The Medicago truncatula Nod factor receptors LYK3 and NFP accumulate in a narrow zone of approximately two cell layers in the nodule apex, where they form heteromeric complexes. This accumulation at the border of the meristem and the infection zone controls bacterial release, and its spatial restriction most likely prevents defense-like responses.
Abstract
Rhizobial Nod factors are the key signaling molecules in the legume-rhizobium nodule symbiosis. In this study, the role of the Nod factor receptors NOD FACTOR PERCEPTION (NFP) and LYSIN MOTIF RECEPTOR-LIKE KINASE3 (LYK3) in establishing the symbiotic interface in root nodules was investigated. It was found that inside Medicago truncatula nodules, NFP and LYK3 localize at the cell periphery in a narrow zone of about two cell layers at the nodule apex. This restricted accumulation is narrower than the region of promoter activity/mRNA accumulation and might serve to prevent the induction of defense-like responses and/or to restrict the rhizobium release to precise cell layers. The distal cell layer where the receptors accumulate at the cell periphery is part of the meristem, and the proximal layer is part of the infection zone. In these layers, the receptors can most likely perceive the bacterial Nod factors to regulate the formation of symbiotic interface. Furthermore, our Förster resonance energy transfer-fluorescence lifetime imaging microscopy analysis indicates that NFP and LYK3 form heteromeric complexes at the cell periphery in M. truncatula nodules.
INTRODUCTION
Legumes have the unique ability to compensate for low levels of nitrogen in the soil by establishing an interaction with rhizobium bacteria. In this interaction, the bacteria are hosted intracellularly in a newly formed organ, the root nodule, where they reduce atmospheric nitrogen into ammonium (Stewart, 1966). The formation of these nodules is initiated upon the perception of signal molecules secreted by rhizobium: specific lipochitooligosaccharides named Nod factors (Lerouge et al., 1990). The role of Nod factor signaling in epidermal infection and the initial steps of nodule organogenesis is well studied (Geurts and Bisseling, 2002; Oldroyd, 2013). However, inside root nodules, rhizobia still produce Nod factors (Marie et al., 1994; Schlaman et al., 1998), but which process they induce and how they are perceived there is unknown.
In the root epidermis, Nod factors are recognized by specific receptors (Geurts et al., 2005). The model legumes Medicago truncatula and Lotus japonicus have two Nod factor receptors: the LysM domain receptor-like kinases NOD FACTOR PERCEPTION (NFP)/NFR5 and LYSIN MOTIF RECEPTOR-LIKE KINASE3 (LYK3)/NFR1 (Limpens et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Smit et al., 2007). In this study, we focus on M. truncatula. LYK3 has an active kinase domain allowing downstream signaling via phosphorylation, whereas an active kinase domain is missing in NFP (Mbengue et al., 2010; Klaus-Heisen et al., 2011). The interaction of these receptors has so far been studied only in the heterologous Nicotiana benthamiana system, where the expression of both L. japonicus or both M. truncatula Nod factor receptors, even in the absence of Nod factors, causes cell death (Madsen et al., 2011; Pietraszewska-Bogiel et al., 2013). In legumes, such cell death is not induced, but how this defense response is avoided is unknown. It has not been possible to study the interaction of the two receptors in legumes, as they could not be visualized.
Nod factor perception at the epidermis induces root hair curling and the formation of an infection thread (an invagination of the plant cell plasma membrane bound by a plant cell wall) in the curl. Simultaneously, cortical cells are mitotically activated, leading to the formation of a nodule primordium. The infection thread grows toward the nodule primordium, and there rhizobia are released into the cytoplasm of the primordium cells (D’Haeze and Holsters, 2002). In M. truncatula, a meristem is then formed at the apex of the primordium, which remains active throughout the lifespan of the indeterminate nodule (Vasse et al., 1990). At this stage, the infection threads grow toward this meristem, and cells derived from the meristem can be penetrated by the infection thread. These infection threads become cell wall-free regions (so-called unwalled droplets) from which bacteria are taken up by the host cell (Rae et al., 1992; Brewin, 2004). During this process, a membrane compartment is formed containing a host-derived membrane and a rhizobium (together named the symbiosome) (Roth and Stacey, 1989). This host membrane forms the symbiotic interface between rhizobia and the host cell. The nodule zone where rhizobia are released from the infection thread and subsequently divide and mature is named the infection zone (Vasse et al., 1990). Maturation of symbiosomes into the nitrogen-fixing state marks the start of the fixation zone. This zone contains cells fully packed with nitrogen-fixing symbiosomes interspersed with specialized uninfected cell types.
Nod factors are still produced by rhizobia in infection threads inside a nodule. However, upon the release of rhizobia, this production stops (Marie et al., 1994). Therefore, we hypothesize that Nod factor perception occurs only in nodule cells that become penetrated by an infection thread, and this triggers the release of rhizobia into the host cytoplasm. However, due to the indeterminate growth of the M. truncatula nodule, all developmental stages are present in a longitudinal section of such a nodule. Accordingly, transient accumulation should be detectable, although it might be restricted to a few cells.
In this study, we show that both M. truncatula Nod factor receptors accumulate in two cell layers that form the border between the meristem and the infection zone and that at least NFP is essential for bacterial release in the infection zone. Our successful visualization of the accumulation of both receptors in the homologous legume background allowed us to show that Nod factor receptor complexes are formed in cells where Nod factor signaling can occur. We further show that ectopic expression of NFP appears to trigger defense-like responses, which suggests that the transient accumulation of the Nod factor receptor(s) is instrumental in avoiding cell death during nodulation.
RESULTS
NFP and LYK3 Accumulate in M. truncatula Nodules
After showing that NFP and LYK3 fluorescent constructs under the control of their native promoters are biologically active by complementation of the corresponding mutant for nodulation (Supplemental Figure 1), we tested whether NFP and LYK3 fused with a fluorescent protein could be detected in nodules 10 to 14 d after inoculation (DAI). As fluorescence markedly decreases during chemical fixation and embedding, hand sections of the transgenic nodules were made and analyzed by confocal microscopy. In nodules expressing fluorescent fusions of LYK3 and NFP, fluorescence was detected in the apex (Figures 1A and 1E; Supplemental Figure 2). To distinguish the fluorescence of the LYK3/NFP fusions from autofluorescence, the fluorescence lifetime (τ) of cyan fluorescent LYK3-mTQ2 and NFP-mTQ2 fusions was determined and compared with the τ measurements of control nodules (expressing an empty vector or yellow fluorescent LYK3-Venus fusion). Fluorescence lifetime imaging microscopy (FLIM) data were subjected to phasor analysis (Supplemental Methods). In short, for each decay curve extracted from an individual region of interest (ROI), we calculated a complex phasor point. The real part and the imaginary part of the phasor points were plotted as a scatterplot together with the semicircle. Phasor points on the semicircle correspond to decay curves with monoexponential decay, and phasor points inside the semicircle correspond to curves with multiexponential decay. Only in nodules expressing LYK3-mTQ2 or NFP-mTQ2 did we observe cyan fluorescence at the cell periphery (Figure 2A, left panel); it was monoexponential and equal to 3.96 ns (confidence interval, 3.93 to 3.98; number of ROIs analyzed [n] = 26 from 11 images) or 3.91 ns (3.87 to 3.94; n = 7 from 6 images), respectively (Figures 2B, left panel, and 2C). These values correlate well with the mTQ2 τ measured in membranes (3.95 to 4.10 ns; Supplemental Methods). In control nodules (expressing an empty vector or LYK3-Venus), cyan autofluorescence was observed in the vacuole lumen but not at the cell periphery (Figure 2A, right panel); it was not monoexponential but was a mixture of at least two lifetimes, of which none was in the range of the mTQ2 τ measured at the periphery of nodule cells (Figure 2D). The average fluorescence lifetime (τav = Στifi, where f is the fraction of a given τ) there was 2.79 ns (2.73 to 2.85; n = 66 from 11 images).
Figure 1.
Localization of the Nod Factor Receptors in Nodules at 10 to 14 DAI.
(A) Typical M. truncatula nodule expressing pLYK3:LYK3-GFP. LYK3 accumulates at the cell periphery in two cell layers (L1 and L2) that form the border between the meristem (L1) and the infection zone (L2).
(B) LYK3-GFP-expressing cell penetrated by an infection thread. Released bacteria (in red) are indicated with arrowheads.
(C) Accumulation of LYK3-GFP at the infection thread membrane and the plasma membrane.
(D) A cell of the infection zone in which an infection thread penetrated and where LYK3-GFP accumulates only at the plasma membrane.
(E) Typical nodule expressing pNFP:NFP-GFP. NFP accumulates at the cell periphery in two cell layers (L1 and L2).
(F) A cell of the infection zone with NFP-GFP accumulation at the plasma membrane and (weaker) at the infection thread membrane.
M marks the position of the nodule meristem, and asterisks mark infection threads. Bars = 10 μm.
Figure 2.
Characterization of τ in Control, LYK3-mTQ2-Expressing, and LYK3-Venus-Expressing Nodules.
(A) Cyan fluorescence intensity image of a nodule transformed with pLYK3:LYK3-mTQ2 (left) or cyan autofluorescence intensity image of a nodule transformed with pLYK3:LYK3-Venus (right; merged with the yellow fluorescent protein intensity image to locate the expression of LYK3-Venus). The arrowhead indicates LYK3-mTQ2 localization at the plasma membrane in the L1/L2 cells. < L indicates cells distal to the L1/L2 cells, and > L indicates cells proximal to L1/L2 cells. Bars = 10 μm.
(B) Lifetime images of (A) based on phasor analysis. The intensity of false colors, representing the lifetimes, was adjusted to match the fluorescence intensity of the cyan (gray scale) fluorescence images.
(C) Phasor plot representing lifetime (phasor) values calculated for ROIs at the cell periphery in nodules transformed with pNFP:NFP-mTQ2 (yellow dots) or pLYK3:LYK3-mTQ2 (red dots). Globally fitted τD values (depicted as color-coded ticks) are represented on the semicircle. Lifetime (phasor) values of vacuolar cyan autofluorescence (in nodules transformed with an empty vector) are depicted as gray dots. Im, imaginary part; Re, real part.
(D) Phasor plot representing lifetime (phasor) values calculated for vacuoles in L1/L2 cells (= L), cells distal to L1 (< L), and/or cells proximal to L2 (> L) in nodules transformed with pNFP:NFP-mTQ2, pLYK3:LYK3-mTQ2, or pLYK3:LYK3-Venus.
Therefore, FLIM and green fluorescent protein (GFP) immunolocalization (Supplemental Figure 2A) show that the fluorescence detected at the cell periphery is specifically derived from the LYK3 and NFP fluorescent fusions.
Vacuolar Localization of the Receptors
Next to the accumulation at the cell periphery, we also detected fluorescence in the vacuole lumen in a broader zone than the L1 and L2 cell layers in nodules expressing NFP or LYK3 fluorescent fusion proteins. These were especially additional cell layers in the infection zone (proximal to the L2 layer), although in the case of NFP fluorescent fusions, accumulation in the vacuole lumen was sometimes also observed in cells distal to the L1 layer (Figure 3B). We assume that this localization represents breakdown of the receptors. Indeed, we noted a correlation between the τav of cyan fluorescence in the vacuole and the location of the cell in nodules expressing LYK3-mTQ2. The τav in vacuoles of the cells distal to the L1 layer was 3.04 ns (2.97 to 3.11; n = 33 from 15 images); in vacuoles of the cells in the L1/L2 layers and of the cells proximal to the L2 layer, the τav = 3.89 ns (3.86 to 3.92; n = 63 from 24 images and n = 25 from 13 images, respectively) (Figure 2D). Convergence of the latter τav values toward the monoexponential mTQ2 τ indicates that these vacuoles contain mTQ2. Similar τav values were measured in vacuoles of the cells in the L1/L2 layers and of the cells proximal to the L2 layer in nodules expressing NFP-mTQ2 (Figure 2D). By contrast, the τav in vacuoles of the cells distal to the L1 layer in nodules expressing LYK3-mTQ2 cluster in the vicinity of the τav from vacuoles of control nodules, indicating that they contain similar autofluorescence lifetimes.
Figure 3.
Colocalization of LYK3 and NFP Fluorescent Fusion Proteins at the Cell Periphery in a M. truncatula Nodule.
(A) to (E) Green and red fluorescence intensity images of LYK3-GFP ([A] and [C]) and NFP-mCherry ([B] and [D]) of nodule cells coexpressing pLYK3:LYK3-GFP and pNFP:NFP-mCherry. (E) shows an overlay of (A) and (B). L1 and L2 indicate the two layers with receptor accumulation at the cell periphery, M marks the position of the meristem, and the box in (E) marks the position of the crop represented in (C) and (D). Bars = 10 μm.
(F) and (G) Fluorescence intensity images of LYK3-mTQ2 (F) and NFP-Venus (G) of nodule cells coexpressing pLYK3:LYK3-mTQ2 and pNFP:NFP-Venus. Bars = 5 μm.
(H) Projection of a z-stack through nodule cells expressing LYK3-GFP. Bar = 5 μm.
NFP and LYK3 Accumulate at the Cell Periphery in Approximately Two Cell Layers at the Border of the Meristem and the Infection Zone
To determine in which cell layers NFP and LYK3 accumulate, median longitudinal sections were analyzed by confocal microscopy. In all nodules (n = 28) expressing LYK3-GFP, GFP fluorescence at the cell periphery was detected in a narrow zone at the apex that consisted of approximately two unordered cell layers (average, 2.2 ± 0.7 [sd]) (Figure 1A). Although these cells resulted from oblique cell divisions and do not form neat tiers, they can be recognized as layers. A similar accumulation in a narrow zone was observed for LYK3-mTQ2, LYK3-Venus, and a stable line expressing pLYK3:LYK3-GFP (Figure 3A; Supplemental Figures 2B and 2C).
The most distal layer where LYK3 accumulated at the cell periphery (layer L1) had no infection threads, and some cells had a morphology indicating that they just ended cytokinesis (Figure 1A). Therefore, we consider L1 to be the last layer of the meristem. In the more proximal layer where LYK3-GFP accumulated at the cell periphery (layer L2), the first infection events were observed. Therefore, L2 is the first layer of the infection zone. Sometimes an additional layer where LYK3 accumulated at the cell periphery occurred either in the meristem or the infection zone.
The accumulation of NFP-GFP in the nodule apex was studied in a similar manner in 10 nodules. NFP localization at the cell periphery was observed in a zone of approximately two cell layers (average, 2.2 ± 0.6) (Figure 1E; Supplemental Figure 2D). Based on cell morphology, these were most likely L1 and L2, which implied that NFP and LYK3 accumulate in the same cell layers. To test this, we introduced pNFP:NFP-mCherry by Agrobacterium rhizogenes root transformation in a stable line expressing pLYK3:LYK3-GFP or coexpressed pLYK3:LYK3-mTQ2 and pNFP:NFP-Venus in the nfp-1 lyk3-1 double mutant. Indeed, we observed colocalization of green and red or cyan and yellow fluorescence signals at the cell periphery in some cells of L1 and L2 layers (Figures 3A to 3G; Supplemental Figure 2E). Additionally, localization of NFP at the cell periphery and vacuole lumen prior to the accumulation of LYK3 was observed occasionally (Figure 3B).
In the case of LYK3 fluorescent fusions, accumulation in puncta of up to 500 nm in diameter was observed in addition to their more uniform localization at the cell periphery (Figure 3H). Because of the optical limitations of confocal imaging, we cannot conclude whether these are vesicles, curved (invaginated) membrane structures, or membrane domains. We could not observe the accumulation of NFP in puncta.
The occurrence of LYK3 and NFP at the cell periphery suggests that they are localized at the plasma membrane. To test this, the receptors were immunolocalized by electron microscopy and anti-GFP immunogold labeling. Indeed, in L1/L2 cells, LYK3-GFP was localized at (Figure 4A) and near (Figure 4B) the plasma membrane, which is consistent with our proposed subcellular localization. It is likely that (part of) the receptor localized near the membrane is present in vesicles. These vesicles are probably involved in endocytosis, leading to degradation of the receptor in the vacuole, but this needs to be demonstrated. Unfortunately, the NFP-GFP levels were too low to allow immunolocalization by electron microscopy.
Figure 4.
Subcellular Localization of LYK3-GFP in M. truncatula Nodules at 10 DAI.
(A) Transmission electron micrograph of LYK3-GFP labeled with anti-GFP antibody and secondary antibody coupled with 10-nm gold particles (indicated with arrowheads) at the plasma membrane of a pLYK3:LYK3-GFP-expressing nodule.
(B) Detail of an infection thread and unwalled droplet showing LYK3-GFP labeled with anti-GFP and gold particles at the unwalled droplet membrane/vesicles (indicated with arrowheads).
The asterisk indicates the plasma membrane. B, released bacteria; CW, cell wall; IT, infection thread. Bar in (A) = 100 nm; bar in (B) = 500 nm.
NFP and LYK3 Localize on Infection Threads before Rhizobia Are Released
In the L1/L2 zone, rhizobia are present intercellularly (in the apoplast) as well as inside the infection threads that have entered L2 cells. It is likely that at both sites rhizobia produce Nod factors, which implies that they may be perceived by receptors present at the plasma membrane and/or at the infection thread membrane. To test this, we analyzed confocal z-stacks of 20 L2 cells expressing LYK3-GFP in which an infection thread was present. In all 20 cells, LYK3-GFP was present at the cell periphery, whereas only in 7 cells was LYK3-GFP also detected on the infection thread (Figures 1B to 1D). Similarly, we studied the subcellular accumulation of NFP-GFP in 21 L2 cells. In all cells, NFP-GFP was detected at both the cell periphery and the infection thread (Figure 1F). Neither NFP nor LYK3 was ever observed on the symbiosome membranes.
We hypothesize that Nod factor signaling takes place at the plasma membrane as well as at the infection thread membrane. Upon release of the bacteria, NFP and LYK3 are removed from these membranes. The absence of LYK3 on most infection threads in L2 cells suggests that the accumulation of LYK3 is slightly more transient than that of NFP.
Oligomerization of LYK3 and NFP in M. truncatula Nodules
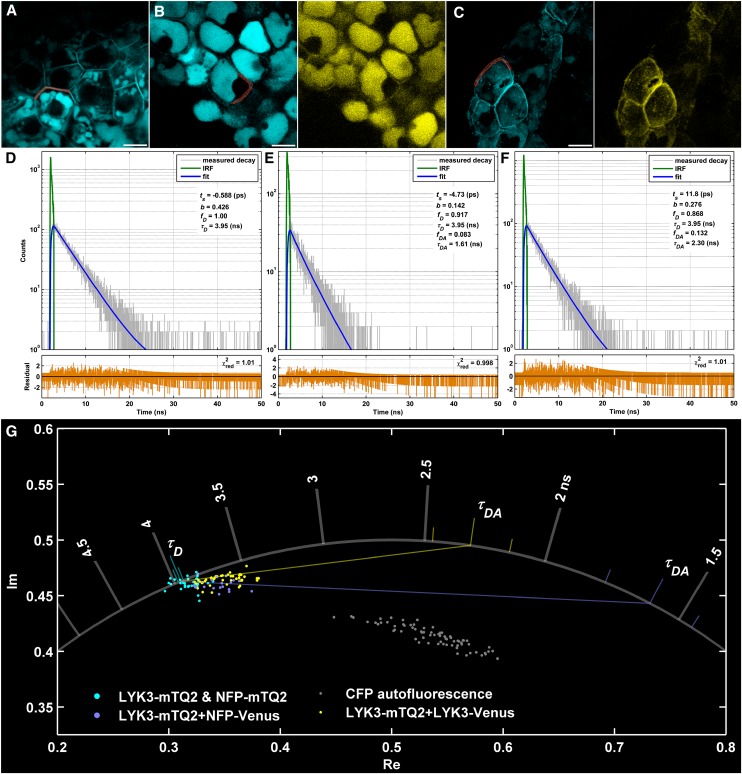
As LYK3 and NFP colocalize, it is possible that they also interact at the molecular level. To test this hypothesis, we cotransformed the lyk3-1 nfp-1 double mutant with pLYK3:LYK3-mTQ2 and pNFP:NFP-Venus and used a Förster resonance energy transfer (FRET)-FLIM technique. FRET caused by the close proximity (not farther apart than 10 nm) of the donor (mTQ2) to the acceptor (Venus), indicating interaction of the fusion proteins (Vogel et al., 2006; Pietraszewska-Bogiel and Gadella, 2011), results in the decrease in the donor fluorescence lifetime (τD). In a situation where equilibrium between interacting and noninteracting proteins is reached, FRET is observed as an appearance of the second donor fluorescence lifetime (τDA) in the fluorescence decay. As a donor-only control, we used the lyk3-1 mutant transformed with pLYK3:LYK3-mTQ2. Fractions and lifetimes were obtained by globally fitting donor-only control and FRET sample decay curves simultaneously. To ascertain whether our FRET-FLIM measurements indicate specific interactions or rather result from aspecific (bystander) FRET, we estimated the concentrations of the receptors at the cell periphery (Supplemental Methods). As both the mean and individual FRET efficiency values calculated for LYK3 and NFP oligomerization (5.8% [confidence interval, 4.5 to 7.4] and 4.6% [3.8 to 5.9] in nodules coexpressing LYK3-mTQ2+LYK3-Venus and LYK3-mTQ2+NFP-Venus, respectively) were higher than the possible (maximal) bystander FRET estimated for the corresponding acceptor concentrations (3%), our FRET-FLIM analysis indicates specific interactions of these receptors in M. truncatula nodules.
In LYK3-mTQ2- or NFP-mTQ2-expressing nodules, the mTQ2 τ measured at the cell periphery was globally fitted with one component, τD = 3.95 ns (3.92 to 3.97; n = 26 + 7, respectively) (Figures 5A and 5D). In accordance with the two-component model (for a mixture of noninteracting/non-FRETting and interacting/FRETting donor fusions), the mTQ2 τ measured at the cell periphery in nodules coexpressing LYK3-mTQ2 and NFP-Venus was fitted with two components: τD = 3.95 ns (3.92 to 3.97) with a fraction (fD) = 92.5% (90 to 94) and τDA = 1.61 ns (1.45 to 1.77) with a fraction (fDA) = 7.5% (6 to 10) (n = 18 from 10 images) (Figures 5B and 5E). Phasor analysis of these measurements showed that the lifetime (phasor) values fell inside the semicircle and clustered along the line connecting the two globally fitted monoexponential lifetimes, τD and τDA (Figure 5G). This confirmed that the complex lifetimes present in the analyzed ROIs were likely a mixture of two lifetimes, similar to those found in the global analysis. Therefore, our results indicate that, on average, minimally 7.5% of LYK3 forms specific heteromeric complexes with NFP at the cell periphery in M. truncatula nodules. Unfortunately, FRET-FLIM analysis for the opposite receptor fusions (i.e., NFP-mTQ2 + LYK3-Venus) was not possible due to too-low levels of accumulation at the cell periphery of the donor fusions.
Figure 5.
NFP and LYK3 Oligomerization at the Cell Periphery in M. truncatula Nodules.
(A) to (C) Examples of cyan and yellow fluorescence intensity images of nodule cells (co)expressing LYK3-mTQ2 (A), LYK3-mTQ2 (in cyan) + NFP-Venus (in yellow) (B), and LYK3-mTQ2 (in cyan) + LYK3-Venus (in yellow) (C). Bars = 10 μm.
(D) to (F) mTQ2 lifetime decay curve (in gray), fit (in blue), and fit residuals (in orange) of the exemplary ROI (in red) presented in (A), (B), and (C), respectively. The τD and τDA were obtained by globally fitting all LYK3+NFP and all LYK3+LYK3 FRET samples; their respective fractions (fD and fDA), background (b), and shift (tS) in relation to the instrumental response function (IRF) are local parameters for the selected ROI. The quality of the fit is indicated by the reduced χ2 value in the residual panels.
(G) Phasor plot representing lifetime (phasor) values for all LYK3-mTQ2 and NFP-mTQ2 donor-only (in cyan), all LYK3+NFP FRET (in purple), and all LYK3+LYK3 FRET (in yellow) measurements. Globally fitted τD and τDA and their confidence intervals (depicted as color-coded ticks) are represented on the semicircle. Phasors of all cyan autofluorescence measurements (from nodules transformed with an empty vector or pLYK3:LYK3-Venus) are depicted as gray dots. Im, imaginary part; Re, real part.
As many receptors function as homomers (Heldin, 1995), we also investigated the homomerization of LYK3. We coexpressed pLYK3:LYK3-mTQ2 and pLYK3:LYK3-Venus in the lyk3-1 mutant. The two lifetimes extracted from the FRET samples were τD = 3.95 ns (3.93 to 3.97) with fD = 86% (81 to 89) and τDA = 2.30 ns (2.14 to 2.47) with fDA = 14% (11 to 19) (n = 40 from 14 images) (Figures 5C and 5F). Again, phasor analysis showed that the lifetime (phasor) values fell inside the semicircle and clustered along the line connecting the τD and τDA (Figure 5G), corroborating the results of global analysis. Therefore, our results indicate that, on average, minimally 14% of LYK3 forms specific homomers (dimers or higher order complexes) at the cell periphery in M. truncatula nodules. Unfortunately, low expression levels of fusion proteins in nfp-1 nodules coexpressing pNFP:NFP-mTQ2 and pNFP:NFP-Venus prohibited an analysis of NFP homomerization.
Additionally, we analyzed the homomerization and heteromerization of LYK3 and NFP fluorescent fusions in a heterologous system of N. benthamiana. To this end, LYK3 and NFP fusions to sYFP2 or mCherry were transiently expressed under the control of a constitutive cauliflower mosaic virus 35S promoter in N. benthamiana leaf epidermal cells (Supplemental Methods and Supplemental Figure 3). There, the apparent FRET efficiency was 5.6% (4.9 to 6.3) for LYK3 homomerization and 4.1% (3.1 to 5.1) for NFP homomerization. Due to the cell death induction upon NFP and LYK3 coexpression in N. benthamiana leaves, a putative heteromerization of NFP with a kinase-inactive LYK3 variant, LYK3 G334E, that did not induce cell death (Pietraszewska-Bogiel et al., 2013), was analyzed instead. The apparent FRET efficiency for this interaction was 2.3% (1.5 to 3.1).
Therefore, our FRET-FLIM studies indicate a specific homomerization of LYK3 (in both species studied) and a specific heteromerization of NFP and LYK3 in M. truncatula nodules.
Function of Nod Factor Receptors in Nodules
As our studies showed that Nod factor receptors are removed from the plasma membrane upon the release of rhizobia into the host cytoplasm, we hypothesized that Nod factor perception triggers the release of rhizobia from infection threads. To test this, we used an RNA interference (RNAi) approach. The challenge was to allow the early symbiotic responses, requiring Nod factor receptors in epidermis and cortex, to occur for the successful formation and infection of a nodule primordium. Therefore, we aimed to specifically reduce the level of the receptors in nodules using the ENOD12 promoter (Pichon et al., 1992; Limpens et al., 2005). This promoter is first activated 3 to 6 h after inoculation and remains active in the meristem and distal part of the infection zone. We expressed a pENOD12:NFP RNAi construct in wild-type M. truncatula and analyzed 10 randomly selected nodules. Eight nodules were markedly smaller than control nodules and had an aberrant infection zone that contained more cell layers with smaller cells in the central tissue (Figure 6). Infection threads did enter the cells, but release of the bacteria was hampered, and the few released bacteria did not develop into mature symbiosomes. In contrast with the block of release in the infection zone, a zone of approximately eight cell layers at the basal part of these nodules showed fully infected cells containing mature symbiosomes. Recently, we showed that these approximately eight basal infected cell layers are derived directly from the reactivated inner cortical root cells and not from the meristem (Xiao et al., 2014). Instead, the small cells of the infection zone in pENOD12:NFP RNAi nodules, where release was blocked, are derived from the meristem. So silencing of NFP results in a block of rhizobium release in cells derived from the meristem.
Figure 6.
Phenotypes of NFP Knockdown in Nodules at 21 DAI.
(A) and (B) A control nodule expressing a DsRED construct.
(C) and (D) A nodule expressing an NFP RNAi hairpin in a nodule-specific manner (pENOD12:NFP RNAi). In this nodule, there are many large infection threads (asterisks) present, and only in rare situations are the bacteria released.
The nodule apex is located at the top of each image. Bars in (A) and (C) = 100 μm; bars in (B) and (D) = 10 μm.
In comparison, the LYK3 RNAi hairpin under the control of the ENOD12 promoter did not block the release (Supplemental Figure 4), although it may be affected, as we saw more infection threads than in control nodules, indicating a slower release process.
Why Is NFP and LYK3 Accumulation Restricted to a Very Narrow Zone?
The coexpression of the Nod factor receptors in N. benthamiana induces a cell death response. Therefore, we hypothesized that the accumulation of NFP and LYK3 at the plasma membrane in only two cell layers is important to avoid a defense response that would hamper nodule development. To test this, we ectopically expressed NFP and LYK3 under the control of the Arabidopsis thaliana UBIQUITIN3 (Ubq) promoter that is active in the nodule apex at a markedly higher level compared with the native NFP or LYK3 promoter and in the infection zone (Limpens et al., 2009). Twelve randomly selected nodules expressing pUbq:NFP at 21 DAI were smaller and contained more uninfected cells in the infection zone compared with control nodules. This could be due to the triggering of defense-like responses against bacteria that hampered their release. The infected cells in all nodules showed signs of premature cell death: symbiosome degradation and the accumulation of yellow/green deposits (Figure 7). Although the identity of the yellow/green deposits is unknown, they might be polyphenols that are induced upon senescence/defense (Kováts et al., 1991) and cause the color change of toluidine blue (Ramalingam and Ravindranath, 1970). Therefore, ectopic expression of NFP appears to trigger defense-like responses in M. truncatula nodules. This supports the hypothesis that a very transient accumulation of NFP is important for successful nodule development.
Figure 7.
Phenotype of the Ectopic Expression of NFP in Nodules at 21 DAI.
Nodule expressing pUbq:NFP. It is smaller, has fewer infected cells in the infection zone, and shows an accumulation of yellow/green deposits. The yellow/green deposits might be polyphenols, which are induced upon senescence/defense and cause a color change in toluidine blue. Bar in (A) = 100 μm; bar in (B) = 50 μm.
In comparison, ectopic expression of LYK3 affected nodule development by reducing bacterial release in the infection zone (Supplemental Figure 5) but did not induce defense-like responses like those we observed with the NFP ectopic expression.
DISCUSSION
Here, we show that the M. truncatula Nod factor receptors, NFP and LYK3, accumulate at the cell periphery, most likely at the plasma membrane, in a narrow zone of approximately two cell layers at the transition of the nodule meristem to the infection zone. At least 7.5% of these receptors are present in heteromeric complexes. NFP is shown to be essential for release of the bacteria from the infection threads into the cytoplasm of cells derived from the nodule meristem. Furthermore, we hypothesize that the accumulation at least of NFP appears strictly regulated in order to prevent the induction of defense-like responses.
NFP and LYK3 accumulate in a narrow zone of approximately two cell layers (L1 and L2) forming the border between the meristem and the infection zone. LYK3 localizes at the plasma membrane and associated vesicles within this zone. The region where NFP and LYK3 accumulate at the cell periphery coincides with the zone where rhizobial Nod genes are active and where, most likely, rhizobia still produce Nod factors (Marie et al., 1994; Schlaman et al., 1998). Therefore, it is very probable that this is the ligand that activates these receptors in the nodule. As Nod factors accumulate in the cell wall and are not translocated to the cytoplasm (Goedhart et al., 2000), receptors at the plasma membrane and infection thread membrane most likely constitute the pool of receptive protein, whereas the vacuole-located receptors most likely represent the fraction of protein targeted for degradation. The Nod factor receptor accumulation at the plasma membrane in a very narrow zone implies that this is a very transient event. As this zone is markedly smaller than the region where NFP and LYK3 are expressed (based on promoter:GUS [Supplemental Figure 1] and in situ [Limpens et al., 2005] data) and because of the accumulation of NFP in the vacuole lumen of cells younger than L1, we hypothesize that the level of receptor is regulated by high protein turnover next to the regulation at the transcriptional level.
This work demonstrates Nod factor receptor (co)localization in root nodules. The same constructs were used to study the Nod factor receptors in the root epidermis with laser-scanning and spinning-disc confocal microscopy. However, we were unable to detect the receptors in the root epidermis (data not shown). This is in contrast with similar studies by Haney et al. (2011) that visualized LYK3-GFP in M. truncatula root hairs but not in nodules. Another difference between the two studies seems to be the transient nature of the signaling. Haney et al. (2011) still detected LYK3 at the infection thread membrane within a root hair even when the tip of this infection thread had already reached the cortical layers. This suggests that the presence of LYK3 at the infection thread membrane is persistent in root hairs, whereas in nodules it appears to be a very transient process.
By an RNAi approach, NFP was shown to be important for the release of rhizobia, and therefore the formation of a symbiotic interface, in cells derived from the meristem. We speculate that LYK3 also might be involved in this process, as it forms a complex with NFP. During early stages of this symbiotic interaction, NFP is essential for all responses, whereas LYK3 is first required for the initiation of infection threads in curled root hairs (Smit et al., 2007). As NFP lacks an active kinase domain, it is very probable that at stages preceding infection thread formation, another LysM domain receptor kinase, LYK3 or its functionally redundant homolog, functions together with NFP (Smit et al., 2007). We speculate that a similar functional redundancy masks the involvement of LYK3 in rhizobium release in the infection zone.
Infected cells present at the base of a M. truncatula nodule are formed by trans-differentiation and the infection of inner cortical/primordial cells (Xiao et al., 2014). Knockdown of NFP did not block the release of rhizobia in these cells, in contrast with cells derived from the meristem. As the trans-differentiation of inner cortical cells precedes the formation of a functional meristem (Xiao et al., 2014), RNAi levels might not yet have reached a sufficient level in the primordial cells before the release of rhizobia was induced. Alternatively, the release could be regulated differently there than in cells of meristem origin. Indeed, an allele of SymRK of pea (Pisum sativum) allows release in the primordium but blocks infection in cells derived from the nodule meristem (Ovchinnikova, 2012). This strongly suggests that release from infection threads in cells derived from the meristem is more stringently regulated than in the nodule primordium. This is in line with the role of SymRK in the release of rhizobia in M. truncatula (Limpens et al., 2005) and Sesbania rostrata (Capoen et al., 2005) and in the interaction of NFR5 and SymRK in L. japonicus (Antolín-Llovera et al., 2014). It would be interesting to see if SymRK interacts on the molecular level with the LYK3-NFP receptor complex we detected inside M. truncatula nodules.
During early stages of symbiotic interaction, Nod factor receptors activate a conserved common symbiotic signaling cascade (Oldroyd, 2013). Components of this pathway also play a role in the release of rhizobia from infection threads (Limpens et al., 2005; Ovchinnikova et al., 2011). Our results support the notion that the Nod factor receptor complex triggers symbiotic interface formation via a signaling cascade that, at least in part, is similar to the common symbiotic signaling pathway. The role of Nod factor signaling in the formation of the symbiotic interface could provide control by the host on the compatibility of rhizobia present in the infection threads. Studies with Parasponia andersonii, belonging to the only nonlegume genus able to establish a nodule symbiosis with rhizobium, indicates that the formation of the symbiotic interface is a primary function of Nod factor signaling. Silencing of the Parasponia NFP ortholog has no effect on infection thread formation in root nodules, but the formation of the symbiotic interface is completely blocked (Op den Camp et al., 2011). The Parasponia-rhizobium symbiosis evolved more recently and represents a rather early form of the rhizobium endosymbiosis. Therefore, we hypothesize that this is one of the most basal host processes regulated by Nod factor receptors and is still maintained in evolutionarily advanced legumes.
The accumulation of both Nod factor receptors in nodules provided, despite its very transient nature, a unique opportunity to study their interaction under physiologically relevant conditions. We showed using FRET-FLIM that NFP and LYK3 indeed form heteromers and that LYK3 is also able to homomerize at the cell periphery, most likely at the plasma membrane, in M. truncatula nodules. In Arabidopsis, for example, the receptor kinases BRI1 and FLS2 homomerize in planta (Wang et al., 2005; Hink et al., 2008; Sun et al., 2012) as well as heteromerize with members of the SERK subfamily (Schulze et al., 2010; Jaillais et al., 2011; Schwessinger et al., 2011). The observed LYK3 homomerization could involve a separate pool of protein or could be realized within higher order protein complexes that also contain NFP.
Our results indicate that the M. truncatula nodule is a good experimental system in which to study the interaction between Nod factor receptors and other proteins. In contrast with the root epidermis, the receptors accumulate here at higher levels that make detection easier. Also, the gradual developmental nature of the M. truncatula nodule makes it a good experimental system to study signaling events despite their transient nature. This allows the comparison of signaling induced by Nod factor receptors in legumes with the observations obtained in heterologous systems. For example, the most striking difference is that simultaneous expression of both receptors in N. benthamiana leaves causes a cell death response (Madsen et al., 2011; Pietraszewska-Bogiel et al., 2013), whereas this is not the case in M. truncatula nodules. This raises the question of whether cell death is avoided due to the very transient nature of Nod factor signaling in nodules. This hypothesis is supported by the ectopic expression of NFP, which appears to trigger defense-like responses in nodules. We speculate that this response is most likely induced by NFP cofunction with another, unknown LysM domain receptor kinase, although LYK3 could perform this function as well. By contrast, the ectopic expression of LYK3 did not result in the triggering of similar defense-like responses. This is consistent with studies in N. benthamiana, where overexpression of LYK3/NFR1 alone also does not trigger cell death (Madsen et al., 2011; Pietraszewska-Bogiel et al., 2013).
NFP is essential for the release of rhizobia from the infection threads, and the accumulation of the receptors is restricted to the cell layers in which this process occurs. Therefore, this strictly regulated accumulation also might be essential to restrict release to one to two cell layers and only occurs at the start of the differentiation of the infected cells.
We hypothesize that the strict regulation of the accumulation of Nod factor receptors is achieved in the following manner. The receptor genes are expressed in a relatively broad zone at the apex of the nodule. This includes cells of the meristem that are not in contact with Nod factors. Nod factor receptors accumulate at the plasma membrane at low, nondetectable levels by which the meristem cells are primed to respond to Nod factors. L1 cells are the first cells that are in contact with Nod factors (secreted by rhizobia located intercellularly). This activates a positive feedback mechanism by which more Nod factor receptors accumulate at the plasma membrane and infection thread membrane. For instance, the mechanism responsible for a high turnover of the receptors might be inhibited, resulting in higher accumulation of a receptive protein and increased signaling. Increased Nod factor signaling triggers the release of the rhizobia from the infection threads but at the same time triggers a negative feedback mechanism by which the receptors are targeted for degradation in the vacuole. In this way, relatively high levels of receptive receptors are restricted to only approximately two cell layers.
METHODS
Constructs, Plant Material, and Transformation
To create the desired entry clones, the desired regions were amplified from genomic DNA using Phusion High-Fidelity DNA Polymerase (Finnzymes) as described in Supplemental Table 1. Binary constructs were generated according to Supplemental Table 2. The generated fluorescent fusion constructs were tested for their biological activity as described in Supplemental Methods. The constructs were transformed by Agrobacterium rhizogenes-mediated hairy root transformation (Limpens et al., 2004) into Medicago truncatula A17, B56 (lyk3-1), C31 (nfp-1), or B56 × C31 (lyk3-1 nfp-1). The transgenic plants were inoculated with Sinorhizobium meliloti 2011 or S. meliloti 2011-mRFP1 (Smit et al., 2005).
To create a stable line, M. truncatula R108 was transformed by Agrobacterium tumefaciens AGL1 carrying pLYK3:LYK3-GFP. Roots from 1-d-old seedlings were cut in an Agrobacterium suspension SH medium (Gamborg et al., 1976) supplemented with 0.5 mg/L BAP, 3 mM MES, 20 mg/L acetosyringone, 400 mg/L l-cysteine, 1 mM DTT, and 0.02% Silvet and left to incubate for 30 min. Root fragments were incubated on cocultivation medium (SH medium supplemented with 0.5 mg/L BAP, 5 mg/L 2,4-D, 3 mM MES, and 20 mg/L acetosyringone) for 2 to 3 d at 25°C in the dark. Transgenic explants were selected on SH medium supplemented with 0.5 mg/L BAP, 5 mg/L 2,4-D, 3 mM MES, 300 mg/L cefotaxime, and 50 mg/L kanamycin and transferred to fresh plates weekly until a callus was formed. Green calluses were transferred to regeneration medium (SH medium supplemented with 20 g/L sucrose, 0.5 mg/L BAP, 0.1 mg/L 1-naphthaleneacetic acid, 10 mg/L AgNO3, 3 mM MES, 300 mg/L cefotaxime, and 50 mg/L kanamycin) and, upon shoot formation, were transferred to SH medium supplemented with 10 g/L sucrose, 0.5 mg/L indole-3-butyric acid, and 300 mg/L cefotaxime to form roots.
For Nicotiana benthamiana transformations, FRET-FLIM imaging, and data analysis in N. benthamiana, see Supplemental Methods.
Structural Analysis and Microscopy
For structural analysis, nodules were selected based on DsRED1 expression using a Leica MZFLIII binocular microscope fitted with an HQ553/30 optical filter (Leica Microsystems). Nodules were fixed overnight in a mixture of 4% paraformaldehyde and 3% glutaraldehyde in 50 mM potassium phosphate buffer, pH 7.4, at 4°C. Roots were dehydrated in ethanol and subsequently embedded in Technovit 7100 (Heraeus Kulzer). Thin 4-μm longitudinal sections were stained with 0.05% toluidine blue and analyzed.
Fluorescence Microscopy
Transgenic roots and nodules were selected based on DsRED1 expression as above. Transgenic nodules were hand-sectioned using double-razor blades, mounted on microscope slides in 0.9% NaCl, and further analyzed on a Zeiss LSM 510 confocal laser-scanning microscope (Carl-Zeiss Axiovert 200 M equipped with a 488-nm argon laser line [GFP] and a 543-nm helium-neon laser line [DsRED1/mRFP1]). The light was guided via an HFT 488/543 primary dichroic mirror through an oil-immersed 40× Plan-Neofluar objective (numerical aperture 1.3) into the sample. Emission was guided via an NFT 545 dichroic mirror, and GFP emission was selectively detected using a 505- to 530-nm BP filter; DsRED1/mRFP1 emission was detected using a 560- to 615-nm BP or 560-nm LP filter. Immunolocalization on thick sections was performed as described in Supplemental Methods.
Time-Domain FLIM Setup
Measurements were performed on an inverted Fluoview 1000 laser-scanning microscope (Olympus). The excitation light of a 440-nm, 20-MHz pulsing laser diode (Picoquant), as controlled by a SepiaII laser driver unit (Picoquant), was attenuated 10 times by a neutral density filter. The light was guided via a D440/514/594 primary dichroic mirror (Chroma) through a water-immersed 60× UPlanS-Apo objective (numerical aperture 1.2) into the sample. The emission light was guided via a pinhole, set at 120 μm, through the Olympus detection box to the fiber output channel. The optical fiber was coupled to a custom-made detection box (Picoquant) containing PDM avalanche photodiodes (MPD). The light was guided into one of the MPDs, where the light was filtered by a 475/45 emission filter (Chroma). The photons were collected for a 256- × 256-pixel image during 150 to 180 s by the SymPhoTime (Picoquant) software. The photon arrival times were recorded by a Picoharp 300 time-correlated single-photon counting system (Picoquant). For global fitting and phasor analysis of time-resolved FLIM data, see Supplemental Methods.
Transmission Electron Microscopy
For electron microscopy, nodules were collected at 10 DAI and prepared according to the conventional method as described previously (Limpens et al., 2009). Thin sections (60 nm) of the same nodule were cut using a Leica Ultracut microtome. Nickel grids with the sections were blocked in 2% BSA in potassium phosphate buffer (pH 7.2) and incubated with polyclonal rabbit anti-GFP antibodies (Molecular Probes) in dilution 1:200. Goat anti-rabbit antibodies coupled with 15-nm gold (BioCell) (1:50 dilution) were used as secondary antibody. Sections were examined using a JEOL JEM 2100 transmission electron microscope equipped with a Gatan US4000 4K×4K camera.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: LYK3 (AY372402.1) and NFP (DQ496250.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Biological Activity of the LYK3 and NFP Fluorescent Fusion Constructs.
Supplemental Figure 2. Accumulation of LYK3 and NFP in the Nodule Apex.
Supplemental Figure 3. NFP and LYK3 Oligomerization at the Plasma Membrane in N. benthamiana Leaf Epidermal Cells.
Supplemental Figure 4. Phenotypes of LYK3 Knockdown in Nodules 21 DAI.
Supplemental Figure 5. Ectopic Expression of LYK3 in Nodules 21 DAI.
Supplemental Table 1. Entry Constructs.
Supplemental Table 2. Destination Constructs.
Supplementary Material
Acknowledgments
A.P.-B. is supported by the European Community Marie Curie Research Training Network Programme (Grant MRTN-CT-2006-035546 NODPERCEPTION) and S.M. and T.B. by Grant ERC-2011-AdG294790.
AUTHOR CONTRIBUTIONS
S.M. performed localization and functional analysis and wrote the article. A.P.-B. performed FRET-FLIM experiments and analysis and wrote the article. E.F. performed electron microscopy. M.P. assisted on FRET-FLIM analysis. M.A.H. assisted on FRET-FLIM analysis. E.L., T.W.J.G., and T.B. wrote the article.
Glossary
- DAI
days after inoculation
- FLIM
fluorescence lifetime imaging microscopy
- ROI
region of interest
- FRET
Förster resonance energy transfer
- RNAi
RNA interference
Footnotes
Online version contains Web-only data.
References
- Antolín-Llovera M., Ried M.K., Parniske M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24: 422–427. [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin N.J. (2004). Plant cell wall remodelling in the Rhizobium–legume symbiosis. Crit. Rev. Plant Sci. 23: 293–316. [Google Scholar]
- Capoen W., Goormachtig S., De Rycke R., Schroeyers K., Holsters M. (2005). SrSymRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 102: 10369–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haeze W., Holsters M. (2002). Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12: 79R–105R. [DOI] [PubMed] [Google Scholar]
- Gamborg O.L., Murashige T., Thorpe T.A., Vasil I.K. (1976). Plant tissue culture media. In Vitro 12: 473–478. [DOI] [PubMed] [Google Scholar]
- Geurts R., Bisseling T. (2002). Rhizobium nod factor perception and signalling. Plant Cell 14 (suppl.): S239–S249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352. [DOI] [PubMed] [Google Scholar]
- Goedhart J., Hink M.A., Visser A.J., Bisseling T., Gadella T.W. Jr. (2000). In vivo fluorescence correlation microscopy (FCM) reveals accumulation and immobilization of Nod factors in root hair cell walls. Plant J. 21: 109–119. [DOI] [PubMed] [Google Scholar]
- Haney C.H., Riely B.K., Tricoli D.M., Cook D.R., Ehrhardt D.W., Long S.R. (2011). Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23: 2774–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C.H. (1995). Dimerization of cell surface receptors in signal transduction. Cell 80: 213–223. [DOI] [PubMed] [Google Scholar]
- Hink M.A., Shah K., Russinova E., de Vries S.C., Visser A.J. (2008). Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys. J. 94: 1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Belkhadir Y., Balsemão-Pires E., Dangl J.L., Chory J. (2011). Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 108: 8503–8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus-Heisen D., Nurisso A., Pietraszewska-Bogiel A., Mbengue M., Camut S., Timmers T., Pichereaux C., Rossignol M., Gadella T.W., Imberty A., Lefebvre B., Cullimore J.V. (2011). Structure-function similarities between a plant receptor-like kinase and the human interleukin-1 receptor-associated kinase-4. J. Biol. Chem. 286: 11202–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kováts K., Binder A., Hohl H.R. (1991). Cytology of induced systemic resistance of cucumber to Colletotrichum lagenarium. Planta 183: 484–490. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Limpens E., Ivanov S., van Esse W., Voets G., Fedorova E., Bisseling T. (2009). Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21: 2811–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Antolín-Llovera M., Grossmann C., Ye J., Vieweg S., Broghammer A., Krusell L., Radutoiu S., Jensen O.N., Stougaard J., Parniske M. (2011). Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 65: 404–417. [DOI] [PubMed] [Google Scholar]
- Marie C., Plaskitt K.A., Downie J.A. (1994). Abnormal bacteroid development in nodules induced by a glucosamine synthase mutant of Rhizobium leguminosarum. Mol. Plant Microbe Interact. 7: 482–487. [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Op den Camp R., Streng A., De Mita S., Cao Q., Polone E., Liu W., Ammiraju J.S., Kudrna D., Wing R., Untergasser A., Bisseling T., Geurts R. (2011). LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova, E. (2012). Genetic Analysis of Symbiosome Formation. PhD dissertation (Wageningen, The Netherlands: Wageningen University). [Google Scholar]
- Ovchinnikova E., et al. (2011). IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol. Plant Microbe Interact. 24: 1333–1344. [DOI] [PubMed] [Google Scholar]
- Pichon M., Journet E.P., Dedieu A., de Billy F., Truchet G., Barker D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietraszewska-Bogiel A., Gadella T.W. (2011). FRET microscopy: From principle to routine technology in cell biology. J. Microsc. 241: 111–118. [DOI] [PubMed] [Google Scholar]
- Pietraszewska-Bogiel A., Lefebvre B., Koini M.A., Klaus-Heisen D., Takken F.L., Geurts R., Cullimore J.V., Gadella T.W. (2013). Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS ONE 8: e65055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Rae A.L., Bonfante-Fasolo P., Brewin N.J. (1992). Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum. Plant J. 2: 385–395. [Google Scholar]
- Ramalingam K., Ravindranath M.H. (1970). Histochemical significance of green metachromasia to toluidine blue. Histochemie 24: 322–327. [DOI] [PubMed] [Google Scholar]
- Roth L.E., Stacey G. (1989). Bacterium release into host cells of nitrogen-fixing soybean nodules: The symbiosome membrane comes from three sources. Eur. J. Cell Biol. 49: 13–23. [PubMed] [Google Scholar]
- Schlaman, H.R.M., Phillips, D.A., and Kondorosi, E. (1998). Genetic organization and transcriptional regulation of rhizobial nodulation genes. In The Rhizobiaceae, H.P. Spaink, A. Kondorosi, and P.J.J. Hooykaas, eds (Dordrecht, The Netherlands: Springer), pp. 361–386. [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Stewart, W.D.P. (1966). Nitrogen Fixation in Plants. (London: Athlone Press).
- Sun W., Cao Y., Jansen Labby K., Bittel P., Boller T., Bent A.F. (2012). Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J., de Billy F., Camut S., Truchet G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172: 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S.S., Thaler C., Koushik S.V. (2006). Fanciful FRET. Sci. STKE 2006: re2. [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. (2014). Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.