The WRKY protein TTG2 is shown to be essential for the activation of the trichome-patterning gene TRY. TTG2 binds to W-boxes in the TRY promoter and enhances the activation by GL1, GL3, and TTG1. Protein interactions of TTG2 and TTG1 suggest the formation of a GL1/GL3/TTG1/TTG2 transcriptional activation complex.
Abstract
Trichome patterning in Arabidopsis thaliana is regulated by a regulatory feedback loop of the trichome promoting factors TRANSPARENT TESTA GLABRA1 (TTG1), GLABRA3 (GL3)/ENHANCER OF GL3 (EGL3), and GL1 and a group of homologous R3MYB proteins that act as their inhibitors. Together, they regulate the temporal and spatial expression of GL2 and TTG2, which are considered to control trichome cell differentiation. In this work, we show that TTG2 is a specific activator of TRY (but not CPC or GL2). The WRKY protein TTG2 binds to W-boxes in a minimal promoter fragment of TRY, and these W-boxes are essential for rescue of the try mutant phenotype. We further show that TTG2 alone is not able to activate TRY expression, but rather drastically enhances the activation by TTG1 and GL3. As TTG2 physically interacts with TTG1 and because TTG2 can associate with GL3 through its interaction with TTG1, we propose that TTG2 enhances the activity of TTG1 and GL3 by forming a protein complex.
INTRODUCTION
Trichome patterning in Arabidopsis thaliana is a well-studied model system for the establishment of a two-dimensional pattern of cell types without reference to already existing positional cues (Pesch and Hülskamp, 2009; Balkunde et al., 2010; Tominaga-Wada et al., 2011; Grebe, 2012). Trichomes in Arabidopsis are large single cells that originate at the basis of young rosette leaves in a regular pattern and become separated from each other by cell divisions of epidermal cells (Hülskamp et al., 1994). Genetic and molecular models explain trichome patterning by a transcriptional network of trichome promoting and repressing genes (Ishida et al., 2008; Pesch and Hülskamp, 2009; Balkunde et al., 2010). Three groups of proteins function as activators: the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) (Koornneef, 1981; Galway et al., 1994; Walker et al., 1999), the R2R3 MYB-related transcription factor GLABRA1 (GL1) (Oppenheimer et al., 1991), and the basic helix-loop-helix (bHLH)-like transcription factors GL3 and ENHANCER OF GL3 (EGL3) (Koornneef et al., 1982; Hülskamp et al., 1994; Payne et al., 2000; Bernhardt et al., 2003; Zhang et al., 2003). Several homologous R3 single repeat MYB genes act in a partially redundant manner as negative regulators of trichome development (Schellmann et al., 2002; Kirik et al., 2004a, 2004b; Wang et al., 2007, 2008, 2010; Tominaga et al., 2008; Wester et al., 2009; Gan et al., 2011). These include TRIPTYCHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, ETC3, TRICHOMELESS1 (TCL1), and TCL2 (Hülskamp et al., 1994; Wada et al., 1997; Schellmann et al., 2002; Kirik et al., 2004a, 2004b; Wang et al., 2007; Gan et al., 2011). The activators form a complex (called MBW) consisting of TTG1, R2R3MYB, and bHLH proteins with TTG1 and the R2R3MYB proteins both binding to the bHLH protein (Payne et al., 2000; Zhang et al., 2003; Zimmermann et al., 2004; Kirik et al., 2005; Digiuni et al., 2008; Gao et al., 2008; Wang and Chen, 2008; Zhao et al., 2008). This complex is considered to be transcriptionally active and is repressed through the binding of a R3MYB to the bHLH protein, which in turn replaces the R2R3MYB (Payne et al., 2000; Bernhardt et al., 2003; Esch et al., 2003). GL2 acts downstream of the activators and inhibitors and regulates the further differentiation of trichome precursor cells (Rerie et al., 1994).
Theoretical modeling of the known genetic and molecular interactions helped to unravel the underlying logic of the gene regulatory network (Benítez et al., 2007, 2008). The network is based on at least two patterning mechanisms each capable of explaining patterning alone (Pesch and Hülskamp, 2009). First, the activators and inhibitors are engaged in a feedback system such that the activators turn on the inhibitors and are themselves downregulated by the inhibitors. Intercellular interactions are mediated by the inhibitors. Second, TTG1 is trapped in incipient trichome cells by GL3 and thereby depleted from the surrounding cells (Bouyer et al., 2008; Balkunde et al., 2011).
In addition to this core machinery, the WRKY transcription factor TTG2 has been implicated in the regulation of trichome patterning. Mutations in TTG2 cause defects in several traits, including trichome patterning, trichome differentiation, proanthocyanidin accumulation, and mucilage production in the seed coat, and the elongation of integument cells (Johnson et al., 2002; Garcia et al., 2005). Consistent with these functions, TTG2 is expressed in leaf blades, trichomes, developing seeds, and non-root hair cells (Johnson et al., 2002). Current data suggest that TTG2 is a downstream gene of TTG1 and, therefore, most likely of the whole patterning network (Johnson et al., 2002; Ishida et al., 2007; Zhao et al., 2008; Gonzalez et al., 2009): TTG2 is not expressed in ttg1 mutants, indicating that its expression depends on TTG1 and it is regulated by R2R3MYB factors. In addition, ttg2 gl2 double mutants have a synergistic phenotype, suggesting that TTG2 regulates trichome cell differentiation together with GL2. It is assumed that trichome cell differentiation involves the regulation of GL2 by TTG2 (Ishida et al., 2007). TTG2 is best known for its regulation of the bHLH gene TRANSPARENT TESTA8 (TT8) in the context of proanthocyanidin regulation in seeds. This pathway is regulated by a MBW complex involving TTG1, a R2R3 MYB protein and bHLH proteins (including TT8). Here, it was shown that TTG2 is important for the expression of TT8 in the endothelium early but not late during seed development (Xu et al., 2013), indicating that TTG2 is involved in the regulation of the temporal and spatial expression of TT8 together with components of the proanthocyanidin regulating MBW complex.
Taken together, while most data point to a role of TTG2 in cell differentiation downstream of the TTG1/R2R3MYB/bHLH system, the latter experiments support the idea that TTG2 can modulate MBW function. To test this possibility, we analyzed the role of TTG2 in trichome formation.
RESULTS
ttg2 Mutants Show Trichome Clusters and a Reduced Trichome Number
In the first report describing the ttg2 mutant phenotype, it was noted that apart from defects in trichome morphology also trichome number is reduced and that the spatial distribution is altered (Johnson et al., 2002). We analyzed the trichome pattern on young leaves on which trichome patterning was still going on and mature leaves. To recognize the earliest trichome stages even before they undergo morphological changes, we used the 5′-GL2:GFP:ER marker line. The overall trichome number was reduced to ∼60% as compared with the wild type in young as well as in older leaves (Table 1). During early leaf development, ∼20% of all trichome initiation sites formed trichome clusters (Figures 1A and 1B, Table 1). On mature leaves, the frequency of trichome clusters dropped to around 10%. This is probably due to an underdevelopment of one of the two neighboring trichomes in the cluster, which cannot be recognized at older stages. This is supported by the observation that one of the two trichomes in a cluster is typically much smaller (Figures 1C and 1D).
Table 1. Analysis of Trichome Number and Cluster Frequency on Leaf Three Using 5′-GL2:GFP:ER to Label Trichomes at All Developmental Stages in the Wild Type and ttg2-1 Mutants (Mean and sd, n = 20).
| Leaf Age | Genotype | Number of TIS | % Cluster |
|---|---|---|---|
| 9 d | Ler | 24.6 ± 3.2 | 0.2 ± 0.9 |
| ttg2-1 | 14.3 ± 1.8 | 20.3 ± 10.9 | |
| 12 d | Ler | 31.4 ± 4.1 | 0.6 ± 1.3 |
| ttg2-1 | 19.8 ± 3.9 | 9.9 ± 5.7 |
TIS, trichome initiation site.
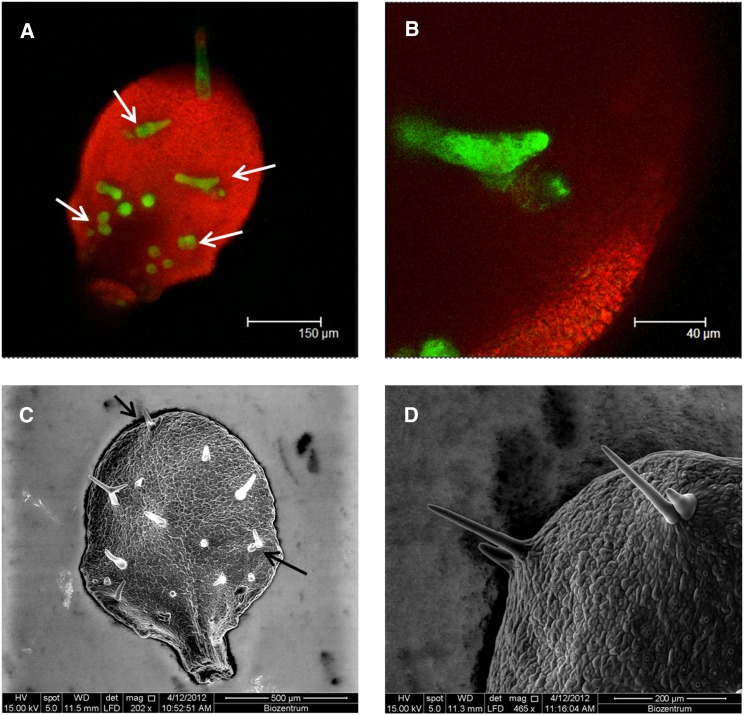
Figure 1.
ttg2 Mutants Exhibit Trichome Clusters.
(A) Confocal laser scanning microscopy image of a young ttg2 leaf of a 5′-GL2:GFP:ER marker line.
(B) Higher magnification from (A) showing trichome clusters.
(C) Scanning electron microscope image of a mature ttg2 mutant leaf.
(D) Higher magnification of two ttg2 mutant trichome clusters. Note that one of the trichomes in a cluster is typically smaller. Arrows mark the position of clusters.
The Expression of TRY, GL2, and CPC Depends on TTG2
The observation that ttg2 mutants show clustered trichomes reminiscent of try mutants prompted us to test whether TTG2 is involved in regulating TRY. Toward this end, we introduced the pTRY:GUS (β-glucuronidase) marker line (Pesch and Hülskamp, 2011) in the ttg2 mutant background. pTRY:GUS expression at the leaf base as well as trichome-specific expression was completely absent, indicating that TTG2 is a positive regulator of TRY (Figures 2A to 2F; Supplemental Figures 1A and 1B). The pTRY:GUS ttg2 plants were crossed with the wild type and the progeny was shown to have normal pTRY:GUS expression. This proved the functionality of the marker construct in the plants and excludes silencing effects in the lines. By contrast, the expression of pCPC:GUS (Figures 2G to 2L; Supplemental Figures 1C and 1D) and pGL2:GUS (Figures 2M to 2R; Supplemental Figures 1E and 1F) was only moderately reduced in ttg2 mutants. This impression was confirmed by quantitative PCR experiments (Supplemental Figure 2) using young leaf stages corresponding to the stages shown in the third column of Figure 2. In these experiments, all three genes were expressed in the ttg2 background, but to a much lesser extent than in the wild type. Together, these data indicate that TTG2 is an activator of the three trichome patterning genes tested and that TRY expression is most severely reduced in ttg2 mutants.
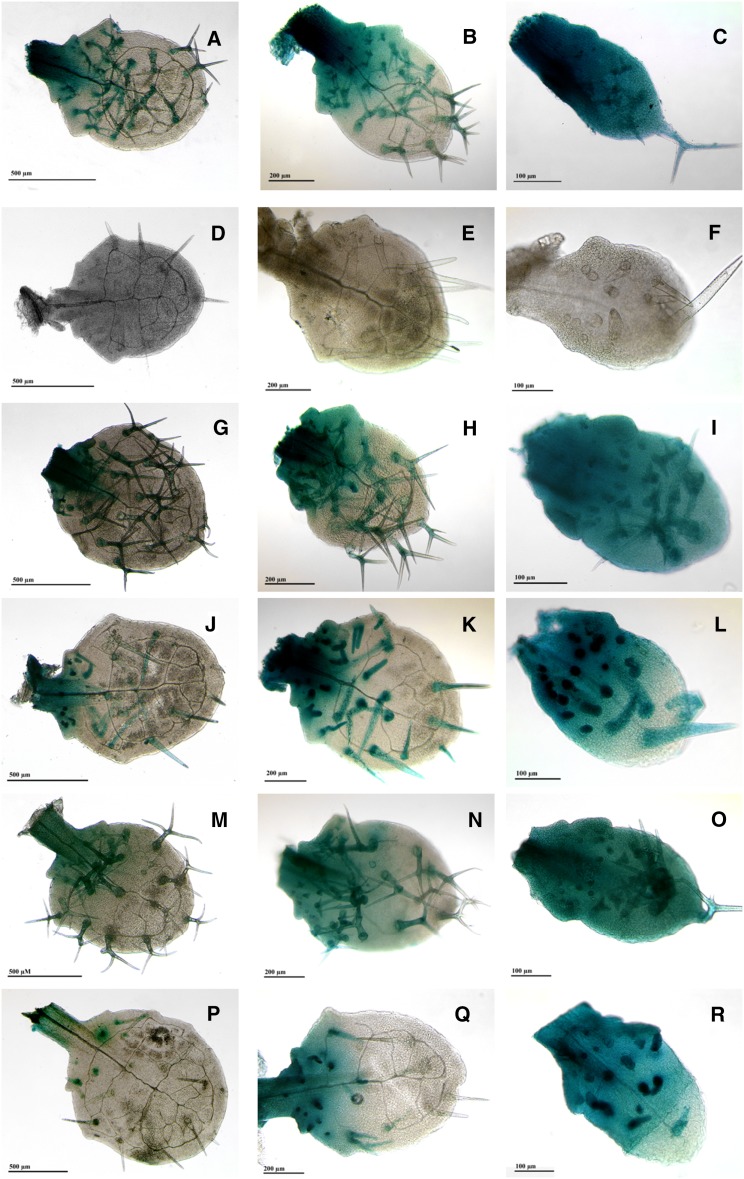
Figure 2.
TRY but Not GL2 or CPC Expression Depends on TTG2.
Expression pattern mediated by the 5′ regulatory regions of GL2, CPC, and TRY in the Ler and ttg2 background as revealed by GUS reporter gene expression. The first column shows leaf number three as an example for a mature leaf. The second column shows leaf number four on which young trichomes are still developing at the base of the leaf. The third column shows leaf number five on which new trichomes are initiated at the base of the leaf. pTRY:GUS Ler ([A] to [C]), pTRY:GUS ttg2 ([D] to [F]), pCPC:GUS Ler ([G] to [I]), pCPC:GUS ttg2 ([J] to [L]), pGL2:GUS Ler ([M] to [O]), and pGL2:GUS ttg2 ([P] to [R]).
WRKY cis-Regulatory Elements Are Important for TRY Expression
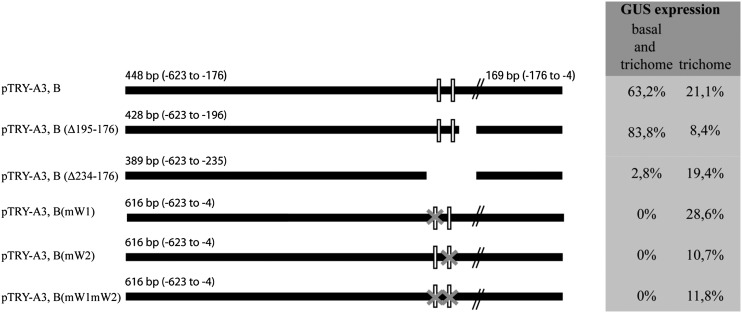
As ttg2 and try mutants share the cluster phenotype, we focused in the following on the role of TTG2 in the transcriptional regulation of TRY. WRKY proteins are transcriptional regulators that bind to their target promoters by interacting with a DNA sequence motif (T)(T)TGAC(C/T) called the W-box (Eulgem et al., 2000). Several W-box motives were found in the TRY promoter as well as in the promoters of its homologs CPC, ETC1, ETC2, ETC3, TCL1, and TCL2 (Supplemental Data Set 1). Further deletions of the minimal TRY promoter fragment pTRY (A3,B) (Pesch and Hülskamp, 2011) were analyzed by promoter:GUS constructs in more than 25 independent T2 lines each to enable a comparison between the different constructs. This study revealed a small DNA stretch that is important for proper expression of TRY in the patterning region (−234 to −196 upstream of the ATG start codon). This region contains two putative W-boxes (Figure 3) that represent possible targets for TTG2. We tested the role of the two W-boxes in TRY expression by substituting the nucleotides GTCAA with CCCGG in each of them or simultaneously in both GUS reporter studies. The minimal wild-type TRY promoter (pTRY-A3,B:GUS) showed the correct TRY expression in >63% of the T2 lines. By contrast, no lines were found to have basal expression when either one of the W-boxes was mutated [pTRY-A3,B(mW1):GUS, pTRY-A3,B(mW2):GUS] (Supplemental Figures 3 and 4) or when both were simultaneously mutated [pTRY-A3, B(mW1mW2):GUS; Supplemental Figure 3]. Expression in incipient trichomes was not found in pTRY-A3,B(mW1mW2):GUS. The number of lines showing expression only in trichomes was not reduced when the W1-box was mutated, but was drastically reduced when the W2-box was destroyed (Figure 3). An additional mutation in the W1-box did not further reduce TRY expression [pTRY-A3, B(mW1mW2):GUS] (Supplemental Figure 3). Together these data indicate that the two W-boxes are essential for the correct expression of TRY.
Figure 3.
Expression Analysis of the TRY Promoter.
Schematic presentation of the TRY promoter and the deletions and mutations of putative WRKY binding sites (W-boxes). The relative position is given with respect to the start codon. The white boxes indicate the relative position of the putative W-boxes. The gray X indicates mutations in the W-boxes. The respective promoter:GUS lines were assayed in more than 25 independent T2 lines for basal epidermal expression and trichome-specific expression. The percentage of T2 lines is given for the two expression categories by distinguishing between lines in which only trichome-specific expression was found or basal expression also was observed.
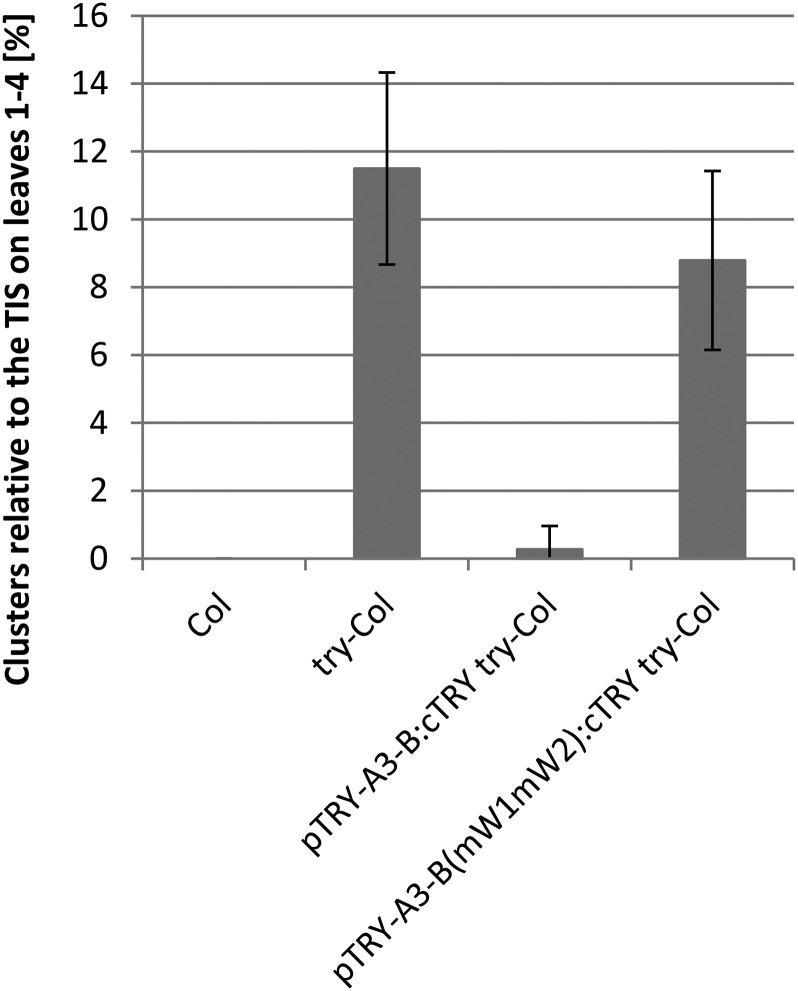
To test whether the W-boxes are functionally relevant, we expressed the coding sequence (CDS) of TRY under the control of the wild-type TRY promoter (pTRY-A3-B:cTRY) and the mutated TRY promoter [pTRY-A3-B(mW1mW2):cTRY]. As shown in Figure 4, the minimal TRY promoter restored the number of clusters to wild-type levels, whereas the promoter carrying the two mutated W-box sites did not.
Figure 4.
Rescue Analysis of Promoters Carrying W-Box Mutations.
The frequency of clusters found at trichome initiation sites (TIS) is compared between wild-type Columbia (Col) (n = 30), try-Col (n = 30) pTRY-A3-B:cTRY try-Col (n = 50) and pTRY-A3-B(mW1mW2):cTRY try-Col (n = 50). pTRY-A3-B:cTRY try-Col was not significantly different from Col (Student’s t test, P < 0.01), whereas pTRY-A3-B(mW1mW2):cTRY try-Col was different from Col. Error bars indicate the sd.
TTG2 Binds to a 71-bp Fragment of the TRY Promoter Containing the Two W-Boxes
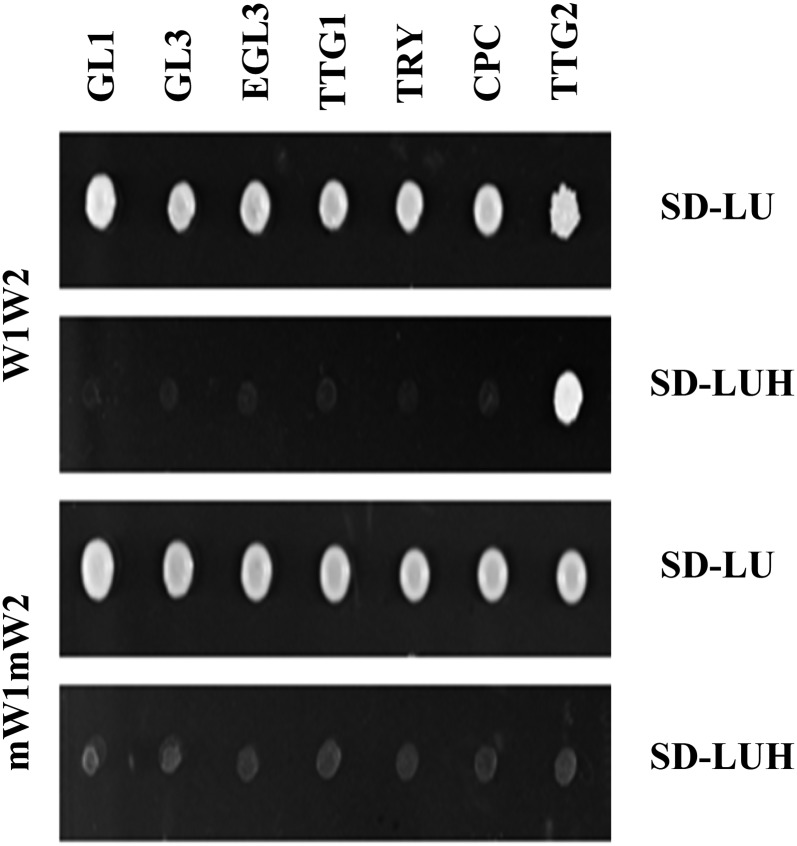
Our findings that TTG2 is required for the activation of TRY expression and that the two WRKY binding sites present in the TRY promoter are important for TRY expression raises the question of whether TTG2 can directly bind to the TRY promoter. To test this, we performed yeast one-hybrid assays using three copies of a 71-bp fragment containing the two W-boxes (Figure 5; Supplemental Figure 4). Colony growth was observed on selection medium when TTG2 was expressed, whereas no interaction was found with GL1, GL3, EGL3, TTG1, TRY, or CPC. As a control we used a 71-bp promoter fragment carrying mutations in the two W-boxes (mW1mW2; Supplemental Figure 4). No binding to the mutated promoter fragment was observed.
Figure 5.
TTG2 Binds to W-Boxes in the 71-bp TRY Promoter Fragment in Yeast One-Hybrid Assays.
A 71-bp fragment of the TRY promoter containing the two W-boxes was used in three tandem copies for yeast one-hybrid assays. Expression of TTG2 but not of GL1, GL3, EGL3, TTG1, TRY, or CPC caused colony growth on selection media (SD-LUH). No binding of TTG2 to the 71-bp fragment was observed when the W-boxes were mutated.
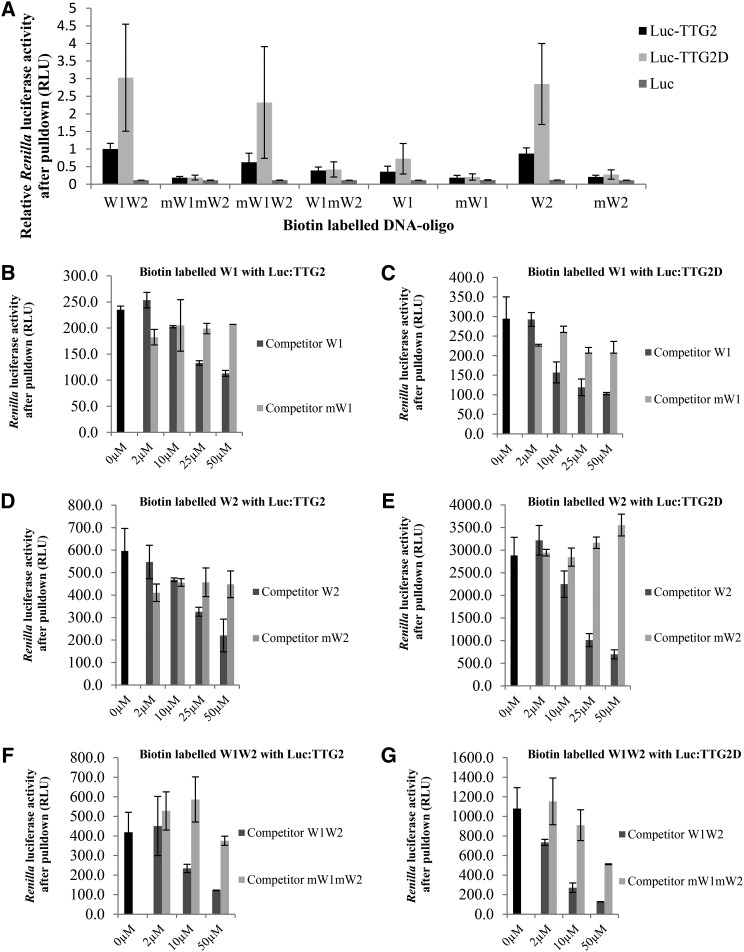
To provide independent evidence for the binding of TTG2 to the TRY promoter fragment, we adapted the ELISA-DNA protein interaction (DPI) assay (Brand et al., 2010) to use Renilla reniformis luciferase fused proteins (called Luc-DPI in the following). In short, biotin-labeled double-stranded short DNA fragments (W1, W2, or W1W2; Supplemental Figure 4) were bound to streptavidin beads in 96-well plates. TTG2 or only the WRKY domain of TTG2 (TTG2D) was expressed in HEK293TN cells as fusions to the luciferase protein (Luc-TTG2 and Luc-TTG2D). The integrity of the Luc-TTG2 and Luc-TTG2D proteins in these experiments was confirmed by immunoblotting using a luciferase antibody as a probe (Supplemental Figure 5). To test the general functionality of Luc-TTG2 and Luc-TTG2D, we designed vectors to express these fusions in planta and confirmed transcriptional enhancing activity for the pTRY:GUS in Arabidopsis cell suspension culture assays (Supplemental Figure 6). The HEK293TN protein lysates were generated. After precipitation of the luciferase-fused proteins through biotin-labeled double-stranded DNA interacting with streptavidin at the bottom of the well, luciferase activity was determined (Figure 6). In this assay, we found that all fragments containing an intact W-box bound with both TTG2 and the TTG2 WRKY domain (Figure 6A). Binding of the TTG2 WRKY domain to the W1W2 fragment was significantly stronger than binding of full-length TTG2. Binding efficiency was similarly strong in W1W2, mW1W2, and W2. W1 and W1mW2 also allowed binding of TTG2 or TTG2D, but the binding was much weaker. These results suggest that the W2-box is bound more strongly than the W1 box. Renilla luciferase alone could not be precipitated with any double stranded DNA oligomer. To further corroborate these data, we performed competitor experiments to demonstrate that DNA fragments without a biotin tag can compete with the interaction of TTG2/TTG2D to the biotin labeled DNA in a dosage-dependent manner and that competitor fragments carrying mutations in the W-box cannot (Figures 6B to 6G). We consistently found a dosage-dependent reduction of TTG2 binding when adding competitor DNA in a range from 0 to 50 μM. Mutated W1 (mW1) and mutated W2 (mW2) fragments exhibited no competition. The mutated mW1mW2 showed a weak competition effect at higher concentration.
Figure 6.
TTG2 Binds to W-Boxes in the 71-bp TRY Promoter Fragment in DPI Assays.
(A) TTG2 and TTG2D (WRKY domain of TTG2) were fused to R. luciferase and precipitated with the 71-bp biotin-labeled fragment containing the W1- and W2-boxes. TTG2/TTG2D binding to intact W-boxes is significantly different to the respective LUC controls (Student’s t test, P < 0.05). TTG2D binds better to the W1W2 fragment than to full-length TTG2 (Student’s t test, P < 0.01). Binding of TTG2/TTG2D to W1W2, mW1W2, and W2 was not significantly different (Student’s t test, P > 0.1). Binding of TTG2/TTG2D to W1 and W1mW2W1 was different to W1W2 (Student’s t test P < 0.01).
(B) and (C) DPI analysis using the W1-box and TTG2 (B) or TTG2D (C). For competition, nonbiotinylated W1 and mutated mW1 were used.
(D) and (E) DPI analysis using the W2-box and TTG2 (D) or TTG2D (E). For competition, nonbiotinylated W2 and mutated mW2 were used.
(F) and (G) DPI analysis using the 71-bp fragment containing the W1- and W2-boxes and TTG2 (F) or TTG2D (G). For competition, nonbiotinylated W1W2 and mutated mW1mW2 were used. Error bars indicate the sd.
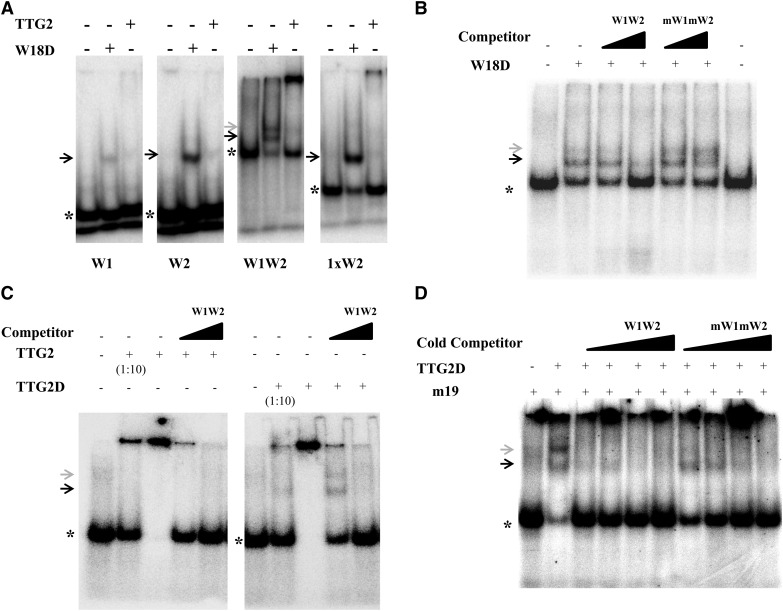
As a third line of experiments demonstrating binding of TTG2 to the TRY promoter fragment, we performed electromobility shift assay (EMSA) (Figure 7). As a positive control, we used the WRKY domain of WRKY18 (W18D), which was demonstrated to strongly bind the parsley (Petroselinum crispum) PR1-1 promoter sequence described as 1xW2 (Rushton et al., 1996; Ciolkowski et al., 2008). W18D showed strong binding to the W1, W2, and W1W2 DNA fragments of the TRY promoter and to the parsley PR1-1 promoter 1xW2. Experiments testing W1W2 showed two higher molecular weight shifted bands that could be diminished by the addition of an excess of nonlabeled W1W2 DNA (Figure 7B).
Figure 7.
TTG2 Binds to W-Boxes in the 71-bp TRY Promoter Fragment in EMSAs.
(A) TTG2 and the WRKY domain from WRKY18 (W18D) were used for EMSA studies on the W1, W2, and W1W2 DNA fragments. The “1xW2” sequence from the parsley Pr1-1 promoter was used as a positive control for WRKY18 binding (Ciolkowski et al., 2008).
(B) W18D binding to the W1W2 DNA fragment and competition with excess competitor DNA.
(C) EMSA testing the binding of TTG2 and TTG2D to the W1W2 DNA fragment. Note that competition with unlabeled W1W2 fragments strongly reduces the high molecular band.
(D) EMSA testing the binding of TTG2D in the presence of the mutated W-Box DNA m19 with and without competitor DNA. Asterisk indicates position of unbound radioactive labeled DNA substrate. The black arrow indicates the lower and the gray arrow the upper shifted band.
TTG2 also showed very weak binding to W1 and W2. Experiments using 1xW2 and W1W2 with TTG2 showed in contrast to W18D a strong shifted band near the gel slot diminishing the amount of unbound free DNA probe capable of entering the gel. However, a faint shifted band also could be observed in the gel using W1W2 and 1xW2 DNA fragments (Figure 7A).
We therefore tested the possibility that the observed band at the gel slot represents multimeric TTG2/DNA aggregations. First, the strength of these high molecular aggregates depends on the relative amount of TTG2. While high TTG2 concentrations can bind virtually all available labeled DNA fragments, a 10-fold dilution of TTG2 recruits only a fraction of the probe (Figure 7C). Second, the addition of specific competitor DNA reduced the recruitment of the DNA in the gel slot. In the case of TTG2D, we observed two shifted bands using lower competitor concentrations whereas binding completely disappeared at higher competitor levels (Figure 7C). Third, to diminish interactions, a mutated W-box fragment designated m19 was used as unspecific DNA. This fragment was previously shown not to interact with WRKY proteins (Ciolkowski et al., 2008). Under these conditions, we again observed two shifted bands with TTG2D to the W1W2 oligomer (Figure 7D). This indicates that additional DNA increases the fraction of released labeled DNA in the gel.
Under optimized conditions comprising TTG2D, W1W2, and unspecific DNA (m19), we could observe that the intensity of the double shifted band could be reduced by increasing levels of nonlabeled W1W2 competitor. The addition of mW1mW2 competitor DNA also reduced the TTG2D/W1W2 interaction, though it was clearly less efficient than the wild-type W1W2 competitor.
The Activation of the TRY Promoter by GL1, GL3, and TTG1 Is Enhanced by TTG2
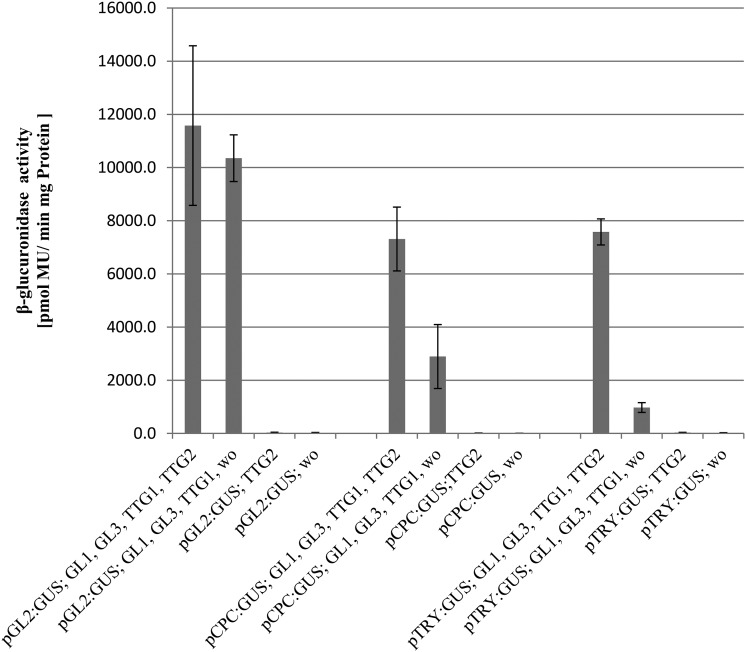
The previous finding that the activation of the TRY promoter depends on GL1, GL3, and TTG1 (Pesch and Hülskamp, 2011) raises the question on the role of TTG2 in transcriptional regulation. Is TTG2 a prerequisite for the function of GL1, GL3, and TTG1 or is TTG2 an enhancer in the presence of the other three proteins? To address this question, we used the Arabidopsis cell suspension culture system that enables a quantitative comparison of the activation of promoter:GUS constructs by different combinations of activating proteins (Figure 8). pTRY:GUS with or without TTG2 showed no GUS activity, indicating that TTG2 alone cannot activate the TRY promoter in this system. Coexpression of GL1, GL3, and TTG1 triggered the activation of the TRY promoter. Additional expression of TTG2 enhanced the activation through GL1, GL3, and TTG1 by ∼10-fold. In contrast, additional expression of TTG2 had no effect on the activation of the GL2 promoter by GL1, GL3, and TTG1. The expression of the CPC promoter was about twofold increased when TTG2 was coexpressed with GL1, GL3, and TTG1.
Figure 8.
TTG2 Enhances the GL1-, GL3-, and TTG1-Dependent Activation of the TRY Promoter.
Arabidopsis cell suspension cultures were transformed with the indicated promoter:GUS constructs along with 35S:cDNAs of GL1, GL3, TTG1, and TTG2. The relative expression levels of the promoter:GUS constructs were determined by GUS assays in three independent experiments. Error bars indicate sd. “wo” indicates the additional expression of the negative control effector construct without CDS fusion to ensure a comparable agrobacteria to cell suspension ratio. The activation of the TRY and CPC promoters by GL1, GL3, and TTG1 are significantly different when TTG2 is additionally expressed (Student’s t test, P < 0.01), whereas no difference was found for the GL2 promoter (Student’s t test, P > 0.1).
Together, these data show that TTG2 exerts its transcriptional activation on the TRY promoter through the enhancement of GL1, GL3, and TTG1 activity.
TTG2 Interacts with TTG1
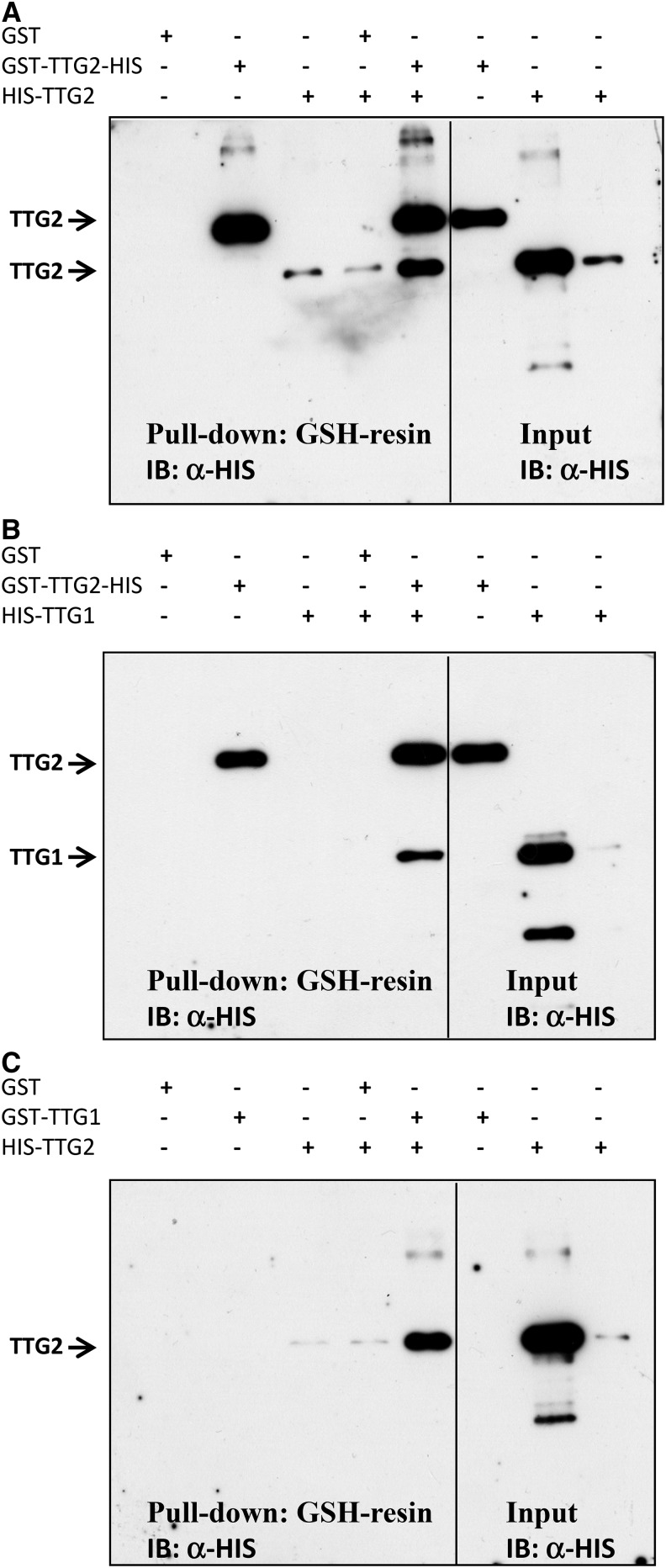
The finding that TTG2 enhances the function of GL1, GL3, and TTG1 suggested to us that TTG2 directly interacts with one or more of the three activator proteins in a higher order transcriptional activating complex. To test this, yeast two hybrid interaction assays were performed with TTG2 fused to the GAL4-activation domain (AD) and to the GAL4-DNA binding domain (DB) (Supplemental Figures 7 and 8). Although we could not test all combinations in both directions because of autoactivation, we consistently found a self-interaction of TTG2 and an interaction with TTG1. No interaction was found with any of the other trichome or root hair patterning genes, including GL1, WER MYB23, GL3, MYC1, TT8, EGL3, TRY, CPC ETC1, ETC2, and ETC3. To confirm the interactions, we expressed TTG1 and TTG2 as HIS and glutathione S-transferase (GST) fusions in Escherichia coli, precipitated the GST fusion proteins, and detected the respective second protein on an immunoblot using a HIS antibody. In these experiments, we observed interactions of TTG2 with itself and with TTG1 in both possible fusion combinations (Figure 9).
Figure 9.
TTG2 Interacts with TTG1.
Bacterially expressed TTG2 and TTG1 proteins fused to GST or a HIS epitope tag were subjected to α-GST pull-down and the proteins detected by immunoblotting (IB) using a HIS antibody.
(A) TTG2-interaction with TTG2.
(B) TTG2 interaction with TTG1 (fused to HIS).
(C) TTG2 (fused to HIS) interaction with TTG1. Note that the GST-TTG2-HIS protein can be distinguished from the HIS-TTG2 protein by size.
TTG2 Requires TTG1 to Interact with GL3
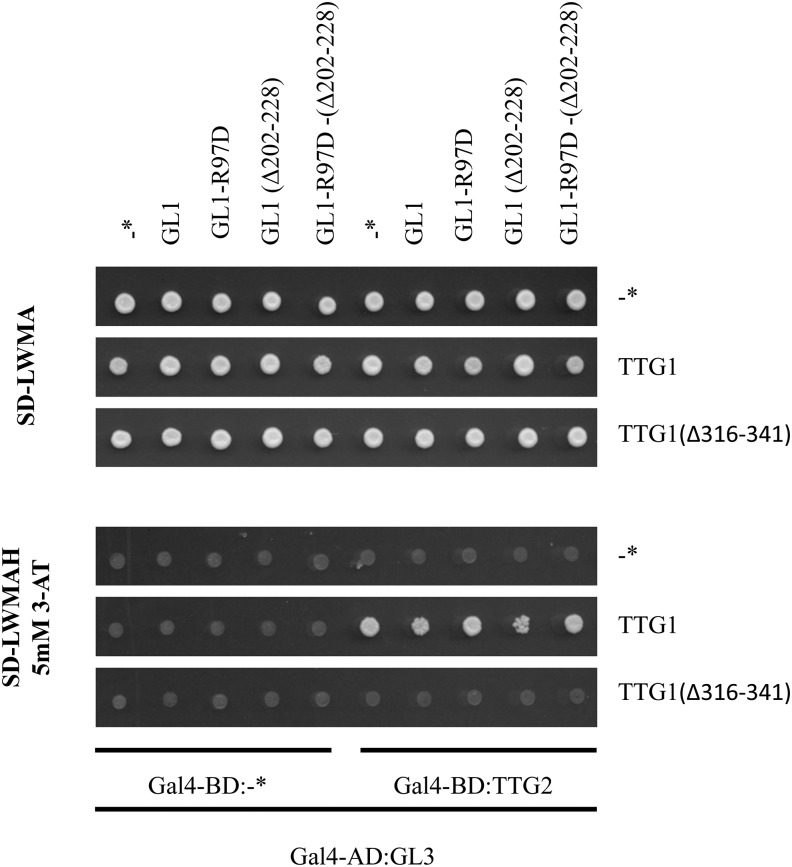
The interaction of TTG2 with TTG1 suggests that TTG2 might be associated with the GL1 GL3 TTG1 protein complex. To test this possibility, we used the yeast four-hybrid system. Gal4-AD-GL3 and Gal4-DB-TTG2 were coexpressed with TTG1 or TTG1∆26 (a mutated version that does not bind to GL3). Coexpression of TTG1 but not of TTG1∆26 mediated the association of AD-GL3 and DB-TTG2 as recognized by growth on interaction plates (Figure 10). This indicates that TTG2 can physically associate with GL3 bridged by TTG1 interacting with both proteins. The ability of TTG1 to mediate the binding of TTG2 to GL3 was not altered by additionally expressing GL1 as a fourth component. Also, the additional expression of mutant versions of GL1, GL1(Δ202-228) (typically used in yeast two-hybrid assays to avoid autoactivation), and GL1-R97D (which does not interact with GL3; Pesch et al., 2013) did not affect the TTG1-mediated binding of TTG2 to GL3 mediated through TTG1 (Figure 10).
Figure 10.
TTG2 Can Interact through TTG1 with GL3.
Yeast four-hybrid interaction assay testing the interaction of GL3-AD and TTG2-BD with and without TTG1. In addition, GL1 and various mutant versions of GL1 were expressed as a fourth component to test whether the addition of GL1 alters the ability of TTG1 to mediate binding of TTG2 to GL3. Asterisk indicates control construct without CDS fusion.
DISCUSSION
Our finding that TTG2 enhances MBW-triggered activation of TRY raised questions that will be discussed below. What is the molecular mechanism by which TTG2 enhances the activator complex? How can TTG2 be integrated in the trichome patterning machinery?
Molecular Mechanism of TTG2-Dependent Transcriptional Activation
TTG2 belongs to a large superfamily of transcription factors containing one or two WRKY domains. WRKY domains are 60 amino acid sequences with a WRKYGQK and a zinc-finger-like motif (Rushton et al., 1995) that can bind through their zinc-finger motif to specific DNA sequences, the W-boxes (de Pater et al., 1996; Eulgem et al., 2000; Rushton et al., 2010; Chi et al., 2013). WRKY proteins may act as transcriptional activators or repressors or may activate and repress in a context-specific manner by binding one or more W-boxes in the respective promoter regions (Rushton et al., 2010). Their activity is thought to be modulated by interactions with other proteins, including VQ containing proteins, mitogen-activated protein kinases, calmodulin, and 14-3-3 proteins (Rushton et al., 2010; Chi et al., 2013).
We have shown that TTG2 can bind to the 71-bp TRY promoter fragment containing two W-boxes in close proximity and to which TTG2 binds with different strength. Whereas the W2-box perfectly matches the typical TTGACC motif, the W1-box motif (TTGACA) was described as a WRKY not binding motif (Ciolkowski et al. 2008). Although the binding of TTG2 and the WRKY18 DNA binding domain (data not shown) to the W1-box was weaker than to the W2-box, our Luc-DPI studies revealed an interaction. This view is supported by our finding that a double shifted band was observed in EMSA studies that can be interpreted as the binding of two proteins to the DNA fragment. The upper band was always weaker, suggesting a weaker binding of the second TTG2 protein.
GUS reporter studies support the importance of both WRKY binding sites because just one of the two WRKY binding sites, W1 or W2, is not sufficient for the correct expression of TRY in trichomes and the basal leaf part. Similar as reported for the Pc-WRKY1 promoter (Eulgem et al., 1999), the tandem arrangement of W-boxes in the TRY promoter could lead to a synergistic effect. Consistent with this hypothesis, the single W-box in the CPC promoter is not sufficient for drastic TTG2-dependent activation although TTG2 can bind to this W-box in yeast one-hybrid assays.
Simultaneous binding of TTG2 proteins to the two W-boxes in the TRY promoter could be additionally strengthened by potential homodimerization of the protein. It is possible that TTG2 proteins interact when binding to the two W-boxes whereby the protein-DNA interaction is reinforced. These bidirectional interaction possibilities could in turn cause the multi-TTG2/DNA aggregates in EMSA studies when using TTG2 instead of the WRKY18 DNA binding domain alone. TTG2 itself contains two WRKY domains and belongs to group1 of the WRKY family (Brand et al., 2013). Studies with the ELISA-DPI technique showed that generally both domains of WRKY33, another group 1 member, could bind DNA. Nevertheless the binding of the cDBD (C-terminal domain) is stronger than of the nDBD (Brand et al., 2013). Whether both WRKY domains of TTG2 can bind to the two W-boxes remains to be investigated.
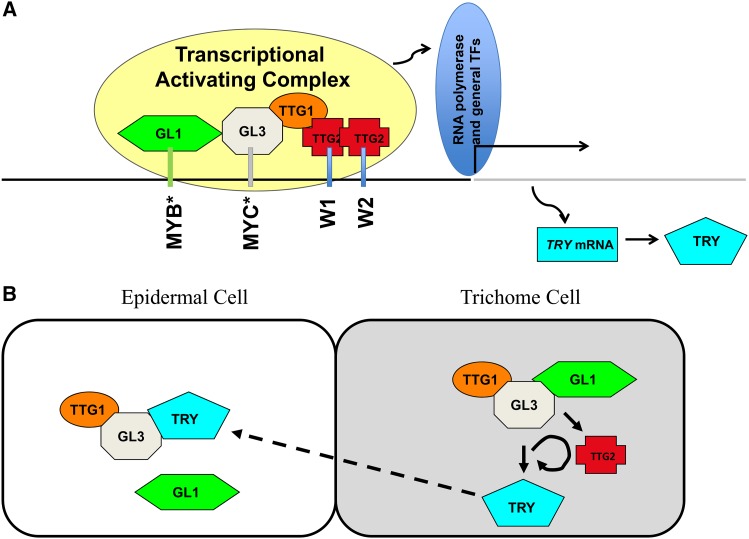
In light of these protein/DNA binding studies, the results are surprising as TTG2 alone cannot activate any of the tested promoters but rather drastically enhances the activity of TTG1, GL3, and GL1 (Figure 11A). As TTG2 interacts with TTG1 and indirectly through TTG1 also with GL3 in yeast four-hybrid assays, we propose that TTG2 forms a protein complex, possibly including a TTG2 dimer, with the MBW proteins for the transcriptional activation of TRY. As the W-boxes are essential, it is conceivable that TTG2 facilitates the targeting of the MBW complex to the correct promoter sequence. It is well possible that a similar mechanism operates on the TT8 promoter (Xu et al., 2013). Here, it was shown that TTG1, MYB5/TT2, and TT8 activate the expression of TT8 in the endothelium. The finding that endothelium expression of TT8 is absent in ttg2 mutants could be explained by an enhancement of the MBW complex similar to that observed for the TRY promoter.
Figure 11.
Model of TTG2 Function
(A) Postulated binding and interaction scheme on the TRY minimal promoter. Black line depicts the minimal TRY-promoter region from −623 to −4. The gray line marks the coding region, including exons und introns. Asterisk indicates predicted MYB and MYC binding sites (Pesch and Hülskamp, 2011).
(B) Role of TTG2 in the regulatory scheme explaining trichome patterning. Arrows indicate activation and blunted bars repression.
[See online article for color version of this figure.]
Role of TTG2 in the Trichome Patterning Gene Network
TTG2 appears to be intimately integrated into the gene regulatory network underlying trichome patterning as it is not only regulated by the trichome patterning genes but also is itself a strong regulator of at least one patterning gene, namely, TRY (Figure 11B). The regulation of TTG2 expression by trichome patterning genes is well documented by showing that ProTTG2:GUS expression is changed in patterning mutants and overexpression lines (Ishida et al., 2007). In support of this, chromatin immunoprecipitation experiments revealed binding of GL1 and GL3 to a proximal TTG2 promoter fragment (Morohashi and Grotewold, 2009). The regulation of patterning genes by TTG2 appears to be quite specific for TRY. On the one hand, TTG2 is absolutely required for the activation of TRY expression in young leaves and ttg2 mutants show a cluster frequency that is similar to that in try mutants. On the other hand, mutations in TTG2 have no strong effect on the expression of CPC or GL2 (Ishida et al., 2007; this article), two other direct target genes of the patterning machinery. Nevertheless, TTG2 moderately regulates the expression of CPC and GL2 as their expression is reduced in the ttg2 mutant background (this article) and in pTTG2:TTG2:SRDX lines (Ishida et al., 2007). In addition, it is possible that TTG2 is involved in its own regulation as pTTG2:TTG2:SRDX lines also show reduced endogenous expression of TTG2, and TTG2 can bind to its own promoter as demonstrated by chromatin immunoprecipitation experiments (Morohashi and Grotewold, 2009).
One important consequence of the current theoretical models is that the feedback loop between TRY and its activators is regulated by TTG2. The activation of TRY by proteins of the MBW complex (Digiuni et al., 2008; Pesch and Hülskamp, 2011) is an essential feature in all models explaining trichome patterning (Pesch and Hülskamp, 2009; Balkunde et al., 2010; Tominaga-Wada et al., 2011; Grebe, 2012). Our data suggest that TTG2 is required for this step by enhancing the MBW-dependent activation of TRY (Figure 11B). What are the consequences when also taking into account that TTG2 itself is regulated by the MBW proteins? Formally, this regulation scheme establishes a nonlinear activation of TRY by the MBW complex such that any level of TTG2 activation translates into a 10-fold higher (according to our cell culture experiments) activation of the TRY promoter. It is difficult to intuitively predict the consequences of this regulation event, but it is conceivable that this may be employed for the fine-tuning or robustness of the system or alternatively that this additional TTG2 loop can be used to link the patterning system to other regulatory inputs forwarded by TTG2.
The analysis of the global expression changes in ttg2 cpc and try cpc mutants revealed similar phenotypes, suggesting that TTG2 promotes TRY expression (Simon et al., 2013). Although less pronounced as found in our study for the trichome system, TRY expression in the root was reduced to 67% of the wild-type expression in ttg2 mutants (Simon et al., 2013). Thus, the role of TTG2 in enhancing the expression of TRY appears to also be relevant in the context of root hair patterning.
METHODS
Plant Lines and Growth Conditions
Plants were grown on soil at 24°C with 16 h daylight. Plant transformations were performed by the floral dip method (Clough and Bent, 1998). The ecotype Landsberg erecta (Ler) was used to produce the promoter GUS reporter lines of the TRY promoter and the try-Col line was used to analyze the rescue efficiency of the TRY promoter lines. The ttg2 mutant was described before (Johnson et al., 2002). The TRY, CPC, and GL2 promoter GUS and GFP-ER reporter lines were created by transformation of either Ler or ttg2-1 mutant plants and analyzed in the F3 generation.
Plant Work
Constructs
The TTG2 CDS, TTG2 (Δ1-80), GL3(Δ1-400), MYB23 CDS, and WER(Δ180-203) CDS were amplified with primers including attB attachment sites from Ler cDNA and cloned in pDONR201 through BP reaction (Supplemental Figure 9). TTG2D (Δ1-158, Δ409-430) containing just the coding regions for the TTG2 WRKY domains with an artificial Stop codon was amplified via PCR. GL2 CDS, GL1 CDS, and WER CDS were PCR amplified subcloned and cloned via EcoRI and NotI, SalI and EcoRI, and SalI and NotI in pENTR1A, respectively. The GUS reporter constructs 5′-GL2:GUS-pAMPAT, 5′-TRY:GUS-pAMPAT, and 5′-CPC:GUS-pAMPAT, pTRY (A3;B) were described before (Pesch and Hülskamp, 2011). The promoter GFP-ER reporter lines were created by LR recombination of 5′-TRY-pAMPAT, 5′-GL2-pAMPAT, and 5′-CPC-pAMPAT with GFP-ER-pDONR201 (Weinl et al., 2005; Wester et al., 2009). The mutated fragments of pTRY (A3) were created by PCR. The pTRY-A3(mW1), pTRY-A3(mW2), and pTRY-A3 (mW1mW2) promoter fragments were produced by combining the respective distal and proximal fragments by fusion PCR followed by BP recombination in pDONR201 and LR recombination with PARB-B. pTRY-A3-B was amplified by PCR and the substituted fragment pTRY-A3-B (mW1mW2) was created by fusing the fragment pTRY (B) to pTRY -A3(mW1mW2) by fusion PCR. The PCR products were recombined in pDONR201, sequenced, and recombined with PARB-TRY-CDS (Pesch and Hülskamp, 2011). pTRY-A3-B-pDONR201 was also recombined in PARB (Supplemental Figure 9). Between 20 and 40 T1 plants were selected, and the GUS staining pattern was analyzed in the next generation after 10 d (Pesch and Hülskamp, 2011) and classified into “basal and trichome” and “only trichome” staining. Cluster frequency in rescue experiments was determined on 50 independent T2 lines.
Light microscopy was done with a Leica MZ16F binocular using the Leica Application Suite Version 3.7.0 or with a Leica DMRE microscope. Image processing was done with Photoshop Elements 7.0. Confocal laser scanning microscopy was performed on the Leica DMRE fluorescence microscope equipped with a TCS-SP2 imaging system (Leica Microsystems). Scanning electron microscopy was done as described previously (Pesch et al., 2013).
Yeast One-Hybrid Assay
The yeast one-hybrid assay was done according to the Matchmaker One-Hybrid System User Manual (Clontech). To create the bait vectors W1W2-pHISi and mW1mW2-pHISi, the promoter regions containing the W1W2 and mW1mW2 were amplified via PCR with different forward and reverse primers attached with EcoRI and KpnI, KpnI and BamHI, and BamHI and XbaI sites, respectively, using pTRY (A3) and pTRY (A3, mW1mW2) as templates. The respective three different fragments were sequentially subcloned in puc18, sequenced, and ligated in pHISi (Invitrogen) via the restriction sites EcoRI and XbaI. All prey vectors based on LR recombination with the destination vector pC-ACT2. TTG2- and EGL3-pC-ACT2 were produced through LR recombination, and all others were already described (Pesch and Hülskamp, 2011). The Saccharomyces cerevisiae strain YM4271 was transformed in two steps first with the bait and second with the different prey vectors through the LiAc transformation method (Gietz et al., 1995) and selected on the respective selective dropout media (SD) lacking uracil or uracil and leucine. Single colonies were tested for interaction on SD medium lacking leucine, uracil, and histidine after 4 d at 37°C. Each interaction was tested four times in parallel in three independent experiments.
Yeast Two-, Three-, and Four-Hybrid Assays
Bait and prey vectors with the respective CDSs were fused to the GAL4 binding domain in pAS2.1 or to GAL4-AD in pC-ACT2 by LR reaction. Transformations were done into the S. cerevisiae strain AH109 (Gietz et al., 1995), selection was performed on synthetic dropout medium (lacking leucine and tryptophan [SD-LW]), and interactions of single colonies were assayed on synthetic dropout interaction medium (lacking leucine, tryptophan, and histidine [SD-LWH] supplemented with 10 or 15 mM 3-aminotriazole] after 3 d at 37°C. Each interaction was monitored three times in parallel in two independent experiments.
For yeast four-hybrid assays, the following constructs were used: pC-ACT2 was recombined with GL3-pDONR201 to fuse Gal4-AD to GL3. The pBRIDGE vector was recombined with pENTR1A-w/o-ccdB, TTG1-pENTR4, and TTG1(Δ316-341)-pENTR4 (Bouyer et al., 2008; Balkunde et al., 2011; Pesch et al., 2013). A HpaI-PstI fragment was removed from pAS-TTG2 and inserted through these restriction sites in pBRIDGE*, pBRIDGE-TTG1, and pBRIDGE-TTG1(Δ316-341). First, the expression of the Gal4 binding domain TTG2 fusion is enabled. Second, the construct pBRIDGE allows the expression of TTG1, TTG1(Δ316-341), or no protein (* empty control fragment) without any fusion under repression by methionine. The third construct, pYEA-GW, was created through cloning the Gateway recombination cassette from pBS-RekA as EcoRV fragment in pYEA after restriction with EcoRI followed by blunting (Pesch and Hülskamp, 2011). In a next step, GL1-pENTR1A, GL1(Δ202-228)-pENTR4, GL1-R97D-pENTR1A, and GL1-R97D-(Δ202-228)-pENTR4 were recombined with pYEA-GW to express the fourth protein without fusion.
The selection was done on dropout medium lacking leucine (selection of pC-ACT2), tryptophan (selection of pBRIDGE-GW), methionine (expression of the unfused protein in pBRIDGE-GW), and adenine (selection of pYEA-GW). Single colonies were plated on interaction medium, which in addition did not contain histidine and was supplemented with 5 mM 3-aminotriazole. Each interaction was monitored two times in parallel in two independent experiments.
Coimmunoprecipitation (GST Pull-Down)
The pGEX2TMGW constructs were created by LR reactions using the TTG1-pENTR4, TTG2-oST-pDONR201 (oST abbreviation for CDS without the stop codon), and pENTR1a-ccdB to express an N-terminal GST fusion of the TTG1protein, an N-terminal GST/C-terminal His fusion of the TTG2 protein, and the GST protein without fusion. pDEST17 was recombined with TTG1-pENTR4 and TTG2-pDONR201 to express N-terminal His-tagged TTG1 and TTG2.
Bacterial cells Bl21DE3RIL (Stratagene) containing the pGEX2TMGW constructs including the TTG1 or TTG2 CDS sequences were grown until an OD600 of 0.5 to 0.8, induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), grown for 4 h, harvested by centrifugation at 7000g for 7 min at 4°C, and lysed (Frangioni and Neel, 1993). Proteins were purified using glutathione sepharose (GE Healthcare) by the batch method (Sambrook and Russell, 2001). The purified GST-tagged proteins were dialyzed against GST binding buffer (50 mM Tris, pH 7.9, 1 mM EDTA, and150 mM NaCl) in Spectra/Por dialysis membranes of MWCO 3,5 (SpectrumLabs). After dialysis in Non-Idet P-40, glycerol and BSA were added to the protein solution to a final concentration of 0.1, 0.1, and 2%. Protein amounts were determined by comparison to BSA standard run on SDS-PAGE, and protein concentrations were measured using the Bradford Reagent from Bio-Rad as explained in the user manual for the reagent.
His-tagged proteins were expressed in Escherichia coli BL21DE3Codon plus (RIL) (Stratagene) containing the IPTG-inducible N-terminal His fusion constructs were grown until an OD600 of 0.5 to 0.8, induced by 0.5 mM IPTG for 3 h, and harvested by centrifugation at 7000g for 7 min at 4°C. Pellets from 50 mL cells were resuspended in 1 mL Tris-based lysis buffer (100 mM NaCl, 50 mM Tris, pH 7.5, 2 mM EDTA,1% Triton X-100, and 200 µg lysozyme [Roth]) and incubated at room temperature for 70 min, sonicated five times for 10 s, and precipitated (the supernatant was kept and run on gel to check if the proteins are in this soluble fraction). Pellets were resuspended two times more in 500 μL of Tris-based lysis buffer, sonicated, and centrifuged as described before. The pellet was resuspended in 250 μL of inclusion body prep solution (100 mM NaHCO3 and 2% SDS, pH 9.1) by sonication until the pellet disappeared and precipitated at 4°C (the prep was analyzed by an SDS- polyacrylamide gel). The supernatant (250 μL) containing the solubilized inclusion bodies and proteins were incubated with 500 μL of Ni-NTA resin in a column for 1 h. The column was washed six times with PBS buffer volume respective to the column volume (137 mM NaCl, 2.7 mM KCl,10 mM Na2HPO4, and 2 mM KH2PO4). Proteins were eluted in 10 fractions each with 150 μL elution buffer (PBS, pH 8, and 20 mM EDTA).
Equal amounts of ∼5 µg of GST-tagged proteins were mixed with His-tagged proteins and incubated for 0.5 h at 4°C on a rotating platform. Then, 100 μL of GSH resin (50% resin and 50% GST binding buffer) was added and the protein resin mixture was further incubated for 0.5 h at 4°C. Resins were washed with 1 mL binding buffer 10 times. Next, 12.5 μL of 5× SDS gel loading buffer (50 mM Tris, pH 6.8, 2% SDS, and 0.1% bromophenol blue) was added and boiled at 95°C for 5 min. Immunoblots were analyzed using anti-His antibodies.
DPI Studies
To fuse the Renilla reniformis luciferase N terminus to TTG2 or to the WRKY-domains of TTG2 (TTG2D), the destination vector pcDNA3-Rluc-GW (Blasche and Koegl, 2013) was recombined by LR reaction with the respective entry clones or with pENTR1A-w/o-ccdB as negative control. Single DNAs were used as complementary strands.
Proteins were transiently coexpressed in HEK293TN cells (BioCat/SBI: LV900A-1) as fusion proteins with the R. reniformis luciferase tag fused to their N termini.
Constructs (9 µg plasmid DNA) were cotransfected into 5 × 106 HEK293TN cells in 10-cm plates using 20 μL of lipofectamine 2000 (Invitrogen). After 48 h, cells were collected in a 15-mL falcon tube and washed two times with 1× PBS by centrifugation (600g, 4°C) and afterwards lysed in 200 μL of ice-cold lysis buffer (20 mM Tris, pH 7.6, 100 mM KCl, and 10 mM DTT) and Protease Inhibitor Cocktail without EDTA (dilution of the stock solution 1:22) with 10 to 15 glass beads (2.5 mm, Biospec No. 11079125) by rigorous shaking for 20 min at 4°C. Subsequently 300 μL lysis buffer was added and lysates were centrifuged (15,000 rpm, 4°C, 10 min). The luciferase activity was measured by the relative light units (RLUs) in white 96-well luminescence plates (Nunc No. 267350) by automatic injection of the assay buffer (1.1 M NaCl, 2.2 mM EDTA, 200 mM K2HPO4/KH2PO4, pH 5.1, 0.44 mg/mL BSA, and 2.5 μM coelenterazine) by a microtiterplate reader (BMG Fluostar Optima). Ten microliters of the lysate with 40 μL PBS was analyzed. The lysate was diluted to obtain an activity of 3000 to 6000 RLUs/10 µL.
To create the short double-stranded DNA fragments, complementary single-stranded DNAs without modification and in addition the forward oligomer with 5′-biotinylation were used. To produce the double-stranded oligomers (ds-bio DNA or ds DNA: W1W2, mW1mW2, mW1W2, W1, mW1, W2, and mW2), complementary single-stranded DNAs (Supplemental Figure 9) were annealed as described previously (Brand et al., 2010).
Sixty microliters (2 pmol) of double-stranded bio DNA in TBST was incubated in Reacti-Bind Streptavidin Coated 96-Well Plates (Pierce Biotechnology) for 2 h at 37°C, washed three times with 300 μL TBST, blocked with 150 μL of α-HIS:HRP antibody blocking reagent (Qiagen) for 30 min, and washed three times with 300 μL TBST. Sixty microliters of the respective luciferase fused protein sample was added and incubated for 1 h at 20°C. The protein solution was removed and washed three times with TBST and four times with PBS. Each well was filled with 50 μL PBS and the luciferase activity (RLU) was determined (Pesch et al., 2013). Competition by nonbiotinylated double-stranded DNAs (wild-type or mutated versions) was achieved by mixing the protein solution with the double-stranded DNA and incubating for 1 h prior to the start of the experiment (2 μL of 1, 5, 12.5, and 50 μM stock solutions).
To determine the full size of the expressed Renilla luciferase fused proteins through immunoblot analysis, transfected HEK293TN cells of a 10-cm dish were lysed with 800 lysis buffer and boiled with 450 μL cracking buffer for 10 min (Bouyer et al., 2008). Fifty microliters of the protein samples was loaded on a SDS gel and afterwards transferred by wet blotting on a nitrocellulose membrane for 16 h at 3°C and 30 V. Immunoreaction was performed using a primary anti-Renilla luciferase antibody (clone 5B11.2; Millipore) and a second anti-mouse-HRP-antibody and detected by chemiluminescence detection with Super Signal West Femto Maximun Sensitivity Substrate (Thermo Scientific) by a luminescent image analyzer (LAS-4000 mini; Fujifilm). Sizes were compared using a size marker (Thermo Scientific PageRuler Prestained Protein Ladder 10-170 kD).
Renilla Luciferase Activity Measurement
Transfected Arabidopsis thaliana cell suspension culture cells, which should express Renilla luciferase, were tested. Five hundred microliters of the suspension were collected by centrifugation at 10 krpm for 5 min and afterwards lysed with lysis buffer (22 mM Tris, pH 7.6, 100 mM KCl, 10 mM DTT, and protease inhibitor) and glass beads and vigorous shaken at 1400 rpm. Protein crude extracts were generated by centrifugation at 13 krpm at 4°C for 20 min. Ten microliters of the lysate was used for luciferase activity measurement (see DPI section).
EMSA
TTG2-pDONR201 and TTG2D-pDONR201 were recombined by LR reaction in pet28a-FrameC to produce an N-terminal His-tag. The WRKY domain of WRKY18 (V164-T244, defined as W18D) was amplified by PCR using cDNA as template and primers attached with SacII and KpnI restriction sites and cloned through these restriction sites in pet11a-His (Birkenbihl et al., 2005).
TTG2 and TTG2D protein expression vectors were transformed in E. coli BL21DE3Codon plus (RIL) (Stratagene) cells and incubated at 37°C in 400 mL Luria-Bertani culture medium supplemented with ampicillin until reaching an adsorbance at 600 nm of 0.8. Cells were induced with 1 mM IPTG overnight at 14°C and collected by centrifugation and frozen in liquid nitrogen. The pellet was solved in lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 10% glycerol, supplemented with Protease Inhibitor Cocktail with EDTA [Roche]) and sonicated. After centrifugation, the supernatant was mixed with prewashed Ni-NTA matrix (Qiagen; as washing buffer serves lysis buffer supplemented with 10 mM Imidazol) and incubated for 1 h. The mixture was washed four times with the washing buffer. Elution was performed with elution buffer (lysis buffer containing 250 mM Imidazol). Protein expression and protein size were checked by a 12% SDS-PAGE and by a protein gel blot with anti-His-antibody. The protein solutions were supplemented with ZnSO4 to a final concentration of 1 mM prior usage in the EMSA studies.
Double-stranded DNA was generated using oligonucleotides (Supplemental Figure 9). Labeling was performed with T4 Polynucleotid Kinase using 10 pmol of single-strand oligonucleotides and freshly ordered 32P-ATP in a 20 μL assay. The 5′-32P-end-labeled upper strands were purified and dissolved in 50 μL TE containing 10 mM MgCl2 and 15 pmol of the respective lower strands and slowly hybridized at 95°C. One microliter of radioactively labeled DNA substrate (∼10,000 counts) was incubated with100 ng/μL of the respective protein or with a dilution to 10 ng/μL (diluted with: 100 mM NaCl, 50 mM Tris, pH 8, 0.1% β-mercaptoethanol, 10% glycerol, 0.1 mM EDTA, and 1 mM ZnSO4), 1 μL of short sequence non binding DNA (m19 [Ciolkowski et al., 2008] and 10 µM), 5 μL sample buffer (100 mM Tris, pH 8, 10 mM EDTA, and 2 mM DTT) and 3 μL loading buffer (40 mM Tris, pH 8, 4 mM EDTA, 25% glycerol, 0.05% bromophenol blue, an 400 µg/mL BSA) to a final volume of 13 µL. In competition experiments, 1 μL of unlabeled (cold) competitor was added. The mixture was incubated for 15 min at room temperature. Samples were loaded on a native 8% (w/v) polyacrylamide gel (40%, 29:1) and run at room temperature at 7 V/cm in 0.5× TBE buffer. Radioactively labeled bands in the gel were visualized on a phosphor imager (FLA-7000; Fujifilm).
RNA Extraction and Real-Time PCR
Ler or ttg2 plants were grown at 23°C under long-day conditions. Forty leaves numbered 3 and 4 of 10-d-old plants were collected and frozen in liquid nitrogen. Total RNA was extracted using the RNeasy Mini Kit (Qiagen). RNA was treated with DNaseI (Thermo Scientific) and quantified by a NanoDrop (ND-XY) spectrophotometer. Five hundred nanograms of RNA was reverse-transcribed (SuperScriptIII; Invitrogen). Quantitative PCRs were performed on an Applied Biosystems 7300 real-time PCR system using POWER SYBR Green PCR-Master Mix (Applied Biosystems). Two biological replicates, each including three technical replicas, were measured. Transcript levels were normalized to 18S rRNA. Primers were used as published before: CPC, GL2 (Morohashi et al., 2007), and TRY (Zhao et al., 2008). Relative RNA levels were calculated according to the comparative Ct(2–ΔΔCt) method.
Cotransfection Assays in Arabidopsis Cell Suspension Culture and GUS Activity Assay
The promoter fragment pTRY from −623 to −4 (containing the upstream region of the CDS used previously in a genomic rescue experiment; Pesch and Hülskamp, 2011) was amplified from genomic DNA of Arabidopsis Ler ecotype and recombined in pDONR201 via BP reaction (Invitrogen). pCPC-pDONR201 from −686 to −158 (containing the 525 bp upstream region of the CDS used previously in a genomic rescue experiment; Wada et al., 1997) was generated in the same manner by BP reaction. The promoter fragment pGL2 from −2132 to −28 bp (Weinl et al., 2005) was amplified with Acc65I and NotI restriction sites and cloned in pENTR1A. To drive expression of the GUS gene under the control of the GL2, CPC, or TRY promoter, the binary plant transformation vector pGWB3i containing an intron within the GUS gene was used (Gigolashvili et al., 2007). Using the LR reaction (Invitrogen), pTRY-pDONR201, pCPC-pDONR201, and pGL2-pENTR1A were recombined with pGWB3i to create pTRY:GUS, pCPC:GUS and pGL2:GUS. The effector constructs (35S:GL3, 35S:TTG1, 35S:GL1, and 35S:TTG2) were also created by LR recombinations of the respective entry clones with pGWB2. For the empty control construct without CDS, pGWB2 was recombined with pENTR1A-w/o-ccdB (Pesch and Hülskamp, 2011). TTG2-pDONR201, TTG2D-pDONR201, and pENTR1a-w/o-ccdB were also recombined with pMDC32-Renilla-attR, which was created by amplifying the Renilla CDS with KpnI and AscI attachments by PCR using pcDNA3-Rluc-GW as template and subsequent cloning in pMDC32.
To quantitatively compare the transcriptional activation of the promoters by different sets of transcription factors, dark-grown Arabidopsis cell suspension cultures were transfected by different combinations of the effector (35S-promoter with the different CDSs in pGWB2) and reporter constructs (pGL2:GUS, pCPC:GUS, or pTRY:GUS in pGWB3i) using the supervirulent Agrobacterium tumefaciens strain LBA4404.pBBR1MCS.virGN54D. To guarantee comparability in one set of experiments several precautions were taken: (1) All Agrobacterium cultures were grown under exactly the same conditions. (2) A combination of the different Agrobacterium cultures for transfection were mixed in advance and then added to each of the three replicates of the cell culture samples. (3) The same amount of Agrobacterium cultures was always used in one set of experiments for transfection. This was done by adding Agrobacterium cultures containing the pGWB2-ccdB-w/o construct without any CDS instead of the respective effectors. The different supervirulent Agrobacterium cultures, in addition to the Agrobacterium strain RK19 containing the antisilencing 19 K protein (Voinnet et al., 1999), were taken from fresh YEB plates, grown overnight in liquid medium with antibiotics, sedimented, and resuspended (at a concentration of 3:1) in cell suspension culture medium (4.3 g/L MS basal salt medium [Sigma-Aldrich], 4 mL vitamin B5 mixture [Sigma-Aldrich], 30 g/L sucrose, pH 5.8, and 1 mg/L 2,4- D). To each transfection sample of 3 mL freshly diluted cell suspension culture, 25 μL of each resuspended Agrobacterium culture was finally added.
After incubating the cell cultures in six-well plates at 22°C and 120 rpm in the dark for 5 d, the cells were harvested by centrifugation, the pellets were frozen in liquid nitrogen, and protein crude extracts were prepared by homogenizing the cell pellets in 500 μL protein extraction buffer (50 mM NaH2PO4/Na2HPO4, pH 7.2, 1 mM EDTA, and 0.1% [v/v] Triton X-100) with glass beads (1.8 to 2.2 mm; Roth) through vigorous shaking at 1400 rpm at 4°C for 20 min. Afterwards the supernatant was cleared by centrifugation (12,000 rpm, 4°C, 20 min).
The crude extracts were diluted 1:10 with the extraction buffer and the protein concentrations were determined according to the manufacturer’s microplate protocol (BCA Protein Assay Kit; Pierce) using sample duplicates. A standard curve for protein concentration was determined using BSA as substrate. GUS activity was determined by measuring the turnover of the 4-methylumbelliferyl β-d-glucuronide substrate for each sample using sample duplicates. For each sample, 200 μL of substrate buffer (1 mM 4-methylumbelliferyl β-d-glucuronide substrate in protein extraction buffer) was mixed with 25 μL of 1:10 diluted protein crude extract and incubated at 37°C in the dark. 4-Methylumbelliferone (4-MU) production was determined fluorometrically (excitation 365 nm; emission 455 nm) for 4 h every 15 min using the Tecan Infinite 200 Titerplate reader and Tecan i-Control 1.4.5.0 software. Different concentrations of 4-MU solved in the extraction buffer were used to generate a standard curve. Finally, the 4-MU production per min was correlated to the total protein amount of the sample. Mean values and standard deviations of the three parallel samples were calculated.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TTG1 (At2g37260), CPC (At2g46410), and TRY (At5g53200).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TRY but Not GL2 or CPC Expression Depends on TTG2.
Supplemental Figure 2. Analysis of TRY, CPC, and GL2 Expression by Real-Time PCR.
Supplemental Figure 3. Expression Analysis of TRY Promoter Fragments Carrying Mutations in W-Boxes.
Supplemental Figure 4. Sequences of Wild-Type and Mutated W-boxes of the TRY Promoter.
Supplemental Figure 5. Integrity and Functionality of Luc-TTG2 and Luc-TTG2D Fusion Proteins.
Supplemental Figure 6. Expression Levels of Promoter:GUS Constructs.
Supplemental Figure 7. Yeast Two-Hybrid Interactions of TTG2-BD with Other Trichome Patterning Proteins.
Supplemental Figure 8. Yeast Two-Hybrid Interactions of TTG2-AD with Other Trichome Patterning Proteins.
Supplemental Figure 9. Primer List.
Supplemental Data Set 1. Analysis of Putative cis-Elements with PLACE Web Signal Scan.
Supplementary Material
Acknowledgments
We thank B. Kernebeck and I. Klinkhammer for excellent technical assistance, H.-P. Bollhagen for help with the scanning electron microscopy, D. Wanke and L. Brand for help with establishing the DPI method, Alex Steffens for help with the quantitative PCR experiments, D. Wanke for thoughtful discussion on WRKY transcription factor structure and functions, and S. Schellmann for critically reading the article. The following materials were kindly provided: pAMPAT-GW from B. Ülker (MPIZ, Cologne), pGWB2 and pGWB3 from T. Nakagawa (Shimane University, Japan), pGWB3i vector from B. Berger and T. Gigolashvili (Botanical Institute of Cologne), pBluescript-GW-RekA from S. Biere (MPIZ, Cologne), TRY, CPC, GL3, TTG1, and entry clones from U. Herrmann, and the pENTR1A-w/o-ccdB from Arp Schnittger. LBA4404.pBBR1MCS.virGN54D was kindly provided by J. Memelink (University of Leiden, The Netherlands) through T. Gigolashvili.
AUTHOR CONTRIBUTIONS
M.P. performed the research, designed the research. B.D. and R.B. performed the research. I.E.S. designed the research. M.H. designed the research and wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- CDS
coding sequence
- DPI
DNA protein interaction
- EMSA
electromobility shift assay
- Ler
Landsberg erecta
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- RLU
relative light unit
- 4-MU
4-methylumbelliferone
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Balkunde R., Pesch M., Hülskamp M. (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 91: 299–321. [DOI] [PubMed] [Google Scholar]
- Balkunde R., Bouyer D., Hülskamp M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048. [DOI] [PubMed] [Google Scholar]
- Benítez M., Espinosa-Soto C., Padilla-Longoria P., Alvarez-Buylla E.R. (2008). Interlinked nonlinear subnetworks underlie the formation of robust cellular patterns in Arabidopsis epidermis: a dynamic spatial model. BMC Syst. Biol. 2: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez M., Espinosa-Soto C., Padilla-Longoria P., Díaz J., Alvarez-Buylla E.R. (2007). Equivalent genetic regulatory networks in different contexts recover contrasting spatial cell patterns that resemble those in Arabidopsis root and leaf epidermis: a dynamic model. Int. J. Dev. Biol. 51: 139–155. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596. [DOI] [PubMed] [Google Scholar]
- Blasche S., Koegl M. (2013). Analysis of protein-protein interactions using LUMIER assays. Methods Mol. Biol. 1064: 17–27. [DOI] [PubMed] [Google Scholar]
- Bouyer D., Geier F., Kragler F., Schnittger A., Pesch M., Wester K., Balkunde R., Timmer J., Fleck C., Hülskamp M. (2008). Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L.H., Fischer N.M., Harter K., Kohlbacher O., Wanke D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41: 9764–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L.H., Kirchler T., Hummel S., Chaban C., Wanke D. (2010). DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Yang Y., Zhou Y., Zhou J., Fan B., Yu J.Q., Chen Z. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6: 287–300. [DOI] [PubMed] [Google Scholar]
- Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Pater S., Greco V., Pham K., Memelink J., Kijne J. (1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24: 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiuni S., Schellmann S., Geier F., Greese B., Pesch M., Wester K., Dartan B., Mach V., Srinivas B.P., Timmer J., Fleck C., Hulskamp M. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch J.J., Chen M., Sanders M., Hillestad M., Ndkium S., Idelkope B., Neizer J., Marks M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Schmelzer E., Hahlbrock K., Somssich I.E. (1999). Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18: 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni J.V., Neel B.G. (1993). Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210: 179–187. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Gan L., Xia K., Chen J.G., Wang S. (2011). Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Gong X., Cao W., Zhao J., Fu L., Wang X., Schumaker K.S., Guo Y. (2008). SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J. Integr. Plant Biol. 50: 906–917. [DOI] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50: 886–901. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Mendenhall J., Huo Y., Lloyd A. (2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325: 412–421. [DOI] [PubMed] [Google Scholar]
- Grebe M. (2012). The patterning of epidermal hairs in Arabidopsis—updated. Curr. Opin. Plant Biol. 15: 31–37. [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misŕa S., Jürgens G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Ishida T., Hattori S., Sano R., Inoue K., Shirano Y., Hayashi H., Shibata D., Sato S., Kato T., Tabata S., Okada K., Wada T. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.S., Kolevski B., Smyth D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268: 506–513. [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55: 389–398. [DOI] [PubMed] [Google Scholar]
- Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485. [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1981). The complex syndrome of ttg mutants. Arabidopsis Information Service 18: 45–51. [Google Scholar]
- Koornneef M., Dellaert L.W.M., van der Veen J.H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. 93: 109–123. [DOI] [PubMed] [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Zhao M., Yang M., Read B., Lloyd A., Lamb R., Grotewold E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145: 736–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493. [DOI] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2009). One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12: 587–592. [DOI] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2011). Role of TRIPTYCHON in trichome patterning in Arabidopsis. BMC Plant Biol. 11: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Schultheiß I., Digiuni S., Uhrig J.F., Hülskamp M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140: 3456–3467. [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Macdonald H., Huttly A.K., Lazarus C.M., Hooley R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 29: 691–702. [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Torres J.T., Parniske M., Wernert P., Hahlbrock K, Somssich I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15: 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Bruex A., Kainkaryam R.M., Zheng X., Huang L., Woolf P.J., Schiefelbein J. (2013). Tissue-specific profiling reveals transcriptome alterations in Arabidopsis mutants lacking morphological phenotypes. Plant Cell 25: 3175–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Sano R., Inoue K., Okada K., Wada T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Ishida T., Wada T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286: 67–106. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto Y.M., Baulcombe D.C. (1999). Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96: 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chen J.G. (2008). Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 49: 1792–1804. [DOI] [PubMed] [Google Scholar]
- Wang S., Barron C., Schiefelbein J., Chen J.-G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185: 387–400. [DOI] [PubMed] [Google Scholar]
- Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J.G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kwak S.H., Zeng Q., Ellis B.E., Chen X.Y., Schiefelbein J., Chen J.G. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882. [DOI] [PubMed] [Google Scholar]
- Weinl C., Marquardt S., Kuijt S.J., Nowack M.K., Jakoby M.J., Hülskamp M., Schnittger A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K., Digiuni S., Geier F., Timmer J., Fleck C., Hülskamp M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496. [DOI] [PubMed] [Google Scholar]
- Xu W., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198: 59–70. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.