Stress-induced nitrate allocation to roots serves as a universal mechanism in response to diverse stresses, although how it perceives the environment remains unknown. This study demonstrates that the ET/JA-NRT1.5/NRT1.8 signaling module functions in this perception to fine-tune nitrate allocation and plant adaptation to the environment.
Abstract
Stresses decouple nitrate assimilation and photosynthesis through stress-initiated nitrate allocation to roots (SINAR), which is mediated by the nitrate transporters NRT1.8 and NRT1.5 and functions to promote stress tolerance. However, how SINAR communicates with the environment remains unknown. Here, we present biochemical and genetic evidence demonstrating that in Arabidopsis thaliana, ethylene (ET) and jasmonic acid (JA) affect the crosstalk between SINAR and the environment. Electrophoretic mobility shift assays and chromatin immunoprecipitation assays showed that ethylene response factors (ERFs), including OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59, bind to the GCC boxes in the NRT1.8 promoter region, while ETHYLENE INSENSITIVE3 (EIN3) binds to the EIN3 binding site motifs in the NRT1.5 promoter. Genetic assays showed that cadmium and sodium stresses initiated ET/JA signaling, which converged at EIN3/EIN3-Like1 (EIL1) to modulate ERF expression and hence to upregulate NRT1.8. By contrast, ET and JA signaling mediated the downregulation of NRT1.5 via EIN3/EIL1 and other, unknown component(s). SINAR enhanced stress tolerance and decreased plant growth under nonstressed conditions through the ET/JA-NRT1.5/NRT1.8 signaling module. Interestingly, when nitrate reductase was impaired, SINAR failed to affect either stress tolerance or plant growth. These data suggest that SINAR responds to environmental conditions through the ET/JA-NRT signaling module, which further modulates stress tolerance and plant growth in a nitrate reductase-dependent manner.
INTRODUCTION
In higher plants, nitrate assimilation occurs by the reduction of nitrate to nitrite, then ammonium, which is then assimilated to glutamate via the glutamine synthase/glutamate oxyglutarate aminotransferase cycle (Crawford, 1995). These metabolic intermediates act as important signaling molecules or precursors of further nitrogen derivatives, which sustain plant growth, development, and responses to biotic and abiotic stresses (Stitt, 1999; Forde and Lea, 2007; Wang et al., 2007; Vidal and Gutiérrez, 2008; Mur et al., 2012). Nitrate assimilation is energy-intensive and occurs preferentially in foliar chloroplasts, where carbon skeletons, energy, and reducing power derived from photosynthesis can be easily accessed. Thus, the direct coupling of nitrate assimilation and photosynthesis in chloroplasts is believed to be energy-efficient and is known as nitrate photoassimilation (Searles and Bloom, 2003), a process adopted by most herbaceous plants and requiring long-distance root-to-shoot transport of nitrate taken up by plant roots (Smirnoff and Stewart, 1985; Andrews, 1986).
Adverse conditions such as low light intensity and heavy metal stresses induce SINAR, in which the assimilation of nitrate in the roots becomes prevalent (Smirnoff and Stewart, 1985; Andrews, 1986; Hernandez et al., 1997) and the direct coordination between nitrate assimilation and photosynthesis decouples, leading to decreased energy efficiency. Despite this loss of efficiency, plants frequently use SINAR and the root assimilation machinery. One explanation is that through SINAR, nitrate assimilation in roots will not compete directly for the reductants and energy generated by photosynthetic electron transport, thus optimizing plant growth in unfavorable environments (Canvin and Atkins, 1974). Thus, elucidation of the mechanisms underlying SINAR might help improve our understanding of nitrogen use efficiency.
SINAR is mediated by two nitrate transporter genes, NRT1.5 and NRT1.8, which encode low-affinity nitrate uptake transporters. NRT1.5 is mainly expressed in root pericycle cells and functions to load nitrate into the xylem, and NRT1.8 is expressed predominantly in xylem parenchyma cells and functions primarily to unload nitrate from the xylem sap (Lin et al., 2008; Li et al., 2010). Under normal conditions, nitrate in roots is loaded into xylem vessels by NRT1.5 and then transferred to aerial tissues, where NRT1.8 unloads it into xylem parenchyma cells. The unloaded nitrate then translocates to mesophyll cells via symplastic diffusion. Under stress conditions, NRT1.5 expression in roots decreases, so much less nitrate is loaded into xylem vessels. Meanwhile, NRT1.8 expression in roots increases strongly, thus unloading nitrate back into the root symplasm. This highly coordinated regulation of NRT1.5 and NRT1.8 fine-tunes nitrate allocation between roots and shoots in response to environmental signals. Moreover, plants with impaired SINAR are more sensitive to cadmium (Cd), salt (Na), and drought stresses (Li et al., 2010; Chen et al., 2012). These observations suggest that SINAR might enhance stress tolerance at the cost of energy efficiency. Interestingly, the coordinated, opposite expression of NRT1.5 and NRT1.8 occurs under diverse stress conditions (Zimmermann et al., 2004), indicating that SINAR might represent a universal response mechanism (Gojon and Gaymard, 2010; Li et al., 2010; Chen et al., 2012), although how it crosstalks with such a wide range of stresses and mediates stress tolerance remains to be determined.
Stresses induce the biosynthesis of ethylene (ET), jasmonic acid (JA), and other stress hormones (Fujita et al., 2006). Once produced, ET binds to and inactivates its receptors, which consequently alleviates the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1)-derived phosphorylation of ETHYLENE-INSENSITIVE2 (EIN2). The C terminus of the nonphosphorylated EIN2 is then cleaved and enters the nuclei, where it activates EIN3/EIN3-Like1 (EIL1) and the downstream transcriptional cascades (Solano et al., 1998; Ju et al., 2012) to mediate a wide range of stress responses (Berrocal-Lobo et al., 2002; Achard et al., 2006; Zhang et al., 2011; Li et al., 2013). Likewise, the stress-induced JA is perceived by CORONATINE INSENSITIVE1 (COI1), which recruits and degrades JASMONATE ZIM-DOMAIN proteins, thus derepressing the downstream transcriptional factors to mediate a series of stress responses (Vijayan et al., 1998; Dombrecht et al., 2007; Kazan and Manners, 2012; Hu et al., 2013). Recent work also showed that ET and JA signaling pathways crosstalk via EIN3/EIL1 and synergistically regulate the expression of a number of downstream genes (Zhu et al., 2011). Other studies further proposed that ET and JA posttranslationally modulate ethylene response factors (ERFs) independent of EIN3/EIL1 (Bethke et al., 2009; Van der Does et al., 2013).
In this study, we provide biochemical, genetic, and physiological evidence to demonstrate that ET and JA signaling pathways crosstalk to fine-tune the SINAR process via the coordinated upregulation of NRT1.8 and downregulation of NRT1.5. Our results further suggest that SINAR mediates the trade-off between stress tolerance and plant growth dependent on the activity of nitrate reductase.
RESULTS
ORA59 Is an Immediate Upstream Regulator of NRT1.8
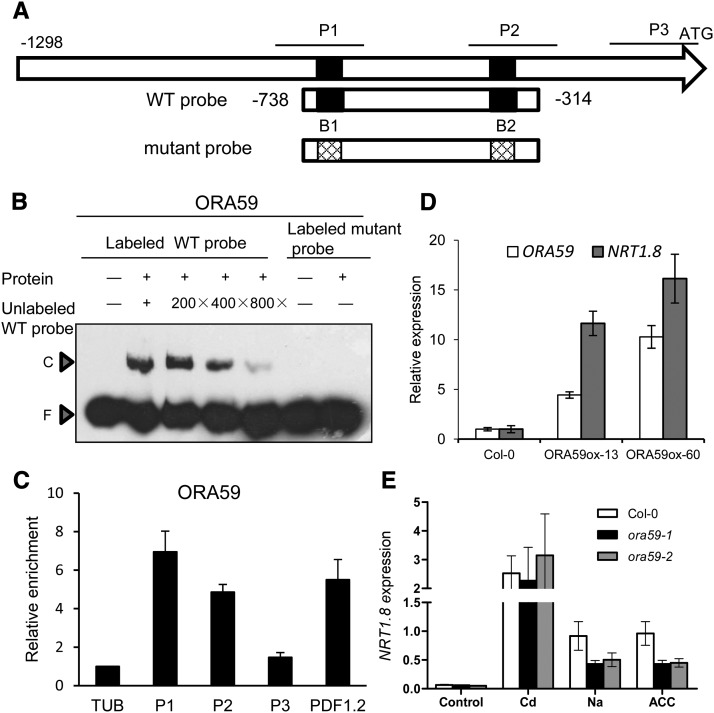
Enhanced NRT1.8 expression was proposed to function as an essential step in the regulation of SINAR (Gojon and Gaymard, 2010; Li et al., 2010). In the effort to identify the upstream regulators that integrate diverse signals to mediate NRT1.8 expression in Arabidopsis thaliana, we noticed that NRT1.8 transcript levels were constitutively elevated in transgenic plants overexpressing OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59; Pré et al., 2008), an ERF family member. To test the idea that ORA59 might be one of the upstream regulators of NRT1.8, the promoter sequence of NRT1.8 was scanned; this identified two putative GCC boxes, B1 (−719 to −713) and B2 (−347 to −341) (Figure 1A), which are known ERF binding motifs (Solano et al., 1998). Gel-shift assays indicated that incubation of the His-ORA59 recombinant protein with a wild-type probe containing this sequence (−738 to −314) resulted in a significant protein-DNA complex. Also, increasing amounts of unlabeled wild-type probe gradually decreased the complex levels, while no complex was observed when His-ORA59 was incubated with labeled mutant probe (Figure 1B). These results indicated that ORA59 binds to the promoter of NRT1.8. Chromatin immunoprecipitation (ChIP) assays using plants overexpressing ORA59 from a 35S:ORA59-TAP construct (ORA59ox) showed significant ORA59 enrichment in the P1 and P2 regions in the NRT1.8 promoter (Figure 1C). Corresponding to the elevated levels of ORA59, NRT1.8 expression was also enhanced in ORA59ox-13 and ORA59ox-60 plants (Figure 1D). These data suggest that ORA59 acts as an immediate upstream regulator of NRT1.8.
Figure 1.
ORA59 Binds to the Promoter of NRT1.8 and Modulates Its Expression.
(A) Schematic diagram of the 1298-bp promoter fragment upstream of the NRT1.8 start codon. P1 (−795 to −634) and P2 (−410 to −233), covering the predicted GCC boxes B1 and B2, as well as P3 (−160 to −7), serving as a negative control, indicate promoter regions subjected to the ChIP-qPCR assay in (C). The wild-type probe used for the EMSA in (B) contains the wild-type GCC boxes (5′-AGCAGCC), while the mutant probe contains the mutagenized GCC boxes (5′-ATCATCC) where G was mutagenized to T.
(B) EMSA determination of complex formation between ORA59 and the NRT1.8 promoter. Wild-type and mutant probes, as indicated in (A), were amplified using the primers listed in Supplemental Table 1. Competition assay for the labeled wild-type probe was performed by adding 200-/400-/800-fold excesses of unlabeled wild-type probe. C, DNA-protein complex; F, free probe.
(C) ChIP assay using IgG beads to precipitate ORA59-TAP protein followed by qPCR detection of P1, P2, and P3 regions as indicated in (A). TUB2 and PDF1.2 were used as internal and positive controls, respectively. Data are means ± sd, n = 3.
(D) Quantitative RT-PCR analysis of NRT1.8 and ORA59 expression in roots of wild-type and ORA59-overexpressing plants grown in hydroponics. Values are means ± sd, n = 3.
(E) Quantitative RT-PCR analysis of NRT1.8 expression in roots of 4-week-old wild-type, ora59-1, and ora59-2 plants treated with 200 μM CdCl2, 150 mM NaCl, or 20 μM ACC for 6 h. NRT1.8 expression levels were normalized to those of SAND.
Values are means ± sd; n = 3.
Given that NRT1.8 is upregulated by various stresses and ORA59 binds to its promoter region, we then determined if ORA59 functions in stress-induced NRT1.8 expression. As shown in Figure 1E, NRT1.8 was induced in the wild-type Columbia-0 (Col-0) when exposed to Cd or Na and the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC), while in the loss-of-function mutants ora59-1 and ora59-2 (Supplemental Figures 1A and 1B), the induction upon treatment with Na and ACC decreased significantly compared with Col-0 (P < 0.05), although changes upon treatment with Cd were not observed (Figure 1E). These results suggest that ORA59 is involved in the upregulation of NRT1.8, but factors other than ORA59 might function redundantly in this induction process, especially when under Cd stress.
ERF1B and ERF104 Function Redundantly with ORA59 to Mediate NRT1.8 Expression
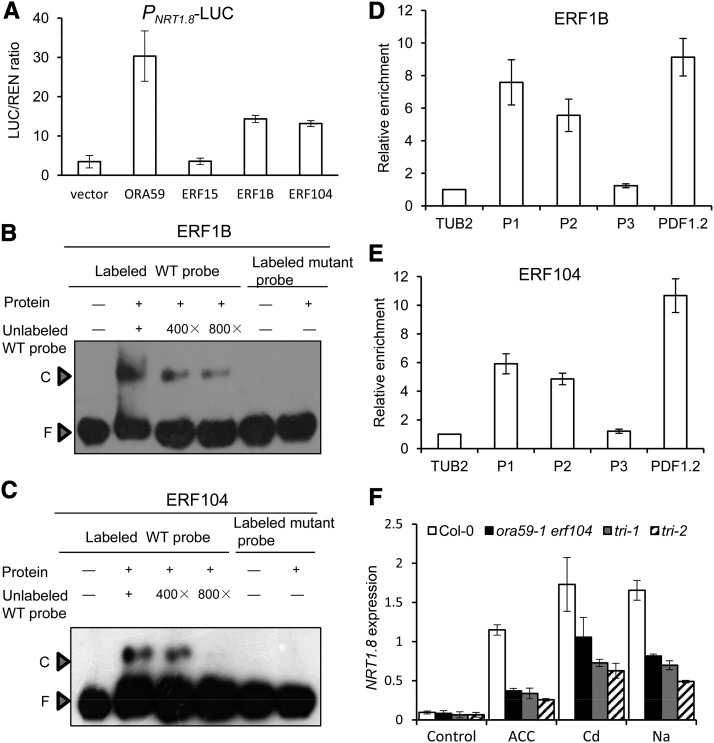
Considering that other factors might regulate NRT1.8 expression, we further characterized another eight reported members of the ERF B-3 subfamily, to which ORA59 belongs (Nakano et al., 2006), including ERF1A (accession number At4g17500, National Center for Biotechnology Information reference sequence NP_567530.4; also known as ATERF1 and referred to as At-ERF#100 by Nakano et al., 2006) and ERF1B (accession number At3g23240, National Center for Biotechnology Information reference sequence NP_188965.1; also known as ERF1 and referred to as At-ERF#092 by Nakano et al., 2006). The results showed that transient overexpression of ORA59, ERF1B, and ERF104 in Arabidopsis protoplasts activated the expression of LUC driven by the NRT1.8 promoter (NRT1.8:LUC; Figure 2A), while overexpression of ERF1A only slightly activated NRT1.8:LUC (Supplemental Figure 1C). Other tested ERFs did not show any significant activation of the NRT1.8 promoter (Figure 2A; Supplemental Figure 1C; P > 0.2). These data suggest that, in addition to ORA59, ERF1B and ERF104 might also participate in the transcriptional regulation of NRT1.8.
Figure 2.
Other ERFs Function Redundantly with ORA59 to Regulate NRT1.8 Expression.
(A) Transient transcriptional activity assay of PNRT1.8:LUC. The PNRT1.8:LUC-35S:REN reporter construct was transiently expressed in Arabidopsis protoplasts together with the control vector, 35S:ORA59, 35S:ERF15, 35S:ERF1B, or 35S:ERF104 effector. REN was used as an internal control. The LUC:REN ratio represents the relative activity of the NRT1.8 promoter.
(B) and (C) EMSA of ERF1B (B) or ERF104 (C) binding to the NRT1.8 promoter regions. Wild-type and mutant probes as indicated in Figure 1A were amplified. Competition for the labeled wild-type probe was performed by adding 400-/800-fold excess of unlabeled wild-type probe. C, DNA-protein complex; F, free probe.
(D) and (E) ChIP assay using IgG beads to precipitate ERF1B-TAP protein (D) or anti-GFP antibody against ERF104-GFP protein (E), followed by qPCR detection of P1, P2, and the negative control P3 regions as indicated in Figure 1A. TUB2 and PDF1.2 were used as internal and positive controls, respectively.
(F) Quantitative RT-PCR analysis of NRT1.8 expression in roots of 4-week-old wild-type, ora59-1 erf104, tri-1, and tri-2 plants exposed to 200 μM CdCl2, 150 mM NaCl, or 20 μM ACC for 6 h. Data were normalized to that of SAND.
Values are means ± sd; n = 3.
Consistent with this, electrophoretic mobility shift assay (EMSA) and ChIP analyses showed that ERF1B and ERF104 bind directly to the GCC boxes in the NRT1.8 promoter (Figures 2B to 2E). In ora59-1 erf104 and the ora59-1 erf104 erf1b triple mutants (tri-1 and tri-2), which showed significantly decreased ERF1B transcript levels (Supplemental Figure 1F), NRT1.8 expression upon treatment with ACC or NaCl was further decreased compared with ora59-1, and significant decrease was also observed for Cd treatment (compare Figures 1E and 2F; P < 0.05). In the ERF104 and ERF1B overexpression lines, NRT1.8 expression was increased (Supplemental Figures 1D and 1E). These results together indicate that ERF1B and ERF104 function redundantly with ORA59 to mediate NRT1.8 expression in response to Cd/Na. Note, however, that NRT1.8 expression in tri-1 and tri-2 was not apparently different from that in ora59-1 erf104 (Figure 2F), which might be attributable to the fact that ERF1B was still detectable in the triple mutant plants (Supplemental Figure 1F), or ERF1B might just play a minor role in Cd/Na-induced NRT1.8 expression.
The Stress-Induced Expression of NRT1.8 Is Mediated by ET/JA Signaling
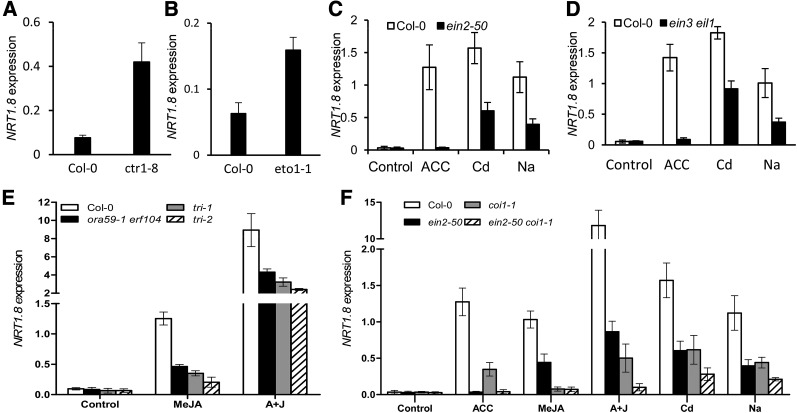
Given that NRT1.8 is greatly induced by ACC (Figure 2F) and that ORA59, ERF1B, and ERF104 are immediate upstream regulators of NRT1.8 (Figures 1 and 2), we questioned to what extent the ET signaling pathway is involved in the SINAR process mediated by NRT1.8. In the ctr1-8 and ethylene overproducing1 (eto1-1) mutant plants, NRT1.8 levels were steadily elevated compared with the wild-type Col-0 (Figures 3A and 3B). Consistent with this, upregulation of NRT1.8 upon Cd/Na treatment decreased in the ET-insensitive mutant ein2-50 compared with Col-0, and its induction by ACC did not occur (Figure 3C). Similar results were obtained in the ET-insensitive ein3 eil1 double mutants (Figure 3D). These data suggest that the ET signaling pathway might be extensively involved in the regulation of NRT1.8 through the major signaling components that have been characterized.
Figure 3.
Regulation of NRT1.8 Expression in ET/JA Signaling Mutants.
NRT1.8 expression levels in the roots of 4-week-old plants were determined by quantitative RT-PCR, treatments were as indicated, and data were normalized to those of SAND.
(A) and (B) NRT1.8 expression in roots of ctr1-8 (A), eto1-1 (B), and Col-0.
(C) and (D) ein2-50 (C), ein3 eil1 (D), and the wild-type control were exposed to 20 μM ACC, 200 μM CdCl2, or 150 mM NaCl for 6 h.
(E) ora59-1 erf104, tri-1, tri-2, and wild-type plants were treated with 50 μM MeJA or 20 μM ACC + 50 μM MeJA (A+J) for 6 h.
(F) ein2-50, coi1-1, ein2-50 coi1-1, and wild-type plants were treated with 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA (A+J), 200 μM CdCl2, or 150 mM NaCl for 6 h.
Values are means ± sd; n = 3 in (A) to (E) and n = 4 in (F).
However, given that the induction of NRT1.8 upon Cd/Na treatment did not decrease to the control level (as observed for the ACC treatment) in the ET signaling mutants (Figures 3C and 3D) and that ORA59 and ERF1B are also regulated by JA (Lorenzo et al., 2003; Pré et al., 2008), we further measured NRT1.8 expression upon treatment with methyl jasmonate (MeJA) or ACC plus MeJA. The results showed that MeJA greatly induced NRT1.8 expression in the wild-type Col-0. Also, the induction upon treatment with ACC plus MeJA was even more dramatic than treatment with either ACC or MeJA alone (Figure 3E), as would be expected for a synergistic effect of ACC and MeJA (Lorenzo et al., 2003; Pré et al., 2008). Moreover, in ora59-1 erf104 double mutants and the tri-1 and tri-2 triple mutants, NRT1.8 expression in response to both treatments was decreased significantly compared with Col-0 (Figure 3E; P < 0.05). These results suggest that ET and JA signaling pathways interact to mediate NRT1.8 expression.
Consistent with this, in ein2-50 and coi1-1 mutants, where ET or JA signaling is essentially prevented, NRT1.8 expression decreased upon treatment with ACC, MeJA, or ACC plus MeJA, and the expression in the double mutant ein2-50 coi1-1 even decreased to levels comparable to those observed under control conditions (Figure 3F). A similar result was obtained for NRT1.8 expression upon Cd/Na treatment, except that the levels in ein2-50 coi1-1 were low but still significant compared with the control condition (Figure 3F; P < 0.01). Comparable expression patterns were also observed for ORA59, ERF1A, ERF1B, and ERF104 (Supplemental Figure 2), the immediate upstream regulators of NRT1.8. These results indicate that ET/JA signaling plays a dominant role in the regulation of NRT1.8 expression, but other mechanisms such as abscisic acid (ABA) signaling might still contribute (Supplemental Figure 3A). Note that ERF104 did not show an apparent response to stress hormones (Supplemental Figure 2D), which might be due to the fact that ERF104 regulation by ACC occurs preferentially at the posttranslational level (Bethke et al., 2009).
Further analysis showed that in ein3 eil1 mutants, ORA59 expression upon treatment with stress hormones or Cd/Na dramatically decreased to the control level (Supplemental Figure 4A), whereas a similar decrease for other ERFs was observed only after treatment with stress hormones (Supplemental Figures 4B to 4D). These results suggest that among the characterized ERFs, ORA59 expression upon Cd/Na treatment is regulated exclusively via EIN3/EIL1, while others are regulated by additional factors. Taking into account the expression pattern of ERFs in ein2-50, coi1-1, and eni2-50 coi1-1 (Supplemental Figure 2), these results further indicate that EIN3/EIL1 act downstream of EIN2 and COI1 to relay signals derived from Cd/Na stresses.
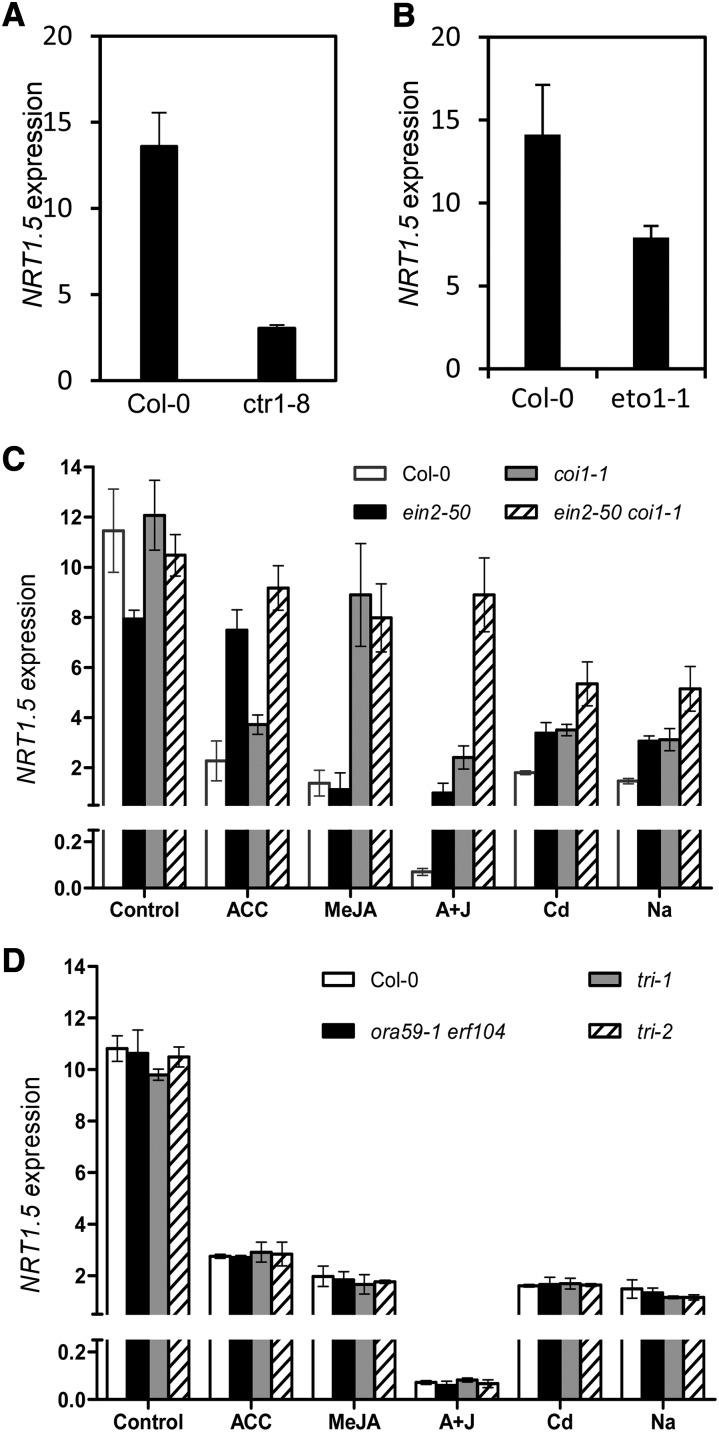
NRT1.5 Inhibition upon Cd/Na Treatments Is Mediated through the ET/JA Signaling Pathway, and EIN3 Acts as an Immediate Upstream Regulator
Given that NRT1.8 and NRT1.5 are coordinately but oppositely regulated by various stresses to mediate nitrate reallocation to roots (Li et al., 2010; Chen et al., 2012), we further determined if the NRT1.5 inhibition under stress conditions was also regulated by ET/JA signaling. As expected, NRT1.5 expression was steadily downregulated in the ctr1-8 and eto1-1 mutants (Figures 4A and 4B). Exposure to Cd/Na as well as stress hormones decreased NRT1.5 expression in the wild-type Col-0, and functional disruption of EIN2 or COI1 significantly alleviated the inhibition of NRT1.5 by the corresponding treatments, although an apparent reciprocal alleviation of NRT1.5 suppression upon ACC or MeJA treatment was not observed in coi1-1 or ein2-50 mutants (Figure 4C). In the ein2-50 coi1-1 double mutant, NRT1.5 expression upon stress hormone treatment was even restored to control levels, and significant restoration was also observed for Cd/Na treatments (Figure 4C), similar to the regulation of NRT1.8, but in an opposite manner (Figure 3F). Interestingly, ABA decreased NRT1.5 expression while salicylic acid did not show any significant effect (Supplemental Figure 3B), as was observed for NRT1.8, and also in an opposite manner (Supplemental Figure 3A). These results together suggest that the coordinated regulation of NRT1.8 and NRT1.5 under stress conditions is predominantly mediated by ET/JA signaling.
Figure 4.
Stresses Inhibit NRT1.5 Expression through ET/JA Signaling.
Four-week-old plants under control conditions or exposed to 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA (A+J), 200 μM CdCl2, or 150 mM NaCl for 6 h as indicated. The expression levels of NRT1.5 were determined by quantitative RT-PCR. Data were normalized to those of SAND. Values are means ± sd; n = 3 to 4.
(A) and (B) NRT1.5 expression in roots of ctr1-8 (A), eto1 (B), and their wild-type control Col-0.
(C) NRT1.5 expression in roots of ein2-50, coi1-1, ein2-50 coi1-1, and Col-0 plants.
(D) NRT1.5 expression in roots of ora59-1 erf104, tri-1, tri-2, and wild-type control plants.
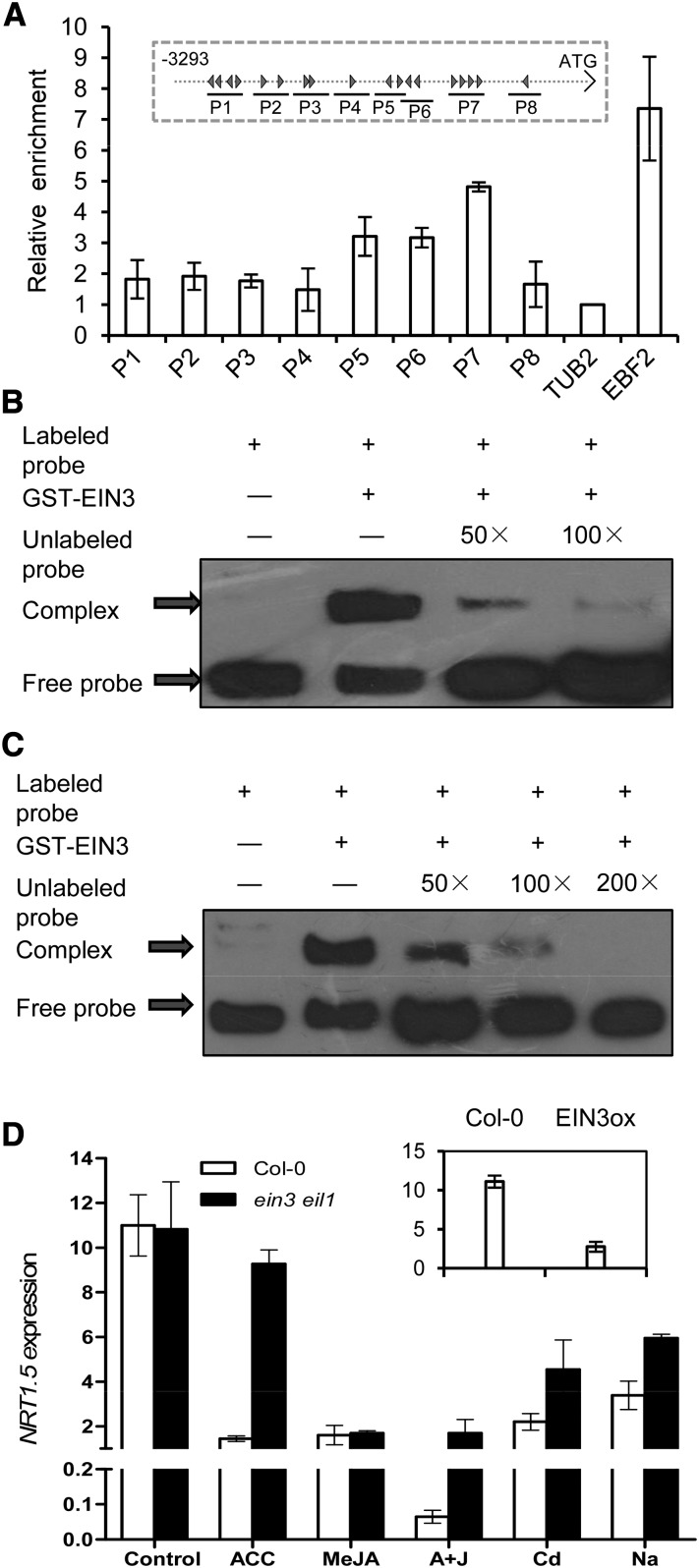
However, further analyses did not show alleviation of the NRT1.5 suppression upon treatments in ora59-1 erf104 double mutants or the triple mutants tri-1 and tri-2 (Figure 4D), indicating that NRT1.5 and NRT1.8 are regulated by different immediate upstream regulators. Consistent with this postulation, scan of the NRT1.5 promoter sequence did not reveal any GCC box, as in the NRT1.8 promoter, but instead identified several EIN3 binding site motifs (Figure 5A, inset). ChIP analysis using 35S:EIN3-GFP/ein3 eil1 plants showed enrichment of EIN3 in the P5, P6, and P7 regions in the NRT1.5 promoter (Figure 5A; P < 0.05 versus TUB2), suggesting that EIN3 binds to the NRT1.5 promoter in vivo. EMSA further indicated in vitro binding of EIN3 to P5 and P7 fragments (Figures 5B and 5C). Consistent with this, suppression of NRT1.5 upon ACC treatment was almost abolished in ein3 eil1 mutants, and the suppression by Cd/Na was also alleviated (P < 0.05), whereas apparent alleviation was not observed upon exposure to MeJA (Figure 5D). NRT1.5 expression decreased in EIN3ox plants (Figure 5D, inset). These results together suggest that EIN3 acts as an immediate upstream regulator of NRT1.5, but the interaction between them appears to be established only when environmental signals are transduced through the ET cascade. Notably, 35S:EIN3-GFP/ein3 eil1 serves as a complementation line and behaves just like a wild-type control (He et al., 2011).
Figure 5.
EIN3 Acts as the Immediate Upstream Regulator of NRT1.5.
(A) ChIP assay using anti-GFP antibody followed by qPCR detection of the enrichment of P1 to P8 regions as indicated in the inset, where a schematic diagram of the 3293-bp promoter fragment upstream of the NRT1.5 start codon is shown and the black arrowheads indicate putative EIN3 binding motifs. Chromatin was isolated from root samples of 4-week-old 35S:EIN3-GFP/ein3 eil1 plants exposed to 20 μM ACC + 50 μM MeJA for 6 h. TUB2 and EBF2 were used as internal and positive controls, respectively. Values are means ± sd; n = 3.
(B) and (C) Representative EMSA for EIN3 binding to the fragments of P5 (B) and P7 (C) in the NRT1.5 promoter.
(D) NRT1.5 expression in roots of 4-week-old wild-type and ein3 eil1 plants treated with 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA (A+J), 200 μM CdCl2, or 150 mM NaCl for 6 h. The inset shows NRT1.5 expression in 6-d-old wild-type Col-0 and EIN3ox seedlings. Data were normalized to those of SAND. Values are means ± sd; n = 3.
The Interaction between Stress Tolerance and Nitrate Reallocation Is Predominantly Coordinated by the ET/JA-NRT1.5/NRT1.8 Signaling Module
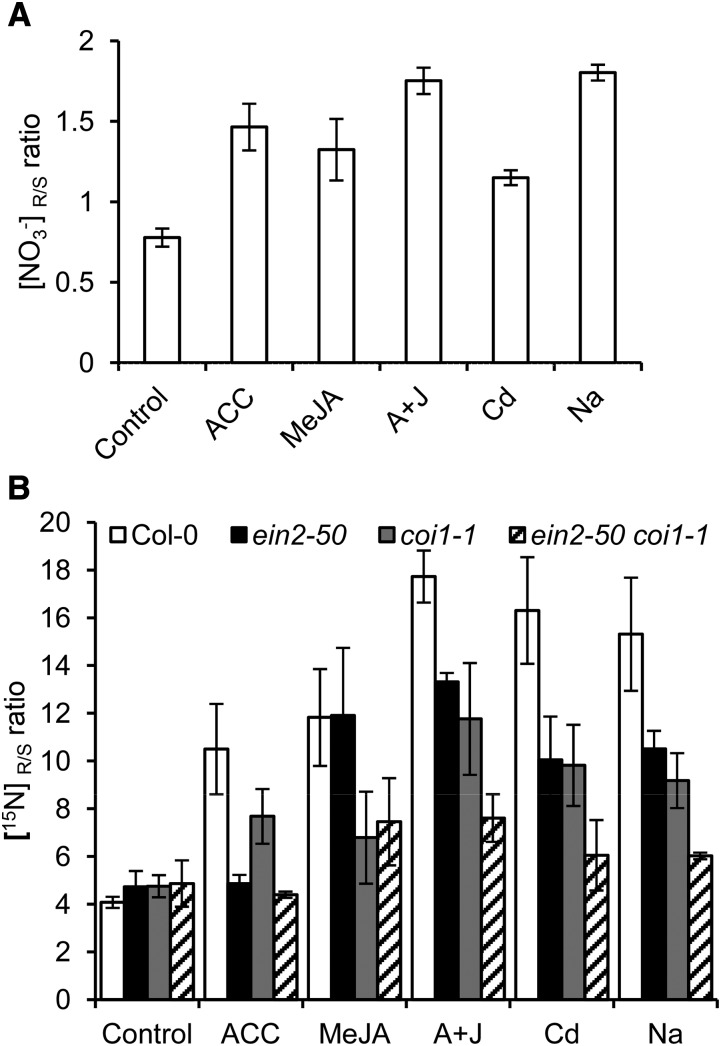
Previous studies revealed that under Cd/Na stresses, NRT1.5 and NRT1.8 respond together to fine-tune SINAR (Li et al., 2010; Chen et al., 2012). Also, our research here further revealed that ET/JA signaling pathways transduce signals to coordinate the regulation of NRT1.5 and NRT1.8 (Figures 1 to 5), indicating that ET/JA would enhance nitrate allocation to roots as Cd/Na do. This hypothesis was tested using the nia1 nia2 double mutant, in which nitrate reduction is almost abolished (Wilkinson and Crawford, 1993), and the results showed that nitrate reallocation to roots increased upon treatment with stress hormones or Cd/Na (Figure 6A; P < 0.05). In ein2-50 and coi1-1 mutants, nitrate allocation to roots upon treatment with stress hormones or Cd/Na decreased correspondingly compared with Col-0 (Figure 6B; P < 0.05). In the ein2-50 coi1-1 double mutant, nitrate reallocation decreased further (Figure 6B), supporting the conclusion that SINAR is predominantly mediated by ET/JA signaling.
Figure 6.
Stress-Induced Nitrate Reallocation Mediated by ET/JA Signaling.
(A) The nia1 nia2 mutant plants were grown hydroponically with NH4HCO3 as the sole nitrogen source. At 4 weeks of age, plants were transferred to 4.5 mM NO3− solution and treated with 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA (A+J), 200 μM CdCl2, or 150 mM NaCl for 12 h. The nitrate ratio between roots and shoots ([NO3−]R/S) was calculated.
(B) Four-week-old Col-0, ein2-50, coi1-1, and ein2-50 coi1-1 plants grown in quarter-strength hydroponics medium were treated with 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA (A+J), 200 μM CdCl2, or 150 mM NaCl for 12 h and then labeled with 3 mM K15NO3 for 30 min. The nitrate ratio between roots and shoots ([15N]R/S) was calculated.
Values are means ± sd; n = 3.
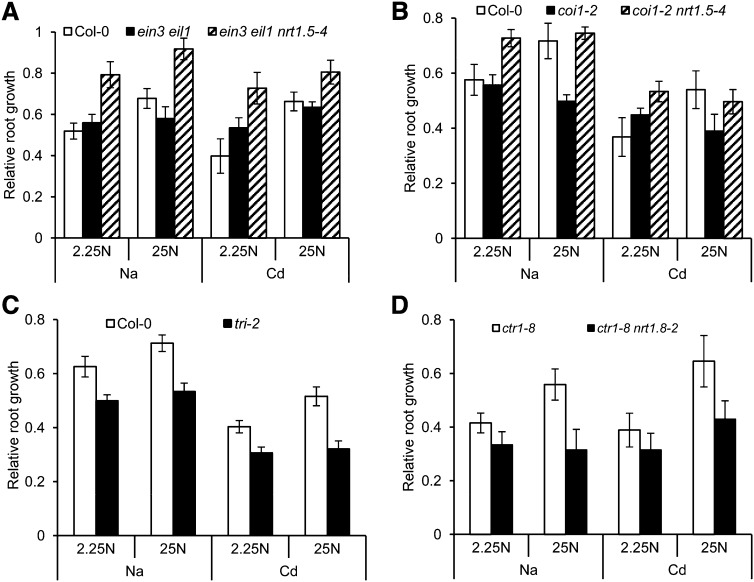
Given that SINAR enhances stress tolerance and that ET/JA signaling mediates SINAR, we further determined the stress tolerance mediated by the interaction between nitrate reallocation and ET/JA signaling. As shown in Figure 7A, when nitrate concentration increased from 2.25 to 25 mM in the medium, enhanced tolerance to Na/Cd was observed in the wild-type Col-0. By contrast, the ET-insensitive ein3 eil1 mutants did not respond. In the triple mutant ein3 eil1 nrt1.5-4, however, steadily enhanced tolerance was observed despite the external nitrate concentration (Figure 7A), which is reasonable, as loss of NRT1.5 function tended to show constitutively promoted nitrate reallocation to roots and stress tolerance (Chen et al., 2012). Similar results were obtained for the JA-insensitive mutant coi1-2 and the double mutant coi1-2 nrt1.5-4 (Figure 7B). The positive relationship between nitrate concentration and Na/Cd tolerance was also suppressed in tri-2 mutants (Figure 7C), which imitate the functional impairment of NRT1.8, thus causing stress sensitivity (Li et al., 2010). In ctr1-8 mutants, enhanced tolerance to Na/Cd was observed with increased nitrate concentration, while the double mutant ctr1-8 nrt1.8-2 showed a decreased response (Figure 7D). These results together suggest that the interaction between stress tolerance and nitrate reallocation is predominantly coordinated by the ET/JA-NRT1.5/NRT1.8 signaling cascade. Note that plant tolerance was quantified here using relative root growth, because ET/JA signaling mutants always show pleotropic effects that might interfere with each other under given conditions, as indicated by the growth of ein3 eil1 and coi1-2 mutants exposed to a combination of Cd and low-level nitrate (Figures 7A and 7B).
Figure 7.
Stress Tolerance Correlates with Nitrate Allocation via the ET/JA-NRT1.5/NRT1.8 Signaling Module.
Four-day-old seedlings germinated on ammonium plates as described in Methods were transferred to medium with either 2.25 mM nitrate (2.25 N) or 25 mM NO3− (25 N) plus CdCl2/NaCl and allowed vertical growth for 6 d ([A] to [C]) or 9 d (D) before the determination of root elongation. Cd, 40 μM CdCl2 in (A) to (C) and 50 μM CdCl2 in (D); Na, 100 mM NaCl in (A) to (C) and 125 mM NaCl in (D). Relative root growth is defined as the ratio of root elongation rates in the presence of Cd/Na against those in the same medium without Cd/Na. Values are means ± sd; n = 7 to 12.
Nitrate Reallocation to Roots Decreases Plant Regular Growth under Nonstressed Conditions
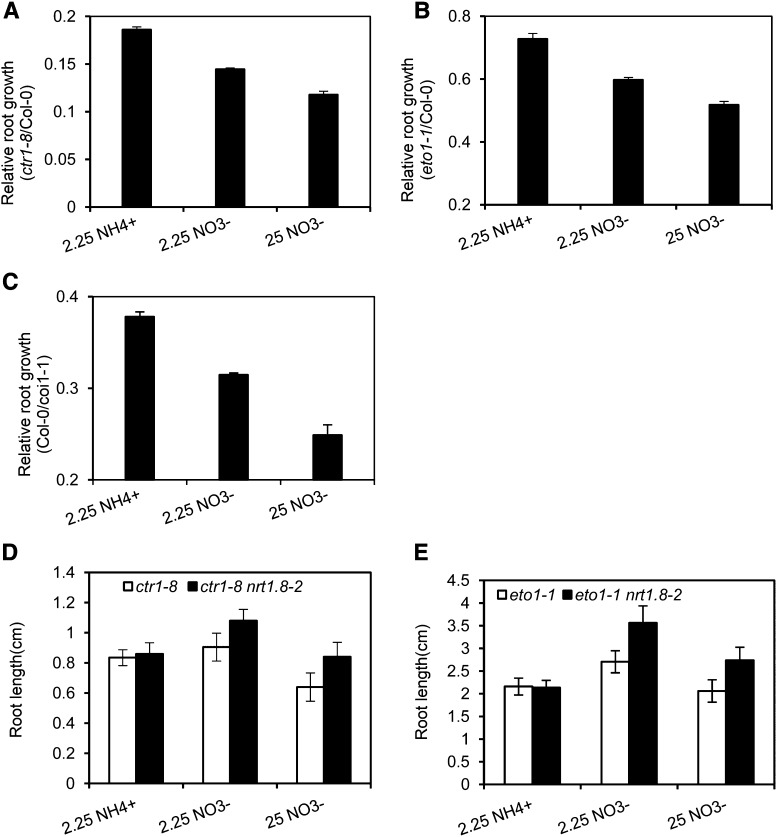
In nrt1.5 mutants, when the nitrate supply increased, stress tolerance was enhanced while plant regular growth was suppressed (Lin et al., 2008; Chen et al., 2012), indicating that nitrate reallocation to roots possibly mediates the trade-off between stress tolerance and plant growth. To test this hypothesis, we used ET/JA signaling-related mutants to mimic nitrate reallocation without exposure to stresses, thus decoupling the two important steps in SINAR. In ctr1-8 mutants, where upregulation of NRT1.8 and downregulation of NRT1.5 were steadily activated (Figures 3A and 4A), suppression of root growth was enhanced when nitrogen was switched from ammonium to nitrate, and further suppression was observed when the nitrate concentration increased (Figure 8A). Consistent data were obtained in eto1-1 mutants (Figure 8B), the MeJA-treated Col-0 plants (Figure 8C), and the EIN3-overexpressing seedlings (Supplemental Figure 5). These results together indicate that nitrate reallocation to roots decreases plant regular growth under nonstressed conditions. Note that the relative growth for JA signaling mutants was defined as Col-0/coi1-1, and, as a constitutive JA response mutant is not available, MeJA was applied to mimic a constitutive JA response in the wild-type Col-0.
Figure 8.
Nitrate Reallocation Mediated by the ET/JA-NRT Signaling Module Reduces Root Growth under the Nonstressed Condition.
Seedlings of ctr1-8, eto1-1, coi1-1, ctr1-8 nrt1.8-2, and eto1-1 nrt1.8-2 mutants and their wild-type control Col-0 were germinated and allowed to grow vertically for 7 to 9 d on medium supplemented with 2.25 mM ammonium (2.25 NH4+), 2.25 mM nitrate (2.25 NO3−), or 25 mM nitrate (25 NO3−), followed by measurement of root growth. Relative root growth of each mutant was calculated by normalizing the data to those of their wild-type control, except for (C).
(A) and (B) Relative root growth of ctr1-8 (A) and eto1-1 (B) against the wild-type Col-0 under different conditions. Values are means ± se; n = 5.
(C) Seeds from heterologous coi1-1(+/−) plants were plated on medium with 10 μM MeJA to screen homozygous seedlings, which together with Col-0 were incubated with different nitrogens plus 10 μM MeJA to mimic the constitutive JA response; correspondingly, the relative growth is defined as the ratio of coi1-1(+/+) to Col-0. Values are means ± se; n = 5.
(D) and (E) Root growth of ctr1-8 and ctr1-8 nrt1.8-2 (D) or eto1-1 and eto1-1 nrt1.8-2 (E) under different conditions. Values are means ± sd; n = 9 to 11.
To further determine how much this process was attributable to the NRT components in the ET/JA-NRT1.8/NRT1.5 signaling pathway, nrt1.8-2 was crossed with ctr1-8 and eto1-1. As expected, similar growth was observed in ctr1-8 and the double mutant ctr1-8 nrt1.8-2 when exposed to ammonium, while increased growth was observed in eto1-1 nrt1.8-2 mutants with nitrate as the nitrogen source (Figure 8D). Although further elevation of nitrate concentration decreased the absolute growth of both ctr1-8 and ctr1-8 nrt1.8-2 mutants, ctr1-8 nrt1.8-2 still showed better growth than ctr1-8 (Figure 8D). Similar results were consistently obtained in eto1-1 nrt1.8-2 and eto1-1 mutants (Figure 8E). Given that nrt1.8 is stress-sensitive (Li et al., 2010), while a cross of nrt1.8-2 with ctr1-8 and eto1-1 increased their regular growth, these observations together support a postulation that the NRT components contribute to the trade-off between stress tolerance and plant growth through nitrate reallocation. Note, though, that the contribution of NRT1.5 was not genetically determined, due to the technical difficulty of alleviating the downregulation of NRT1.5 in ctr1-8 and eto1-1 mutants. Nevertheless, the conclusion should stand as indicated by other lines of evidence and previous studies (Figures 4, 7A, and 7B; Lin et al., 2008; Chen et al., 2012).
SINAR Functions in Mediating Plant Growth and Stress Tolerance Require Nitrate Reductase
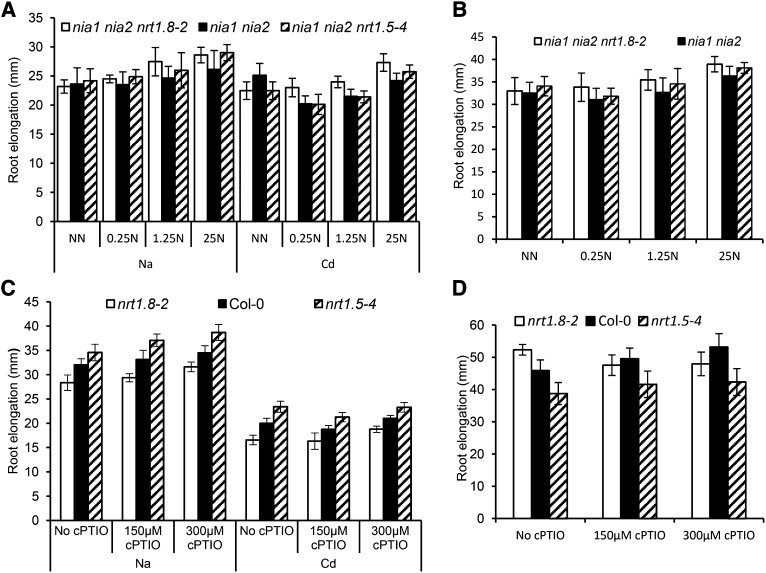
Nitrate reallocation was proposed to act as a signal to mediate stress tolerance (Gojon and Gaymard, 2010; Li et al., 2010; Chen et al., 2012), and here it was further identified to balance stress tolerance and plant growth (Figures 7 and 8). To address the question of whether nitrate, nitrite, nitric oxide (NO), or another nitrogen metabolite is perceived as the signal for nitrate reallocation, nrt1.8-2 and nrt1.5-4 were crossed to nia1 nia2, where typical nitrate reallocation was induced upon treatment with Na/Cd (Supplemental Figures 6A to 6D). However, the results unexpectedly showed that the nia1 nia2 nrt1.8-2 and nia1 nia2 nrt1.5-4 triple mutants responded to Na/Cd treatments similarly to the nia1 nia2 double mutants (Figure 9A; P > 0.2), in contrast with what we observed in nrt1.8-2 and nrt1.5-4 with normal nitrate reductase function (Figure 7; Chen et al., 2012). Furthermore, the previously observed differential plant growth in response to nitrate reallocation (Figure 8; Lin et al., 2008; Chen et al., 2012) was also impaired in the nia1 nia2 nrt1.8-2 and nia1 nia2 nrt1.5-4 mutants under nonstressed conditions (Figure 9B; P > 0.1). These results suggest that nitrate reallocation functions to mediate stress tolerance and plant growth, dependent on nitrate reductase.
Figure 9.
Nitrate Reallocation Mediates Stress Tolerance and Plant Growth Dependent on Nitrate Reductase.
(A) and (B) Stress response and regular growth of nrt1.8-2 and nrt1.5-4 in the nia1 nia2 background. Arabidopsis seeds were germinated on 2.25 mM ammonium medium for 4 d, transferred to the same ammonium medium spiked with various levels of nitrate (NN, no nitrate; 0.25N, 0.25 mM KNO3; 1.25N, 1.25 mM KNO3; 25N, 25 mM KNO3) and 40 μM CdCl2 or 100 mM NaCl (A) or no stress treatment (B), and allowed vertical growth for 6 d before the determination of root elongation.
(C) and (D) Stress response and regular growth of nrt1.8-2 and nrt1.5-4 treated with a NO scavenger. Four-day-old seedlings were transferred from 2.25 mM ammonium medium to 25 mM nitrate medium with (C) or without (D) the combined treatment of Cd/Na (Cd, 40 μM CdCl2; Na, 100 mM NaCl) and cPTIO and allowed vertical growth for 6 d before the determination of root elongation.
Values are means ± sd; n = 7.
Given that nia1 nia2 mutants show a substantial decrease in nitrate reduction and thus in subsequent assimilates and derivatives, we wondered if NO might play a role in SINAR signal perception, as proposed (Gojon and Gaymard, 2010). However, when the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) was applied, the differential response to Na/Cd was still observed in nrt1.8-2 and nrt1.5-4 mutants compared with Col-0 (Figure 9C; P < 0.01). When under nonstressed conditions, consistent phenotypes were observed (Figure 9D). These results appeared to indicate that NO might not serve as the signal molecule in SINAR signal perception. Note, though, that nrt1.8-2 showed enhanced growth under control conditions (no cPTIO), as expected, and application of cPTIO appeared to impair its growth (Figure 9D), which might be due to the interaction between cPTIO toxicity and the increased stress sensitivity in nrt1.8-2.
DISCUSSION
Recent studies indicated that SINAR serves as a universal mechanism in response to diverse stresses (Li et al., 2010; Chen et al., 2012). However, how SINAR responds to the environment remains unknown. This study demonstrates that, in response to Cd/Na treatment, the ET/JA signaling pathways are activated to mediate EIN3 and its induction of NRT1.5, as well as several ERFs including ORA59, which further induce NRT1.8, thus establishing a dynamic coordination of the essential components NRT1.5 and NRT1.8 in SINAR. Our data also revealed that SINAR mediates stress tolerance and plant growth, dependent on nitrate reductase.
The ET/JA-NRT Signaling Module Presents a Primary Mechanism to Coordinate Crosstalk between SINAR and the Environment
Stresses induce the biosynthesis of ET and JA (Fujita et al., 2006), which further activate downstream signaling pathways to mediate a wide range of stress responses (Vijayan et al., 1998; Berrocal-Lobo et al., 2002; Achard et al., 2006; Dombrecht et al., 2007; Zhang et al., 2011; Kazan and Manners, 2012; Hu et al., 2013; Li et al., 2013). A recent study further discovered that ET and JA signaling pathways crosstalk via EIN3/EIL1 and function synergistically (Zhu et al., 2011). In our research, we showed that ET/JA signaling might function conservatively, via known signaling components, to regulate the communication between SINAR and the environment (Figure 10). One point of supporting evidence is that in the mutants of major ET or JA signaling components, such as ctr1-8, ein2-50, and coi1-1, NRT1.8/NRT1.5 expression altered correspondingly as expected (Figures 3 to 5), and this alteration was enhanced in the double mutant ein2-50 coi1-1, where both ET and JA signaling are essentially blocked (Figures 3F and 4C). Further supporting evidence came from the results that NRT1.8 induction upon ET in coi1-1 or upon JA in ein2-50 still occurred (Figure 3F) and that nitrate reallocation to coi1-1 roots upon ACC or to ein2-50 roots upon JA was still significant (Figure 6B), in line with previous observations that the expression of ERF1B and ORA59 in response to JA or ET requires intact ET and JA signaling pathways (Lorenzo et al., 2003; Pré et al., 2008). These data support the model that ET/JA signaling is involved in Cd/Na-mediated SINAR via important, well-characterized components (Figure 10; Zhao and Guo, 2011; Zhu et al., 2011). Note that in ein3 eil1, the inhibition of NRT1.5 upon JA treatment was not alleviated as it was upon ET treatment (Figure 5D), suggesting that the stress-derived JA signaling cascade mediates NRT1.5 expression via new component(s) other than EIN3/EIL1, the integrator of JA and ET signals (Figure 10; Zhu et al., 2011; Kazan and Manners, 2012).
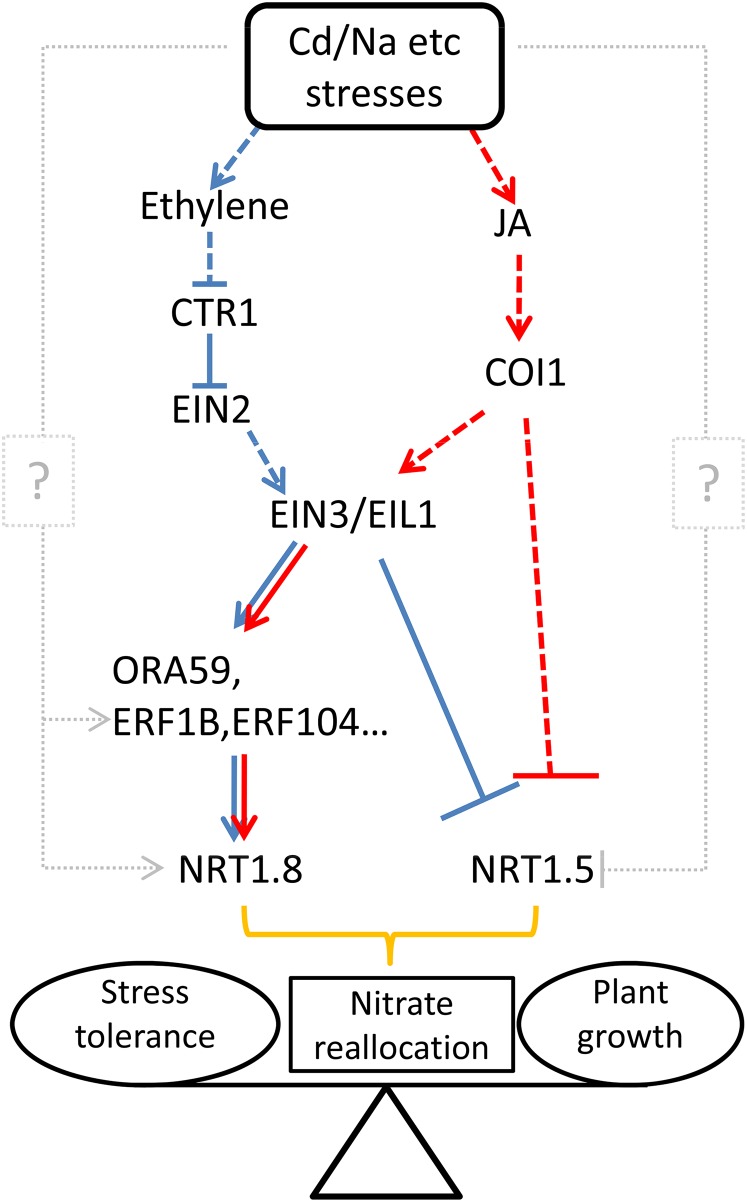
Figure 10.
A Simplified Model for the Interaction between ET/JA Signaling and Nitrate Reallocation to Mediate Plant Adaptation to the Environment.
Stresses such as cadmium or salt induce the production of ET and JA, which further activates the ET and JA signaling pathways, respectively. EIN3/EIL1 acts to converge signals transduced by ET and JA cascades, and further transduces the signal down to ERFs, which induce NRT1.8 expression by binding to its promoter, or EIN3/EIL1 decodes signals via the ET pathway by binding to the promoter of NRT1.5 and suppresses its expression. The JA pathway also participates in the downregulation of NRT1.5 via COI1. The coordinated upregulation of NRT1.8 and downregulation of NRT1.5 fine-tunes nitrate reallocation to roots, which modulates the balance between stress tolerance and plant growth. Blue lines display the route for signals going through the ET signaling pathway, while red lines indicate those through the JA signaling pathway. Dashed lines indicate steps not shown or possible unidentified components. Gray dotted lines and question marks indicate alternative pathways/components that contribute, but to a much lesser extent, in the regulation of NRT1.5 and NRT1.8.
Moreover, although NRT1.8/NRT1.5 expression upon ET/JA was not induced in corresponding ET/JA signaling mutants, a significant regulation upon Cd/Na treatment still occurred (Figures 3 and 4), indicating that alternative signaling pathway(s) might work with ET/JA signaling to mediate SINAR. The results that NRT1.8 and NRT1.5 were coordinately regulated by ABA supported this postulation (Supplemental Figure 3). However, given that nitrate reallocation to roots of ein2-50 coi1-1 under Cd/Na exposure was restored to levels comparable to those under control conditions (Figure 6B), the alternative pathway(s) might have a subtle contribution. Note that we do not exclude the possibility that nitrate transporters other than NRT1.5/NRT1.8 might contribute, but they are likely to be controlled by ET/JA signaling (Figure 6B).
Furthermore, given that SINAR was proposed as a universal adaptive response due to the highly coordinated NRT1.5/NRT1.8 expression under diverse conditions (Gojon and Gaymard, 2010; Li et al., 2010; Chen et al., 2012) and that ET/JA signaling acts genetically upstream of NRT1.5/NRT1.8 to mediate SINAR under Cd/Na stresses, another concern is if ET/JA-NRT signaling represents a common mechanism in the regulation of SINAR. This possibility was only investigated in a limited set of conditions here, but the fact that ET/JA are produced under a wide range of stresses might suggest so (Glazebrook, 2005; Maksymiec et al., 2005; Arteca and Arteca, 2007; Cao et al., 2007; Zhang et al., 2011; Ismail et al., 2012; Poltronieri et al., 2013), as ET/JA production would eventually activate the downstream signaling cascades, although new components specific to certain stimuli might exist, as was observed for the JA-mediated NRT1.5 expression (Figure 10).
How SINAR Balances Plant Growth and Stress Tolerance
Previous studies proposed that reallocated nitrate per se might act as a signal to coordinate responses (Li et al., 2010). A more recent study further excluded the possibility of a nutritional role by substituting nitrate with ammonium and provided evidence supporting the signal hypothesis (Chen et al., 2012). However, when investigating the stress response in nia1 nia2 nrt1.8-2 and nia1 nia2 nrt1.5-4 mutants, where nitrate reduction is fundamentally disrupted, we surprisingly found that they responded to Cd/Na similarly and did not show any apparent difference from the control nia1 nia2 (Figures 9A and 9B). These observations appear to be contradictory to the hypothesis that nitrate per se might be the signal molecule.
Gojon and Gaymard (2010) proposed an alternative hypothesis where NO serves as the signal molecule, because Cd promotes NO synthesis in roots (Besson-Bard et al., 2009), and nitrate reductase represents an important way to synthesize NO. However, application of the NO scavenger cPTIO did not alter the varied growth rates or differential responses of nrt1.8-2 and nrt1.5-4 mutants to Cd/Na (Figures 9C and 9D), implying that NO is unlikely to be the signal molecule. In this respect, we hypothesize that Glu/Gln or other nitrogen assimilates might be the candidate signaling molecules, as they are known to function as signaling molecules in mediating various biological processes (Stitt, 1999; Walch-Liu et al., 2006; Forde and Lea, 2007; Szabados and Savouré, 2010). A more likely model is that nitrate per se does act as the signal molecule to mediate the balance between stress tolerance and plant growth, but its function requires the activity of nitrate reductase.
SINAR Is a Dynamic Mechanism to Modulate Plant Growth and Environmental Adaptation
SINAR decouples the direct correlation between nitrate assimilation and photosynthesis; thus, it is believed to decrease energy use efficiency and evolutionary competitiveness (Canvin and Atkins, 1974; Smirnoff and Stewart, 1985; Andrews, 1986). One major concern about this phenomenon is what exactly the physiological importance is. Recent studies discovered that when SINAR was impaired, plants became sensitive to Cd stress (Li et al., 2010), while promotion of this process enhanced tolerance to diverse stresses (Li et al., 2010; Chen et al., 2012), indicating that SINAR decreases energy efficiency but increases stress tolerance. This study further discovered that in mutant plants with enhanced SINAR, regular root growth decreased under nonstressed conditions (Figures 8A to 8C; Supplemental Figure 5) and vice versa (Figures 8D and 8E). These observations support a model in which SINAR partitions energy within plants in response to environmental cues, thus allowing a dynamic balance between plant growth and stress tolerance (Figure 10).
ET/JA signaling regulates the trade-off between plant growth and stress tolerance (Achard et al., 2003, 2006; Yang et al., 2012), and here we showed that SINAR also regulates this and functions downstream of the ET/JA signaling cascade (Figures 1 to 8). This suggests that SINAR contributes significantly to the overall balancing mechanism mediated by ET/JA, although the exact contribution might remain to be determined due to the technical difficulties in concomitantly modulating NRT1.5 and NRT1.8 expression. It is worth noting that plant growth in this context was evaluated only with roots, considering the pleiotropic effects of ET/JA signaling mutants, but the conclusion is likely also applicable to shoot growth, mainly because nrt1.5-2 mutants showed dramatic growth reduction in the whole plant (Lin et al., 2008).
Additionally, given that nitrate assimilation requires a lot of energy, an intriguing strategy to improve nitrogen use efficiency is to block SINAR, thus enhancing nitrate photoassimilation. This strategy is of special importance in China, where eutrophication is widespread (Ju et al., 2009); however, considering the frequency of environmental stress, a more plausible strategy might be to enhance the basal long-distance root-to-shoot nitrate transport, retaining the SINAR mechanism but decreasing it to a minimal level that will require further study to be determined. This hypothesis is supported by the growth promotion in nonstressed ctr1-8 nrt1.8-2 or eto1-1 nrt1.8-2 mutants (Figures 8D and 8E), where the ET/JA-derived SINAR was partially impaired by nrt1.8. Considering that NRT1.8/NRT1.5 are two essential components in SINAR, further promotion of plant growth might be obtained by constitutive elevation of NRT1.5 and concomitant disruption of NRT1.8 in roots.
In conclusion, we revealed that the ET/JA-NRT signaling module is involved in the regulation of SINAR and that EIN3/EIL1 act as pivotal signal integrators to modulate NRT1.5 and NRT1.8 expression. Components other than EIN3/EIL1 might exist in the signaling pathway to relay the stress-initiated JA signal, and SINAR mediates plant adaption to environments in a nitrate reductase-dependent manner.
METHODS
Plant Materials
Arabidopsis thaliana mutant or transgenic lines were gifts from colleagues (ein2-50, coi1-1, ein3 eil1, coi1-2, nia1 nia2, EIN3ox, ctr1-8, eto1-1, and 35S:EIN3-GFP/ein3 eil1) or generated in our laboratory (nrt1.8-2 and nrt1.5-4) as described (Chen et al., 2012). The mutant lines ora59-1 (CS405772), ora59-2 (CS855464), and erf104 (SALK_057720C) were ordered from the ABRC and the European Arabidopsis Stock Centre. All double or triple mutant lines were generated by crossing the single mutants and subsequent PCR genotyping, except that an additional screen with 50 μM MeJA in plate medium was performed to identify the coi1-2 nrt1.5-4 homozygote (Xu et al., 2002) or 50 μM MeJA plus 10 μM ACC for ein2-50 coi1-1.
DNA Constructs and Transformation into Plants
To make the ERF1B-RNAi construct, a cDNA fragment specific to ERF1B was amplified using the PCR primers listed in Supplemental Table 1, and the fragment was further introduced into the modified binary vector pFGC5941 (a gift from Hai Huang) by a two-step cloning strategy, according to the instructions (http://www.chromdb.org/rnai/vector_info.html). The resulting construct, ERF1B-RNAi, was then transformed into ora59-1 erf104 to generate the ora59-1 erf104 erf1b triple mutants tri-1 and tri-2. To make the 35S:ORA59-TAP and 35S:ERF1B-TAP constructs, the coding sequences of ORA59 and ERF1B were amplified and introduced into the pCambias1300-TAP vector, as described (Luo et al., 2012). The 35S:ERF104-GFP construct was generated by introducing green fluorescent protein (GFP) into the vector pCambias1300 and then fusing the coding sequence of ERF104 to the 5′ terminus of GFP. All constructs were transformed into the indicated plants by the floral dip method (Clough and Bent, 1998). Transgenic lines with 3:1 segregation on selection plates were used for further isolation of homozygotes with strong expression of target genes.
Growth Conditions and Phenotyping
Arabidopsis plants were generally grown in quarter-strength minimal medium or hydroponics as described (Gong et al., 2003). At 4 weeks of age, plants were treated with the indicated hormones or stresses and tissues were sampled and subjected to further assays. Alternatively, the minimal medium was supplemented with elevated nitrate [24 mM KNO3, 0.5 mM Ca(NO3)2] or substituted nitrogen sources [2.25 mM ammonium:1.125 mM (NH4)2 succinate, 1.25 mM KCl, 0.5 mM CaCl2]. Surface-sterilized seeds were plated on the above media as indicated and incubated at 4°C for 4 d, then allowed to grow vertically at 22°C for 7 to 9 d before root elongation was measured. Or, as indicated, Arabidopsis seeds were germinated on minimal medium-containing plates supplemented with 2.25 mM ammonium for 4 d, transferred to medium with the indicated nitrogen plus CdCl2/NaCl, and allowed to grow vertically for the indicated time before the determination of root elongation.
RT-PCR and Real-Time Quantitative RT-PCR
Total RNA from plants grown under the indicated conditions was prepared using TRIzol reagent (Invitrogen). First-strand cDNA synthesis, RT-PCR, and quantitative RT-PCR were performed as described previously (Li et al., 2010). The primers used in these assays are listed in Supplemental Table 1, and the expression levels were normalized to those of the SAND control.
Protein Purification and EMSA
Fragments of ORA59, ERF1B, and ERF104 coding sequences were cloned into the vector pET-30a(+), and the resulting constructs were further transformed into Escherichia coli BL21(DE3) pLysS. Recombinant proteins were induced by isopropyl β-d-1-thiogalactopyranoside and purified using Ni-NTA agarose (Qiagen). For EIN3, a truncated cDNA fragment representing the EIN3 DNA binding domain (amino acids 1 to 314) was cloned into pGEX 4T-1, and the recombinant protein was purified by glutathione S-transferase-agarose affinity chromatography (GE Healthcare).
EMSA was performed using the Light Shift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. All DNA fragments used as wild-type probes were amplified using the PCR primers listed in Supplemental Table 1, then labeled with biotin or left unlabeled and used as competitors. Mutant probes were generated using the Multipoints Mutagenesis Kit (Takara).
Transient Transcriptional Activity Assay
A 1298-bp promoter fragment upstream of the NRT1.8 start codon (ATG) was cloned into pGreen II 0800 to generate the reporter plasmid, and the coding sequences of selected ERF genes were cloned into pGreen II 62-SK to make effector plasmid (Hellens et al., 2005). Once transformed with 12 μg of effector plasmids and 3 μg of reporter plasmids, the Arabidopsis protoplasts were incubated at 20 to 25°C overnight and then harvested and lysed (Yoo et al., 2007). Firefly luciferase (LUC) and Renilla luciferase (REN) activities were assayed using the dual-luciferase reporter assay system (Promega).
ChIP-Quantitative PCR
At 4 weeks of age, root samples from hydroponically grown transgenic plants (ERF-overexpressing or 35S:EIN3-GFP/ein3 eil1 lines) with or without treatments (20 μM ACC + 50 μM MeJA for 6 h) were fixed in formaldehyde crosslinking solution. ChIP was performed as described previously (Aggarwal et al., 2010) using anti-GFP antibody (for ERF104-GFP or EIN3-GFP protein fusions; Invitrogen) or IgG beads (for ERF1B-TAP or ORA59-TAP protein fusions; GE Healthcare). The enriched DNA fragments were subjected to quantitative PCR (qPCR) determination using the primers listed in Supplemental Table 1. qPCR was performed using SYBR Premix Ex Taq (Takara) according to the manufacturer’s instructions, and relative enrichment of fragments was calculated by comparing samples treated with or without antibodies. TUB2 was used as an internal control.
Determination of Nitrate Content
The nia1 nia2 plants were grown hydroponically with ammonium as the sole nitrogen source; the original 1.25 mM KNO3 and 0.5 mM Ca(NO3)2 in the hydroponic solution (Gong et al., 2003) were replaced with 2.25 mM NH4HCO3, 1.25 mM KCl, and 0.5 mM CaCl2. At 4 weeks of age, plants were subjected to treatments as indicated and tissues were sampled. Nitrate was extracted and determined as described (Li et al., 2010).
Analysis of Root-to-Shoot Nitrate Allocation Using 15NO3−
Four-week-old Col-0, ein2-50, coi1-1, and ein2-50 coi1-1 plants grown in quarter-strength hydroponics medium were treated with 20 μM ACC, 50 μM MeJA, 20 μM ACC + 50 μM MeJA, 200 μM CdCl2, and 150 mM NaCl for 12 h, washed in 0.1 mM CaSO4 for 1 min, labeled for 30 min in 3 mM K15NO3 medium (99.15% atom excess of 15N; pH 6.0), and washed again in 0.1 mM CaSO4 for 1 min (Wang and Tsay, 2011). The root and shoot tissues were sampled separately and dried at 80°C for 3 d. 15N contents were determined using a continuous-flow isotope ratio mass spectrometer coupled to a carbon nitrogen elemental analyzer (Vario EL III/Isoprime; Elementar).
Statistical Analysis
Two-tailed Student’s t tests were performed. Differences were deemed significant at P < 0.05 and extremely significant at P < 0.01.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NRT1.5 (At1g32450), NRT1.8 (At4g21680), ERF1B/ERF1 (At3g23240), ORA59 (At1g06160), ERF104 (At5g61600), ERF1A/ATERF1 (At4g17500), ERF2 (At5g47220), ERF6 (At4g17490), ERF13 (At2g44840), ERF15 (At2g31230), EIN3 (At3g20770), EIL1 (At2g27050), CTR1 (At5g03730), EIN2 (At5g03280), COI1 (At2g39940), ETO1 (At3g51770), NIA1 (At1g77760), NIA2 (At1g37130), SAND (At2g28390), and TUB2 (At5g62690). Additional sequence data are available in Supplemental Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ERF Family Members Function Redundantly in the Regulation of NRT1.8 Expression.
Supplemental Figure 2. ERF Expression upon Stress/Hormone Treatment in the ET/JA Mutants.
Supplemental Figure 3. NRT1.5 and NRT1.8 Expression in Response to Salicylic Acid or ABA Treatment.
Supplemental Figure 4. The Expression of ERFs in the ein3 eil1 Mutant in Response to Different Treatments.
Supplemental Figure 5. Root Growth of EIN3-Overexpressing Lines under Nonstressed Conditions.
Supplemental Figure 6. Nitrate Reallocation upon Stresses in nrt1.8 and nrt1.5 Mutants in the nia1 nia2 Background.
Supplemental Table 1. List of Primer Sequences.
Supplementary Material
Acknowledgments
We thank Chi-Kuang Wen (Shanghai Institutes for Biological Sciences), Hongwei Guo (Peking University), Chuanyou Li (Chinese Academy of Sciences), Daoxin Xie (Tsinghua University), and Nigel Crawford (University of California San Diego) for kindly providing the seeds of ein2-50, ctr1-8, eto1-1, EIN3ox, ein3 eil1, 35S:EIN3-GFP/ein3eil1, coi1-1, coi1-2, and nia1 nia2; Hai Huang (Shanghai Institutes for Biological Sciences) and Roger P. Hellens (Mt. Albert Research Centre) for the vectors of pFGC5941, pGreen II 0800, and pGreen II 62-SK; Jingli Hou and Li Zhang (Instrumental Analysis Center, Shanghai Jiao Tong University) for helping with 15N analysis; Chi-Kuang Wen for careful reading of the article; and Yan-Lei Fu for helping with ChIP analysis. This work was supported by the National Science Foundation of China (Grants 31325003 and 31121063), the Ministry of Agriculture of China (Grant 2014ZX08009-003-005), and the SA-SIBS scholarship program.
AUTHOR CONTRIBUTIONS
J.G. and G.Z. designed the research. G.Z. and H.Y. performed the research. J.G. and G.Z. analyzed the data and wrote the article.
Glossary
- SINAR
stress-initiated nitrate allocation to roots
- ET
ethylene
- JA
jasmonic acid
- ChIP
chromatin immunoprecipitation
- Col-0
Columbia-0
- ACC
1-aminocyclopropane-1-carboxylic acid
- EMSA
electrophoretic mobility shift assay
- MeJA
methyl jasmonate
- ABA
abscisic acid
- NO
nitric oxide
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- qPCR
quantitative PCR
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- Achard P., Vriezen W.H., Van Der Straeten D., Harberd N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal P., Vaites L.P., Kim J.K., Mellert H., Gurung B., Nakagawa H., Herlyn M., Hua X., Rustgi A.K., McMahon S.B., Diehl J.A. (2010). Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. (1986). The partitioning of nitrate assimilation between root and shoot of higher plants. Plant Cell Environ. 9: 511–519. [Google Scholar]
- Arteca R.N., Arteca J.M. (2007). Heavy-metal-induced ethylene production in Arabidopsis thaliana. J. Plant Physiol. 164: 1480–1488. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32. [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Gravot A., Richaud P., Auroy P., Duc C., Gaymard F., Taconnat L., Renou J.P., Pugin A., Wendehenne D. (2009). Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 149: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin D.T., Atkins C.A. (1974). Nitrate, nitrite and ammonia assimilation by leaves: Effect of light, carbon dioxide and oxygen. Planta 116: 207–224. [DOI] [PubMed] [Google Scholar]
- Cao W.H., Liu J., He X.J., Mu R.L., Zhou H.L., Chen S.Y., Zhang J.S. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Z., Lv X.F., Li J.Y., Yi H.Y., Gong J.M. (2012). Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159: 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crawford N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7: 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B.G., Lea P.J. (2007). Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 58: 2339–2358. [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Gojon A., Gaymard F. (2010). Keeping nitrate in the roots: An unexpected requirement for cadmium tolerance in plants. J. Mol. Cell Biol. 2: 299–301. [DOI] [PubMed] [Google Scholar]
- Gong J.M., Lee D.A., Schroeder J.I. (2003). Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10118–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., et al. . (2011). A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L.E., Garate A., Carpena-Ruiz R (1997). Effects of cadmium on the uptake, distribution and assimilation of nitrate in Pisum sativum. Plant Soil 189: 97–106. [Google Scholar]
- Hu P., Zhou W., Cheng Z., Fan M., Wang L., Xie D. (2013). JAV1 controls jasmonate-regulated plant defense. Mol. Cell 50: 504–515. [DOI] [PubMed] [Google Scholar]
- Ismail A., Riemann M., Nick P. (2012). The jasmonate pathway mediates salt tolerance in grapevines. J. Exp. Bot. 63: 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., et al. . (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X.T., Xing G.X., Chen X.P., Zhang S.L., Zhang L.J., Liu X.J., Cui Z.L., Yin B., Christie P., Zhu Z.L., Zhang F.S. (2009). Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. USA 106: 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31. [DOI] [PubMed] [Google Scholar]
- Li J.Y., et al. . (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.H., Kuo H.F., Canivenc G., Lin C.S., Lepetit M., Hsu P.K., Tillard P., Lin H.L., Wang Y.Y., Tsai C.B., Gojon A., Tsay Y.F. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Bernard D.G., Balk J., Hai H., Cui X. (2012). The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24: 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksymiec W., Wianowska D., Dawidowicz A.L., Radkiewicz S., Mardarowicz M., Krupa Z. (2005). The level of jasmonic acid in Arabidopsis thaliana and Phaseolus coccineus plants under heavy metal stress. J. Plant Physiol. 162: 1338–1346. [DOI] [PubMed] [Google Scholar]
- Mur L.A.J., Mandon J., Persijn S., Cristescu S.M., Moshkov I.E., Novikova G.V., Hall M.A., Harren F.J.M., Hebelstrup K.H., Gupta K.J. (2012). Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 5: pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltronieri, P., Taurino, M., De Domenico, S., Bonsegna, S., and Santino, A. (2013). Activation of the jasmonate biosynthesis pathway in roots in drought stress. In Climate Change and Plant Abiotic Stress Tolerance, N. Tuteja and S.S. Gill, eds (Weinheim, Germany: Wiley-VCH), pp. 325–342. [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M.J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles P.S., Bloom A.J. (2003). Nitrate photo-assimilation in tomato leaves under short-term exposure to elevated carbon dioxide and low oxygen. Plant Cell Environ. 26: 1247–1255. [Google Scholar]
- Smirnoff N., Stewart G.R. (1985). Nitrate assimilation and translocation by higher plants: Comparative physiology and ecological consequences. Physiol. Plant. 64: 133–140. [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2: 178–186. [DOI] [PubMed] [Google Scholar]
- Szabados L., Savouré A. (2010). Proline: A multifunctional amino acid. Trends Plant Sci. 15: 89–97. [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M.C., Rodenburg N., Pauwels L., Goossens A., Körbes A.P., Memelink J., Ritsema T., Van Wees S.C.M., Pieterse C.M.J. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Gutiérrez R.A. (2008). A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11: 521–529. [DOI] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C.A., Cook R.J., Browse J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P., Liu L.H., Remans T., Tester M., Forde B.G. (2006). Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol. 47: 1045–1057. [DOI] [PubMed] [Google Scholar]
- Wang R., Xing X., Crawford N. (2007). Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 145: 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Y., Tsay Y.F. (2011). Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J.Q., Crawford N.M. (1993). Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. 239: 289–297. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCFCOl1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. . (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang L., Li Z., Quan R., Li G., Wang R., Huang R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Guo H.W. (2011). Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4: 626–634. [DOI] [PubMed] [Google Scholar]
- Zhu Z., et al. . (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.