This work explores the intriguing roles of Mediator subunit CYCLIN-DEPENDENT KINASE8 (CDK8) in plant immune responses to fungal infection. CDK8 regulates jasmonate-responsive gene expression and cuticle development via interactions with MEDIATOR COMPLEX SUBUNIT25 and transcription factor WIN1, respectively, while other interactions suggest evolutionary conservation of the Mediator kinase module.
Abstract
CYCLIN-DEPENDENT KINASE8 (CDK8) is a widely studied component of eukaryotic Mediator complexes. However, the biological and molecular functions of plant CDK8 are not well understood. Here, we provide evidence for regulatory functions of Arabidopsis thaliana CDK8 in defense and demonstrate its functional and molecular interactions with other Mediator and non-Mediator subunits. The cdk8 mutant exhibits enhanced resistance to Botrytis cinerea but susceptibility to Alternaria brassicicola. The contributions of CDK8 to the transcriptional activation of defensin gene PDF1.2 and its interaction with MEDIATOR COMPLEX SUBUNIT25 (MED25) implicate CDK8 in jasmonate-mediated defense. Moreover, CDK8 associates with the promoter of AGMATINE COUMAROYLTRANSFERASE to promote its transcription and regulate the biosynthesis of the defense-active secondary metabolites hydroxycinnamic acid amides. CDK8 also interacts with the transcription factor WAX INDUCER1, implying its additional role in cuticle development. In addition, overlapping functions of CDK8 with MED12 and MED13 and interactions between CDK8 and C-type cyclins suggest the conserved configuration of the plant Mediator kinase module. In summary, while CDK8’s positive transcriptional regulation of target genes and its phosphorylation activities underpin its defense functions, the impaired defense responses in the mutant are masked by its altered cuticle, resulting in specific resistance to B. cinerea.
INTRODUCTION
The plant transcriptome is significantly reprogrammed in response to pathogen infection (Hahlbrock et al., 2003). The expression of genes related to hormone signaling pathways, the cell wall, cuticle modification processes, and the synthesis of defense metabolites, as well as pathogenesis-related (PR) genes, increases upon attempted pathogen infection (Dixon, 2001; Ausubel, 2005; Jones and Dangl, 2006; Boller and Felix, 2009; Fu and Dong, 2013). Precise transcriptional regulation of a battery of genes encoding diverse molecules in these plant processes determines plant resistance or susceptibility (Somssich and Hahlbrock, 1998). Plant transcriptional regulators and coregulators of plant immunity have been described (Moore et al., 2011). Recent studies link Mediator to transcriptional processes underpinning plant immunity to bacterial and fungal infection (Canet et al., 2012; Chen et al., 2012; Wathugala et al., 2012; Zhang et al., 2012; An and Mou, 2013; Lai et al., 2014). Mediator is an evolutionarily conserved eukaryotic multiple-protein complex that recruits RNA polymerase II (RNAP II) to specific promoters by either linking activators and repressors or recruiting transcription factors (TFs) to RNAP II (Kornberg, 2005; Malik and Roeder, 2005). In yeast (Saccharomyces cerevisiae) and animal cells, the core Mediator is subdivided into the head, middle, and tail modules (Guglielmi et al., 2004; Chadick and Asturias, 2005) and a fourth and separable kinase module consisting of CYCLIN-DEPENDENT KINASE8 (CDK8), C-type cyclin (CycC), MEDIATOR COMPLEX SUBUNIT12 (MED12), and MED13 (Borggrefe et al., 2002; Andrau et al., 2006).
Previous studies in yeast and metazoan cells suggested that CDK8 is mainly a negative regulator of transcription. First, yeast CDK8 (Srb10) was originally identified from a genetic screen for RNAP II C-terminal domain suppressors (Nonet and Young, 1989). Second, CDK8 could inhibit transcription in vitro probably because the kinase module association with core Mediator prevented the interaction between core Mediator and RNAP II (Elmlund et al., 2006). Other kinase module subunits, including CycC, MED12, and MED13, are also required for the repressive function of the kinase module, as the loss-of-function mutants of those four subunits displayed similar phenotypes, especially in response to nutrient deprivation and heat shock in yeast (Carlson, 1997; Knuesel et al., 2009a). Increasing evidence also demonstrates that CDK8 serves as a coactivator of transcription (Nemet et al., 2014) in many pathways, including p53-dependent transcription (Donner et al., 2007a, 2007b; Beckerman et al., 2009), the serum response network (Donner et al., 2010), the Transforming Growth Factor β signaling pathway (Alarcón et al., 2009; Knuesel et al., 2009b), and thyroid hormone-dependent transcription (Belakavadi and Fondell, 2010). Moreover, the kinase activity of CDK8 seems to be essential for its biological functions. For example, CDK8 positively affected TF Gal4-dependent transcription by directly phosphorylating Gal4 at the critical site Ser-699 (Hirst et al., 1999; Rohde et al., 2000). Disruption of this phosphorylation suppressed the induction of Gal4-dependent transcription (Ansari et al., 2002). Several other CDK8 phosphorylation substrates have also been identified. Recombinant CDK8 was able to phosphorylate the C-terminal domain of RNAP II and TFIIH as well as histone H3, MED13, and some general TFs (Hengartner et al., 1998; Rickert et al., 1999; Meyer et al., 2008; Knuesel et al., 2009b), but how those phosphorylation events contribute to the biological function of CDK8 is still largely unknown.
Purification of the core Mediator from Arabidopsis thaliana suspension cells (Bäckström et al., 2007) demonstrated that most Mediator components are present in the Arabidopsis complex as in other organisms. However, the kinase module was missing, in accordance with the transient and reversible association between the kinase module and core Mediator observed in other systems (Andrau et al., 2006). A number of Mediator subunits play critical roles in plant defense and development. MED21 is the first Mediator subunit implicated in resistance to necrotrophic pathogens based on the increased susceptibility of the MED21 RNA interference lines to Botrytis cinerea and Alternaria brassicicola (Dhawan et al., 2009). Subsequently, MED8, MED16, MED15, and MED25 were also implicated in resistance to necrotrophic pathogens (Canet et al., 2012; Chen et al., 2012; Wathugala et al., 2012; Zhang et al., 2012; An and Mou, 2013). Two independent studies reported that MED16 regulates both salicylic acid (SA)-mediated systemic acquired resistance and jasmonic acid (JA)/ethylene-mediated plant defense (Wathugala et al., 2012; Zhang et al., 2012). MED25 integrates signals from biotic and abiotic stresses and also contributes to the control of flowering time, hormone signaling, stress tolerance, and plant defense responses to necrotrophic pathogens. Consistent with its multiple functions, MED25 interacts with diverse TFs, including ERF1, ORA59, MYC2, MYC3, MYBs, WRKYs, and many ERF/AP2s (Çevik et al., 2012).
The kinase module has also been suggested to function in plant development. Arabidopsis CDK8 (CDKE1) was first identified as the mammalian and yeast CDK8 homolog (van de Peppel et al., 2005), later named HUA ENHANCER3, and is known to regulate floral organ identity (Wang and Chen, 2004). CDK8 interacts with Arabidopsis LEUNIG, a transcription corepressor, which in turn interacts with HISTONE DEACETYLASE19, a regulator of JA-dependent defense responses, as well as MED14 (Gonzalez et al., 2007). More recently, studies on Arabidopsis regulator of alternative oxidase1, which carries a mutation in the CDK8 gene, revealed that CDK8 regulates mitochondrial retrograde signaling in response to H2O2 and cold stress (Ng et al., 2013). The other kinase module components MED12 and MED13 are required for proper embryo axis development (Gillmor et al., 2010; Ito et al., 2011). The med13 mutant enhances auxin responses (Ito et al. 2011). In addition, the Arabidopsis genome contains two cyclin C-type genes, but no clear biological function has been assigned to the two cyclins (Menges et al., 2005).
In this work, we describe the functions of the Arabidopsis Mediator subunit CDK8 in plant defense and its underlying molecular and biochemical mechanisms. CDK8 regulates the expression of defensin genes, including PDF1.2 and several ETHYLENE RESPONSE TRANSCRIPTION FACTORS (ERFs) known for their disease resistance functions. Consistent with this observation, CDK8 is required for ERF1- and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59)-dependent activation of PDF1.2 expression. These data, coupled with the interaction between CDK8 and MED25, implicate CDK8 in the regulation of plant immunity through a JA-dependent pathway. CDK8 also regulates resistance to A. brassicicola through direct transcriptional regulation of AGMATINE COUMAROYLTRANSFERASE (AACT1), which is critical for the biosynthesis of hydroxycinnamic acid amides (HCAAs), secondary metabolites known for their functions in fungal resistance. Intriguingly, despite the loss of critical defense responses, the cdk8 mutant shows enhanced resistance to another necrotrophic pathogen, B. cinerea, possibly attributed to the increased cuticle permeability and altered cuticle structure in the cdk8 mutant. These features of plant cuticles have previously been associated with enhanced resistance to B. cinerea (Kurdyukov et al., 2006; Bessire et al., 2007; Chassot et al., 2007). CDK8 interacts with WAX INDUCER1 (WIN1), an ERF family protein known to regulate cuticular wax biosynthesis, thus implicating CDK8 in cuticle development, which is consistent with the observed changes in cuticle structure and permeability of the cdk8 mutant. Moreover, expression of CDK8 that is defective in phosphorylation activity failed to rescue the A. brassicicola susceptibility of the cdk8 mutant, while the resistance to B. cinerea was restored to the wild-type level, suggesting the kinase-dependent and independent functions of CDK8. CDK8 strongly interacts with two CycCs, suggesting an evolutionarily conserved structure of the kinase module in plants. Two other kinase module mutants, med12 and med13, exhibit disease responses and increased cuticle permeability similar to that of the cdk8 mutant, underlining the common functions and structural conservation of the kinase module. Taken together, our data shed light on multiple regulatory roles of the Mediator CDK8 module in plant defense and development.
RESULTS
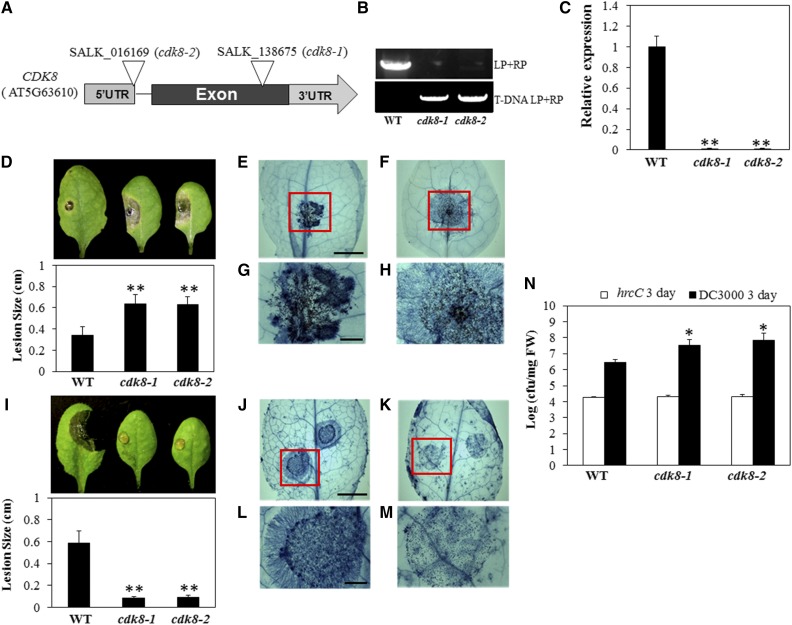
CDK8 Mediates Responses to Fungal and Bacterial Pathogens
We have previously described the functions of Mediator subunits MED21 and MED18 in plant defense responses to necrotrophic pathogens (Dhawan et al., 2009; Lai et al., 2014). In a parallel reverse genetic screen covering mutants of Mediator subunits, we determined that two homozygous mutant alleles of CDK8 from the SALK T-DNA insertion collection were susceptible to A. brassicicola (Figure 1). PCR analysis confirmed that both cdk8-1 and cdk8-2 carry homozygous T-DNA insertions in the CDK8 gene (Figures 1A and 1B), and quantitative RT-PCR (RT-qPCR) revealed that the expression of CDK8 is abolished in the two mutant lines (Figure 1C). The primer sequences used to verify the T-DNA insertion in CDK8 and to determine gene expression are listed in Supplemental Data Set 1. When inoculated with A. brassicicola, both cdk8 mutant lines exhibited symptoms of increased susceptibility to the fungus (Figure 1D). At 4 d after inoculation (DAI), leaves of wild-type plants had only very restricted disease lesions at the inoculation sites characteristic of Arabidopsis wild-type responses to this fungus, in contrast with cdk8 mutant plants, which had significantly larger disease lesions surrounding the inoculation sites. Fungal growth was analyzed in inoculated plants to determine whether the enhanced disease symptoms in the mutant were accompanied by increased fungal growth. Trypan blue staining to observe fungal growth patterns clearly revealed that A. brassicicola proliferated significantly more quickly on leaves of the cdk8 mutant at 48 h after inoculation, with hyphal growth extending beyond the inoculation sites (Figures 1E to 1H; Supplemental Figures 1C and 1D).
Figure 1.
The cdk8 Mutant Displays Increased Susceptibility to A. brassicicola and P. syringae but Resistance to B. cinerea.
(A) Genomic structure of the CDK8 gene and positions of the T-DNA insertion in cdk8 mutant alleles. UTR, untranslated region.
(B) Verification of cdk8 T-DNA insertion mutants by genomic PCR. LP, T-DNA left border genomic primer; RP, T-DNA right border primer.
(C) RT-qPCR data showing loss of CDK8 expression in cdk8 mutants. Relative transcript levels were normalized with Arabidopsis ACT2. The normalized expression level of the wild type was set to 1. Error bars indicate se (n = 3). Two independent biological replicates were performed. Significance between the mean values was analyzed statistically (Student’s t test, **P < 0.01).
(D) A. brassicicola disease symptoms (top panel) and disease lesion size (bottom panel) in the wild type and the cdk8 mutant at 4 DAI.
(E) and (F) Trypan blue staining of A. brassicicola-inoculated leaves of the wild type (E) and the cdk8 mutant (F) 48 h after inoculation. Bar = 4 mm.
(G) and (H) Closeups of the disease lesion areas from (E) and (F). Bar = 1 mm.
(I) B. cinerea disease symptoms (top panel) and disease lesion size (bottom panel) in the wild type and the cdk8 mutant at 4 DAI.
(J) and (K) Trypan blue staining of the wild type (J) and the cdk8 mutant (K) 36 h after inoculation with B. cinerea. Bar = 4 mm.
(L) and (M) Closeups of the disease lesion areas from (J) and (K). Bar = 1 mm.
(N) Bacterial growth in wild-type and cdk8 mutant plants. Leaves of 5-week-old plants were infiltrated with bacterial suspension (OD600 = 0.0005). At 3 DAI, leaf discs were collected and bacterial growth was quantified and expressed in colony-forming units (cfu). FW, fresh weight.
The disease assay was performed by drop-inoculation of B. cinerea on leaves of soil-grown plants or A. brassicicola on detached leaves. The average lesion sizes are mean values ± se from four independent replicates (n = 40). A minimum of 10 leaves for each genotype were used for each biological replicate, and the disease assay was repeated at least four times with similar results. All of the data were statistically analyzed. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
We then challenged cdk8 and wild-type plants by drop-inoculation with a conidial suspension of B. cinerea as described previously (Mengiste et al., 2003). B. cinerea infection caused obvious disease symptoms, with larger and expanding disease lesions beyond the inoculation site in the wild-type plants at 4 DAI (Figure 1I; Supplemental Figure 1A). However, in cdk8 mutants, disease symptoms were restricted to the site of inoculation without any noticeable spread of disease symptoms. Similarly, B. cinerea spray inoculation confirmed that both cdk8 mutant lines exhibited enhanced resistance compared with the wild-type plants (Supplemental Figure 1B). The contrasting responses of the cdk8 mutants to these two closely related necrotrophic fungi were unexpected in light of previous reports that show largely similar host response mechanisms. In contrast with the responses to A. brassicicola, the growth of B. cinerea was significantly restricted on leaves of the cdk8 mutant compared with the wild type at 36 h after inoculation (Figures 1J to 1M; Supplemental Figure 1D). Leaves of the wild type and the cdk8 mutant were also stained with trypan blue after spray inoculation with B. cinerea, which also demonstrated that the growth of B. cinerea was attenuated on the cdk8 mutant (Supplemental Figure 1C). The germination of conidia was not inhibited, but later stages of fungal growth were significantly restricted.
We also investigated the responses of cdk8 mutants to the bacterial pathogen Pseudomonas syringae virulent strain DC3000 and the nonpathogenic strain DC3000 hrcC, which is defective in the type III secretion system. This secretion system is used by plant pathogenic bacteria to deliver effector proteins into plant cells. Three days after inoculation with P. syringae DC3000, both cdk8 mutant lines supported significantly more bacterial growth than wild-type plants, revealing the susceptibility of the cdk8 mutant (Figure 1N). No differences were observed after inoculation with the hrcC strain. These data suggest that CDK8 is also a positive regulator of bacterial resistance similar to its function in A. brassicicola resistance.
CDK8-Dependent Arabidopsis Transcriptome Revealed by RNA Sequencing
RNA sequencing (RNA-seq) analyses of mock- or B. cinerea-inoculated plants were conducted to further understand the genome-wide regulatory impact of CDK8 in pathogen-induced transcriptional changes and to identify CDK8-regulated target genes. B. cinerea and A. brassicicola trigger similar transcriptional changes (van Wees et al., 2003; AbuQamar et al., 2006; Lai and Mengiste, 2013); thus, we conducted the RNA-seq experiment from B. cinerea-inoculated samples. Reads per kilobase of exon model per million mapped reads values for each sample of the RNA-seq analysis were calculated from the count data as described recently (Lai et al., 2014). Pathogen-induced or -suppressed genes with at least 2-fold changes and false discovery rate values (P ≤ 0.05) were selected. The Venn diagram in Supplemental Figure 2A summarizes the number of genes that were differentially expressed in CDK8-dependent and independent manners. Moreover, genes belonging to different functional categories, including abiotic and biotic stress, transcription, lipid and secondary metabolism, hormone response, and signaling, were closely analyzed because their expression was altered in response to infection. The heat map in Supplemental Figure 2B shows differentially expressed genes in the different functional categories. Compared with wild-type plants at 36 h after B. cinerea infection, a number of genes encoding TFs, including ERF/AP2, MYB, WRKY, and bHLH family genes, were differentially expressed in cdk8 in response to infection (Supplemental Data Set 2), implying that CDK8 is a critical regulator with broader impact on gene regulation. In addition, we found that the expression of defense marker PDF genes was significantly reduced in the cdk8 mutant, which is consistent with the observed susceptibility to A. brassicicola but does not correlate with the resistance to B. cinerea.
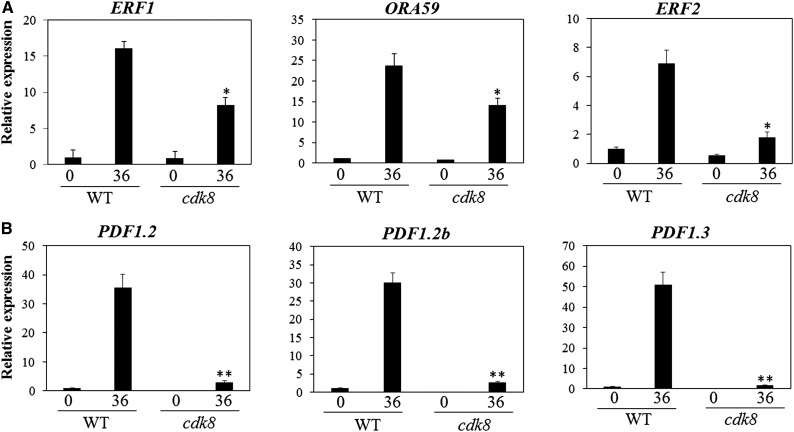
A number of studies have shown that the necrotrophic pathogens B. cinerea and A. brassicicola trigger similar defense responses (Laluk and Mengiste, 2011; Lai and Mengiste, 2013; Lai et al., 2014). Therefore, we investigated the expression of several defense genes in wild-type and cdk8 plants after inoculation with the two necrotrophic pathogens to link the disease response phenotype with changes in gene expression. Analysis of RT-qPCR data confirmed the CDK8-dependent expression of ERF1, ORA59, and ERF2, which was significantly suppressed in the cdk8 mutant (Figure 2A). The expression of plant defensin genes PDF1.2, PDF1.2b, and PDF1.3, which are also ERF target genes, was also significantly reduced in cdk8 (Figure 2B). Similarly, the expression of ORA59, PDF1.2, and PDF1.3 was compromised in the cdk8 mutant in response to A. brassicicola, despite the contrasting phenotypes (Supplemental Figure 3). Thus, the differences in the responses of cdk8 to B. cinerea and A. brassicicola are not linked to the pathogen-induced expression of these defense genes.
Figure 2.
CDK8-Dependent Expression of ERF and Plant Defensin Genes.
(A) Induced expression of genes encoding the TFs ERF1, ORA59, and ERF2 is reduced in the cdk8 mutant.
(B) Induced expression of defensin genes is dependent on CDK8.
Total RNAs were extracted from 5-week-old plants grown in soil before and after B. cinerea inoculation. Relative transcript levels were normalized with Arabidopsis ACT2. The normalized expression level of the wild type at 0 h was set to 1. Error bars indicate se (n = 3). Three independent biological replicates were performed. Significance of differences between the mean values was analyzed statistically (Student’s t test, *P < 0.05, **P < 0.01).
CDK8 and MED25 Are Required for TF-Dependent Activation of PDF1.2
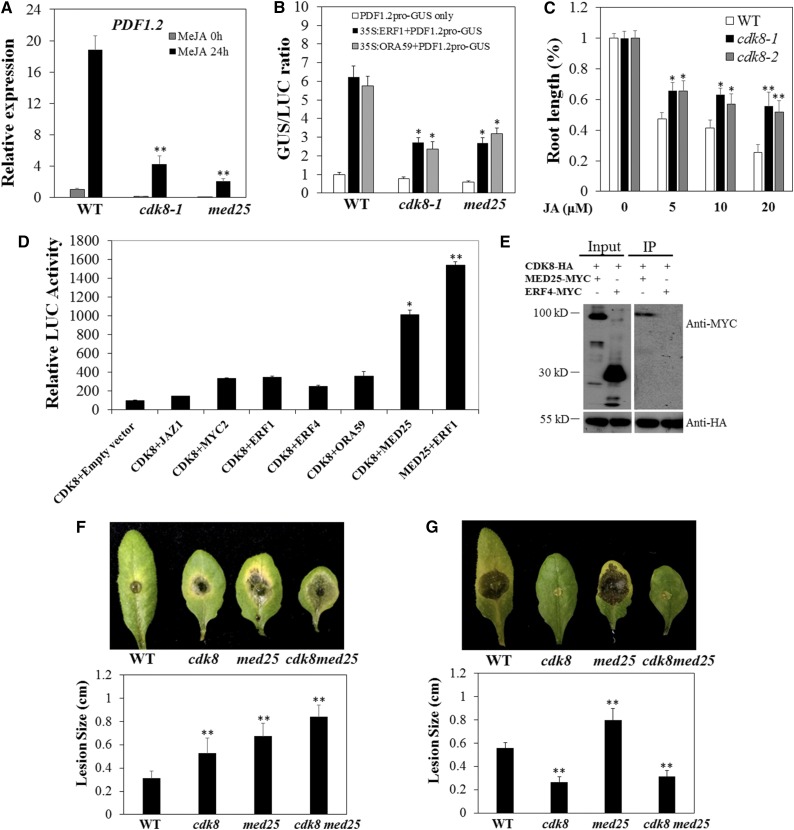
From an analysis of gene expression data, we observed a clear reduction of PDF1.2 expression in the cdk8 mutant. After methyl jasmonate (MeJA) treatment, PDF1.2 was significantly induced in wild-type plants but remained at extremely low levels in cdk8, similar to the response of med25 mutants (Figure 3A; Kidd et al., 2009). These data suggest that both CDK8 and MED25 are required for the induced expression of PDF1.2. The TFs ERF1 and ORA59 activate PDF1.2 expression (Lorenzo et al., 2003; Pré et al., 2008). To determine the role of CDK8 in PDF1.2 gene expression, we performed transactivation assays. The activation of PDF1.2 by ERF1 and ORA59 requires functional MED25 (Çevik et al., 2012). To test whether CDK8 is required for the coactivation of PDF1.2, we coexpressed a PDF1.2 promoter-β‑glucuronidase (GUS) reporter with effector construct 35S:ERF1/ORA59-MYC or the empty vector in Arabidopsis protoplasts with cauliflower mosaic virus 35S-driven, full-length luciferase (LUC) coexpressed as an internal control. When PDF1.2 pro-GUS and the empty vector alone were coexpressed, the GUS activities from the PDF1.2 promoter were similar in the wild type and the cdk8 and med25 mutants (Figure 3B). On the other hand, coexpression of the effector plasmids 35S:ERF1-MYC and 35S:ORA59-MYC, run in parallel, noticeably enhanced the PDF1.2 pro-GUS activity in wild-type protoplasts, which confirms previously reported observations (Çevik et al., 2012). In particular, the average induction of PDF1.2 pro-GUS activity in med25 and cdk8 protoplasts was significantly reduced compared with that of the wild type (Figure 3B), which is consistent with a previous report (Kidd et al., 2009). Protein gel blot analysis was performed to confirm the equivalent transformation efficiency in different protoplasts (Supplemental Figure 4). These results indicate that CDK8 and MED25 are required for the ERF1- or ORA59-dependent activation of PDF1.2. Thus, CDK8 is a coactivator in the regulation of PDF1.2. Moreover, seedling growth responses to MeJA revealed that both cdk8 mutant lines were less sensitive to a range of JA concentrations than the wild type (Figure 3C), further suggesting that MED25 and CDK8 function in the JA response pathway.
Figure 3.
Interactions between CDK8 and MED25 Promote the Expression of PDF1.2.
(A) Reduced MeJA-induced expression of PDF1.2 in cdk8 and med25 mutants. Gene expression was determined by RT-qPCR with the Arabidopsis ACT2 used for normalization. The expression level of PDF1.2/ACT2 in the mock-treated wild type was set at 1. Error bars indicate se (n = 3).
(B) CDK8 and MED25 are required for ERF1/ORA59-mediated PDF1.2 activation. The activity of the reporter gene construct PDF1.2 pro-GUS was normalized to the full-length LUC construct used as an internal control. Relative GUS:LUC activity ratios (fold change) are mean values from three independent biological replicates (n = 6). Error bars indicate se.
(C) Root growth responses of cdk8 to MeJA. Wild-type and cdk8 seeds were germinated on half-strength Murashige and Skoog medium. Root lengths were measured at 7 d after transferring to medium containing MeJA at the indicated concentrations. Results are mean values ± se from two independent replicates (n = 50).
(D) Interaction between CDK8 and MED25 revealed by split-luciferase complementation assay. Equal amounts of purified plasmids were transiently coexpressed in Arabidopsis protoplasts with substrate luciferin. The LUC activities are mean values from three biological replicates, and error bars indicate se (n = 3).
(E) CDK8 and MED25 interact in Co-IP assays in Arabidopsis protoplasts. MED25-MYC, but not ERF4-MYC, was present in CDK8-HA-precipitated complex. The experiment was repeated three times.
(F) A. brassicicola disease symptoms and disease lesion size in the wild type, cdk8, med25, and cdk8 med25 double mutant.
(G) B. cinerea disease symptoms and disease lesion size in the wild type, cdk8, med25, and cdk8 med25 double mutant.
The disease assays and lesion size measurements were performed as described in Figure 1 and were repeated at least three times. The lesion sizes are mean values ± se from at least 20 disease lesions. The lesion sizes and photographs are from 4 DAI. All of the data were statistically analyzed. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
[See online article for color version of this figure.]
Arabidopsis CDK8 Interacts with MED25
Analysis of gene expression demonstrated that the transcription of PDF1.2 and other plant defensin genes was significantly reduced in cdk8 mutants in mock-, pathogen-, or MeJA-treated plants. Thus, CDK8 was hypothesized to play roles in either recruiting ERF/AP2 domain TFs to the PDF1.2 promoter or interacting with some important factors in the JA pathway. With this rationale, we cloned the genes encoding the TFs ERF1, ORA59, ERF4, MYC2, JAZ1, and MED25 and performed a split-luciferase complementation assay to explore the potential interaction between these proteins and CDK8. CDK8 was fused to N-terminal luciferase (nLuc), and other TFs and MED25 were fused to C-terminal luciferase (cLuc). After coexpression, there was only a background level of LUC activity from coexpression of CDK8-nLuc with the empty cLuc vector, which was used as a negative control (Figure 3D). The coexpression of CDK8-nLuc with JAZ1, MYC2, ERF1, or ORA59-cLuc resulted in slightly stronger LUC activity (only ∼2-fold higher) than that of the negative control. By contrast, there was significantly stronger LUC activity from the coexpression of CDK8-nLuc and MED25-cLuc (∼5- to 6-fold higher), indicating that CDK8 directly interacts with MED25. Recently, MED25 was shown to physically interact with 18 TFs, including ERF1, ORA59, MYC2, and WRKYs (Çevik et al., 2012). Therefore, we also constructed MED25-nLuc and coexpressed it with ERF1-cLuc as a positive control, which exhibited strong LUC activity, confirming the efficiency of our split-luciferase complementation assays. Moreover, the interaction between CDK8 and MED25 was verified in coimmunoprecipitation (Co-IP) assays in Arabidopsis protoplasts. MED25-MYC was immunoprecipitated by CDK8-HA but ERF4-MYC was absent in CDK8-HA precipitates, confirming the specific interaction between CDK8 and MED25 (Figure 3E).
To determine the genetic interaction between MED25 and CDK8, we generated a cdk8 med25 double mutant by crossing cdk8-1 and med25. The cdk8 med25 double mutant displayed increased susceptibility to A. brassicicola, significantly more than that of the single mutants, at 3 DAI (Figure 3F), indicating that both CDK8 and MED25 may affect disease responses in more than one pathway. Interestingly, the double mutant exhibited increased resistance to B. cinerea similar to that of the cdk8-1 single mutant, despite the susceptibility of the med25 single mutant to B. cinerea (Figure 3G). These data suggest that the cdk8 mutation is epistatic to med25 with respect to B. cinerea resistance but may affect overlapping as well as distinct pathways with respect to A. brassicicola resistance.
Transcriptional Regulation of PDF1.2 by CDK8
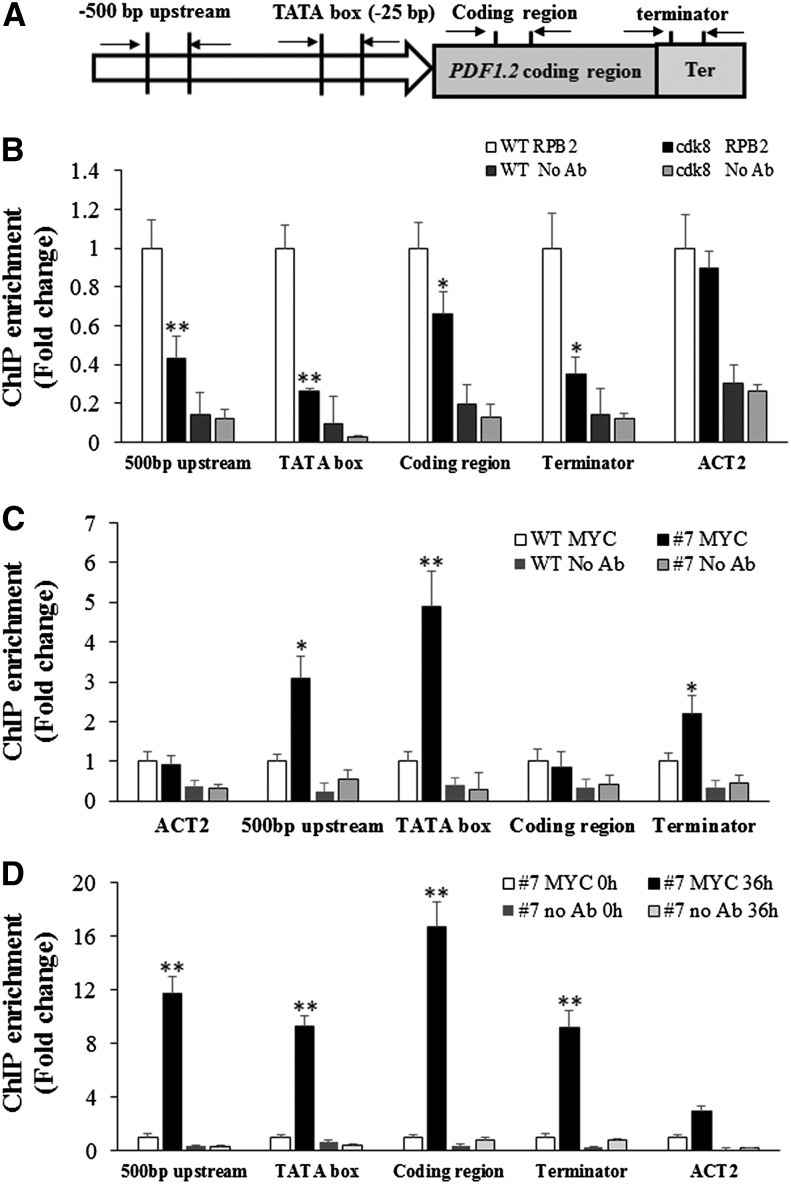
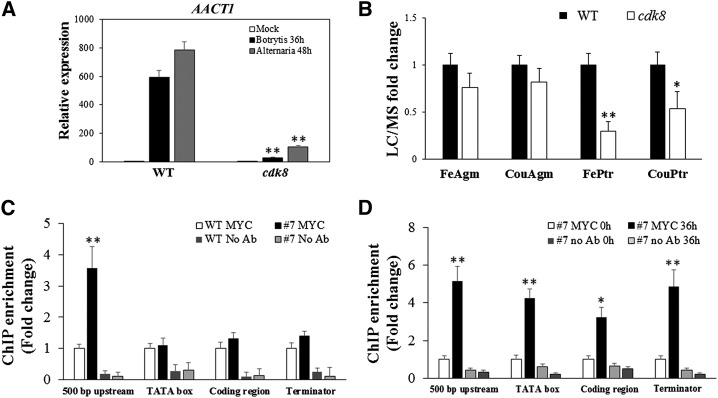
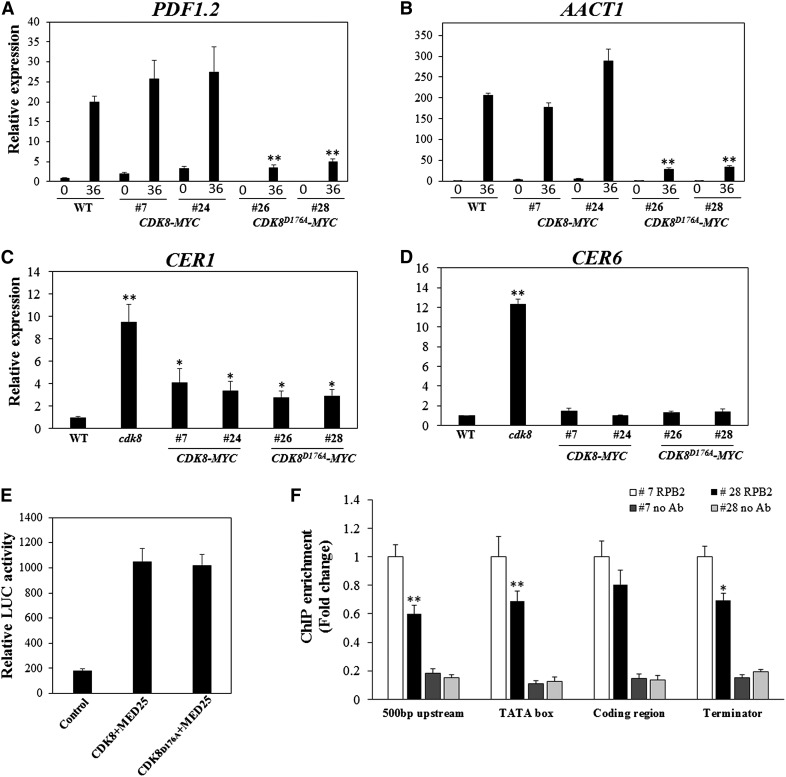
To shed light on the molecular mechanisms of the regulation of PDF1.2 expression by CDK8, we sought to determine whether CDK8 is required for the recruitment of RNAP II to the promoter regions of PDF1.2. Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay was conducted using antibodies raised against the C-terminal domain of RNAP II (anti-RPB2) and quantitative PCR primers designed at different positions, including 500-bp upstream, TATA box (the RNAP II binding site), and coding and terminator regions of PDF1.2 (Figure 4A). The recruitment of RNAP II to all four PDF1.2 regions was significantly lower in the cdk8 mutant than in the wild type after MeJA treatment, especially for the TATA box of the PDF1.2 promoter region. The recruitment of RNAP II to Arabidopsis ACTIN2 (ACT2), which was used as an endogenous control, was similar in the wild type and the cdk8 mutant (Figure 4B). From these findings, we infer that CDK8 contributes to the transcription of PDF1.2 through its role in RNAP II recruitment. In parallel, the cdk8;35S:CDK8-MYC transgenic plants were used for ChIP-qPCR experiments to determine the association of CDK8 with target genes. The association of CDK8-MYC with the 500-bp upstream, TATA box, and terminator sites of the PDF1.2 promoter region were enhanced compared with wild-type plants in mock-treated plants (Figure 4C). The enrichment of CDK8-MYC was at least 2- to 5-fold higher in these regions. By contrast, there was no difference at the PDF1.2 coding region and the endogenous ACT2 control. We then investigated whether pathogen infection would enhance the enrichment of CDK8 at the PDF1.2 promoter. The recruitment of CDK8 to the four PDF1.2 regions tested was significantly enhanced by B. cinerea infection, suggesting that CDK8 directly regulates the expression PDF1.2 in response to pathogen attack (Figure 4D).
Figure 4.
CDK8 Associates with the Regulatory Regions of the PDF1.2 Gene.
(A) Schematic showing the positions of PDF1.2 gene primers used for ChIP-qPCR.
(B) CDK8 is required for MeJA-induced RNAP II recruitment to the PDF1.2 promoter. ChIP-qPCR results are shown with PDF1.2 gene-specific primers. Chromatin was extracted from wild-type and cdk8 seedlings 1 h after treatment with 100 μM MeJA and then precipitated with anti-RPB2 antibody (Abcam) or only IgG (negative control with no antibody [No Ab]). The RNAP II recruitment at Arabidopsis ACT2 was used as a control because its expression is independent of CDK8. The ChIP-qPCR data show that RNAP II recruitment to PDF1.2 500-bp, TATA box, and coding and terminator regions decreased compared with the wild type.
(C) CDK8 associates with the PDF1.2 promoter. Chromatin was extracted from 5-week-old wild-type and cdk8;35S:CDK8-MYC transgenic plants and then precipitated with anti-MYC antibody (Abcam) or only IgG (No Ab).
(D) Recruitment of CDK8 to the PDF1.2 promoter is significantly enhanced by B. cinerea infection. Chromatin was extracted from 5-week-old cdk8;35S:CDK8-MYC #7 transgenic plants that were mock-inoculated or B. cinerea spray-inoculated. ChIP was performed with anti-MYC antibody (Abcam) or only IgG (No Ab). The CDK8 recruitment to 500-bp upstream, TATA box, and coding and terminator regions of PDF1.2 was determined by quantitative PCR using primers at different positions of the PDF1.2 gene as shown in (A).
Error bars in (B) to (D) indicate se (n = 3). Two biological replicates were performed with similar results for each ChIP-qPCR experiment. The significance of differences in mean values is marked by asterisks (Student’s t test, *P < 0.05, **P < 0.01).
CDK8 Is Required for the Accumulation of HCAAs
The RNA-seq analysis identified many genes that are significantly induced by B. cinerea in a CDK8-dependent manner. Among these, the AACT1 gene, which catalyzes the last step of HCAA biosynthesis, was significantly induced by B. cinerea or A. brassicicola infection, but its expression remained at significantly lower levels in the cdk8 mutant (Figure 5A; Supplemental Data Set 3). The HCAAs p-coumaroylagmatine, feruloylagmatine, p-coumaroylputrescine, and feruloylputrescine are important secondary metabolites that accumulate in response to infection and are known to contribute to resistance to A. brassicicola (Muroi et al., 2009). Liquid chromatography-mass spectrometry (LC-MS) analysis revealed that p-coumaroylputrescine and feruloylputrescine accumulated at significantly lower levels in the cdk8 mutant (Figure 5B). Consistently, the Arabidopsis mutants in the AACT1 gene exhibited increased susceptibility to B. cinerea (Supplemental Figure 5). Previous data show that the aact1 mutant has reduced accumulation of HCAAs and enhanced susceptibility to A. brassicicola (Muroi et al., 2009).
Figure 5.
CDK8 Is Required for the Accumulation of Arabidopsis HCAAs.
(A) Expression of the AACT1 gene in response to B. cinerea and A. brassicicola. RNA was extracted from mock-, B. cinerea-, or A. brassicicola-inoculated leaves. The experiment was repeated three times. Error bars indicate se (n = 3).
(B) Detection of HCAAs from rosette leaves of 5-week-old Arabidopsis wild-type and cdk8 plants at 48 h after inoculation with A. brassicicola. The data are mean values ± se from two biological replicates (n = 4). FerAgm, feruloylagmatine; CouAgm, p-coumaroylagmatine; FerPtr, feruloylputrescine; CouPtr, p-coumaroylputrescine.
(C) CDK8 is specifically recruited to a 500-bp region upstream of the AACT1 promoter region.
(D) Recruitment of CDK8 to the AACT1 gene is enhanced by B. cinerea infection.
In (C) and (D), chromatin was extracted from 5-week-old cdk8;35S:CDK8-MYC #7 transgenic plants that were mock- or B. cinerea-inoculated. Error bars indicate se (n = 3). ChIP-qPCR experiments were repeated two times with similar results. No Ab, negative control with no antibody.
The data were statistically analyzed. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
To determine whether the transcriptional regulation of AACT1 by CDK8 is direct or indirect, we studied the association of CDK8 with the AACT1 gene by ChIP-qPCR using primers designed based on the regulatory and coding regions of AACT1. CDK8 associated with the 500-bp upstream region but not the TATA box of the AACT1 promoter (Figure 5C). After inoculation with B. cinerea, the associations of CDK8 at the 500-bp upstream, TATA box, and coding and terminator regions were all significantly increased (Figure 5D), indicating that the association of CDK8 with these regions contributes to the regulation of AACT1 transcription in response to pathogen infection.
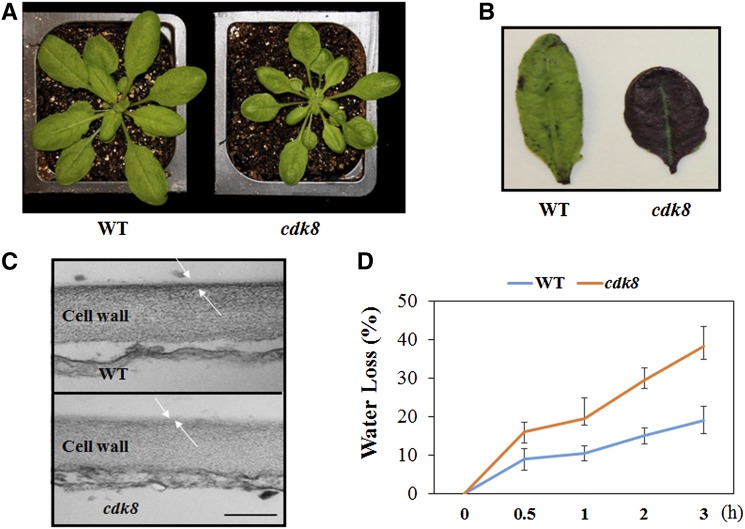
The cdk8 Mutant Has Altered Cuticle Structure
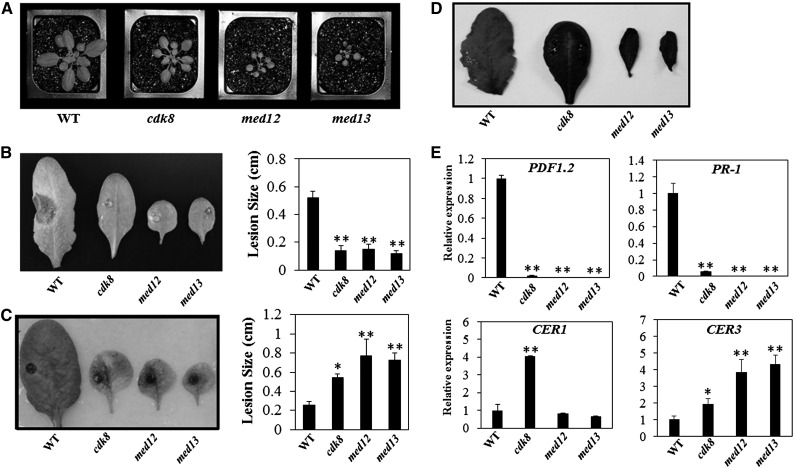
Changes in cuticle composition and structure are known to alter plant defense responses (Bessire et al., 2007; Reina-Pinto and Yephremov, 2009; L’Haridon et al., 2011). We noticed that the leaves of cdk8 mutants are particularly glossy, which is reminiscent of altered cuticle structure (Figure 6A; Supplemental Figure 6). Toluidine blue staining revealed that leaves of the cdk8 mutant stained more quickly than leaves of the wild type (Figure 6B), suggesting increased permeability in the cdk8 mutant. After inspection of the cuticle by transmission electron microcopy, we observed that the cuticle layer is thinner in the cdk8 mutant than in wild-type plants (Figure 6C). Accordingly, relative to wild-type plants, the percentage water loss is higher in the cdk8 mutant and those plants wilt more quickly, as predicted from the changes in the cuticle structure of the mutant (Figure 6D). The expression of genes implicated in cuticle and cuticular wax biosynthesis was studied to explore the potential mechanisms underlying the altered cuticle structure in the cdk8 mutant. The expression of several cuticle wax biosynthesis genes, including ECERIFERUM1 (CER1) and CER6, was significantly increased in the cdk8 mutant (Supplemental Figure 7). Overexpression of CER1 has been shown to cause increased susceptibility to bacterial and fungal pathogens (Bourdenx et al., 2011), which is consistent with the susceptibility of the cdk8 mutant to A. brassicicola and P. syringae DC3000.
Figure 6.
The cdk8 Mutant Has Increased Cuticle Permeability and a Reduced Cuticular Layer.
(A) Leaves of the cdk8 mutant (right) showing the glossy phenotype.
(B) Enhanced cuticle permeability of cdk8 leaves (right) revealed by toluidine blue staining.
(C) Transmission electron microscopic images of wild-type and cdk8 mutant leaf epidermal cells. The white arrows mark cutin. Bar = 200 nm.
(D) The cdk8 mutant shows enhanced water loss. Water loss was expressed as the percentage of initial fresh weight. Values are averages from 20 leaves for each of three independent experiments. Values shown are means ± se.
Previous studies have shown that increased cuticle permeability is associated with enhanced resistance to B. cinerea (Bessire et al., 2007). Thus, the increased permeability of the cdk8 mutant may contribute to the resistance of cdk8 to B. cinerea. The growth phenotype and permeability of the cuticle of the cdk8 med25 double mutant were also analyzed (Supplemental Figure 8). The cuticle permeability of the med25 cdk8 double mutant was significantly increased relative to the med25 single mutant but was comparable to the cdk8 mutant (Supplemental Figure 8B). These data, together with the B. cinerea resistance of the double mutant, suggest that the compromised defense responses in med25 is overcome by the enhanced cuticle permeability, resulting in resistance to B. cinerea.
CDK8 and MED25 Interact with the Ethylene Response TF WIN1
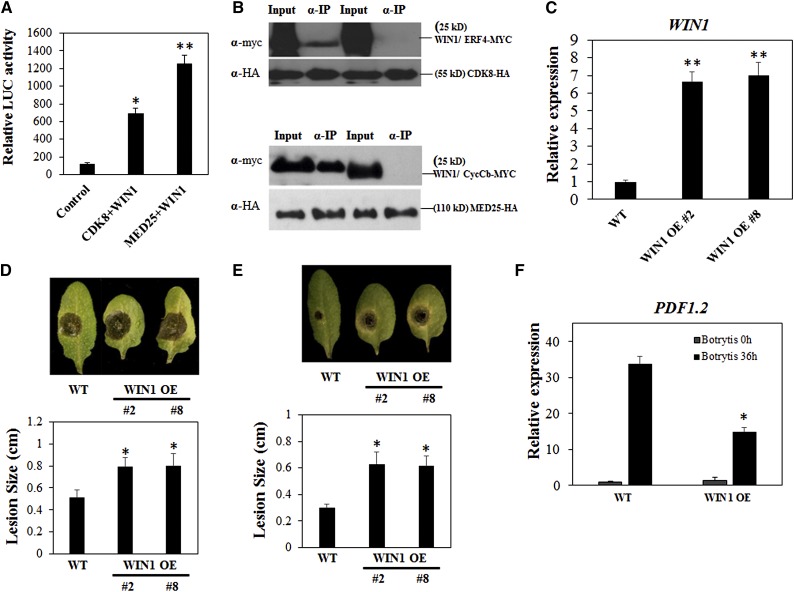
It was previously reported that overexpression of the TF WIN1 increases cuticle thickness (Broun et al., 2004; Kannangara et al., 2007) and results in susceptibility to B. cinerea (Sela et al., 2013), which contrasts with the phenotypes of the cdk8 mutant. These observations suggest that CDK8 and WIN1 have overlapping functions. Therefore, we tested the possible interaction between CDK8 and WIN1, a protein belonging to the ERF family of TFs. The coexpression of CDK8-nLuc and WIN1-cLuc led to ∼5-fold higher LUC activity than the negative control, suggesting a physical interaction (Figure 7A). Co-IP experiments in Arabidopsis protoplasts confirmed that WIN1-MYC but not ERF4-MYC was immunoprecipitated by CDK8-HA (Figure 7B). The interaction between CDK8 and MED25 further raised the possibility of MED25 and WIN1 being in the same complex with CDK8. Interestingly, the WIN1-MED25 interaction was stronger than WIN1-CDK8, because the coexpression of WIN1-cLuc and MED25-nLuc resulted in more than 10-fold higher LUC activity than the negative control (Figure 7A). These data show that MED25 interacts with other ERF/AP2 domain TFs besides ERF1 and ORA59. Co-IP experiments also demonstrated that WIN1-MYC was precipitated by MED25-HA but not by CycCb, which was used as a control (Figure 7B).
Figure 7.
CDK8 and MED25 Both Interact with the TF WIN1.
(A) CDK8 and MED25 interact with WIN1 in a split-luciferase complementation assay. The LUC activities are mean values from three biological replicates, and error bars indicate se (n = 3).
(B) WIN1 and CDK8/MED25 interact in a Co-IP assay.
(C) The expression of WIN1 in two independent transgenic WIN1 overexpression lines.
(D) B. cinerea disease symptoms and disease lesion size in the wild type and WIN1 overexpression lines at 4 dpi.
(E) A. brassicicola disease symptoms and disease lesion size in the wild type and WIN1 overexpression lines at 4 dpi.
(F) Expression of the PDF1.2 gene in the wild type and WIN1 overexpression lines after mock or B. cinerea inoculation.
Two independent WIN1 overexpression lines (WIN1 OE #2 and WIN1 OE #8) were used in these studies. The significance of differences between mean lesion sizes was analyzed. Values shown are means ± se. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
We generated WIN1 overexpression (WIN1 OE) lines by transforming 35S:WIN1-HA into wild-type plants. As confirmed by RT-qPCR, the expression of WIN1 was more than 6-fold higher in WIN1 OE lines 2 and 8 plants relative to the wild-type plants (Figure 7C). Next, we tested the WIN1 OE lines for responses to A. brassicicola and B. cinerea infection. Leaves of WIN1 OE lines 2 and 8 plants had larger disease lesions at 4 d after B. cinerea drop-inoculation, suggesting enhanced susceptibility (Figure 7D). Similarly, leaves of WIN1 OE lines 2 and 8 plants exhibited increased susceptibility to A. brassicicola at 4 DAI (Figure 7E). The enhanced susceptibility to the two fungal pathogens was accompanied by significantly reduced expression of PDF1.2 (Figure 7F). Thus, it appears that WIN1 actually affects defense through its function in the cuticle as well as other distinct pathways. It is noteworthy that WIN1 functions in fatty acid biosynthesis and most likely has more functions than just cuticle development (Kannangara et al., 2007). Furthermore, to determine whether WIN1 is a major component of the CDK8-regulated pathway, we expressed WIN1-HA in the cdk8 mutant background (cdk8;WIN1-HA). The cdk8;WIN1-HA plants showed disease lesions similar to wild-type plants at 4 DAI (Supplemental Figure 9). By contrast, the cdk8 mutant exhibited resistance without detectable disease symptoms. This genetic evidence further supports the notion that the resistance to B. cinerea in the cdk8 mutant is due to the altered cuticle profile linked to the loss of CDK8 functions.
The Phosphorylation Activity of CDK8 Is Required for the Suppression or Activation of Defense
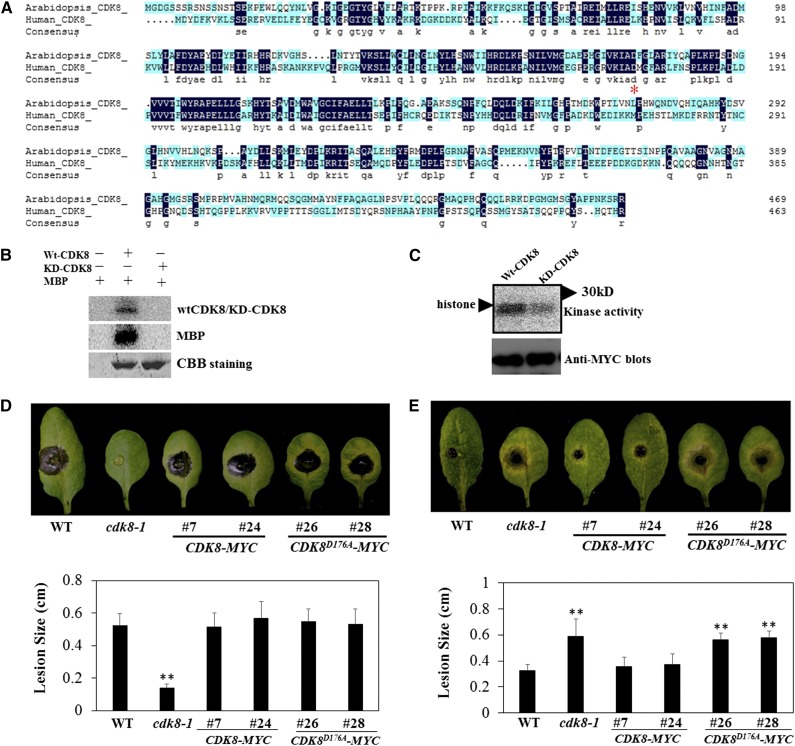
CDK8 is the only component of Mediator with known enzymatic functions. Therefore, we sought to determine the biological functions of CDK8 enzymatic activity. Sequence comparisons show that Arabidopsis CDK8 shares significant sequence identity with human CDK8 (Figure 8A). From previous studies of human CDK8, the single amino acid substitution of aspartic acid to alanine (D173A) was sufficient to nullify the kinase activity of human CDK8 (Knuesel et al., 2009b). The same residues are conserved in other animal and plant species (Supplemental Figure 10). Therefore, we generated Arabidopsis CDK8 with alanine substitution at position 176 (CDK8D176A) corresponding to the human CDK8 173D, and the recombinant protein was produced as a glutathione S-transferase (GST) fusion. Based on in vitro kinase assays, the wild-type CDK8 displayed autophosphorylation as well as phosphorylation of myelin basic protein (MBP), a commonly used artificial kinase substrate. Nonetheless, the CDK8D176A mutant resulted in a complete loss of autophosphorylation and MBP phosphorylation activities (Figure 8B). Subsequently, we cloned CDK8 and CDK8D176A into a plant transformation vector and transformed the constructs into the cdk8 mutant, generating cdk8;35S:CDK8-MYC and cdk8;35S:CDK8D176A-MYC transgenic plants (Supplemental Figure 11). Immunocomplex kinase assays with anti-MYC conjugated beads revealed that immunoprecipitates from 35S:CDK8-MYC phosphorylate histones, whereas the immunoprecipitates of 35S:CDK8D176A-MYC plants showed much weaker kinase activity, which is consistent with the data from bacterially expressed recombinant protein (Figure 8C).
Figure 8.
CDK8 Functions in Kinase-Dependent and Independent Manners.
(A) Amino acid sequence comparison between Arabidopsis and human CDK8 proteins. The conserved aspartic acid at amino acid position 176 (176D) of CDK8, which is critical for its kinase activity, is marked by the red asterisk.
(B) In vitro kinase assay showing that substitution of 176D to alanine nullifies the kinase activity of Arabidopsis CDK8. GST-CDK8 recombinant protein displayed autophosphorylation as well as phosphorylation of substrate MBP. However, GST-CDK8D176A recombinant protein lost both autophosphorylation and MBP phosphorylation activities. The experiment was repeated two times with similar results. CBB, Coomassie Brilliant Blue.
(C) Immunocomplex kinase assays showing the phosphorylation of histones by CDK8. Kinase assay was performed using protein extracts from cdk8;35S:CDK8-MYC #7 and cdk8;35S:CDK8D176A-MYC #26 transgenic seedlings.
(D) Ectopic expression of CDK8-MYC or CDK8D176A-MYC restores wild-type levels of B. cinerea responses in the cdk8 mutant.
(E) CDK8-MYC but not CDK8D176A-MYC plants restore wild-type responses to A. brassicicola.
In (D) and (E), the disease assays were performed as described in Figure 1, and disease lesions were recorded at 4 DAI. Data are mean values ± se (n = 20), and statistical analysis was conducted to determine the significant differences of values (Student’s t test, **P < 0.01).
To determine whether the kinase activity of CDK8 contributes to its defense functions, the transgenic plants cdk8;35S:CDK8-MYC and cdk8;35S:CDK8D176A-MYC were tested for their disease responses. At 4 DAI, larger disease lesions were observed in wild-type, cdk8;35S:CDK8-MYC, and cdk8;35S:CDK8D176A-MYC lines after inoculation with B. cinerea, suggesting that both CDK8 constructs restored the B. cinerea disease responses to wild-type levels (Figure 8D). On the other hand, cdk8;35S:CDK8-MYC transgenic lines showed wild-type levels of resistance to A. brassicicola, indicating that ectopic expression of CDK8 rescued the susceptibility of the cdk8 mutant to this fungus. However, the 35S:CDK8D176A-MYC lines were as susceptible as the cdk8 mutant to A. brassicicola, indicating that the kinase activity of CDK8 is necessary for resistance to A. brassicicola but not for responses to B. cinerea (Figure 8E).
At the molecular level, the expression of PDF1.2 and AACT1 was rescued in the cdk8;35S:CDK8-MYC lines but not in the cdk8;35S:CDK8D176A-MYC lines (Figures 9A and 9B), indicating that the kinase activity of CDK8 is essential for the regulation of PDF1.2 and AACT1 by CDK8. On the other hand, the expression of both wax biosynthesis genes CER1 and CER6 was restored to wild-type levels in cdk8;35S:CDK8-MYC and cdk8;35S:CDK8D176A-MYC transgenic lines (Figures 9C and 9D), indicating a CDK8 kinase-independent regulation.
Figure 9.
The Kinase Activity of CDK8 Is Required for Target Gene Expression.
(A) to (D) PDF1.2 (A), AACT1 (B), CER1 (C), and CER6 (D) gene expression levels in wild-type, cdk8;35S:CDK8-MYC, and cdk8;35S:CDK8D176A-MYC transgenic plants. Lines 7 and 24 are two independent transgenic lines expressing 35S:CDK8-MYC, whereas lines 26 and 28 are 35S:CDK8D176A-MYC lines. Error bars represent se (n = 3).
(E) The kinase-dead CDK8 mutation does not affect the CDK8-MED25 interaction in split-luciferase complementation assays. Error bars represent se (n = 3).
(F) RNAP II recruitment to the PDF1.2 promoter is dependent on the kinase activity of CDK8. Reduced RNAP II recruitment is shown in cdk8;35S:CDK8D176A-MYC transgenic plants compared with cdk8;35S:CDK8-MYC plants after B. cinerea infection. Error bars indicate se (n = 3). ChIP-qPCR experiments were repeated two times with similar results. No Ab, negative control with no antibody.
Statistical analysis was conducted to determine differences in mean values. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
The roles of the kinase activity of CDK8 in its interaction with functional partners and in RNAP II recruitment were further studied. To explore whether the kinase activity of CDK8 affects the CDK8-MED25 interaction, we cloned CDK8D176A in the nLuc vector and coexpressed it with MED25-cLuc. The LUC activities from the coexpression of CDK8-nLuc or CDK8D176A with MED25-cLuc were almost the same, implying that the CDK8-MED25 interaction is not dependent on kinase activity (Figure 9E). To better understand whether the RNAP II recruitment to the PDF1.2 promoter is affected by its kinase activity, cdk8;35S:CDK8D176A-MYC and cdk8;35S:CDK8-MYC plants were spray-inoculated with B. cinerea and used for chromatin immunoprecipitation (ChIP) assays. RNAP II recruitment to the PDF1.2 promoter regions was significantly reduced in the lines expressing kinase-dead CDK8, suggesting that kinase activity is required for RNAP II recruitment to the PDF1.2 promoter (Figure 9F).
med12 and med13 Mutants Display Phenotypes Similar to Those of the cdk8 Mutant
CDK8, CycC, MED12, and MED13, together, constitute the kinase module of Mediator in other eukaryotic organisms (Knuesel et al., 2009a). In parallel with the cdk8 mutant, we characterized SALK T-DNA insertion alleles of MED12 and MED13 for plant growth and pathogen responses (Figure 10; Supplemental Figure 12). The cdk8, med12, and med13 mutants exhibit reduced overall plant stature, glossy leaves, and late flowering (Figure 10A; Supplemental Figure 12B). The med12 and med13 mutants displayed enhanced resistance to B. cinerea but increased susceptibility to A. brassicicola, mirroring the disease response phenotypes of cdk8 (Figures 10B and 10C), which suggested that subunits of the kinase module share similar functions in regulating plant responses to necrotrophic fungi. Moreover, toluidine blue staining of cdk8, med12, and med13 mutants revealed enhanced permeability relative to the wild type. When 0.05% toluidine blue was applied, leaves of cdk8, med12, and med13 mutant plants stained much more quickly and strongly than wild-type leaves (Figure 10D). These observations suggest that the kinase module may regulate cuticle development in a coordinated manner.
Figure 10.
Mediator Kinase Module Components MED12 and MED13 Mediate Defense Responses.
(A) Growth phenotypes of 4-week-old wild-type, cdk8, med12, and med13 mutant plants.
(B) B. cinerea disease symptoms and disease lesion sizes in the wild type and Mediator kinase module mutants.
(C) A. brassicicola disease symptoms and disease lesion sizes in the wild type and Mediator kinase module mutants.
(D) Enhanced cuticle permeability of the wild type and Mediator kinase module mutants revealed by toluidine blue staining.
(E) Relative expression of defense and cuticular wax genes in the wild type and Mediator kinase module mutants.
The disease assays in (B) and (C) were conducted as described in Figure 1. The lesion sizes are mean values ± se from at least 20 disease lesions, and the data were analyzed with Student’s t test (*P < 0.05, **P < 0.01).
In addition to the disease phenotypes and increased leaf permeability noted among kinase module mutants, gene expression data suggest that CDK8, MED12, and MED13 regulate the expression of some of the same target genes, including PDF1.2 and PR1 (Figure 10E). The expression of both defense-related genes was significantly reduced in cdk8, med12, and med13 mutants. Interestingly, CER3 was upregulated in all mutants, while CER1 was upregulated only in cdk8 (Figure 10E), indicating that MED12 and MED13 may also have functions different from those of CDK8.
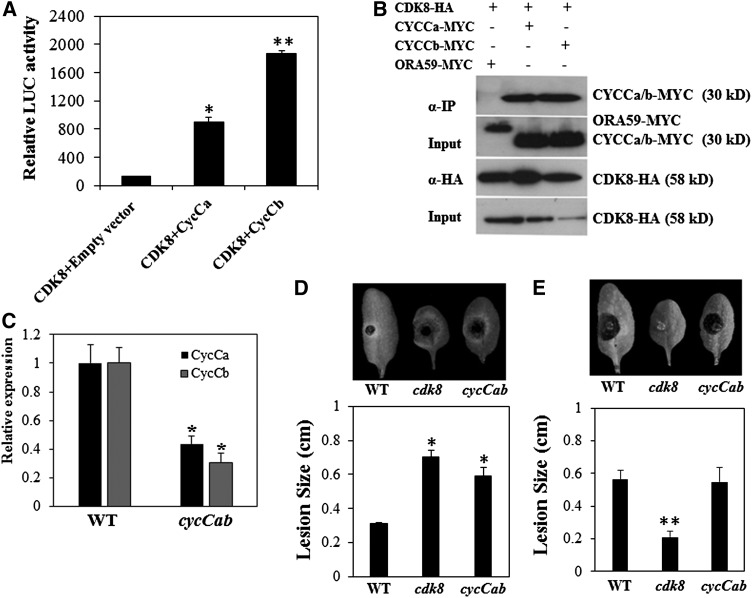
CDK8 Physically Interacts with Two CycCs
In yeast and mammalian cells, Mediator kinase modules contain a CycC that interacts with CDK8 (Borggrefe et al., 2002; Andrau et al., 2006). The Arabidopsis genome contains two CycC genes, CycCa (At5g48630) and CycCb (At5g48640) (Menges et al., 2005). In split luciferase complementation assays to determine interactions between CDK8 and CycCa/b, the coexpression of CDK8-nLuc and CycCa/CycCb-cLuc resulted in significant LUC activities, which averaged ∼7- and 10-fold higher than the negative control, respectively (Figure 11A). The interaction of CDK8 and CycCb was stronger than that of CDK8 and CycCa based on the LUC reconstitution signal. In addition, Co-IP assays confirmed the direct interactions between CDK8 and CycCa/b. Both CycCa-MYC and CycCb-MYC were successfully precipitated by CDK8-HA, whereas the negative control ORA59-MYC was not precipitated (Figure 11B). These data suggest that the CycC-CDK8 interaction is evolutionarily conserved in Arabidopsis, yeast, and mammalian cells.
Figure 11.
Arabidopsis CDK8 Interacts with Two CycCs.
(A) Strong interactions between CDK8 and two CycCs in split-luciferase complementation assays. Data are mean values ± se from three independent experiments. Error bars indicate se (n = 3).
(B) CDK8 interacts with CycCa and CycCb in Co-IP assays. The experiment was repeated at least three times.
(C) Reduced expression of CycCa and CycCb genes in the cycCab mutant allele. The cycCab allele (SALK_039400C) carries a T-DNA insertion between the CycCa and CycCb genes disrupting the expression of both genes. CycCa and CycCb are tightly linked and transcribed in the same orientation.
(D) A. brassicicola disease symptoms and disease lesion sizes in wild-type, cdk8, and cycCab mutant plants.
(E) B. cinerea disease symptoms and disease lesion sizes in wild-type, cdk8, and cycCab mutant plants.
The disease assays in (C) and (D) were conducted as described in Figure 1. The lesion sizes are mean values ± se from at least 20 disease lesions. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
The disease phenotypes of cycC mutants were examined to determine the defense functions of the two cyclin genes. Unlike cdk8, med12, and med13 mutants, the single cycCa and cycCb mutants showed wild-type levels of disease responses (Supplemental Figure 13), which implied redundant functions of the two CycC genes. The two genes are linked, which precluded the possibility of generating double mutants. However, we identified a T-DNA insertion homozygous line (SALK_039400C) designated cycCab in which both CycCa and CycCb are downregulated due to the insertion of the T-DNA in the small intergenic region between the two genes (Figure 11C). The cycCab mutant showed increased susceptibility to A. brassicicola compared with wild-type plants, with larger disease lesions at 4 DAI, similar to the cdk8 mutant, which is consistent with their physical interaction (Figure 11D). The cycCab mutants displayed no altered responses to B. cinerea, unlike the resistance in the cdk8 mutant.
DISCUSSION
In yeast and metazoan cells, CDK8 defines the kinase module of Mediator, which also contains MED12, MED13, and CycC (Borggrefe et al., 2002; Andrau et al., 2006). Although increasing evidence has demonstrated that CDK8 is a critical regulator of transcription in metazoan and yeast cells, the function of plant CDK8 remains largely unknown. In this work, we show that Arabidopsis CDK8 has both negative and positive roles in the regulation of plant responses to pathogens, and we provide insights into the molecular and biochemical functions of CDK8 in plant immunity. Overall, we provide extensive data supporting the complex function of CDK8 and the kinase module of Mediator. These include (1) genetic data revealing the contrasting defense functions of CDK8 to related necrotrophic fungi sharing similar mechanisms of pathogenesis, virulence, and modes of nutrition; (2) epispastic interactions between resistance based on altered cuticle layer and JA-mediated immune responses; (3) the impact of the enzymatic activity of CDK8 and its biological function, particularly in response to fungal infection; (4) the molecular interactions between CDK8 and other Mediator subunits (MED25, cyclins) as well as the ERF/AP2 family TF WIN1, shedding light on the various mechanisms of CDK8 function; and (5) CDK8 regulates the accumulation of secondary metabolite HCAAs through association with AACT1, which mediates a critical biosynthetic step.
CDK8 Positively Regulates JA Responses and PDF1.2 and AACT1 Gene Expression
Analysis of loss-of-function alleles of CDK8 revealed that CDK8 contributes to JA-mediated defense responses and regulates the synthesis of the secondary metabolite HCAAs, which are essential for plant resistance to fungal infection. ChIP data demonstrated that CDK8 positively regulates PDF1.2 and AACT1 transcription by associating with their promoter regions. These findings suggest a positive role for CDK8 in the regulation of gene expression. Furthermore, CDK8 interacts with the multifunctional Mediator subunit MED25, which is known to regulate JA responses (Kidd et al., 2009). MED25 interacts with diverse TFs, including MYC2, ERF1, and ORA59 (Çevik et al., 2012; Chen et al., 2012), which are all implicated in plant defense against fungi. MYC2 is known to suppress resistance to B. cinerea, while ERF1 and ORA59 contribute to resistance (Berrocal-Lobo et al., 2002; Lorenzo et al., 2004; Pré et al., 2008). The interaction data indicate that CDK8 functions in concert with MED25 to regulate the expression of JA-responsive genes. CDK8 may be recruited by ERF1/ORA59 through MED25 to the PDF1.2 promoter. This is consistent with the contributions of MED25 and CDK8 to the ERF1- and ORA59-dependent activation of PDF1.2 expression as well as the association of CDK8 with the PDF1.2 promoter.
It was also recently suggested that plants regulate the transcription of specific genes through the combined actions of subsets of Mediator subunits (Hemsley et al., 2014). For example, RNAP II recruitment to C-repeat binding factor-responsive cold-regulated genes requires MED16, MED2, and MED14 subunits. Genes inducible by darkness also required MED16, but they required a different combination of Mediator subunits for their expression from those of genes induced by cold. Similarly, the Arabidopsis yellow and sensitive to iron-deficiency1 (yid1) mutant, which harbors a mutation in the MED16 gene, accumulates reduced amounts of iron. MED16 interacted with MED25, which implicated MED25 in the regulation of iron homeostasis. This role of MED25 was attributed to the interaction with EIN3 and EIL1, two TFs that function in ethylene signaling that were previously associated with the regulation of iron homeostasis (Yang et al., 2014). The transcriptome in yid1 and med25 mutants was significantly affected by iron deficiency. The transcription of marker genes for iron homeostasis and iron uptake was reduced in the yid1 and med25 mutants under iron-deficient conditions. Thus, MED16 and MED25 coordinately regulate iron homeostasis in Arabidopsis. These findings illustrate the complex functions of Mediator subunits through combinatorial interactions with other Mediator subunits and non-Mediator proteins. To date, MED16 has been implicated in defense, iron homeostasis, and abiotic stress tolerance (Wathugala et al., 2012; Yang et al., 2014). Similarly, CDK8 may also have different functions through interactions with MED25, cyclins, and WIN1 and possible interactions with other Mediator subunits and TFs.
The Differential Impact of CDK8 on Responses to Necrotrophic Fungal Species Is Linked to Its Function in Cuticle Development
The plant cuticle is a structured layering of long-chain soluble waxes comingled within a cutin polyester matrix that serves as the primary physical barrier against pathogens and prevents nonstomatal water loss (Bessire et al., 2007). Despite the positive regulatory role of CDK8 in the expression of PDF1.2 and AACT1 genes and in the accumulation of HCAAs, the cdk8 mutant is resistant to B. cinerea. We attribute this to the likely loss of plant cues specifically required for the virulence of B. cinerea, which may have masked the defects in other pathways. We also cannot preclude the possible loss of other susceptibility factors regulated by CDK8, besides changes in the cuticle profile, which are specifically required for B. cinerea. Many pathogens are dependent on host susceptibility factors or virulence targets to cause disease. Alternatively, cdk8 may accumulate compounds that have specific inhibitory effects on B. cinerea but not on A. brassicicola. However, based on current observations by many laboratories on the correlation between loss of cuticle integrity and resistance, we suggest that the resistance of cdk8 is also linked to its altered cuticle.
The function of CDK8 in cuticle development is supported by genetic and molecular data. Leaves of the cdk8 mutant exhibit glossy and smooth appearances reminiscent of changes in the cuticle. Furthermore, the cdk8 mutant has enhanced cuticle permeability and reduced cuticle thickness. Importantly, CDK8 interacts with WIN1, an ERF family TF that is known to modulate the wax profile of the Arabidopsis cuticle by directly binding to the LACS2 promoter (Kannangara et al., 2007). LACS2 encodes a member of the long-chain acyl-CoA synthases involved in cutin biosynthesis in Arabidopsis (Schnurr et al., 2004). Interestingly, WIN1 overexpression results in increased expression of several wax biosynthesis genes, increased alkane levels, and susceptibility to fungal infection (Broun et al., 2004; Sela et al., 2013). Due to the lack of appropriate loss-of-function alleles of the WIN1 gene, we could not make direct comparisons of the win1 and cdk8 mutants. However, overexpression of CDK8 restores susceptibility to B. cinerea to at least wild-type levels, and WIN1 overexpression confers susceptibility. Similarly, WIN1 overexpression enhances cuticle thickness (Broun et al., 2004), but the cdk8 mutation reduced cuticle thickness. These data of contrasting phenotypes in the gain-of-function WIN1 and loss-of-function cdk8 suggest shared functions of WIN1 and CDK8 consistent with their physical interactions. Intriguingly, the expression of CER1 and CER6, implicated in cuticle biosynthesis pathways, is significantly high in the cdk8 mutant, but the biological significance of this is unclear. Despite the reported correlation between loss of cuticle integrity and resistance to B. cinerea in many Arabidopsis mutants, the exact mechanisms are still unclear. This intriguing correlation is also debatable, as there are mutants that have permeable cuticles but do not show altered pathogen responses (Voisin et al., 2009).
CDK8 Functions in Both Kinase-Dependent and Independent Manners
Arabidopsis CDK8 shows phosphorylation activities consistent with previous data in yeast and mammalian cells (Nelson et al., 2003; Knuesel et al., 2009b). Interestingly, our data suggest that CDK8 could function in kinase-dependent and independent manners. The expression of kinase-dead CDK8 failed to rescue the attenuated PDF1.2 and AACT1 transcription as well as the A. brassicicola susceptibility of the cdk8 mutant. By contrast, the negative regulation of B. cinerea resistance by CDK8 is independent of its kinase functions, because the kinase-dead CDK8 restored the responses to B. cinerea to wild-type levels. Consistent with the phenotypes, the kinase-dead CDK8 also restored the expression of CER1 and CER6, two cuticular wax biosynthesis genes, to wild-type levels.
Overlapping Functions of CDK8 Module Subunits in Arabidopsis Defense and Development
All four subunits of the kinase module of Mediator CDK8 (i.e., MED12, MED13, and the two CycCs) share defense functions. For example, all aspects of the cdk8 mutant appear to be less severe forms of the med12 and med13 mutants, thus adding credence to the common functions of the kinase module of Mediator. cdk8, med12, and med13 mutants displayed similar morphological and disease phenotypes as well as increased cuticle permeability. Similarly, PDF1.2, PR1, and CER3 show similar patterns of gene expression in the three kinase module mutants. CER1 is upregulated only in cdk8, but not in med12 and med13, suggesting that MED12 and MED13 also have some separate functions. Our study also demonstrates strong physical interactions between CDK8 and two Arabidopsis CycCs, suggesting the conserved partnership of CDK8-CycC throughout eukaryotes. Consistent with this physical interaction, the cycCab mutant, in which both CycCa and CycCb are downregulated, exhibited enhanced susceptibility to A. brassicicola. The detailed mechanism has yet to be determined, but these data further support the complexity of CDK8 function.
cdk8 Identifies Differences in the Pathogenesis of B. cinerea and A. brassicicola
The contrasting responses of cdk8, med12, and med13 mutants to the necrotrophic fungi B. cinerea and A. brassicicola was unexpected but suggest that the two pathogens might rely on different plant cues for activating their virulence. The two fungi are known to share related pathogenesis strategies, and plants activate similar immune responses against these pathogens (Lai and Mengiste, 2013). Most Arabidopsis mutants identified to be susceptible to B. cinerea also exhibit susceptibility to A. brassicicola, with the exception of some ethylene pathway mutants, which show unaltered responses to A. brassicicola (van Wees et al., 2003). The genetic data suggest that CDK8 is a negative regulator of immune responses to B. cinerea. However, resistance to B. cinerea in the cdk8 mutant is linked to changes in the cuticle profile, which is epistatic to the functions of CDK8 in regulating secondary metabolites and plant defensins. Resistance due to cuticle permeability was previously attributed to faster activation of defense, faster mobility of defense compounds to the infection site (Bessire et al., 2007), efficient perception of cutin monomers in the absence of intact cuticle, and attenuated fungal virulence due to loss of plant-derived cues (Chassot et al., 2007). In a comprehensive study involving many Arabidopsis mutants with altered cuticles and having permeable cuticles, resistance in cuticle mutants was suggested to be independent of the cuticle composition per se but due to the overactivation of defense (Voisin et al., 2009). In cdk8, changes in cuticle integrity may have resulted in reduced activation of virulence in B. cinerea but not A. brassicicola. Histological observation of infected tissue through trypan blue staining also clearly showed that B. cinerea has attenuated virulence and fungal growth that is significantly retarded on the cdk8 mutant. Consistent with this, many Arabidopsis mutants with permeable cuticles, including long-chain acyl CoA synthase2, are resistant to B. cinerea but not susceptible to A. brassicicola. The disease response of the cdk8 med25 double mutant suggests that the cdk8 mutation is epistatic to med25 with respect to B. cinerea resistance but synergistic with respect to A. brassicicola resistance and JA responses. Despite the extreme susceptibility of med25 to B. cinerea, cdk8 med25 is resistant to B. cinerea, which further strengthens the observation that the altered cuticle in cdk8 overrides defects in other plant immune responses.
Mediator and Plant Immunity
Many recent reports implicate plant Mediator in the regulation of gene expression underlying various physiological processes, including hormone responses, flowering time, and biotic and abiotic stress tolerance (Canet et al., 2012; Chen et al., 2012; Wathugala et al., 2012; Zhang et al., 2012; An and Mou, 2013). Additional functions will likely be discovered as research in plant Mediator expands. The challenge, however, is to dissect the sequence of molecular and biochemical actions of Mediator as it relates to other regulators, including TFs. Also, the posttranscriptional regulation of Mediator subunits will be an interesting layer to study, as that adds complexity and functional diversity to the functions of the complex. Most Mediator subunits are implicated in defense, most likely, because pathogen response may be an obvious phenotypic read out for Mediator functions in other fundamental cellular and molecular processes. Direct defense function was recently provided for a Mediator subunit when it was discovered that Mediator is a target of pathogen virulence. The Hyaloperonospora arabidopsidis nucleus-localized effector HaRxL44 interacts with MED19a, resulting in the degradation of MED19a in a proteasome-dependent manner. HaRxL44 interferes with Mediator function by degrading MED19, shifting the balance of defense transcription from SA-responsive defense to JA/ethylene signaling and enhancing susceptibility to biotrophs by attenuating SA-dependent gene expression (Caillaud et al., 2013).
In conclusion, this work demonstrates the impacts of the kinase module of the Mediator complex on the regulation of a major group of plant defense molecules, secondary metabolites, and defense-linked TFs as well as the development of the plant cuticle. The distinct responses of CDK8 in plant defense to related pathogens are the consequences of its regulatory impacts on cuticle development on the one hand and plant defensins and secondary metabolites on the other. The cdk8 mutant displays unique immune responses that combine susceptibility to A. brassicicola and P. syringae with resistance to B. cinerea. Thus, CDK8 provides a powerful genetic tool for dissecting differences in plant immunity to the two related necrotrophs.
METHODS
Plant Materials, Growth Conditions, and Disease Assays
All mutant and transgenic Arabidopsis thaliana lines used in the studies described here are in the Columbia-0 (Col-0) ecotype background unless indicated otherwise. cdk8-1 (SALK_138675), cdk8-2 (SALK_016169), med12 (SALK_108241), med13 (SALK_018056), med25 (SALK_129555C), aact1 (SALK_097380), cycCa (SAIL_102_B02), cycCb (SALK_053291C), and cycCab (SALK_039400C) were obtained from the ABRC. The T-DNA insertions were confirmed by genomic DNA-PCR using the primers listed in Supplemental Data Set 1.
For root growth response assay to JA, Arabidopsis seeds were surface-sterilized and incubated at 4°C for 2 d before plating on half-strength Murashige and Skoog medium containing 0.8% agar and 1% sugar with the indicated concentration of MeJA. Then, the seeds were germinated under continuous light at 22°C for 7 to 10 d.
Plant growth conditions, fungal growth media, and fungal disease assays were as described previously (Dhawan et al., 2009; Laluk and Mengiste, 2011). In brief, plants were grown in growth chambers at 24°C, 70% relative humidity, and a 12-h-light/12-h-dark cycle. For Botrytis cinerea assay, plants were grown for 5 weeks in growth chambers under the above conditions and then inoculated with a conidial suspension of 250,000 to 300,000 spores/mL in 1% Sabouraud Maltose Broth buffer. For Alternaria brassicicola assay, at least 10 fully expanded adult leaves were excised and placed in plastic trays lined with wet filter paper for each biological replicate. Leaves were inoculated with 500,000 spores/mL in 1% Sabouraud Maltose Broth buffer. The transparent trays with leaves were sealed to maintain high humidity. Disease lesion diameters were measured after 3 to 6 d. All disease assays were repeated at least four times.
Trypan Blue Staining
Trypan blue stock solution contains 10 g of phenol, 10 mL of lactic acid, 10 mL of glycerol, 10 mL of distilled water, and 40 mg of trypan blue (Sigma-Aldrich). Trypan blue working solution is made by mixing 2 volumes of 96% ethanol and 1 volume of trypan blue stock solution. After standard inoculation with conidial suspensions, the detached leaves were incubated in boiling trypan blue working solution for 1 min. Then, the leaves were transferred to wells of cell culture plates and immersed in chloral hydrate solution (250%, w/v) until chlorophyll was removed. The stained leaves were then observed with a Leica imaging system or the photographs were taken using a 10×/0.25 numerical aperture objective on a Nikon Microphot SA wide-field fluorescence microscope equipped with a CCD camera (ORCA-ER C4742-95; Hamamatsu Photonics).
Molecular Cloning
The full-length coding sequences of CDK8, MED25, WIN1, ERF1, ORA59, ERF4, MYC2, JAZ1, CycCa, CycCb, and other genes described in our studies were amplified by PCR from wild-type Col-0 Arabidopsis cDNA using the primers listed in Supplemental Data Set 1. For Gateway-based cloning, fresh PCR products were first cloned into pENTER vector (Invitrogen) and then transferred to destination vectors that contained N/C-terminal fragments of LUC via LR reactions (according to the Invitrogen manual). For Co-IP, the indicated genes were transferred to pHBT95-HA/MYC vectors from sequenced templates via transfer PCR (Erijman et al., 2011).
The CDK8D176A point mutation was generated by overlapping PCR with the primers listed in Supplemental Data Set 1. Full-length sequence of CDK8 and CDK8D176A were cloned into pGEX-4T-2 for recombinant protein expression and in vitro kinase assay and subsequently inserted into the binary vector pBA-MYC for generating transgenic plants. All constructs were verified by sequencing.
Generation of Arabidopsis Transgenic Plants and Double Mutant
The constructs with the expression cassettes 35S:CDK8-MYC and 35S:CDK8D176A-MYC were transformed into cdk8-1 mutant plants by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Primary transformants were selected for Basta resistance, and CDK8 protein expression levels were determined by protein gel blotting with anti-MYC antibody (1:5000 dilution; Abcam). Transgenic lines with a 3:1 (resistant:sensitive to Basta) segregation ratio were selected, and at least three homozygous lines were identified in the T3 generation. The med25 cdk8 double mutant was generated by crossing, and homozygous double mutant individuals were identified by genomic DNA-PCR.
RNA Extraction, Reverse Transcription, and RT-qPCR
Total RNA was extracted from Arabidopsis leaves or seedlings with Trizol reagent according to the manufacturer’s instructions (Sigma-Aldrich). After RNase-free DNase I treatment (Ambion), total RNA was reverse-transcribed into cDNA (New England Biolabs). RT-qPCR was performed with a SYBR Green kit (Bio-Rad) according to the manufacturer’s instructions using gene-specific primers with Arabidopsis ACT2 as an endogenous reference for normalization. A minimum of three technical replicates of the RT-qPCR assay were used for each sample with at least three biological replicates. Primers used are listed in Supplemental Data Set 1.
RNA-seq and Analysis
Five-week-old wild-type (Col-0) and cdk8-1 plants grown on soil were mock-inoculated or inoculated with B. cinerea. The leaves were collected at 36 h after inoculation for RNA isolation. The RNA-seq data analysis and sequence quality control were performed according to Lai et al. (2014). Gene expression levels were determined by reads per kilobase per million reads. Genes with more than 2-fold changes (P value and false discovery rate ≤ 0.05) in cdk8 compared with wild-type plants were selected as differentially expressed genes. The differentially expressed gene expression patterns in the wild type and cdk8 were presented as a heat map using the R statistical computing package.
Split-Luciferase Complementation Assays
Transient expression in Arabidopsis protoplasts was conducted as described previously (Yoo et al., 2007; Zhu et al., 2010). All plasmids used in this assay were purified with the Qiagen Plasmid Maxi Kit or the Qiagen Plasmid Midi Kit. Ten micrograms of purified plasmid of each construct was used for protoplast transformation. After coexpression, the substrate LUC was added and incubated in the dark for at least 1 h. The recombinant LUC activity was measured using a Wallac VICTOR2 plate reader.
Co-IP
Purified plasmids were transiently expressed in Arabidopsis protoplasts as described (Yoo et al., 2007; Zhu et al., 2010). After transformation, protoplasts were harvested and suspended in 1 mL of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% [v/v] Triton X-100, and 1× protease inhibitor cocktail from Sigma-Aldrich plus 1 mM phenylmethylsulfonyl fluoride) and then centrifuged at 12,000 rpm for 10 min at 4°C. Fifty microliters of the supernatant served as input, and the rest of the supernatant was incubated with 20 μL of prewashed monoclonal anti-HA agarose antibody (Sigma-Aldrich) at 4°C for at least 4 h with gentle rotation. The agarose was washed four times with lysis buffer and boiled in 50 μL of 1× SDS loading buffer for 5 min. Protein samples were then subjected to protein gel blot analysis and detected with anti-HA or anti-MYC antibody (1:5000 dilution; Abcam).
Transcriptional Activation Assay
Arabidopsis protoplasts were isolated as described (Yoo et al., 2007; Zhu et al., 2010). Protoplasts were cotransformed with 35S:full-length LUC for an internal control, PDF1.2 promoter-GUS, and effector plasmid construct 35S:ERF1-MYC or 35S:ORA59-MYC, which were described previously (Çevik et al. 2012). After overnight transformation, protoplasts were suspended on ice in 20 μL of lysis buffer containing 2.5 mM Tris-phosphate, pH 7.8, 1 mM DTT, 2 mM DACTAA, 10% glycerol, and 1% Triton X-100. Protoplast lysates mixed with 20 μL of LUC substrate (Promega) were measured with a plate reader for LUC activity. At the same time, 5 μL of protoplast lysate was mixed with 10 μL of 4-methylumbelliferyl β-d-glucuronide substrate buffer (10 mM Tris-HCl, pH 8, 1 mM 4-methylumbelliferyl β-d-glucuronide, and 2 mM MgCl2) for at least 1 h at 37°C, and the reaction was stopped by the addition of 100 μL of 0.2 M Na2CO3. GUS activity was also measured using a plate reader. GUS activities were normalized by internal LUC activities from the same sample, and at least three biological replicates were analyzed.
In Vitro and Immunocomplex Kinase Assays
Recombinant GST-CDK8 or GST-CDK8D176A was incubated in 25 μL of kinase reaction buffer (50 mM Tris, pH 7.5, 1 mM DTT, 10 mM MgCl2, 1 µM ATP, and 5 µCi of [γ-32P]ATP) with 1 µg of MBP at room temperature for 30 min. The reaction was stopped by the addition of SDS sample buffer, and the mixtures were then subjected to SDS-PAGE. Fourteen-day-old cdk8;35S:CDK8-MYC or cdk8;35S:CDK8D176A-MYC transgenic seedlings were used for the immunoprecipitation kinase assay. After immunoprecipitates were washed with 1 mL of kinase reaction buffer (50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, and 1 mM DTT), kinase assays were performed at room temperature for 30 min in buffer containing 25 mM ATP, 1 µM ATP, 5 µCi of [γ-32P]ATP, and histone. After electrophoresis on a 12% SDS gel, the phosphorylated histone was visualized by autoradiography.
Transmission Electron Microscopy
For transmission electron microscopy analysis, the ninth leaves from 4-week-old plants were fixed by the microwave method as described previously (Dhawan et al., 2009). The primary fixation was performed with buffer containing 2% paraformaldehyde, 2.5% glutaraldehyde, and 0.1 M potassium phosphate buffer, pH 6.8. Secondary fixation was performed with buffer containing 1% OsO4 and 1.5% K3Fe(CN)6. After fixation, samples were dehydrated in an ethanol series and propylene oxide. Samples were embedding in polymerized resin. Ultrathin sections were viewed with an FEI/Philips CM-100 transmission electron microscope.
Measurement of HCAAs by LC-MS
Wild-type and cdk8 mutant plants were inoculated with A. brassicicola, and at 48 h after inoculation, ∼200 mg of Arabidopsis leaves was extracted with 2 mL of methanol overnight at 25°C. HCAAs were analyzed with a Waters 600 LC-MS instrument (Agilent QQQ 6460 system coupled to a 1200 series liquid chromatography pump and autosampler) as described (Muroi et al., 2009).
ChIP-qPCR
ChIP assays were performed according to Saleh et al. (2008) with minor modifications. Basically, 1 g of 14-d-old cdk8;35S:CDK8-MYC seedlings or 4 g of 30-d-old leaves of wild-type, cdk8;35S:CDK8-MYC, or cdk8;35S:CDK8D176A-MYC transgenic plants were used for each assay. Chromatin was isolated and sonicated. One hundred microliters of immunoprecipitate was diluted 10 times with nuclear lysis buffer and incubated at 4°C overnight with anti-MYC or anti-RPB2 antibody (Abcam) or rabbit IgG as a negative control. The protein-chromatin complexes were then captured by salmon sperm DNA/protein A agarose (Millipore). After washing and reverse cross-linking, the precipitated DNA was purified with a PCR clean-up kit. RT-qPCR was performed with negative control (IgG-precipitated DNA) and input control (the chromatin before precipitation). The ChIP enrichment was normalized to the input and then converted to fold change. All primers are listed in Supplemental Data Set 1.
Toluidine Blue Staining and Water Loss Measurement
Fully expanded adult leaves were excised and submerged in 0.05% toluidine blue solution. The stain was allowed to incubate up to 1 h at room temperature. At least eight leaves from each genotype were tested, and the staining experiments were repeated at least three times. For water loss experiments, leaves of 5-week-old plants were detached and left at room temperature, and the weight (∼20 leaves) was recorded. Water loss was expressed as the percentage of initial fresh weight.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: CDK8 (AT5G63610), MED12 (AT4G00450), MED13 (AT1G55325), CYCCa (AT5G48630), CYCCb (AT5G48640), MED25 (AT1G25540), ERF1 (AT3G23240), ORA59 (AT1G06160), AACT1 (AT5G61160), WIN1 (AT1G15360), ERF2 (AT5G47220), ERF4 (AT3G15210), CER1 (AT1G02205), CER2 (AT4G24510), CER3 (AT5G57800), CER4 (AT4G33790), CER6 (AT1G68530), CER7 (AT3G60500), LTP3 (AT5G59320), and KCS1 (AT1G01120).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Disease Symptoms and Fungal Growth Revealed by Trypan Blue Staining of B. cinerea-Inoculated Leaves.
Supplemental Figure 2. Venn Diagram and Heat Map Presenting Differential Expression of Genes in the Wild Type and cdk8 after Mock or B. cinerea Inoculation.
Supplemental Figure 3. The Expression of PDF1.2, ORA59, and PDF1.3 in the Wild Type and cdk8 in Response to A. brassicicola.
Supplemental Figure 4. Protein Gel Blot Confirming Equal Protein Amounts Used for Transactivation Assays.
Supplemental Figure 5. The Expression of Cuticle Wax Biosynthesis Genes in the Wild Type and cdk8 Mutant.
Supplemental Figure 6. The CDK8 Target Gene AACT1 Is Required for Resistance to B. cinerea.
Supplemental Figure 7. The Glossy Leaf Surface of cdk8 Mutant Compared with Wild-Type Plants.
Supplemental Figure 8. Growth and Cuticle Permeability of cdk8 and med25 Single and Double Mutants.
Supplemental Figure 9. B. cinerea Disease Symptoms of cdk8;WIN1-HA Plants.
Supplemental Figure 10. Amino Acid Sequence Comparisons between Full-Length Plant, Human, Drosophila, and Mouse CDK8 Proteins.
Supplemental Figure 11. Protein Gel Blot Showing Plants Expressing Wild-Type and Kinase-Dead CDK8 Protein.
Supplemental Figure 12. Arabidopsis Mutants med12 and med13 in CDK8 Kinase Module Have Altered Growth.
Supplemental Figure 13. Disease Symptoms in cycCa and cycCb Single Mutants after Inoculation with B. cinerea and A. brassicicola.
Supplemental Data Set 1. Primer Sequences Used in This Study.
Supplemental Data Set 2. Differential Expression of Transcription Factors in cdk8 Compared with Wild-Type Plants after B. cinerea Infection.
Supplemental Data Set 3. List of Differentially Expressed Genes in cdk8 Compared with the Wild Type from RNA-seq Analysis.
Supplementary Material
Acknowledgments
We thank Yang Zhao, Yueh-Ju Hou, and Xing Lu (Department of Horticulture and Landscape, Purdue University) and Volkan Çevik (John Innes Centre, Sainsbury Laboratory) for plasmid vectors. We thank Dylan J. Taatjes (Colorado State University) for valuable advice in designing kinase-dead CDK8 constructs and Chao Cai (Biology Department, Purdue University) for assistance with the imaging of fungal growth in inoculated plants. This work was supported by the National Science Foundation (Grant IOS-1050095), the Next-Generation BioGreen 21 Program (Grant PJ00949503), Rural Development Administration, Republic of Korea, and the National Research Foundation of Korea (Grant 2013R1A2A1A01005170).
AUTHOR CONTRIBUTIONS
Y.Z. conducted most of the experiments. Y.Z. and T.M. designed most of the experiments and directed the project. C.M.S. performed the initial mutant screen that identified the cdk8 and cyclin mutants. F.F. and J.T. performed the analyses of RNA-seq data. P.W. conducted the kinase assays. T.M., J.-K.Z., D.-J.Y., and S.Y.L. analyzed the data and wrote the article.
Glossary
- TF
transcription factor
- SA
salicylic acid
- JA
jasmonic acid
- HCAA
hydroxycinnamic acid amide
- RT-qPCR
quantitative RT-PCR
- DAI
days after inoculation
- RNA-seq
RNA sequencing
- MeJA
methyl jasmonate
- ChIP
chromatin immunoprecipitation
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
- LC-MS
liquid chromatography-mass spectrometry
- Co-IP
coimmunoprecipitation
- Col-0
Columbia-0
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- AbuQamar S., Chen X., Dhawan R., Bluhm B., Salmeron J., Lam S., Dietrich R.A., Mengiste T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48: 28–44. [DOI] [PubMed] [Google Scholar]
- Alarcón C., et al. (2009). Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Mou Z. (2013). The function of the Mediator complex in plant immunity. Plant Signal. Behav. 8: e23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau J.C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M.G., van de Peppel J., Werner M., Holstege F.C. (2006). Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22: 179–192. [DOI] [PubMed] [Google Scholar]
- Ansari A.Z., Koh S.S., Zaman Z., Bongards C., Lehming N., Young R.A., Ptashne M. (2002). Transcriptional activating regions target a cyclin-dependent kinase. Proc. Natl. Acad. Sci. USA 99: 14706–14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979. [DOI] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729. [DOI] [PubMed] [Google Scholar]
- Beckerman R., Donner A.J., Mattia M., Peart M.J., Manley J.L., Espinosa J.M., Prives C. (2009). A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 23: 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belakavadi M., Fondell J.D. (2010). Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol. Cell. Biol. 30: 2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32. [DOI] [PubMed] [Google Scholar]
- Bessire M., Chassot C., Jacquat A.C., Humphry M., Borel S., Petétot J.M., Métraux J.P., Nawrath C. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26: 2158–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Borggrefe T., Davis R., Erdjument-Bromage H., Tempst P., Kornberg R.D. (2002). A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277: 44202–44207. [DOI] [PubMed] [Google Scholar]
- Bourdenx B., Bernard A., Domergue F., Pascal S., Léger A., Roby D., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P., Poindexter P., Osborne E., Jiang C.Z., Riechmann J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J.D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet J.V., Dobón A., Tornero P. (2012). Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. (1997). Genetics of transcriptional regulation in yeast: Connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13: 1–23. [DOI] [PubMed] [Google Scholar]
- Çevik V., Kidd B.N., Zhang P., Hill C., Kiddle S., Denby K.J., Holub E.B., Cahill D.M., Manners J.M., Schenk P.M., Beynon J., Kazan K. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick J.Z., Asturias F.J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30: 264–271. [DOI] [PubMed] [Google Scholar]
- Chassot C., Nawrath C., Métraux J.P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49: 972–980. [DOI] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A. (2001). Natural products and plant disease resistance. Nature 411: 843–847. [DOI] [PubMed] [Google Scholar]
- Donner A.J., Ebmeier C.C., Taatjes D.J., Espinosa J.M. (2010). CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A.J., Hoover J.M., Szostek S.A., Espinosa J.M. (2007a). Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle 6: 2594–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A.J., Szostek S., Hoover J.M., Espinosa J.M. (2007b). CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J., Holmberg S., Hebert H., Gustafsson C.M. (2006). The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. USA 103: 15788–15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erijman A., Dantes A., Bernheim R., Shifman J.M., Peleg Y. (2011). Transfer-PCR (TPCR): A highway for DNA cloning and protein engineering. J. Struct. Biol. 175: 171–177. [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Gillmor C.S., Park M.Y., Smith M.R., Pepitone R., Kerstetter R.A., Poethig R.S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Bowen A.J., Carroll T.S., Conlan R.S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27: 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.M., Holstege F.C., Werner M. (2004). A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K., Bednarek P., Ciolkowski I., Hamberger B., Heise A., Liedgens H., Logemann E., Nürnberger T., Schmelzer E., Somssich I.E., Tan J. (2003). Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc. Natl. Acad. Sci. USA 100 (suppl. 2): 14569–14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A., Hurst C.H., Kaliyadasa E., Lamb R., Knight M.R., De Cothi E.A., Steele J.F., Knight H. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner C.J., Myer V.E., Liao S.M., Wilson C.J., Koh S.S., Young R.A. (1998). Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2: 43–53. [DOI] [PubMed] [Google Scholar]
- Hirst M., Kobor M.S., Kuriakose N., Greenblatt J., Sadowski I. (1999). GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3: 673–678. [DOI] [PubMed] [Google Scholar]
- Ito J., Sono T., Tasaka M., Furutani M. (2011). MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol. 52: 539–552. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kannangara R., Branigan C., Liu Y., Penfield T., Rao V., Mouille G., Höfte H., Pauly M., Riechmann J.L., Broun P. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M.T., Meyer K.D., Bernecky C., Taatjes D.J. (2009a). The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M.T., Meyer K.D., Donner A.J., Espinosa J.M., Taatjes D.J. (2009b). The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 29: 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235–239. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Nawrath C., Bär S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Métraux J.P., Yephremov A. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Mengiste T. (2013). Genetic and cellular mechanisms regulating plant responses to necrotrophic pathogens. Curr. Opin. Plant Biol. 16: 505–512. [DOI] [PubMed] [Google Scholar]
- Lai Z., Schluttenhofer C.M., Bhide K., Shreve J., Thimmapuram J., Lee S.Y., Yun D.J., Mengiste T. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5: 3064. [DOI] [PubMed] [Google Scholar]
- Laluk K., Mengiste T. (2011). The Arabidopsis extracellular UNUSUAL SERINE PROTEASE INHIBITOR functions in resistance to necrotrophic fungi and insect herbivory. Plant J. 68: 480–494. [DOI] [PubMed] [Google Scholar]
- L’Haridon F., Besson-Bard A., Binda M., Serrano M., Abou-Mansour E., Balet F., Schoonbeek H.J., Hess S., Mir R., Léon J., Lamotte O., Métraux J.P. (2011). A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30: 256–263. [DOI] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Donner A.J., Knuesel M.T., York A.G., Espinosa J.M., Taatjes D.J. (2008). Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 27: 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.W., Loake G.J., Spoel S.H. (2011). Transcription dynamics in plant immunity. Plant Cell 23: 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi A., Ishihara A., Tanaka C., Ishizuka A., Takabayashi J., Miyoshi H., Nishioka T. (2009). Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 230: 517–527. [DOI] [PubMed] [Google Scholar]
- Nelson C., Goto S., Lund K., Hung W., Sadowski I. (2003). Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421: 187–190. [DOI] [PubMed] [Google Scholar]
- Nemet J., Jelicic B., Rubelj I., Sopta M. (2014). The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 97: 22–27. [DOI] [PubMed] [Google Scholar]
- Ng S., et al. (2013). Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 288: 3449–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M.L., Young R.A. (1989). Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Pinto J.J., Yephremov A. (2009). Surface lipids and plant defenses. Plant Physiol. Biochem. 47: 540–549. [DOI] [PubMed] [Google Scholar]
- Rickert P., Corden J.L., Lees E. (1999). Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18: 1093–1102. [DOI] [PubMed] [Google Scholar]
- Rohde J.R., Trinh J., Sadowski I. (2000). Multiple signals regulate GAL transcription in yeast. Mol. Cell. Biol. 20: 3880–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schnurr J., Shockey J., Browse J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D., Buxdorf K., Shi J.X., Feldmesser E., Schreiber L., Aharoni A., Levy M. (2013). Overexpression of AtSHN1/WIN1 provokes unique defense responses. PLoS ONE 8: e70146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich I.E., Hahlbrock K. (1998). Pathogen defence in plants: A paradigm of biological complexity. Trends Plant Sci. 3: 86–90. [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T.T., van Leenen D., Holstege F.C. (2005). Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522. [DOI] [PubMed] [Google Scholar]
- van Wees S.C., Chang H.-S., Zhu T., Glazebrook J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin D., Nawrath C., Kurdyukov S., Franke R.B., Reina-Pinto J.J., Efremova N., Will I., Schreiber L., Yephremov A. (2009). Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet. 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chen X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala D.L., Hemsley P.A., Moffat C.S., Cremelie P., Knight M.R., Knight H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195: 217–230. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ou B., Zhang J., Si W., Gu H., Qin G., Qu L.J. (2014). The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77: 838–851. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang Y., Li R., Song X., Wang Q., Huang S., Jin J.B., Liu C.M., Lin J. (2010). Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 61: 223–233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.