GC cell lines. HOTAIR gene expression was downregulated dramatically after HOTAIR knockdown. (c) Effect of HOTAIR suppression on MKN45 and KATOIII cell proliferation as assessed by the MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide) assay. (d) Cell cycle analysis demonstrated that the G0/G1 fraction increased after HOTAIR knockdown. Each value represents the mean ± SE. *P < 0.05.
In order to determine whether siRNA transfection of HOTAIR has biological consequences in controlling cell proliferation in human cancer cell lines, we analyzed the effect of HOTAIR–siRNA on cell proliferation by MTT assays and cell cycle analysis in transfected cell lines. Cell proliferation was significantly reduced following HOTAIR knockdown, in comparison with mock-transfected cells (siControl) as measured by a MTT proliferation assay (Figure 2c). Cell cycle analysis revealed that the G0/G1-phase fraction was significantly increased after HOTAIR knockdown (Figure 2d).
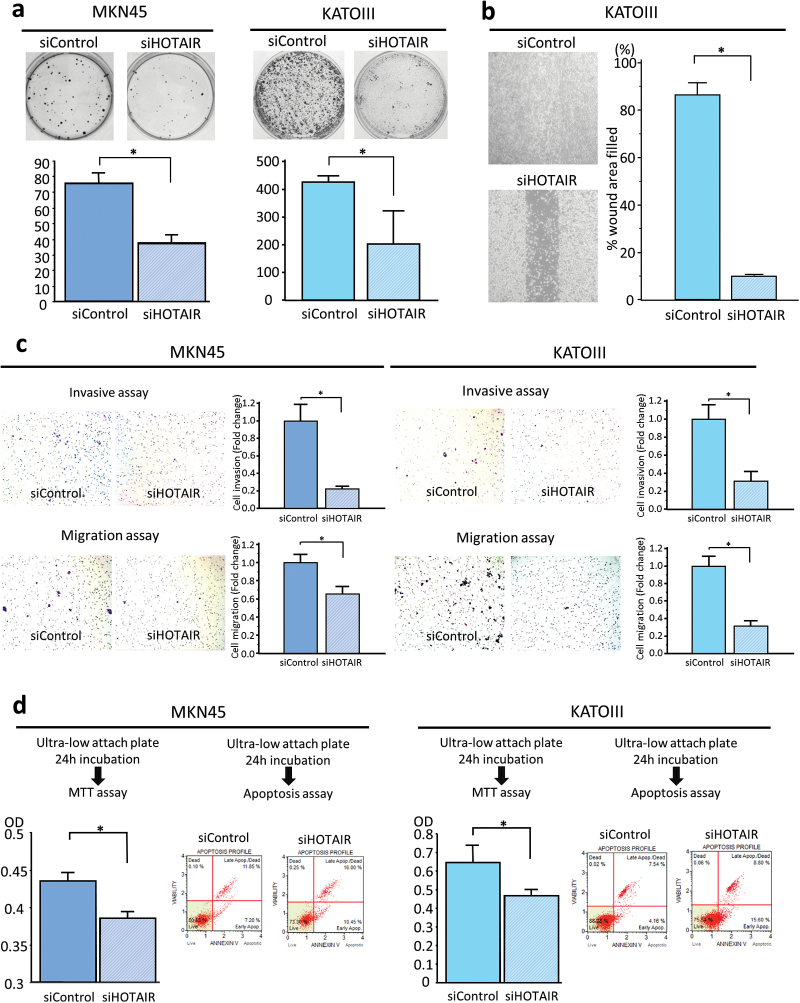
Next, to examine the therapeutic effect of HOTAIR knockdown on the colony-forming ability of single cells plated in vitro, we performed colony formation assays. GC cells treated with siHOTAIR resulted in significantly reduced number of colonies compared with siControl transfected cells (Figure 3a).
Fig. 3.
HOTAIR downregulation causes inhibition of colony-forming ability, migration/invasion, and anoikis resistance in GC cell lines. (a) Colony formation assay. The number of colonies with >50 cells were counted after 10 days incubation. (b) Migration scratch assay to investigate the migratory potential of cultured cells treated with or without HOTAIR knockdown. Cell monolayers were scratched with a pipette tip and imaged 48h after wound formation. (c) Cell invasion and migration assay using Matrigel-coated transwell membranes (upper panel representative picture of invasive assay, bottom panel representative picture of migration chamber, average counts from four random microscopic fields). Each value represents the mean ± SE. *P < 0.05. (d) Anoikis assay to investigate the anoikis resistance of MKN45 and KATOIII cells after HOTAIR siRNA knockdown. Following induction of anoikis for 24h, the number of viable floating cancer cells in low attachment plates was calculated by MTT assay (left panel) and apoptosis rates were measured by annexin V staining (right panel). Apoptotic cells were calculated as upper right (UR) and lower right (LR).
To determine whether silencing HOTAIR inhibited cell migration and invasion, in vitro migration and invasion assays were performed. First, a wound-healing scratch assay was performed to compare the migratory potential of GC cells transfected with HOTAIR siRNA versus control siRNA. As illustrated in Figure 3b and 3c, HOTAIR siRNA transfection of MKN45 and KATOIII GC cells resulted in significantly diminished invasive and migratory potential compared with cells transfected with control siRNA. All of these results suggest that HOTAIR might be intimately involved in the pathogenesis of GC by enhancing the rates of cell growth, colonogenic survival and the invasive and migratory potential of GC cells.
HOTAIR induces anoikis resistance in gastric cancer cells
Anoikis induces apoptosis by loss of cell adhesion (34), and resistance to anoikis is considered a necessary feature of cancer cells during dissemination and metastasis (35). Because our data revealed that HOTAIR overexpression is an independent risk factor for peritoneal dissemination, we hypothesized that one of the novel functions of HOTAIR might be to provide resistance to anoikis in advanced stages of gastric carcinogenesis. To interrogate whether HOTAIR silencing induces anoikis, we incubated GC cells with or without HOTAIR knockdown treatments in anchorage-independent culture using an ultra-low attachment plate. Given the importance of HOTAIR function in GC cells, we examined cell survival using multiple methods following the induction of anoikis. Annexin V staining and flow cytometry demonstrated significantly more apoptosis in both MKN45 and KATOIII cells after HOTAIR siRNA transfection compared with control siRNA-transfected cells. In addition, we used the MTT assay to evaluate the number of viable GC cells floating in low-attachment plates. GC cell lines with siRNA knockdown of HOTAIR showed a decreased number of viable GC cells, which was significantly lower than observed in siControl-transfected cells (Figure 3d).
To further understand the biological consequences of key HOTAIR-dependent target genes in GC cells, we evaluated the expression of 10 genes (ABL2, SNAI1, LAMB3, LMAC2, JAM2, PCDH10, PCDHB5, GDF15, MX1 and OAS1), as they are known to be regulated by HOTAIR in other types of cancer, in HOTAIR siRNA and mock transfected cells (28,36). Inhibition of HOTAIR resulted in changes in the expression of several of its target genes (LAMB3, GDF15 and OAS1) in both MKN45 and KATOIII cells (Supplementary Figure S1, available at Carcinogenesis Online). These observations are consistent with recent reports on HOTAIR-mediated gene expression in other types of cancers (36,37), highlighting that this lncRNA may drive the malignant phenotype in gastric neoplasia by altering the expression of its target genes.
Downregulation of HOTAIR expression suppresses gastric carcinogenesis in xenograft and peritoneal metastasis mouse models
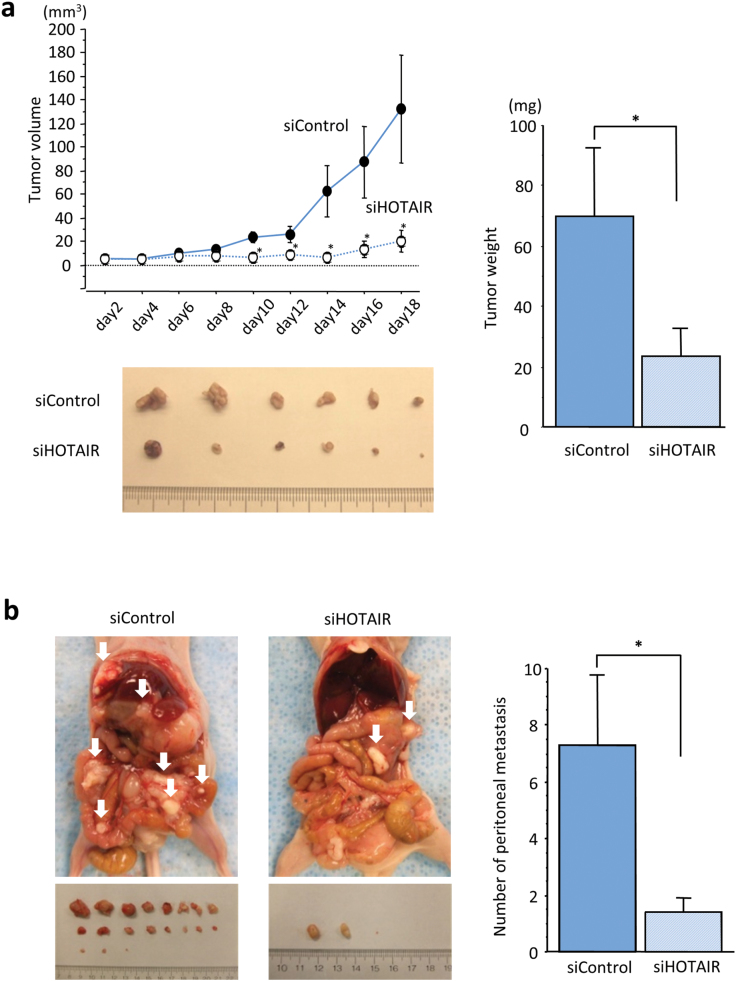
To assess whether knockdown of HOTAIR suppresses the tumorigenicity or formation of peritoneal dissemination in GC, we administered MKN45 cells transfected with either HOTAIR siRNA or control siRNA, subcutaneously or intraperitoneally into nude mice (3 × 106 cells per mouse). In the xenograft mouse model, 18 days post-transfection, tumor volume and weight in cells transfected with HOTAIR siRNA were significantly decreased compared with control siRNA transfected cells (Figure 4a). Likewise, in mice where cells were injected into the peritoneum, numbers of peritoneal tumors following HOTAIR siRNA transfection were significantly fewer than in mice transfected with control siRNA (Figure 4b).
Fig. 4.
Functional analysis of HOTAIR using multiple in vivo models. Effect of HOTAIR knockdown in MKN45 on the xenograft model (a) was assessed by evaluating tumor volume and weight compared with controls. (b) HOTAIR inhibition suppressed the growth of MKN45-induced peritoneal dissemination in the peritoneum compared with negative controls. Seven mice were used in each group and each value represents the mean ± SE. *P < 0.05.
These data using multiple in vivo models further validate our in vitro results and data obtained from the analysis of clinical specimens, further highlighting the possibility that therapies targeting HOTAIR might be attractive for the development of novel treatment for GC patients.
Discussion
Accumulating evidence suggests the importance of lncRNAs in various human cancers (28,30,31,36), but the associations between specific lncRNAs and a mechanistic role in driving GC, especially peritoneal metastasis, has been unclear. In this study, we interrogated the expression patterns of two key cancer-related lncRNAs in GC, determined its association with peritoneal metastasis, characterized their functional roles in gastric neoplasia and made several novel observations. First, we have demonstrated that two metastasis-related lncRNAs, MALAT1 and HOTAIR, were upregulated in primary GC tissues compared corresponding adjacent normal mucosa, and moreover, their expression significantly correlated with peritoneal metastasis. Second, HOTAIR expression in primary tissues was found to be an independent prognostic and risk factor for peritoneal metastasis in GC patients. Third, inhibition of HOTAIR impaired invasion and migration of GC cells as well as proliferation, cycle progress and anoikis resistance in GC cells. Fourth, we were able to successfully validate in-vitro results in two animal models; knockdown of HOTAIR reduced the tumor growth in the xenograft model and inhibited the formation of metastatic nodules in a peritoneal carcinomatosis model.
In recent years, several lncRNAs have been identified and their involvement in human carcinogenesis is currently being ascertained. Since metastasis is the principal cause of cancer-related deaths, there is a growing interest in the identification and understanding of metastasis-associated lncRNAs in human malignancies. One such lncRNA, MALAT1 was originally discovered to be a marker for metastasis development in early stage lung adenocarcinoma (14), and more recently it was shown to be a metastasis-related marker in squamous cell carcinoma of the lung and hepatocellular carcinoma (26,38). HOTAIR has also been described as a metastasis-associated lncRNA and has been shown to be associated with disease progression and prognosis in several types of cancer (28,29,39). A key finding of our study is that these two metastasis-associated lncRNAs were expressed at significantly higher levels in GC tissues than in corresponding adjacent normal mucosa. Moreover, elevated expression of these lncRNAs correlated significantly with peritoneal metastasis in GCs. These results were consistent with our hypothesis and previous studies in various other type of cancer; however, logistic regression analysis of our study illustrated that high levels of HOTAIR in GC was an independent risk factor for peritoneal metastasis, whereas high expression of MALAT1 was compromised by other factors. Furthermore, we showed that elevated expression of HOTAIR was an independent prognostic factor in GC patients. Although another recent study suggest that high expression of HOTAIR is a predictor of poor prognosis in GC patients (40), this previous research included a very small group of GCs and was unable to determine the strong correlation of HOTAIR with prognosis and/or peritoneal metastasis. Multivariate analysis in our study was clearly illustrate that HOTAIR was the only independent marker for predicting patient survival, and emerged as a strongest predictive marker for peritoneal metastasis in the same cohort of GC tissues as described previously (32). From the clinical standpoint, in spite of few advances made for the treatment of peritoneal dissemination, this type of metastasis remains the most frequent and life-threatening form in GC patients. Moreover, it is difficult to diagnose the peritoneal metastasis because individual peritoneal lesions are usually too small to be evaluated accurately by using CT scans, and therefore availability of predictive biomarkers for peritoneal metastasis is critical in the overall management of patients with advanced GC. In this scenario, our data are significant as these suggest that quantification of HOTAIR expression in primary tumor might assist in risk stratification for GC patients that are at risk for disease progression to peritoneal metastasis.
Our study establishes that high expression of HOTAIR is significantly correlated with peritoneal metastasis, and more importantly, there was a trend for its correlation with well-established disease progression factors such as the T-classification and lymph node metastasis. These data suggest that HOTAIR is intimately involved in disease progression, and could be a candidate for targeted therapy in GC. To further understand the biological function of HOTAIR in GC progression, we investigated the ability of HOTAIR to modify malignant cellular behaviors in cell lines through siRNA knockdown experiments. This works demonstrates that silencing HOTAIR inhibits invasion and migration in GC cells. Recent evidence suggests that knockdown of HOTAIR suppressed the proliferation caused by Go/G1 cell cycle arrest in various types of the cancer cells (37,41,42). These previous reports combined with our present data suggest that HOTAIR plays critical roles in the malignant potential in GC.
Interaction with extracellular matrix components activates intracellular pro-survival signaling pathways while disruption of cell-matrix interactions induces apoptotic cell death, via ‘anoikis’ (34). Resistance to anoikis enhances anchorage-independent growth during tumor dissemination and metastasis (35). In addition, recent reports suggest that the acquisition of resistance to anoikis plays an important role in peritoneal dissemination of gastric and ovarian cancer (32,43). In this study, we discovered that knockdown of HOTAIR significantly induced anoikis in vitro and inhibited the establishment of peritoneal metastasis in vivo. Recently, Gupta et al. (28) demonstrated that HOTAIR overexpression promoted anchorage-independent colony growth in soft agar using breast cancer cell lines, Peritoneal metastasis of GC occurs through a multi-step process consisting of invasion into the serosa from intraluminal epithelial sites, detachment from the primary tumor, movement into the peritoneal cavity, attachment to the distant peritoneum and finally proliferation. There is a high degree of consistency between the prior data and this study, which used different methods of HOTAIR alteration, all of which is consistent with the results of our clinical study drawn from a large number of patient samples. Collectively, this suggests that HOTAIR might be directly involved in driving peritoneal metastasis, particularly in the mechanisms involved in detachment from the primary tumor into the peritoneal cavity, which may be a target for preventive therapeutics.
In conclusion, a systematic assessment of metastasis-associated lncRNAs demonstrates novel evidence for the role of HOTAIR in the clinical progression of GC. The present study highlights that HOTAIR expression may serve as potentially important disease biomarker for the identification of high-risk GC patients. Moreover, we have data supporting a mechanistic role for HOTAIR in GC progression, employing both in in vitro and in vivo studies. Therefore, we propose that this lncRNA could be useful clinically as a diagnostic and prognostic indicator in GC patients, and might be a novel therapeutic target for developing more robust and targeted therapeutic regimens.
Supplementary material
Supplementary Materials and methods, Table S1 and Figure S1 can be found at http://carcin.oxfordjournals.org/
Funding
Grant in Aid for Scientific Research from Takeda science foundation, Japan to Y.O.; National Cancer Institute (R01 CA72851), National Institutes of Health, Baylor Research Institute to A.G. and C.R.B.; Charles A Sammons Cancer Center, Baylor University Medical Center to A.G.
Supplementary Material
Acknowledgements
The authors thank Margaret Hinshelwood from the Department of Scientific Publications for careful proofreading and editing of this manuscript. Study concept and design (Y.O., A.G.); provision of samples (Y.T., K.T., Y.I., Y.M., M.K.); acquisition of data (Y.O., S.T., A.G.); analysis and interpretation of data (Y.O., Y.T., K.H., A.G.); statistical analysis (Y.O., Y.T., A.G.); drafting of the manuscript (Y.O., C.R.B., A.G.).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- GC
gastric cancer
- HOTAIR
HOX-Antisense Intergenic RNA
- lncRNAs
long non-coding RNAs
- MALAT1
Metastasis-Associated Lung Adenocarcinoma Transcript 1
- siRNA
small interfering RNA.
References
- 1. Jemal A., et al. (2011). Global cancer statistics. CA Cancer J. Clin., 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Yoo C.H., et al. (2000). Recurrence following curative resection for gastric carcinoma. Br. J. Surg., 87, 236–242. [DOI] [PubMed] [Google Scholar]
- 3. Sasako M., et al. ; Japan Clinical Oncology Group. (2008). D2 lymphadenectomy alone or with para-aortic nodal dissection for gastric cancer. N. Engl. J. Med., 359, 453–462. [DOI] [PubMed] [Google Scholar]
- 4. Ishizone S., et al. (2006). Efficacy of S-1 for patients with peritoneal metastasis of gastric cancer. Chemotherapy, 52, 301–307. [DOI] [PubMed] [Google Scholar]
- 5. Thomassen I., et al. (2014). Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int. J. Cancer, 134, 622–628. [DOI] [PubMed] [Google Scholar]
- 6. Carninci P., et al. (2005). The transcriptional landscape of the mammalian genome. Science, 309, 1559–1563. [DOI] [PubMed] [Google Scholar]
- 7. Kapranov P., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- 8. Rinn J.L., et al. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem., 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mercer T.R., et al. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet., 10, 155–159. [DOI] [PubMed] [Google Scholar]
- 10. Ponting C.P., et al. (2009). Evolution and functions of long noncoding RNAs. Cell, 136, 629–641. [DOI] [PubMed] [Google Scholar]
- 11. Guttman M., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 458, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carninci P. (2008). Non-coding RNA transcription: turning on neighbours. Nat. Cell Biol., 10, 1023–1024. [DOI] [PubMed] [Google Scholar]
- 13. Wang X., et al. (2011). The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb. Perspect. Biol., 3, a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ji P., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031–8041. [DOI] [PubMed] [Google Scholar]
- 15. Tripathi V., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell, 39, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang L., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell, 147, 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin R., et al. (2007). A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene, 26, 851–858. [DOI] [PubMed] [Google Scholar]
- 18. Luo J.H., et al. (2006). Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology, 44, 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamada K., et al. (2006). Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci., 97, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y., et al. (2009). Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res. Treat., 118, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu C., et al. (2011). MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol., 39, 169–175. [DOI] [PubMed] [Google Scholar]
- 22. Fellenberg J., et al. (2007). Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod. Pathol., 20, 1085–1094. [DOI] [PubMed] [Google Scholar]
- 23. Guo F., et al. (2010). Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim. Biophys. Sin. (Shanghai), 42, 224–229. [DOI] [PubMed] [Google Scholar]
- 24. Ying L., et al. (2012). Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst., 8, 2289–2294. [DOI] [PubMed] [Google Scholar]
- 25. Tano K., et al. (2010). MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett., 584, 4575–4580. [DOI] [PubMed] [Google Scholar]
- 26. Schmidt L.H., et al. (2011). The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol., 6, 1984–1992. [DOI] [PubMed] [Google Scholar]
- 27. Woo C.J., et al. (2007). HOTAIR lifts noncoding RNAs to new levels. Cell, 129, 1257–1259. [DOI] [PubMed] [Google Scholar]
- 28. Gupta R.A., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Z., et al. (2011). Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol., 18, 1243–1250. [DOI] [PubMed] [Google Scholar]
- 30. Li D., et al. (2013). Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am. J. Pathol., 182, 64–70. [DOI] [PubMed] [Google Scholar]
- 31. Kogo R., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res., 71, 6320–6326. [DOI] [PubMed] [Google Scholar]
- 32. Okugawa Y., et al. (2013). Brain-derived neurotrophic factor/tropomyosin-related kinase B pathway in gastric cancer. Br. J. Cancer, 108, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chisholm K.M., et al. (2012). Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One, 7, e47998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frisch S.M., et al. (1994). Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol., 124, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eccles S.A., et al. (2007). Metastasis: recent discoveries and novel treatment strategies. Lancet, 369, 1742–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim K., et al. (2013). HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene, 32, 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X., et al. (2013). Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer, 109, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai M.C., et al. (2012). Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol., 29, 1810–1816. [DOI] [PubMed] [Google Scholar]
- 39. Nie Y., et al. (2013). Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci., 104, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Z.Y., et al. (2013). Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci., 9, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J.X., et al. ; Chinese Glioma Cooperative Group. (2013). HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro. Oncol., 15, 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Z., et al. (2013). The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21 (WAF1/CIP1) expression. PLoS One, 8, e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simpson C.D., et al. (2008). Anoikis resistance and tumor metastasis. Cancer Lett., 272, 177–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.