Summary
We systematically examined common genetic variants in the 9p21 region and risk of eight cancers, based on GWAS data deposited in dbGaP. A number of SNPs were associated with multiple cancers, which are not confined to the CDKN2/MTAP cluster.
Abstract
The chromosome 9p21 region has been implicated in the pathogenesis of multiple cancers. We analyzed 9p21 single nucleotide polymorphisms (SNPs) from eight genome-wide association studies (GWAS) with data deposited in dbGaP, including studies of esophageal squamous cell carcinoma (ESCC), gastric cancer (GC), pancreatic cancer, renal cell carcinoma (RCC), lung cancer (LC), breast cancer (BrC), bladder cancer (BC) and prostate cancer (PrC). The number of subjects ranged from 2252 (PrC) to 7619 (LC). SNP-level analyses for each cancer were conducted by logistic regression or random-effects meta-analysis. A subset-based statistical approach (ASSET) was performed to combine SNP-level P values across multiple cancers. We calculated gene-level P values using the adaptive rank truncated product method. We identified that rs1063192 and rs2157719 in the CDKN2A/2B region were significantly associated with ESCC and rs2764736 (3′ of TUSC1) was associated with BC (P ≤ 2.59 × 10−6). ASSET analyses identified four SNPs significantly associated with multiple cancers: rs3731239 (CDKN2A intronic) with ESCC, GC and BC (P = 3.96 × 10− 4); rs10811474 (3′ of IFNW1) with RCC and BrC (P = 0.001); rs12683422 (LINGO2 intronic) with RCC and BC (P = 5.93 × 10− 4) and rs10511729 (3′ of ELAVL2) with LC and BrC (P = 8.63 × 10− 4). At gene level, CDKN2B, CDKN2A and CDKN2B-AS1 were significantly associated with ESCC (P ≤ 4.70 × 10− 5). Rs10511729 and rs10811474 were associated with cis-expression of 9p21 genes in corresponding cancer tissues in the expression quantitative trait loci analysis. In conclusion, we identified several genetic variants in the 9p21 region associated with the risk of multiple cancers, suggesting that this region may contribute to a shared susceptibility across different cancer types.
Introduction
Genetic variants associated with risk of cancers at multiple sites have been observed (1–3). Of note, germline variants in the 9p21 region have been associated with the risk of several cancers. In addition, somatic mutations and deletions of the 9p21 region have been observed in a variety of tumors, underscoring an important role of this genomic region in cancer etiology (4–8). This region harbors a cluster of interferon (IFN) genes, several tumor suppressor genes, including cyclin-dependent kinase inhibitors 2A (CDKN2A), CDKN2B, CDKN2B-AS1 [also known as ANRIL (antisense non-coding RNA in the INK4 locus)] and MTAP and other genes with a less well-established role in cancer, including ELAVL2, TUSC1, MOB3B, C9orf72, LINGO2 and ACO1. CDKN2A was the first high-risk melanoma-predisposition gene identified, and rare genetic mutations in CDKN2A have been identified in familial melanoma kindreds (9,10). In genome-wide association studies (GWAS) of individual cancers, genetic polymorphisms in the CDKN2A/2B-AS1/2B/MTAP gene cluster have been associated with melanoma (11,12), basal cell carcinoma (13), glioma (14–16), lung squamous cell carcinoma (LUSC) (17), breast cancer (BrC) (18), nasopharyngeal cancer (19) and glaucoma (20).
Prior reports have provided some insights into a possible common role of the CDKN2A/2B-AS1/2B/MTAP gene cluster in multiple cancers (11–20). However, evidence for other 9p21 genes in cancer has been sparse. Thus, combining results of existing GWAS may aid the discovery of novel genetic variants with pleiotropic effects, even if their associations with any individual cancer may not surpass a genome-wide significance threshold. However, few studies have systematically investigated the association of common genetic variants with risk of multiple tumors (1–3), partly because of the challenges in combining results from multiple studies using standard meta-analysis approaches when an individual single nucleotide polymorphism (SNP) has an effect in only a subset of cancers or is associated with different cancers in opposite directions. Using a meta-analysis approach that can account for subset-specific and bidirectional effects of individual variants (21), we previously identified several SNPs that were associated with the risk of multiple cancers using data from studies participating in a National Cancer Institute (NCI) iSelect project (2). However, most cancers included in the iSelect analysis were rare cancers and had moderate sample sizes. In addition, the iSelect platform provided only limited SNP coverage. To expand the examination of the 9p21 region, we analyzed SNPs in this region from eight large cancer GWAS, which were deposited in the database of Genotype and Phenotype (dbGaP) (22). The eight cancers included are esophageal squamous cell carcinoma (ESCC), gastric cancer (GC), renal cell carcinoma (RCC), pancreatic cancer (PanC), lung cancer (LC), BrC, bladder cancer (BC) and prostate cancer (PrC). The studies of ESCC and GC consisted of only Chinese, whereas subjects for other cancers were predominantly Caucasians. Three cancers (ESCC, GC and RCC) were also studied in the previous iSelect analysis, but the number of cases included in GWAS was more than doubled for GC and RCC, and nearly doubled for ESCC.
Methods
Study populations
At the time when we requested data from dbGaP (http://www.ncbi.nlm.nih.gov/gap), GWAS data from 12 cancer studies were available. We excluded four studies because of small sample size (two studies with total number of cases and controls <500), lack of controls or unavailable principle component analysis (PCA) data (we only had access to SNPs for the 9p21 region and therefore cannot regenerate PCA). In the final analysis, we included eight cancers, all of which are part of the NCI-Cancer Genetic Markers of Susceptibility (CGEMS, http://dceg.cancer.gov/research/how-we-study/genomic-studies/cgems-summary) project, including upper gastrointestinal cancer (ESCC and GC, dbGaP accession number phs000361), PanC (consent group 1, phs000206), RCC (phs000351, only the NCI scan part available in dbGaP), LC (consent group 1, phs000336), BrC (phs000147), BC (phs000346) and PrC (phs000207) (Table I). Details on the subjects in the genome-wide scanning phase of these GWAS have been described previously (23–30). For ESCC and GC, additional subjects were genotyped using the same platform as used in the initial scanning (23,31), and data from these subjects were provided by the study principal investigator (P.R.T.). ESCC and GC share the same control subjects (n = 2111). Overlapping subjects, predominantly shared controls, were also present for most of the other six cancers (Supplementary Table S1, available at Carcinogenesis Online).
Table I.
Description of the eight cancer GWAS included in this analysisa
| Cancer type (dbGaP study accession) | Cases (n) | Controls (n) | Ethnicity | Source of participants (substudies) | No. of SNPsb | Covariates |
|---|---|---|---|---|---|---|
| ESCC (phs000361. v1.p1) | 1942 | 2111 | Asian | One case–control and one case-only study: Shanxi (China); one cohort study: NITs (China) | 2272 | Age, sex and substudy |
| GC (phs000361.v1.p1) | 1758 | 2111 | Asian | One case–control and one case-only study: Shanxi (China); one cohort study: NITs (China) | 2267 | Age, sex and substudy |
| BrC (phs000147. v1.p1) | 1145 | 1142 | Caucasian | One cohort study: NHS (USA) | 2470 | Age and three PCs |
| LC (phs000336.v1.p1) | 3782 | 3837 | Caucasian | Three cohort studies: ATBC (Finland), CPS II (USA) and PLCO (USA) | 2451 | Age, sex, substudy and four PCs |
| RCC (phs000351. v1.p1) | 1311 | 3424 | Caucasian | Three cohort studies: ATBC (Finland), CPS II (USA) and PLCO (USA); one case– control study: USKC (USA) | 2600 | Sex, substudy and two PCs |
| BC (phs000346.v1.p1) | 2811 | 3241 | Caucasian | Two case–control studies: SBCS (Spain) and NEBCS-ME, VT (USA); two cohort studies: ATBC (Finland) and CPS II (USA) | 3674 | Age, sex, substudy and 10 PCs |
| PrC (phs000207. v1.p1) | 1151 | 1101 | Caucasian | One cohort study: PLCO (USA) | 2509 | Age and 10 PCs |
| PanC (phs000206. v3.p2) | 2440 | 2452 | Caucasian, Asian | 12 cohort studies (USA, unless noted otherwise): ATBC (Finland), CLUE II, CPS II, EPIC, HPFS, NHS, NYU- WHS, PHS, PLCO, SMWHS (China), WHI and WHS; two case–control studies (USA): Mayo and MDA. | 2517 | Age, sex, substudy and eight PCs |
CGEMS, Cancer Genetic Markers of Susceptibility; CLUE, Give Us a Clue to Cancer and Heart Disease Study; CPS, the American Cancer Society Cancer Prevention Study; dbGaP, database of Genotype and Phenotype; EPIC, European Prospective Investigation Into Cancer and Nutrition Study; HPFS, Health Professionals Follow-up Study; Mayo, Mayo Clinic Molecular Epidemiology Case–Control Study; MDA, MD Anderson Cancer Center; NEBCS-ME, VT, New England Bladder Cancer Study-Maine, Vermont; NHS, Nurses’ Health Study; NIT, Nutrition Intervention Trial; NYU-WHS, The New York University Women’s Health Study; PHS, Physicians Health Study; PC, principal components; SBCS, Spanish Bladder Cancer Study; Shanxi, Shanxi UGI Cancer Genetic Project; SMWHS, Shanghai Men’s and Women’s Health Study; USKC, National Cancer Institute United States Kidney Cancer Study; WHI, Women’s Health Initiative; WHS, Womens’ Heath Study.
aData on ESCC and GC were provided by the study principal investigator (P.R.T.) which included additional subjects after the initial GWAS scan. Other discrepancies between numbers here and those shown in dbGaP are due to the limit of the authors’ data access or the applied quality control criteria.
bSNPs without deviation from Hardy–Weinberg equilibrium (P > 10−7) in controls, with minor allele frequency >0.01 in cases and controls combined, and with completion rate >0.90 in cases and controls combined.
Selection of genes and SNPs and quality control
Genotyping data for SNPs located in 35 genes (including their flanking areas of 20kb upstream and 10kb downstream) and intergenic regions comprising the chromosome 9p21 region from 21 067 104 to 32 440 834 (hg18/build 36) were extracted. These genes include IFNB1, IFNW1, IFNA21, IFNA4, IFNA7, IFNA10, IFNA16, IFNA17, IFNA14, IFNA5, KL HL9, IFNA6, IFNA13, IFNA2, IFNA8, IFNA1, IFNE1, MTAP, CDKN2A, CD KN 2B-AS1, CDKN2B, DMRTA1, ELAVL2, TUSC1, C9orf82, PLAA, IFT74, LRRC19, TEK, C9orf11, MOB3B, IFNK, C9orf72, LINGO2 and ACO1. Genotyping methods were described previously (23–30). For each study separately, we excluded SNPs with minor allele frequency <0.01 in the case–control combined dataset, completion rate <0.90, or deviation from Hardy–Weinberg equilibrium among controls (P < 10− 7). Subjects in Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial in the BC study had a generally low completion rate for all 9p21 SNPs (1055 out of 2573 subjects had completion rate <55%), and we therefore excluded all PLCO subjects from the BC study. After quality control filters, the number of evaluable SNPs ranged from 2267 in GC to 3674 in BC (Table I). No SNPs mapped to IFNA16 for ESCC, GC, PanC, RCC and LC, so only 34 genes were analyzed for these cancers.
Imputation analysis of SNPs in the 9p21 region
Genome-wide imputation data were available for three cancers (ESCC, PanC and LC) and we evaluated imputed SNPs in the 9p21 region for these three cancer outcomes. Imputation was conducted separately for each scan using IMPUTE2 software version 2.2.2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html) and version 3 of the 1000 Genomes Project data (http://www.1000genomes.org/) as the reference set. First, the genomic coordinates were lifted over from NCBI human genome build 36 to build 37 using the UCSC lift over tool (http://hgdownload.cse.ucsc.edu/downloads.html). The few loci that failed to be lifted over were also excluded from the imputation. Second, the strand of the inference data was aligned with the 1000 Genomes data by simple allele state comparison or allele frequency matching for A/T and G/C SNPs. We excluded those A/T or G/C SNPs with minor allele frequency >0.45 before any imputation was conducted. We implemented a 4 Mb sliding window to impute across the genome, resulting in 744 jobs running in parallel on the National Institutes of Health BIOWULF cluster (http://biowulf.nih.gov/). A prephasing strategy with SHAPEIT software version 1 (http://www.shapeit.fr/) was adopted to improve the imputation performance. The phased haplotypes from SHAPEIT were fed directly into IMPUTE2. Imputed loci with imputed R2 <0.3 or minor allele frequency <0.01 were excluded from further association analysis.
Statistical analyses
Individual SNP-level analyses were conducted for each cancer separately. An unconditional logistic regression model was initially fitted to calculate the odds ratio and 95% confidence interval (CI) for one minor allele of each SNP in a log-additive model using the R software. We adjusted for the same covariates as in the original GWAS publications or as suggested by study principal investigators (Table I ) (23–30).
For several cancers (LC, BC, PanC and RCC), we had data from substudies conducted in different countries. We evaluated the heterogeneity across substudies for the top 20 SNPs identified from each cancer using the Q statistic. Significant heterogeneity (P heterogeneity < 0.10) was found only for a small subset of SNPs in BC and RCC and none in PanC. However, in LC, 9 of the 20 selected SNPs were heterogeneous across substudies (data not shown). To examine whether the extensive heterogeneity was influenced by different smoking exposures in the different substudies, we obtained detailed smoking data from study investigators (D.A., N.E.C., S.M.G. and M.T.L.); however, the adjustment for smoking variables (smoking status, duration and pack-years) and the analyses restricting to male current smokers only or male current/former smokers [Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) only had male current smokers] did not appreciably reduce the heterogeneity (data not shown). For SNPs showing significant heterogeneity (P heterogeneity < 0.10), we further calculated the effect of SNPs in each individual substudy using logistic regression and combined substudies-specific SNP effects using a random-effect meta-analysis.
We evaluated the linkage disequilibrium (LD) between SNPs using Haploview 4.2. After excluding SNPs with pairwise r 2 ≥ 0.80 in controls, a Bonferroni-corrected significance level for the single SNP analysis was calculated based on the number of SNPs and outcomes studied [P < 3.65 × 10− 6, 0.05/13 712 SNPs (the total number of SNPs in all eight cancers combined), footnote of Table II]. The PanC study included a small proportion of Chinese subjects (n = 113). We conducted a sensitivity analysis by excluding these subjects and obtained similar results (data not shown). We therefore included all subjects in subsequent analyses.
Table II.
SNPs in the 9p21 region significantly associated with individual cancers after Bonferroni correctiona
| Gene | SNP (major, minor allele) | Location | MAF: cases, controls | OR (95% CI) | P |
|---|---|---|---|---|---|
| BC | |||||
| 3′ of TUSC1 | rs2764736 (T, C) | 25057731 | 0.015, 0.027 | 0.36 (0.26–0.49) | 5.29×10−11 |
| ESCC | |||||
| 3′ UTR of CDKN2B | rs1063192 (T, C) | 21993367 | 0.235, 0.194 | 1.29 (1.16–1.44) | 2.45×10−6 |
| CDKN2B-AS1 intronic | rs2157719 (A, G) | 22023366 | 0.141, 0.108 | 1.38 (1.21–1.58) | 2.59×10−6 |
MAF, minor allele frequency.
aSNPs significant after Bonferroni correction for multiple comparisons (P < 3.65×10−6), ranked by P values. The P values and odds ratios (ORs) (per one minor allele) were calculated from random-effect meta-analysis (LC) or logistic regression models (other cancers), adjusting for cancer-specific covariates.
For 39 SNPs that were associated with at least one cancer at P < 0.001, or at least two cancers at P < 0.01 (Supplementary Table S2, available at Carcinogenesis Online), we assessed their associations with multiple cancers using the standard random-effect meta-analytic model (PROC GLIMMIX, SAS 9.2) and a subset-based model, which combines SNP-level associations using the association analysis based on subset (ASSET) implemented in R (21). We did not find significant heterogeneity in effect estimates for these SNPs across substudies of LC, BC, PanC and RCC. Overlapping subjects in different outcomes were accounted for in all meta-analyses. The same reference allele was used for each SNP across the eight cancers, selected as the minor allele for the outcome with the most significant observed P value. ASSET explores subsets of cancers for the presence of true association signals (21). For a subset S of k cancers, ASSET defines Z(S) as test statistic for meta-analyses of the k cancers, for which the weight was determined by the sample size for one outcome relative to the total sample size in the subset. ASSET exhaustively tests all possible subsets and reaches the maximum of the subset-specific Z(S). Two-sided ASSET was used to allow for effects in opposite directions. In our analysis, the significance for meta-analyses was set at P < 0.0013 (0.05/39).
We applied the adaptive rank-truncated product approach available in an R package, to combine association signals for all SNPs within a gene and calculate gene-level associations (1000000 resamplings) (32), for each cancer respectively. This method takes into account the number of SNPs within one gene. The significance level for gene-level analysis was defined as P < 1.79 × 10− 4 [0.05/(35 genes × 8 studies)].
We conducted single SNP-level analysis for imputed SNPs for ESCC (96 238 SNPs), PanC (102 799 SNPs) and LC (106 837 SNPs), respectively. A Bonferroni-corrected significance level for the single SNP analysis was calculated based on the number of SNPs and outcomes studied (P < 1.63 × 10− 7, 0.05/305 874 imputed SNPs). For 200 imputed SNPs that were associated with at least one cancer at P < 0.001, or at least two cancers at P < 0.01, we assessed their associations with multiple cancers using ASSET and significance level was set at P < 2.50 × 10− 4 (0.05/200). We examined the LD between the imputed SNPs significantly associated with multiple cancers and the genotyped SNPs significant for multiple cancers in Table III, based on 1000 Genome data.
Table III.
SNPs with P value <0.001 in two-sided subset search or random-effect meta-analysesa
| rs10811474 | rs3731239 | rs10511729 | rs12683422 | |
|---|---|---|---|---|
| Location | 3′ of IFNW1 | CDKN2A intronic | 3′ of ELAVL2 | LINGO2 intronic |
| Effect alleleb | G | C | G | T |
| Meta-analysis | ||||
| Random-effect P c | 0.005 | 0.19 | 0.16 | 0.64 |
| Two-side subset searchd | ||||
| Combined P | 0.008 | 5.21×10 −4 | 0.006 | 6.99×10 −4 |
| P for positive effect | 0.001 | 3.96×10 −4 | 1.00 | 0.11 |
| Subsets with positive association | BrC, RCC | ESCC, GC, BC | — | — |
| P for negative effect | 0.89 | 0.12 | 7.93×10 −4 | 5.93×10 −4 |
| Subsets with negative association | — | — | LC, BrC | RCC, BC |
| Association with individual cancerse, OR (P) | ||||
| ESCC | 1.05 (0.31) | 1.33 (7.50×10 −6) | 1.04 (0.41) | — |
| GC | 1.04 (0.37) | 1.20 (0.007) | 1.08 (0.13) | — |
| PanC | 1.01 (0.83) | 0.95 (0.21) | 1.02 (0.62) | 0.88 (0.33) |
| RCC | 1.18 (9.19×10 −4) | 1.02 (0.66) | 0.95 (0.33) | 0.64 (2.86×10 −4) |
| LC | 1.08 (0.03) | 1.04 (0.22) | 0.89 (0.001) | 4.13 (0.11) |
| BrC | 1.21 (0.001) | 0.90 (0.08) | 0.83 (0.004) | NA |
| BC | 0.99 (0.78) | 1.09 (0.03) | 0.99 (0.79) | 0.71 (0.009) |
| PrC | 0.99 (0.87) | 0.94 (0.29) | 1.01 (0.88) | NA |
NA, not applicable; OR, odds ratio.
aSNPs were ordered by the chromosome location. Significant P for meta-analysis (P < 0.05/39 = 0.0013) or for analysis in individual outcomes (P < 0.01) was marked in bold.
bThe minor allele in the cancer outcome with the most significant P value.
cRandom-effect meta-analysis was conducted using GLIMMIX, accounting for overlapping subjects.
dResults were from ASSET, a subset-based association analysis for combining SNP-based results, accounting for overlapping subjects.
eORs and P values per one effect allele, obtained by random-effect meta-analysis (for LC) or unconditional logistic regression (for other cancers) adjusting for cancer-specific covariates.
SNP function annotation
To explore whether any of the significant SNPs identified might have potential regulatory functions in the specific cancer-associated tissues, we used custom tracks on the UCSC Genome browser (http://genome.ucsc.edu) to screen Roadmap and ENCODE data for each implicated SNP region for evidence of regulatory relevance (33,34), such as overlapping with chromatin marks, CpG site methylation and transcription factor binding motifs. We also used the online tools HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) and RegulomeDB (http://regulome.stanford.edu) as a complementary analysis to confirm the location of each SNP in relation to annotated protein-coding genes and/or non-coding RNA genes (footnotes of Supplementary Table S3, available at Carcinogenesis Online).
For SNPs that were significantly associated with individual cancer (Table II) or multiple cancers (Table III and Supplementary Table S4, available at Carcinogenesis Online), we performed expression quantitative trait loci (eQTL) analysis to assess the impact of 9p21 SNPs on cis-expression levels in corresponding cancer tissues, based on The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/) (35). We downloaded the total expression estimated based on the RNA-seq data of tumor samples using RSEM software (36) and TCGA germline SNP data derived from blood DNA, which are based on the Genome-Wide Human SNP Array 6.0 platform. For each tumor site, we performed eQTL analysis for each 9p21 gene-SNP pair. The genetic association was evaluated using a linear regression adjusting for the top five PCA scores derived from SNP genotyping data to control for population stratification and top three PCAs derived from expression levels to remove potential artifacts such as batch effects. For each gene, we further calculated the average of the segmented copy number scores based on TCGA Level 3 data and average methylation levels for CpG probes annotated to be related with the gene. The association between SNPs and the gene was then further adjusted for the average copy number alteration scores and methylation levels as described by Li et al. (37). Several genotyped (rs3731239 and rs12683422) or imputed SNPs (rs7874405, rs2383607 and rs180733044) significantly associated with multiple cancers were not included in the TCGA data. None of the significant SNPs for individual cancer (Table II) was included in the TCGA data. We therefore only examined rs10511729 [in tissues of BrC (n = 702), lung adenocarcinoma (LUAD, n = 426) or LUSC (n = 362)] and rs10811474 [in BrC tissues only (n = 702), since no information on RCC tissues was available].
Results
SNP-level analysis for individual cancers
Information on the design of each GWAS, number of subjects, SNPs and available covariates is summarized in Table I and Supplementary Table S1, available at Carcinogenesis Online. Based on random-effect meta-analysis for LC and logistic regression for all other cancers, five SNPs were significant after Bonferroni correction for individual cancers (P ≤ 2.59 × 10− 6), including rs2764736 (3′ of TUSC1) and rs1502895 (5′ of LINGO2) for BC, rs1063192 (3′ UTR of CDKN2B) and rs2157719 (CDKN2B-AS1 intronic) for ESCC and rs7033375 (5′ of C9orf72) for RCC. However, rs1502895 and rs7033375 showed significant heterogeneity across substudies of BC and RCC, respectively (P heterogeneity < 0.10), and became non-significant when accounting for heterogeneity using a random-effect meta-analysis (P = 0.50 and P = 0.21, respectively). We therefore did not include these SNPs in subsequent meta-analyses. Table II shows the remaining three SNPs that were significant (Table II and Supplementary Figure S1, available at Carcinogenesis Online).
Meta-analysis across multiple cancers
For 39 SNPs with P < 0.001 for at least one cancer or with P < 0.01 for at least two cancers (Supplementary Table S2, available at Carcinogenesis Online), we conducted meta-analyses and found significant associations for 4 SNPs based on ASSET (P ≤ 0.0013), which were not significant by random-effect meta-analyses (Table III and Supplementary Figure S1, available at Carcinogenesis Online). Specifically, the C allele of rs3731239 (CDKN2A intronic) was associated with increased risk of ESCC, GC and BC (P = 3.96 × 10− 4); the G allele of rs10811474 (3′ of IFNW1) was associated with increased risk of RCC and BrC (P = 0.001); the T allele of rs12683422 (LINGO2 intronic) was associated with a decreased risk of RCC and BC (P = 5.93 × 10− 4) and the G allele of rs10511729 (3′ of ELAVL2) was associated with a decreased risk of LC and BrC (P = 8.63 × 10− 4).
Gene-level analysis
In the gene-level analysis, three genes (CDKN2A, CDKN2B-AS1 and CDKN2B) were significantly associated with the risk of ESCC (P ≤ 4.70 × 10− 5). None of the 9p21 genes examined showed significant associations with other cancers (Table IV).
Table IV.
Gene-level P values with number of SNPs for 9p21 genes in eight cancersa
| Gene | Starting | Ending | ESCC | GC | PanC | RCC | LC | BrC | BC | PrC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | P | No. | P | No. | P | No. | P | No. | P | No. | P | No. | P | No. | P | |||
| IFNB1 | 21067104 | 21087943 | 4 | 0.66 | 4 | 0.80 | 4 | 0.79 | 4 | 0.02 | 4 | 0.30 | 5 | 0.14 | 7 | 0.12 | 5 | 0.63 |
| IFNW1 | 21120631 | 21152144 | 7 | 0.15 | 7 | 0.28 | 8 | 0.72 | 8 | 0.36 | 8 | 0.68 | 8 | 0.23 | 10 | 0.14 | 8 | 0.54 |
| IFNA21 | 21145636 | 21176659 | 3 | 0.95 | 3 | 0.81 | 3 | 0.31 | 4 | 0.98 | 3 | 0.23 | 3 | 0.55 | 8 | 0.82 | 3 | 0.24 |
| IFNA4 | 21166693 | 21197670 | 5 | 0.95 | 5 | 0.52 | 5 | 0.09 | 7 | 0.92 | 4 | 0.48 | 5 | 0.90 | 8 | 0.50 | 5 | 0.36 |
| IFNA7 | 21181468 | 21212204 | 4 | 0.94 | 4 | 0.50 | 4 | 0.09 | 5 | 0.91 | 3 | 0.47 | 4 | 0.90 | 7 | 0.57 | 4 | 0.36 |
| IFNA10 | 21186180 | 21217142 | 3 | 0.93 | 3 | 0.49 | 3 | 0.08 | 4 | 0.90 | 3 | 0.47 | 4 | 0.90 | 7 | 0.57 | 4 | 0.36 |
| IFNA16 | 21196372 | 21227310 | — | — | — | — | — | — | — | — | — | — | 1 | 0.74 | 4 | 0.84 | 1 | 0.75 |
| IFNA17 | 21207242 | 21238221 | 1 | 0.44 | 1 | 0.85 | 1 | 0.14 | 2 | 0.69 | 1 | 0.44 | 1 | 0.97 | 4 | 0.41 | 1 | 0.74 |
| IFNA14 | 21219201 | 21249978 | 3 | 0.64 | 2 | 0.82 | 3 | 0.26 | 4 | 0.86 | 3 | 0.58 | 3 | 0.24 | 5 | 0.45 | 3 | 0.27 |
| IFNA5 | 21284686 | 21315255 | 7 | 0.91 | 7 | 0.56 | 7 | 0.04 | 9 | 0.95 | 6 | 0.89 | 6 | 0.98 | 10 | 0.97 | 7 | 0.75 |
| KLHL9 | 21311018 | 21345429 | 6 | 0.83 | 6 | 0.24 | 6 | 0.02 | 6 | 0.21 | 6 | 0.27 | 6 | 0.52 | 10 | 0.62 | 6 | 0.82 |
| IFNA6 | 21330317 | 21360886 | 3 | 0.54 | 3 | 0.22 | 3 | 0.01 | 4 | 0.14 | 3 | 0.15 | 3 | 0.31 | 11 | 0.89 | 3 | 0.58 |
| IFNA13 | 21347423 | 21378075 | 3 | 0.08 | 3 | 0.05 | 3 | 0.03 | 4 | 0.78 | 3 | 0.79 | 3 | 0.42 | 12 | 0.92 | 3 | 0.80 |
| IFNA2 | 21364254 | 21395396 | 6 | 0.13 | 6 | 0.09 | 6 | 0.04 | 6 | 0.86 | 6 | 0.89 | 6 | 0.57 | 12 | 0.80 | 6 | 0.93 |
| IFNA8 | 21379146 | 21410184 | 6 | 0.54 | 6 | 0.23 | 9 | 0.24 | 9 | 0.27 | 9 | 0.88 | 8 | 0.56 | 14 | 0.89 | 9 | 0.60 |
| IFNA1 | 21410440 | 21441315 | 4 | 0.56 | 4 | 0.21 | 5 | 0.71 | 5 | 0.10 | 4 | 0.53 | 5 | 0.82 | 6 | 0.29 | 5 | 0.51 |
| IFNE1 | 21460839 | 21492312 | 7 | 0.75 | 7 | 0.008 | 7 | 0.24 | 6 | 0.006 | 4 | 0.18 | 5 | 0.33 | 10 | 0.96 | 6 | 0.45 |
| MTAP | 21772635 | 21865969 | 16 | 0.04 | 16 | 0.81 | 20 | 0.62 | 19 | 0.17 | 19 | 0.08 | 21 | 0.22 | 42 | 0.02 | 20 | 0.94 |
| CDKN2A | 21947751 | 22004490 | 15 | 2.70×10 −5b | 15 | 0.06 | 16 | 0.56 | 15 | 0.88 | 15 | 0.09 | 15 | 0.52 | 23 | 0.11 | 15 | 0.52 |
| CDKN2B-AS1 | 21964790 | 22121093 | 31 | 4.70×10 −5b | 31 | 0.22 | 32 | 0.43 | 31 | 0.70 | 30 | 0.13 | 30 | 0.92 | 46 | 0.16 | 31 | 0.35 |
| CDKN2B | 21982902 | 22019312 | 10 | 1.80×10 −5b | 10 | 0.36 | 11 | 0.40 | 10 | 0.88 | 10 | 0.05 | 10 | 0.91 | 15 | 0.07 | 10 | 0.36 |
| DMRTA1 | 22416840 | 22452472 | 10 | 0.47 | 10 | 0.28 | 10 | 0.32 | 10 | 0.70 | 10 | 0.19 | 10 | 0.24 | 15 | 0.42 | 10 | 1.00 |
| ELAVL2 | 23670103 | 23836063 | 51 | 0.07 | 51 | 0.39 | 53 | 0.03 | 52 | 0.94 | 51 | 0.84 | 49 | 0.92 | 67 | 0.99 | 50 | 0.94 |
| TUSC1 | 25656387 | 25688856 | 5 | 0.77 | 5 | 0.13 | 6 | 0.85 | 6 | 0.98 | 6 | 0.64 | 6 | 0.80 | 8 | 0.14 | 6 | 0.40 |
| C9orf82 | 26820683 | 26902725 | 8 | 0.64 | 8 | 0.52 | 6 | 0.88 | 9 | 0.86 | 9 | 0.73 | 9 | 0.39 | 20 | 0.34 | 9 | 0.32 |
| PLAA | 26884518 | 26957139 | 6 | 0.77 | 6 | 0.17 | 10 | 0.50 | 10 | 0.74 | 9 | 0.99 | 10 | 0.79 | 23 | 0.37 | 10 | 0.17 |
| IFT74 | 26926371 | 27062931 | 10 | 0.41 | 10 | 0.41 | 14 | 0.54 | 17 | 0.83 | 17 | 0.71 | 17 | 0.81 | 32 | 0.57 | 19 | 0.31 |
| LRRC19 | 26973586 | 27015670 | 1 | 0.39 | 1 | 0.16 | 2 | 0.39 | 2 | 0.31 | 2 | 0.42 | 2 | 0.38 | 7 | 0.43 | 3 | 0.11 |
| TEK | 27079147 | 27230172 | 72 | 0.51 | 72 | 0.28 | 84 | 0.26 | 86 | 0.76 | 84 | 0.48 | 83 | 0.17 | 110 | 0.69 | 84 | 0.46 |
| C9orf11 | 27264667 | 27307137 | 19 | 0.79 | 19 | 0.96 | 22 | 0.46 | 21 | 0.54 | 20 | 0.15 | 21 | 0.24 | 31 | 0.77 | 21 | 0.45 |
| MOB3B | 27305207 | 27539850 | 63 | 0.87 | 63 | 0.36 | 73 | 0.01 | 73 | 0.10 | 69 | 0.18 | 69 | 0.41 | 105 | 0.26 | 69 | 0.60 |
| IFNK | 27494312 | 27526496 | 8 | 0.97 | 8 | 0.90 | 8 | 0.75 | 8 | 0.55 | 8 | 0.40 | 8 | 0.66 | 14 | 0.97 | 8 | 0.16 |
| C9orf72 | 27526544 | 27583476 | 20 | 0.60 | 20 | 0.52 | 19 | 0.81 | 19 | 0.71 | 18 | 0.41 | 17 | 0.89 | 28 | 0.98 | 18 | 0.10 |
| LINGO2 | 27928528 | 28729303 | 239 | 0.74 | 240 | 0.19 | 269 | 0.05 | 274 | 0.31 | 258 | 0.87 | 261 | 0.10 | 344 | 0.49 | 263 | 0.29 |
| ACO1 | 32354601 | 32440834 | 19 | 0.02 | 19 | 0.05 | 24 | 0.68 | 24 | 0.46 | 23 | 0.50 | 24 | 0.88 | 35 | 0.21 | 25 | 0.03 |
aGenes were ordered by the chromosome location and those with P < 0.01 were marked in bold.
bSignificant after multiple comparison adjustment [P < 0.05/(35 genes × 8 studies)] = 1.79×10−4.
SNP function annotation based on ENCODE data
Based on ENCODE data, several SNPs such as rs3731239 (CDKN2A intronic), rs10511729 (3′ of ELAVL2) and rs2157719 (CDKN2B-AS1 intronic) overlapped with weak but potential enhancers in one or more cancer-associated tissues suggesting a potential regulatory function for the SNPs in these tissues (Figure 1 and Supplementary Table S3, available at Carcinogenesis Online). Interestingly, in addition to rs2157719, which maps to the antisense RNA gene CDKN2B-AS1, rs10511729 (3′ of ELAVL2) and rs2764736 (3′ of TUSC1) were also located proximal to non-coding RNA genes (Supplementary Table S3, available at Carcinogenesis Online), and in some instances much closer to a non-coding RNA gene than the previously annotated protein-coding gene.
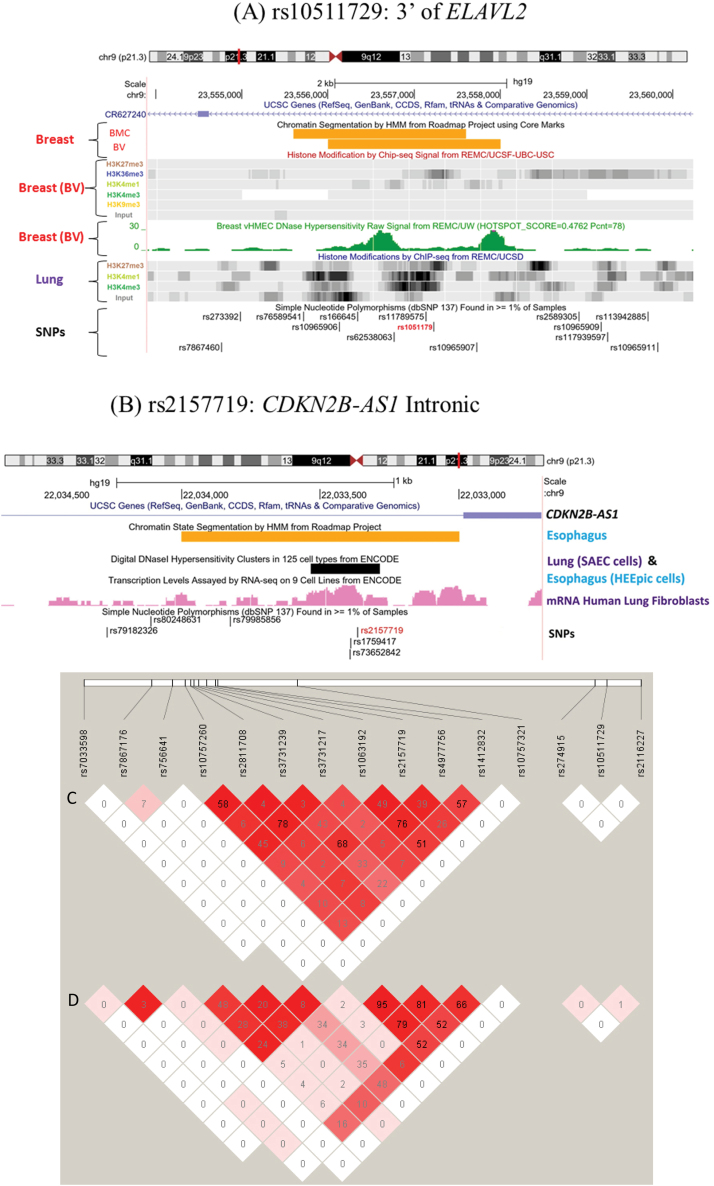
Fig. 1.
SNP function annotation using UCSC Genome Browser and LD structure of rs10511729 and rs2157719. (A) UCSC Genome Browser image of 3′ region of ELAVL2 on human assembly hg19 showing 9p21 region, UCSC Genes, Roadmap Chromatin State Segmentation by a Hidden Markov Model (HMM) and core marks for Roadmap breast myoepithelial cells and breast variant human mammary epithelial cells, histone marks H3K27me3, H3K36me3, H3K4me1, H3K4me3 and H3K9me3 in Bv cells, DNaseI sites in Bv cells, histone marks H3K27ac, H3K4me1, H3K4me3 and H3K9me3 in Roadmap lung cells and dbSNP 137 track on the left hand scale. The SNP rs10511729 is colored red. The HMM utilizes computationally integrated ChIP-seq data for 5/6 core marks (H3K27me3, H3K36me3, H3K4me1, H3K4me3, H3K9ac, H3K9me3) + H3K27ac with the associated yellow segment color indicating a weak enhancer annotation for the overlapping SNP region in normal breast cells. The overlapping SNP region is also bordered by two separate DNaseI peaks in normal breast epithelial cells. In normal lung cells, histone marks (H3K27ac, H3K4me1 and H3K4me3) which are associated with active enhancers were also observed for the overlapping region, suggesting rs10511729 is contained within a potential regulatory region in breast and lung tissues. (B) UCSC genome Browser image of intronic region of CDKN2B-AS1 on human assembly hg19 showing 9p21 region, ENCODE/GENCODE V17 gene annotation, UCSC Genes, Roadmap Chromatin State Segmentation by a HMM for Roadmap normal esophageal cells, DNaseI sites/cluster for 8/125 ENCODE cell lines, RNA-seq in ENCODE normal Human Lung Fibroblasts and dbSNP 137 on the right hand scale. The SNP rs2157719 is colored red. The associated yellow segment color for HMM indicates a weak enhancer annotation for the overlapping SNP region in Roadmap normal esophageal cells. The rs2157719 containing region also overlaps with a strong DNaseI cluster in a number of ENCODE cells lines including small airway epithelial cells (SAEC) and esophageal epithelial cells (HEEpic) cells suggesting rs2157719 is contained within a potential regulatory region in esophageal and lung (bronchiole) tissues. (C) The LD structures of rs10511729 and rs2157719 among controls of Chinese in the ESCC study. Only SNPs with P value <0.001 for at least one cancer or P value <0.01 for at least two cancers, that locate <500kb apart from rs10511729 and rs2157719, were analyzed. (D) The LD structures of rs10511729 and rs2157719 among controls of Caucasian in the LC study. Only SNPs with P value <0.001 for at least one cancer or P value <0.01 for at least two cancers, that locate less than 500kb apart from rs10511729 and rs2157719, were analyzed. The LD structures of these two SNPs among controls of Caucasians in the BrC study are very similar. BMC, breast human mammary epithelial cells; BV, breast myoepithelial cells.
Analysis using imputed SNPs
We had the SNP imputation data for three cancers (ESCC, PanC and LC). To identify additional 9p21 SNPs associated with multiple cancers, we examined these imputed SNPs but did not find any single imputed SNP that was significantly associated with any individual cancer after Bonferroni correction. Meta-analyses using ASSET found significant associations for three imputed SNPs (Supplementary Table S4, available at Carcinogenesis Online), including rs7874405 (intronic CDKN2A, with C allele associated with decreased risk of ESCC and PanC, P = 1.49 × 10− 4), rs2383607 (3′ of TUSC1, with C allele correlated with decreased risk of ESCC and LC, P = 1.80 × 10− 4) and rs180733044 (intergenic LINGO2-ACO1, with T allele correlated with increased risk of ESCC and LC, P = 2.44 × 10− 4). We did not find evidence for high LD between these SNPs and the genotyped SNPs significant for multiple cancers in Table III, based on 1000 Genome data.
eQTL analysis
In eQTL analyses based on TCGA data, we found that the G allele of rs10511729, which was negatively associated with LC and BrC risk, was significantly associated with increased expression of TUSC1 (P = 0.006) and IFNA14 (P = 0.02) in BrC, increased expression of CDKN2B in LUSC (P = 0.02) and LUAD (P = 0.03) tissues and increased expression of LRRC19 (P = 0.049) in LUSC tissues, but was associated with decreased ACO1 expression (P = 0.02) in LUAD. For rs10811474, which was associated with RCC and BrC, we found that the G allele was significantly associated with increased IFNW1 expression in BrC (P = 0.03) (there is no TCGA data available for RCC). Similar results were obtained with the further adjustment of copy number alteration and methylation within the same tissue (Supplementary Table S5, available at Carcinogenesis Online).
Discussion
In this study, we systematically examined the associations between common genetic variants in 35 9p21 genes and risk of eight cancers by a subset-based meta-analysis using ASSET. We found that a number of SNPs were associated with multiple cancers, further highlighting the role of this region in the shared susceptibility and etiology of various cancers. Our data also suggest that the identified associations might not be confined to the CDKN2A/2B-AS1/2B/MTAP cluster; rather, that different genes/subregions may be associated with different cancer outcomes.
We previously conducted a similar meta-analysis evaluating up to 203 9p21 SNPs and the risk of multiple cancers based on selected SNPs, from a customized platform, iSelect (2). The associations we found in that study were restricted to subsets containing ESCC and colorectal cancer, which were the only two cancers with >1000 cases. Thus, these findings could reflect a lack of power for cancers with smaller sample sizes. The current GWAS meta-analysis was based on much larger sample sizes (range 2252–7619 cases for each cancer) and vastly improved SNP coverage (2267–3674 SNPs). In addition, most cancers in the current analysis, including BC, BrC, PanC, PrC and LC, were not included in the previous analysis which focused primarily on rare malignancies.
The protein-coding genes CDKN2A and CDKN2B and non-coding antisense RNA gene CDKN2B-AS1 lie in a region that is frequently mutated, deleted or hypermethylated in tumor tissues and are among the loci most consistently associated with multiple chronic diseases in GWAS (10,38,39). We observed that all three genes in the CDKN2A/2B-AS1/2B cluster and two SNPs (rs1063192, 3′ UTR of CDKN2B, and rs2157719, CDKN2B-AS1 intronic) were significantly associated with ESCC in both gene- and SNP-level analyses. These SNPs were previously associated with glaucoma and glioma in Caucasians (14,16,20). One GWAS conducted in China found that rs1412829 (CDKN2B-AS1 intronic), which is in high LD (r 2 > 0.8) with rs2157719, was associated with nasopharyngeal cancer (19). Particularly, a number of risk SNPs for multiple chronic diseases in this gene cluster, such as rs1063192, was associated with allelic-specific expression of CDKN2B/2A/2B-AS1 (38,39).
The region containing rs2157719 appears to be in an active, but weak enhancer in normal esophageal cells as well as an open chromatin conformation (strong DNaseI site) in cultured esophageal epithelial cells (HEEpic), suggesting a potential regulatory function for this SNP in esophageal tissues. Similarly, rs3731239 (CDKN2A intronic), which has been associated with increased risk of ESCC (Chinese) and endometrial cancer (Caucasian) in our previous report (2) and with ESCC (Chinese), GC (Chinese) and BC (Caucasians) in our analysis, appeared to be in a region overlapping a weak and poised enhancer in normal stomach mucosa (and possibly esophagus) based on Roadmap histone data tracks. Consistent with previous reports, these data suggest that SNPs in the CDKN2A/2B-AS1/2B cluster may modulate disease susceptibility primarily through regulating expression levels of genes in the cluster.
Other genes in the 9p21 region have less established roles in cancer development compared to CDKN genes; however, several, such as ACO1, ELAVL2 and TUSC1, are involved in cellular functions or pathways that are relevant to cancer (10,40–44). For example, ELAVL2 is involved in cell growth and differentiation and increased expression has been implicated in neuroblastoma and teratocarcinoma cells (42,43). TUSC1 has been suggested to be a tumor suppressor gene in LC (44). Unlike previous individual cancer GWAS in which most significant associations were confined to the CDKN-MTAP region, the current analysis revealed associations for several SNPs in a number of non-CDKN genes and intergenic regions. For example, rs2764736 (3′ of TUSC1), which is located proximal to a non-coding RNA gene, was associated with BC risk. The G allele of rs10511729 (3′ of ELAVL2) was significantly associated with decreased risk of LC and BrC in ASSET analysis. This SNP is located in an active but weak enhancer in normal breast tissues and overlaps with H3K4me1 and H3K27me3 histone marks in normal lung tissues. Using BrC TCGA data, we found that rs10511729 G allele was correlated with increased expression of TUSC1 (BrC), IFNA14 (BrC), CDKN2B (LUSC and LUAD) and LRRC19 (LUSC), as well as decreased expression of ACO1 in LUAD, suggesting that this SNP may have an important regulatory function in these tissues. Similarly, the G allele of rs10811474 in the 3′ of IFNW1, which was associated with increased risk of BrC and RCC, has the potential to form a CpG site in the DNA context and may also alter Sp100 transcription factor binding. Indeed, the eQTL analysis using TCGA data also revealed significantly elevated IFNW1 expression by G allele in BrC. Our ASSET meta-analysis found significant SNPs with heterogeneous effects in different cancers. Our findings support a complex genetic susceptibility associated with cancer etiology implicated by this chromosome region and highlight the importance of comprehensively evaluating genes located in the 9p21 region in future genetic studies.
Notably, we found SNPs associated with BC even at genome-wide significance threshold that were not revealed in the original GWAS publications (26,28), which may be partly due to different subjects included in the original GWAS and our analysis (we excluded PLCO subjects in BC). Our study also raised an issue of heterogeneity among substudies. For LC, considerable heterogeneity in SNP effects existed among substudies, especially between ATBC (Finland) and the two US substudies [Cancer Prevention Study (CPS) II and PLCO], and was not resolved after further adjusting for smoking variables and principal components and confining subject to male smokers. The issue of heterogeneity requires attention in future studies involving multiple countries and centers.
Strengths of our study include the use of a subset-based method (ASSET) designed to combine association signals that may only manifest in a specific subset or have different directions across studies. This approach has better power in some settings compared to standard meta-analyses (21). In addition to SNP-level analyses, we also calculated gene-level associations. Further, we adjusted for the same covariates that were included in the original GWAS whenever possible and for additional variables that were not included in dbGaP such as principal components and smoking variables for LC, which we obtained from study investigators.
Our study also has several limitations. First, our analysis was based on the available GWAS data deposited in dbGaP, which covers only a small subset of cancer GWAS published in the literature and may have limited the power in identifying common genetic mechanisms for different cancers. However, we aimed to use the largest number of studies we could get from dbGaP, and the GWAS included were all the eligible data we can get when we initiated the study. In addition, studies included in this analysis varied by design, population ethnicity and SNP coverage and had overlapping subjects which posed challenges for combining data. We addressed the across-cancer heterogeneity using a subset-based meta-analysis, accounting for the overlapping subjects. Because ESCC and GC studies consisted of only Chinese subjects, whereas other cancers were predominantly among Caucasians, the subset-based meta-analysis may have been limited in its ability to identify shared susceptibility loci between upper gastrointestinal cancers (ESCC and GC) and other cancers. However, since Asian populations have high incidences of ESCC and GC, including these studies may provide important insights for the evaluation of a variety of different cancers. Second, we did not have SNP imputation data for all cancers, which may have limited the power for finding additional loci in meta-analyses. However, the secondary analysis for three cancers with imputed SNPs did not find other significant regions, beyond the regions that were shown significant in the primary analysis (CDKN2A/2B-AS1/2B/MTAP cluster, ELAVL2/TUSC1, LINGO2/ACO1 and IFNW1 region). Third, because of the need to control for a large number of overlapping subjects across studies, our results likely represent the most conservative estimates of SNP effects in multiple cancers. Fourth, there is growing evidence of heterogeneity within a single cancer site such as tumor subtypes defined by histology, gene expression, or major exposures. However, our study based on the public database is limited by sample size and relevant subtype information to address this issue.
In conclusion, our analyses based on multiple cancer GWAS identified several susceptibility loci in the 9p21 region that were associated with a number of different cancers, suggesting shared genetic susceptibility. On the other hand, variants in different genes/regions also appear to be associated with distinct outcomes, indicating heterogeneous cancer susceptibility associated with this chromosomal region. Further studies integrating GWAS data with those obtained from detailed genomic analyses in multiple tumor tissues such as TCGA would be helpful in understanding the complex genetic regulation of the 9p21 region in the development of each specific cancer outcome.
Supplementary material
Supplementary Tables S1–S5 and Figure S1 can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program of the National Institute of Health (NIH) ; National Cancer Institute; Division of Cancer Epidemiology and Genetics.
Supplementary Material
Acknowledgements
We are grateful to Drs David J.Hunter (Harvard School of Public Health), Mark P.Purdue (DCEG), Nathaniel Rothman (DCEG), Jonine D.Figueroa (DCEG) and Montserrat Garcia-Closas (the Institute of Cancer Research, UK) for their support and suggestions for quality control and data analyses. We thank William Wheeler and Jane Wang, Information Management Services, Inc., Calverton, MD, for their assistance in data analysis. W.-Q.L. and X.R.Y. conceived and designed the study. W.-Q.L., R.M.P., S.B., J.L., X.X., X.D. and N.C. conducted data analysis. W.-Q.L. and X.R.Y. wrote the manuscript. J.S., F.G., Z.W., M.Y., N.H., P.R.T., D.A., N.E.C., S.M.G., L.A., S.J.C., M.T.L., M.A.T., A.M.G. and X.R.Y. contributed significantly to data acquisition. W.-Q.L., R.M.P., P.L.H., A.M.G. and X.R.Y. critically revised the manuscript for important intellectual content. W.-Q.L. and X.R.Y. supervised this study. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ASSET
association analysis based on subset
- ATBC
Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study
- BC
bladder cancer
- BrC
breast cancer
- ESCC
esophageal squamous cell carcinoma
- GC
gastric cancer
- GWAS
genome-wide association studies
- LC
lung cancer
- LD
linkage disequilibrium
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- PanC
pancreatic cancer
- PCA
principle component analysis
- PLCO
Prostate, Lung, Colon and Ovarian Cancer Screening Trial
- PrC
prostate cancer
- RCC
renal cell carcinoma
- SNP
single nucleotide polymorphism
- TCGA
The Cancer Genome Atlas.
References
- 1. Chung C.C., et al. (2011). Current status of genome-wide association studies in cancer. Hum. Genet., 130, 59–78. [DOI] [PubMed] [Google Scholar]
- 2. Gu F., et al. (2013). Common genetic variants in the 9p21 region and their associations with multiple tumours. Br. J. Cancer, 108, 1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jin G., et al. (2012). Genetic variants at 6p21.1 and 7p15.3 are associated with risk of multiple cancers in Han Chinese. Am. J. Hum. Genet., 91, 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia-Closas M., et al. ; Australian Ovarian Cancer Management Group; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer. (2008). Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet., 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu N., et al. (2004). High frequency of CDKN2A alterations in esophageal squamous cell carcinoma from a high-risk Chinese population. Genes. Chromosomes Cancer, 39, 205–216. [DOI] [PubMed] [Google Scholar]
- 6. Hustinx S.R., et al. (2005). Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod. Pathol., 18, 959–963. [DOI] [PubMed] [Google Scholar]
- 7. Schmid M., et al. (2000). A methylthioadenosine phosphorylase (MTAP) fusion transcript identifies a new gene on chromosome 9p21 that is frequently deleted in cancer. Oncogene, 19, 5747–5754. [DOI] [PubMed] [Google Scholar]
- 8. Grady B., et al. (2001). Frequently deleted loci on chromosome 9 may harbor several tumor suppressor genes in human renal cell carcinoma. J. Urol., 166, 1088–1092. [PubMed] [Google Scholar]
- 9. Goldstein A.M. (2004). Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum. Mutat., 23, 630. [DOI] [PubMed] [Google Scholar]
- 10. Yang X.R., et al. (2010). Associations of 9p21 variants with cutaneous malignant melanoma, nevi, and pigmentation phenotypes in melanoma-prone families with and without CDKN2A mutations. Fam. Cancer, 9, 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bishop D.T., et al. (2009). Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet., 41, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amos C.I., et al. ; GenoMEL Investigators; Q-Mega Investigators; AMFS Investigators. (2011). Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet., 20, 5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stacey S.N., et al. (2009). New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet., 41, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrensch M., et al. (2009). Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet., 41, 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajaraman P., et al. (2012). Genome-wide association study of glioma and meta-analysis. Hum. Genet., 131, 1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shete S., et al. (2009). Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet., 41, 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timofeeva M.N., et al. ; Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team. (2012). Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet., 21, 4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turnbull C., et al. ; Breast Cancer Susceptibility Collaboration (UK). (2010). Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet., 42, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bei J.X., et al. (2010). A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet., 42, 599–603. [DOI] [PubMed] [Google Scholar]
- 20. Osman W., et al. (2012). A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum. Mol. Genet., 21, 2836–2842. [DOI] [PubMed] [Google Scholar]
- 21. Bhattacharjee S., et al. ; GliomaScan Consortium. (2012). A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet., 90, 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mailman M.D., et al. (2007). The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet., 39, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abnet C.C., et al. (2010). A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet., 42, 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amundadottir L., et al. (2009). Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet., 41, 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purdue M.P., et al. (2011). Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet., 43, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landi M.T., et al. (2009). A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunter D.J., et al. (2007). A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet., 39, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rothman N., et al. (2010). A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet., 42, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeager M., et al. (2007). Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet., 39, 645–649. [DOI] [PubMed] [Google Scholar]
- 30. Petersen G.M., et al. (2010). A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet., 42, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W.Q., et al. (2013). Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis, 34, 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu K., et al. (2009). Pathway analysis by adaptive combination of P-values. Genet. Epidemiol., 33, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenbloom K.R., et al. (2013). ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res., 41, D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meyer L.R., et al. (2013). The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res., 41(Database issue), D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Research Network. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li B., et al. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q., et al. (2013). Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 152, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cunnington M.S., et al. (2010). Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet., 6, e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson A.D., et al. (2013). Resequencing and clinical associations of the 9p21.3 region: a comprehensive investigation in the Framingham heart study. Circulation, 127, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chow A., et al. (2010). Molecular characterization of human homologs of yeast MOB1. Int. J. Cancer, 126, 2079–2089. [DOI] [PubMed] [Google Scholar]
- 41. Ghosh M.C., et al. (2008). Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc. Natl Acad. Sci. USA 105, 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antic D., et al. (1999). ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev., 13, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gupta A., et al. (2006). Cellular retinoic acid-binding protein II is a direct transcriptional target of MycN in neuroblastoma. Cancer Res., 66, 8100–8108. [DOI] [PubMed] [Google Scholar]
- 44. Shan Z., et al. (2013). TUSC1, a putative tumor suppressor gene, reduces tumor cell growth in vitro and tumor growth in vivo . PLoS One, 8, e66114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.