The FAMA gene that confers stomatal identity also specifies the fate of myrosin idioblasts that together with guard cells, which are also myrosin cells, make up the “mustard oil bomb” that reduces plant predation via the glucosinolate-myrosinase system.
Abstract
Crucifer shoots harbor a glucosinolate-myrosinase system that defends against insect predation. Arabidopsis thaliana myrosinase (thioglucoside glucohydrolase [TGG]) accumulates in stomata and in myrosin idioblasts (MIs). This work reports that the basic helix-loop-helix transcription factor FAMA that is key to stomatal development is also expressed in MIs. The loss of FAMA function abolishes MI fate as well as the expression of the myrosinase genes TGG1 and TGG2. MI cells have previously been reported to be located in the phloem. Instead, we found that MIs arise from the ground meristem rather than provascular tissues and thus are not homologous with phloem. Moreover, MI patterning and morphogenesis are abnormal when the function of the ARF-GEF gene GNOM is lost as well as when auxin efflux and vesicular trafficking are chemically disrupted. Stomata and MI cells constitute part of a wider system that reduces plant predation, the so-called “mustard oil bomb,” in which vacuole breakage in cells harboring myrosinase and glucosinolate yields a brew toxic to many animals, especially insects. This identification of the gene that confers the fate of MIs, as well as stomata, might facilitate the development of strategies for engineering crops to mitigate predation.
INTRODUCTION
Crucifers harbor an injury-induced defense pathway termed a “mustard oil bomb,” a glucosinolate-myrosinase system that reduces predation by forming products toxic to microbes and insects. Myrosinase comprises a family of glucosinolate hydrolases present at high levels in many Brassicaceae species (Rask et al., 2000). In Arabidopsis thaliana, myrosinase is a synonym for thioglucosidase, which is also referred to as thioglucoside glucohydrolase (TGG) (Xue et al., 1992; Chadchawan et al., 1993). Six myrosinase genes have been identified in Arabidopsis, of which two, TGG1 and TGG2, are strongly expressed in the shoot, with TGG3 likely to be a pseudogene (Andréasson et al., 2001; Zhang et al., 2002). The functions of the three other myrosinase genes, TGG4, TGG5, and TGG6, are still not well described. TGG4 and TGG5 encode functional myrosinases and appear to be expressed specifically in roots, while TGG6 shows expression only in pollen and does not appear to harbor myrosinase activity (Andréasson et al., 2001; Kissen et al., 2009). In Brassica napus and Sinapis alba seeds, myrosinase is found in myrosin cells in the form of water-soluble myrosin grains located in protein storage bodies in cotyledons and in the embryonic axis (Bones et al., 1991). Plant myrosinases and glucosinolates are synthesized and stored separately in adjacent cells termed myrosin cells and S-cells, respectively (Eriksson et al., 2002; Kissen et al., 2009; Ahuja et al., 2010). During predation or unnatural cell breakage, myrosinase can hydrolyze glucosinolate from damaged plant tissues yielding a glucose molecule and an unstable glucone. The latter is quickly transferred to either a thiocyanate, an isothiocyanate, or to a nitrile, all of which are toxic to insects and microorganisms (Wittstock and Halkier, 2002). B. napus plants that lack myrosinase activity due to the ablation of myrosin cells were more actively fed upon by animals, consistent with reduced toxicity (Borgen et al., 2010). In addition to plant defense, myrosinases contribute to counteracting diabetes, heart disease, and cancer (Halkier and Gershenzon, 2006).
Crucifers contain two types of myrosin cells that inhibit predation, guard cells (GCs) in stomata and specific cells reported to be located in the phloem that have been termed “phloem idioblasts” (Andréasson et al., 2001; Husebye et al., 2002). Stomata, which regulate gas exchange between the shoot and the environment, are present in nearly all plant taxa, bryophytes and above. Many aspects of stomatal development are well defined, including patterning and division regulation in the cell lineage (Pillitteri and Torii, 2012). Dynamic changes of auxin activity in stomatal lineage stem cells result from auxin transport and signaling that enforce stomatal morphology and patterning (Le et al., 2014). The final stage of stomatal development is regulated via a master basic helix-loop-helix (bHLH) transcription factor FAMA that confers guard cell fate and ensures that an oval guard mother cell (GMC) divides only once symmetrically, thus forming a pair of mature guard cells (Hachez et al., 2011). FAMA is strongly expressed in late GMCs and young guard cells, but not in mature stomata (Ohashi-Ito and Bergmann, 2006).
“Phloem idioblasts” differ in size and morphology from adjacent cells (Kissen et al., 2009). These cells are reported to be localized throughout the shoot in the abaxial phloem parenchyma (Andréasson et al., 2001; Husebye et al., 2002). Recently, the loss-of-function of CONTINUOUS VASCULAR RING1 (COV1), which encodes a protein localized to the trans-Golgi network, was found to increase the number of these idioblasts (Shirakawa et al., 2014). Similarly, the Q-SNARE protein, so called VACUOLAR MORPHOGENESIS PROTEIN3 (VAM3) that contributes to the regulation of vesicular transport of vacuolar proteins, also normally restricts idioblast number (Ueda et al., 2006).
However, relatively little is otherwise known about how these idioblasts develop, including the genes that confer their fate, as well as where these idioblasts originate in planta. Here, we report that these cells originate from the ground meristem rather than the procambium and thus refer to them as myrosin idioblasts (MIs) instead of phloem idioblasts. In addition, we show that FAMA as well as the E1728 enhancer trap that both mark GC fate are also expressed in developing as well as in mature MIs. Importantly, this work demonstrates that FAMA is required for MI fate as well as TGG expression. In addition, we report that MI shape and distribution are regulated by intercellular auxin transport as well as by vesicular trafficking.
RESULTS
Guard Cell Fate Markers Are Expressed in Myrosin Idioblasts
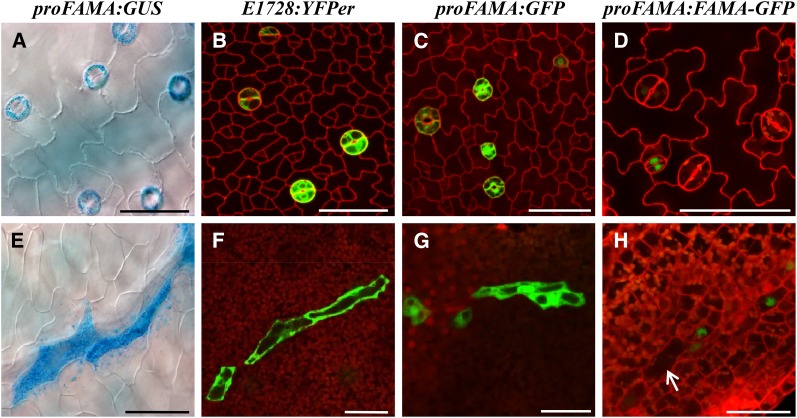
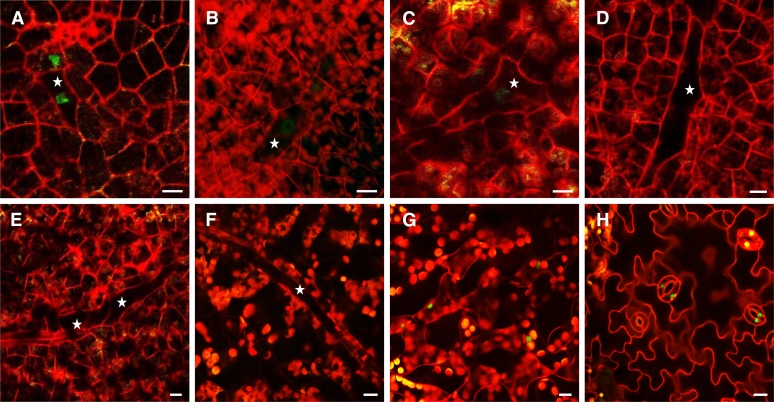
Stomatal-related reporter gene transcriptional fusions, such as ProFAMA:GFP, ProFAMA:GUS (β-glucuronidase), as well as the E1728:YFPer enhancer trap, have been shown to be expressed in GMCs and in young GCs (Ohashi-Ito and Bergmann, 2006). In addition to their expression during stomatal development (Figures 1A to 1C), we found that all of these markers were also expressed in long, isolated, and irregularly shaped cells (Figures 1E to 1G). MIs first appear in Arabidopsis development in cotyledons of nearly mature seeds (Supplemental Movie 1 and Supplemental Figure 16A) and then became located near the vasculature throughout the shoot in both vegetative and reproductive parts, including petioles, leaves, sepals, petals, and carpels (Supplemental Figure 1).
Figure 1.
GC Markers Are Also Expressed in MIs.
(A) GCs show epidermal ProFAMA:GUS expression.
(B) GCs exhibit E1728:YFPer expression.
(C) Epidermal ProFAMA:GFP expression in GMCs and GCs.
(D) Young GCs harbor ProFAMA:FAMA-GFP expression.
(E) MIs express ProFAMA:GUS near a leaf vein in the same field shown in the epidermis in (A).
(F) Three MIs harboring E1728:YFPer expression near leaf veins.
(G) MIs with ProFAMA:GFP expression (near leaf veins; data not shown).
(H) Young MIs near leaf veins exhibit nuclear ProFAMA:FAMA-GFP expression. Arrow indicates a large and older MI that no longer emits fluorescence.
All samples were taken from first rosette leaves 10 to 15 DAG. Images were captured using confocal microscopy except for bright-field microscopy ([A] and [E]). Cell walls were visualized in red using PI staining and confocal microscopy. (A) to (D) show the leaf epidermis, whereas (E) to (H) indicate MI cells in the leaf interior. Bars = 50 μm.
The expression patterns of two major myrosinase genes, TGG1 and TGG2, as well as the distribution of myrosin cells, have been described previously for Arabidopsis leaves, inflorescences, sepals, petals, and gynoecia (Barth and Jander, 2006). TGG1 is expressed in stomata as well as in MIs, but TGG2 is expressed exclusively in MIs (Barth and Jander, 2006). To determine the identity of this elongated cell type, expression patterns driven by the FAMA and E1728 reporters were compared with those of the myrosin cell-type-specific marker, a TGG1-GUS transcriptional fusion (ProTGG1:GUS). In leaves transformed with all three reporters, FAMA and E1728 were expressed in these two distinct and isolated cell types that are also marked by TGG1 expression (Supplemental Figure 2). Thus, FAMA, E1728, and TGG1 are all coexpressed in both types of myrosin cells, i.e., MIs as well as guard cells.
Notably, unlike the transcriptional fusion, a ProFAMA:FAMA-GFP translational fusion (referred to hereafter as FAMAtrans; Lee et al. 2014), was only expressed strongly in nuclei of young, but not in mature MIs (Figures 1D and 1H; Supplemental Figures 3D to 3F and 16B and Supplemental Movie 2). The absence of FAMA expression from mature MIs parallels the lack of ProFAMA:FAMA-GFP fluorescence from mature GCs and also parallels its presence just before and after GMC symmetric division (Supplemental Figure 3). These results indicate that FAMA acts before MIs mature and also suggest that FAMA expression is negatively and posttranscriptionally regulated during MI maturation.
Myrosin Idioblasts Originate from the Ground Tissue Meristem
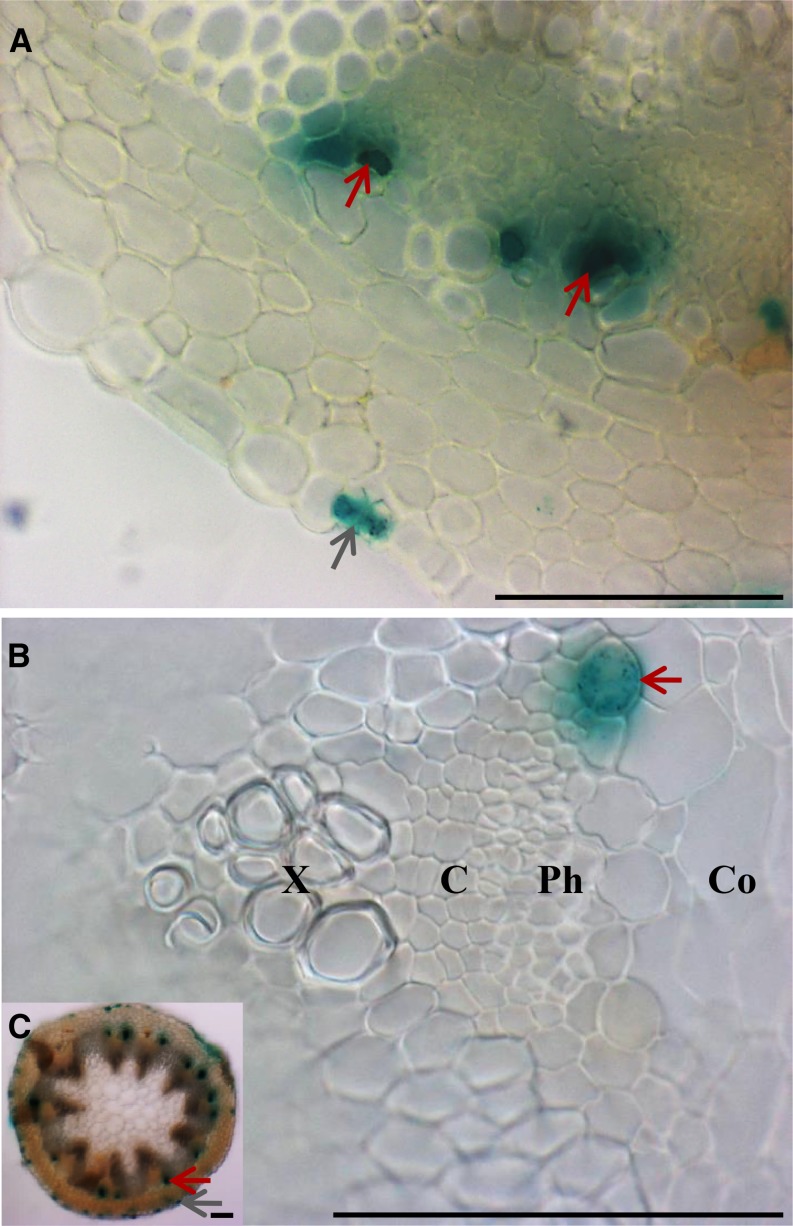
Myrosin idioblasts are reported to be located in Arabidopsis vascular bundles and thus to originate from the procambium (Husebye et al., 2002). Leaf primordia develop from the three types of primary tissue meristems (protoderm, procambium, and the ground meristem) with the protoderm differentiating into the epidermis and the ground tissue meristem producing the mesophyll (Esau, 1965). Vascular network patterning in leaves starts from selected ground cells that then form the procambium. Procambial cells elongate, divide, and then differentiate into xylem and phloem in vascular bundles, while vascular bundle sheath cells originate from the ground tissue (Zhang et al., 2002; Donner et al., 2009). However, these reports did not define whether MIs arise from the ground tissue meristem or the procambium. To identify the tissue system that produces MIs, the distribution of GUS staining driven by the FAMA promoter was analyzed in cross sections of primary inflorescence stems. Consistent with the previous reports, Figure 2 shows that mature MIs are located in the vicinity of the phloem and the cortex (Kissen et al., 2009).
Figure 2.
FAMA Promoter-Driven GUS (ProFAMA:GUS) Expression in MIs in Stems.
Red and gray arrows indicate GUS expression in MIs and GCs, respectively. X, xylem; C, vascular cambium; Ph, phloem; Co, cortex. All three images were from hand sections of stems 2 weeks after bolting and were captured using light microscopy. Bars = 0.1 mm.
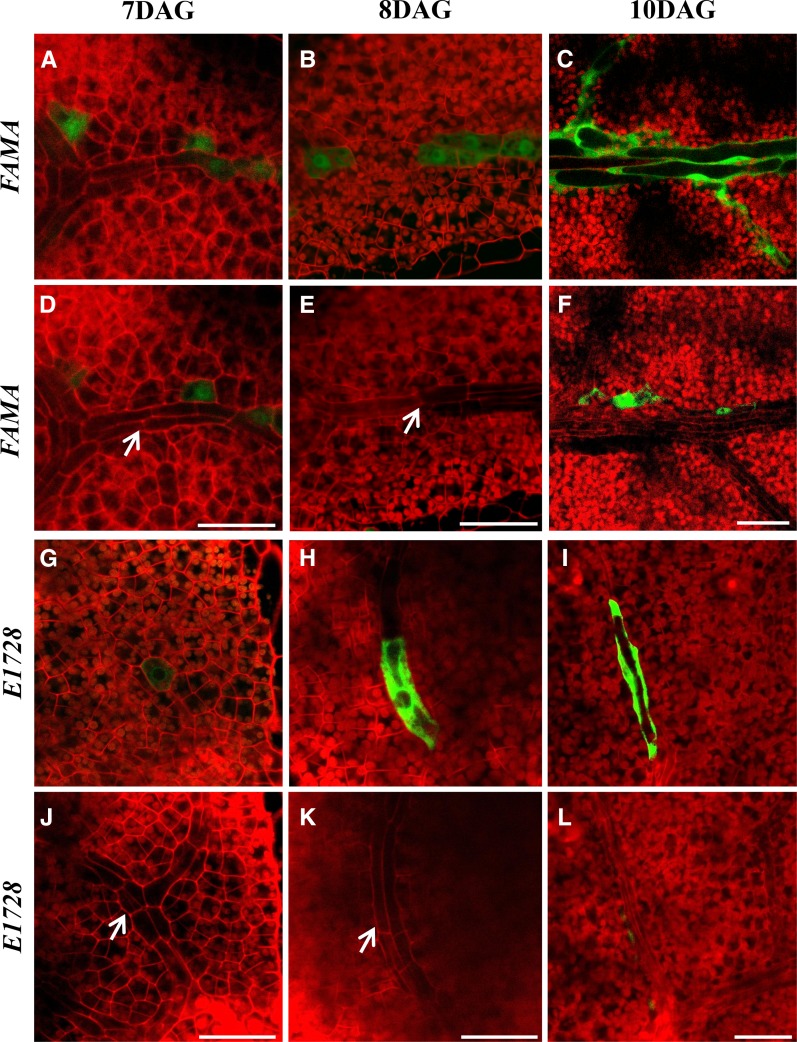
During the vascular development of leaf primordia, procambial cells and young MIs form at roughly comparable times (Figure 3; Supplemental Movies 3 to 6 and Supplemental Figures 16C to 16F). Procambial cells elongate and proliferate during early shoot growth and then differentiate (Scarpella et al., 2010). FAMA and E1728 expression first appeared in small cells that were part of the ground meristem (Figures 3A and 3G; Supplemental Movie 3 and Supplemental Figure 16C), as shown by their shorter length compared with adjacent and still elongating procambial cells (Figures 3D and 3J). Young MIs can form singly or adjacent (Figures 3A and 3G). Following the enlargement of young MIs (Figures 3B and 3H), and the subjacent initial differentiation of the vasculature (Figures 3E and 3K), MIs then matured into irregularly shaped and isolated Idioblasts (Figures 3C and 3I) positioned next to abaxial vascular tissue (Figures 3F and 3L). Cell fate markers for the procambium were also analyzed. The location of FAMA expression in juvenile rosette leaves was then compared with that of markers of the elongating procambium, i.e. fluorescence from reporters of the transcription factor gene Arabidopsis thaliana HOMEOBOX8 (At-HB8), as well as from the enhancer trap Q0990 (Scarpella et al., 2010). The localization of the MI expression markers FAMA and E1728 (Supplemental Figures 4E to 4H and 16D to 16F and Supplemental Movies 4 to 6) differed from that of the procambial markers At-HB8 (Supplemental Figures 4A, 4B, and 17A and Supplemental Movie 7) and Q0990 (Supplemental Figures 4C, 4D, 17B, and 17C and Supplemental Movies 8 and 9). These data show that while MIs are located close to the phloem, they likely instead derive from the ground tissue meristem rather than the procambium. Thus, the term “phloem idioblast” is a misnomer and this cell type should instead be referred to just as “myrosin idioblasts.”
Figure 3.
Comparison of Stages of MI and Vascular Development Shown Using Two Different MI Markers.
Confocal images taken 7, 8, and 10 DAG from regions below the epidermis near the leaf tip in first leaves. Pairs of vertical images (e.g., [A] and [D], and [I] and [L]) show the same field focused on either the MI layer ([A] to [C] and [G] to [I]) or the subjacent procambial/provascular tissues ([D] to [F] and [J] to [L]). The images in (G) and (J) are from Supplemental Movie 6. The arrows in (D), (E), (J), and (K) indicate the procambium. “FAMA” denotes the transcriptional ProFAMA:GFP fusion. “E1728” refers to expression from the E1728:YFPer construct. All images were captured using confocal microscopy. The red signal is from PI staining. Bars = 50 μm.
MIs in mature rosette leaves become 5 to 6 times longer than surrounding spongy mesophyll cells, and their diameter was often greater than adjacent and narrow phloem cells (Supplemental Figure 5B). Mature MIs harbored a spindle-shaped nucleus (Supplemental Figure 5A), as well as large vacuoles (Supplemental Figure 5C) containing soluble myrosinases (Ueda et al., 2006). MI shape varied, and these cells were found either singly or in contact (Supplemental Figures 5D to 5I), as opposed to the more continuous arrangement of, e.g., spongy mesophyll or developing vasculature.
FAMA Is Required for Myrosin Cell Fate and TGG Expression
FAMA has been previously shown to confer stomatal cell fate, but no other role for this bHLH transcription factor has been reported for Arabidopsis (Pillitteri and Torii, 2012). To investigate the function of FAMA in MI development, the phenotypes of a fama loss-of-function mutant (fama-1 allele; Ohashi-Ito and Bergmann, 2006) were evaluated (Supplemental Figure 6K). This mutant fails to accumulate FAMA transcripts and instead of stomata produces abnormal clusters of small and narrow cells that lack guard cell fate and morphology (Ohashi-Ito and Bergmann, 2006). Compared with bolting wild-type plants (Supplemental Figure 6A), fama-1 mutants arrested growth as pale and stunted plants that only contain seven to eight small leaves before bolting and that later harbored small and infertile flowers (Supplemental Figures 6B and 6J). Whereas a rich vein network, visualized using dark-field microscopy, is present in the wild type (Supplemental Figure 6D), leaves of the same age in fama-1 contained only several veins in primary growth, such as a mid-vein and first and second loop veins (Supplemental Figure 6E). Thus, this mutant displays severe defects in growth and vascular development.
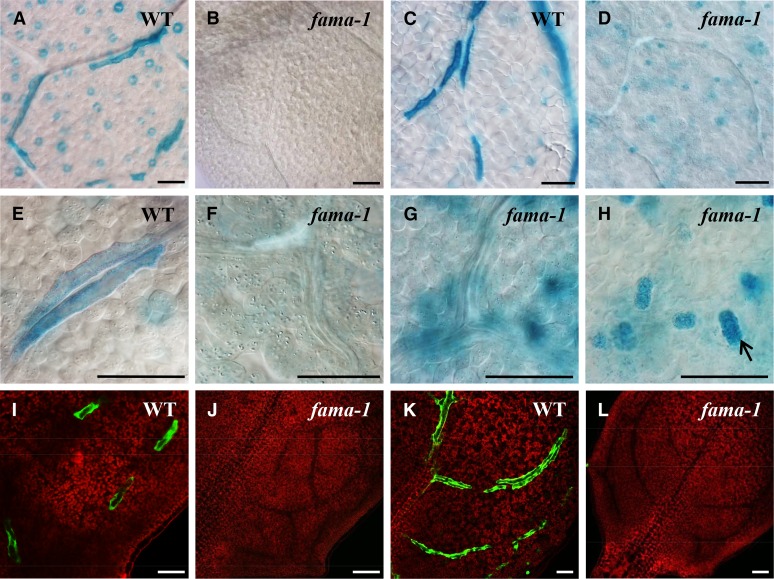
To further define fama phenotypes during MI development, three MI markers, FAMA, E1728, and TGG1, were transformed into wild-type and fama-1 mutant plants. In contrast to reporter-based expression in MIs in wild-type rosette leaves (Figure 4; Supplemental Figure 6G), in fama-1, FAMA promoter activity was only detected in clusters of narrow cells located in place of stomata (Figure 4; Supplemental Figure 6H). This mutant completely lacked any recognizable and definable MIs near procambial strands or the phloem (Figure 4) throughout the shoots, including mature leaves, inflorescence stems, as well as flowers (Supplemental Figures 6H to 6J). By contrast, the tgg1 tgg2 double mutant harbored MIs (as well as stomata) that were normally distributed (Supplemental Figures 7G and 7K).
Figure 4.
Loss-of-Function Phenotypes of GCs and MIs in fama-1 Leaves.
(A) Wild type showing ProTGG1:GUS expression in stomata and in MIs.
(B) Whole mount of leaf showing that fama-1 lacks ProTGG1:GUS expression.
(C) Wild type with ProFAMA:GUS staining in MIs.
(D) fama-1 lacks MIs. Note absence of stained linearly arranged cells. However, ProFAMA:GUS is expressed in clusters of cells (that are not stomata) but that are produced by the stomatal lineage. The roughly circular outline derives from the vasculature.
(E) A higher magnification showing ProTGG1:GUS expression in two adjacent MIs.
(F) Absence of ProTGG1:GUS staining resulting from the abrogation of MI fate by the loss of FAMA function. The elongated cells are part of the phloem.
(G) Note the absence of elongated, ProFAMA:GUS-expressing MIs in fama-1. The globular structures expressing GUS are out-of-focus epidermal cell clusters shown in (H).
(H) ProFAMA:GUS is expressed in clusters of cells (e.g., at arrow) that arise from excess symmetric divisions in GMCs in fama-1, but these cells do not form stomata.
(I) Expression of the E1728:YFPer marker in MIs.
(J) E1728:YFPer expression is absent in fama-1, which lacks MIs.
(K) Wild type showing ProFAMA:GFP fluorescence in elongated MIs.
(L) ProFAMA:GFP expression is absent from fama-1, which lacks MIs.
Images from (A) to (H) are from 4-week-old fifth to sixth rosette leaves using bright-field microscopy. (I) to (L) show confocal microscopy images of 2-week-old, third rosette leaf hydathode regions. Red fluorescence in (I) to (L) derives from PI staining in cell walls as well as from the autofluorescence of chloroplasts. Bars = 50 μm.
Because the fama mutant abolishes the presence of stomata as well as MIs, we asked whether mutations in the bHLH genes speechless (spch) and mute that also disrupt guard cell development (Supplemental Figures 7H and 7I; MacAlister et al., 2007) affect the presence or distribution of MIs. In contrast to fama, neither the spch-1 nor mute mutants altered MI development or distribution (Supplemental Figures 7L, 7M, 18A, and 18B and Supplemental Movies 10 and 11). Using ProSPCH:SPCH-GFP and ProMUTE:MUTE-GFP translational fusions, we found that neither SPCH nor MUTE was expressed outside the epidermis (Supplemental Movies 12 and 13 and Supplemental Figures 18C and 18D). And at the level of the whole plant, each mutant (spch and mute) was pale and stunted and had a reduced leaf vasculature compared with the wild type (Supplemental Figures 7A to 7F). MI development was not disrupted in two different types of stomatal mutants that harbor extra adjacent stomata, i.e., in four lips (flp-7 allele) and the closely related myb88 mutant (Lai et al., 2005) or in the double mutant (Supplemental Figures 7J and 7N).
Additionally, the complementation of fama-1 mutant plants with a ProFAMA:FAMA-GFP translational fusion (Lee et al., 2014) that restores stomata, also resulted in normal plant growth (e.g., a complete leaf vascular network was present) and reproduction as well as the presence of MIs (Supplemental Figures 6C, 6F, and 8D to F). Thus, the loss of FAMA function blocks the conferral of MI fate as well as MI expansion and morphogenesis.
Both types of myrosin cells accumulate the myrosinase TGG1, while MIs, but not guard cells, contain high concentrations of TGG2 (Barth and Jander, 2006). Quantitative RT-PCR analyses of 3-week-old shoot tissues from the wild type, both the fama-1 allele as well as the tgg1-3 tgg2-1 double mutant eliminated the transcription of both the TGG1 and TGG2 (Supplemental Figures 7O, 7P, 8B, and 8C). By contrast, FAMAtrans fama-1 plants, as well as FAMAtrans, spch-1, and mute plants, harbored low transcript levels of TGG1 and TGG2 (Supplemental Figures 7O, 7P, 8B, and 8C). Moreover, a second fama mutant allele, i.e., Salk_049126 (Supplemental Figure 9F), which was also pale and stunted and lacked stomata (Supplemental Figures 9A to 9C), also abolished the expression of both TGG transcripts (Supplemental Figures 9D and 9E). Thus, unlike other stomata-related regulators such as spch, mute, and the flp-7 myb88 double mutant, only FAMA activity leads to the transcription and activity of myrosinases, as well as MI differentiation in Arabidopsis.
FAMAtrans plants were previously shown to exhibit a gain of function, stoma-in-stoma phenotype in mature rosette leaves, as evidenced by the repeated loss and resetting of GC fate (Supplemental Figures 10B to 10D; Lee et al., 2014). By contrast, no comparable phenotype was found with respect to MIs, i.e., MIs did not form within preexisting idioblasts (Supplemental Figure 10A). However, the expression levels of TGG1 and TGG2 were reduced over 50% (Supplemental Figures 8B and 8C).
Characterization of Auxin Activity during Myrosin Idioblast Cell Development
Auxin accumulation in stomatal precursor cells and in the procambium leads to the differentiation of stomata and the vasculature as well as their correct patterning (Donner et al., 2009; Le et al., 2014). To determine whether MI development also involves auxin-mediated transcription, auxin activity during their development was monitored using auxin input and output markers (Brunoud et al., 2012). Transcription driven by the DR5 promoter has been shown to represent auxin-mediated output activity (Vernoux et al., 2011). YFP fluorescence from ProDR5rev:3XVENUS-N7 (Heisler et al., 2005) was analyzed during MI development. The YFP signal was high in nuclei in small and developing MIs (Figures 5A and 5B; Supplemental Movie 14 and Supplemental Figure 19A), but decreased during MI expansion (Figures 5C and 5D), indicating that auxin-mediated transcriptional output activity is strongest in early MIs. This result is similar to the decrease in DR5 activity that takes place after GMCs divide symmetrically and by the apparent lack of this signal in mature GCs (Le et al., 2014).
Figure 5.
Auxin Output and Input Activity in MIs of Developing and Mature Rosette Leaves.
(A) ProDR5:3XVENUS-N7 fluorescence (star) marks nuclei in two adjacent young MIs among immature tissues in a first leaf 7 DAG.
(B) Auxin-DR5 output activity is detected in nuclei of a young, elongating MI (star). This image from Supplemental Movie 14.
(C) As in (B), except later stage of MI expansion/development that shows little or no MI fluorescence.
(D) DR5 expression is absent in an older and larger MI.
(E) MI layer from 9 DAG seedling showing absence of Pro35S:DII-3XVENUS-N7 activity. Stars indicate elongating MIs.
(F) No DII fluorescence was detected in an MI layer from a 30 DAG plant. Star marks a differentiated MI. Cell walls and chloroplasts visualized via red fluorescence.
(G) Leaf spongy mesophyll cells exhibit DII-3XVENUS-N7 fluorescence.
(H) Leaf lower epidermis showing fluorescence from DII-3XVENUS-N7 in the nucleus of mature GCs.
The samples were taken from 12 DAG first leaves in (B) to (D) and 30 DAG plants in (F) to (H). All stars mark MIs. Cell walls visualized with PI. Bars = 10 μm.
In addition, expression of the auxin-related input marker DII-VENUS is repressed when local indole-3-acetic acid (IAA) levels become elevated (Brunoud et al., 2012). Pro35S:GFP expression was used as a control to estimate relative fluorescence intensity by comparing the epidermis to deeper tissues such as the vasculature (Supplemental Figure 11). In contrast to DR5, DII signal levels were relatively high during GC maturation (Figure 5H), consistent with the presence of low auxin levels and activity late in stomatal development (Le et al., 2014). However, DII-VENUS (Pro35S:DII-3XVENUS-N7) was only expressed in mature mesophyll cells surrounding leaf veins (Figure 5G), while no expression was detected during MI elongation and maturation (Figures 5E and 5F). These findings indicate that there is no significant decrease in local auxin levels during MI development. In addition, the expression patterns of these two markers (DR5 and DII) differ between the two types of myrosin cells.
Polar Auxin Transport and Vesicular Trafficking Regulate MI Patterning and Morphogenesis
Auxin efflux can be blocked chemically with the synthetic phytotropin 1-N-naphthylphthalamic acid (NPA) (Morris et al., 2010). In addition, the fungal toxin Brefeldin A (BFA) binds directly to multiple ARF-GEFs as well as to coat adaptors that are required for forming and transporting vesicles (Tanaka et al., 2014). BFA inhibits GNOM activity and blocks auxin efflux across the cell membrane (Steinmann et al., 1999; Geldner et al., 2001). The Arabidopsis GNOM gene encodes an ADP Ribosylation Factor-GTP Exchange Factor (ARF-GEF), a GTPase/small G-protein necessary for vesicle trafficking and cargo selection (Donaldson and Jackson, 2000). Both chemical inhibitors, as well as mutant gnom alleles, have been shown to induce numerous developmental defects including disrupting the continuity of vascular networks, as well as the formation of stomata abnormally in contact (Kleine-Vehn et al., 2009; Le et al., 2014).
To further characterize possible relationships between MI development and auxin efflux-related signaling, seedlings were treated separately with NPA and BFA. Both NPA and BFA reduced seedling growth and caused leaves to become small and round (Supplemental Figures 12A to 12C), but these chemicals affected MI and stomatal patterning in different ways. As shown by the GC and MI reporter markers E1728 and FAMA, MIs normally form a loose network associated with the vasculature (Figures 6A to 6D and 6M). However, NPA application caused MIs and vascular tissues to redistribute into leaf margins and petioles, with both MIs and the vasculature become radially arranged (Figures 6E, 6F, and 6N). After germination on NPA-containing medium, TGG1 transcript levels in 12 d after germination (DAG) seedlings were significantly upregulated, while those of TGG2 were unchanged (Figures 6P and 6Q). However, NPA did not appear to affect the morphology of GCs and MIs, even though the leaves became abnormally rounded (Figures 6G and 6H).
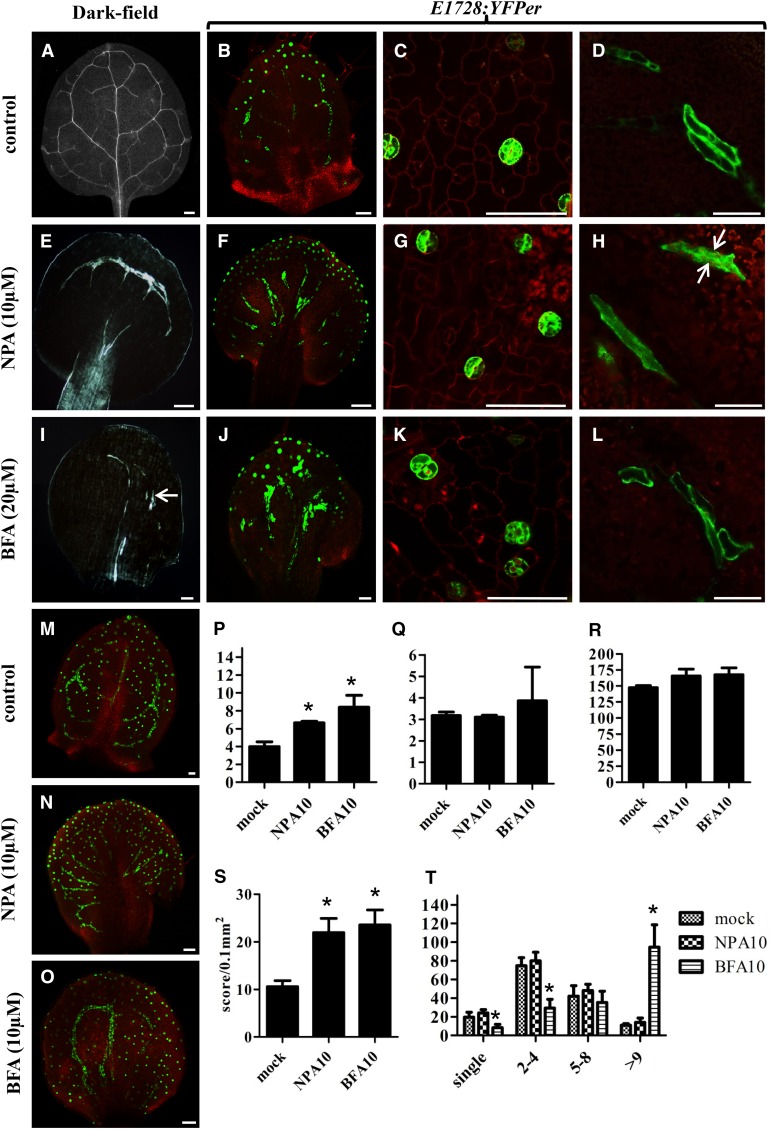
Figure 6.
Disruption of MI, Vascular, and Stomatal Patterning by the Auxin Pathway-Related Chemical Inhibitors NPA and BFA.
(A) to (O) MIs were visualized using ProFAMA:GFP fluorescence. Figures in (A) to (L) show 8 DAG first leaves grown on half-strength Murashige and Skoog medium, some of which contained the chemicals specified. (M) to (O) were as above except from 12 DAG third to fourth rosette leaves. All images in (A) to (O) were captured using confocal microscopy. Red fluorescence represents PI staining as well as autofluorescence from chloroplasts. Bars = 50 μm.
(A) Normal vascular patterning, dark-field microscopy.
(B) Normal distribution of stomata (dots in optical plane focused on stomata) as well as of linearly shaped MIs located deeper in the leaf visualized with E1728:YFPer fluorescence.
(C) and (D) Normal distribution and patterning of stomata and MIs, respectively.
(E) NPA (10 μM) collapses and radializes vascular distribution in the leaf margin and petiole. Dark-field microscopy.
(F) NPA radializes MI patterning between the leaf margin and the petiole.
(G) and (H) NPA does not significantly disrupt stomatal or MI morphology. Arrows in (H) mark two MIs in contact, a grouping also found in untreated plants.
(I) BFA (20 μM) shortens and disrupts the vasculature (arrow). Dark-field microscopy.
(J) BFA induces MI swelling and increases the number of MIs in contact.
(L) BFA induces MI swelling (cf. to length-to-width ratio of MIs in [D]).
(M) Normal distribution of stomata (dots) and MIs (lines) visualized with ProFAMA:GFP fluorescence.
(N) NPA (10 μM) radializes MI distribution between the leaf margin and petiole. MIs visualized by ProFAMA:GFP fluorescence.
(O) BFA (10 μM) increases the number of MIs in contact.
(P) Data from quantitative RT-PCR showing TGG1 transcript levels in 14 DAG mock, NPA, and BFA-treated plants. TGG1 expression was normalized to that of ACTIN2. Unpaired t tests were used to analyze statistical differences; significance at the level of P < 0.05 is indicated by an asterisk. Bars represent means of three replicates ± sd.
(Q) As in (P) except for TGG2 transcript levels.
(R) Total numbers of MIs scored from 12 DAG first leaves.
(S) Quantification of MI density in12 DAG first leaves. y axis indicates the number of MIs scored per leaf area (0.1 mm2). Each value is the mean calculated from five leaves in different plants from three biological replicates ± sd. Asterisks indicate significant differences compared with controls (mock treatment) analyzed by unpaired t tests (P < 0.01).
(T) Number of MIs in contact scored from leaves 12 DAG. x axis indicates the number of one or more MIs in direct contact. Each column represents the mean calculated from five first leaves using the ProFAMA:GFP MI marker and confocal microscopy. Bars represent the means from three replicates ± sd. Asterisks indicate statistical significance as analyzed by unpaired t tests (P < 0.01).
Treatment with NPA and BFA increases MI density by reducing leaf expansion (Figures 6M to 6O and 6S). BFA (but not NPA) generates discontinuities in the leaf vasculature (Figures 6I and 6J), and BFA also causes many MIs to form in direct contact (Figures 6O and 6T). Moreover, BFA induced GC and MI swelling (Figures 6K and 6L; Le et al., 2014). Using quantitative RT-PCR, only TGG1 transcript levels increased significantly, whereas those of TGG2 remained unchanged in 12 DAG seedlings treated with BFA (Figures 6P and 6Q). However, neither NPA nor BFA affected the total number of MIs in leaves or their location with respect to the vasculature (abaxial to leaf veins) (Figure 6R).
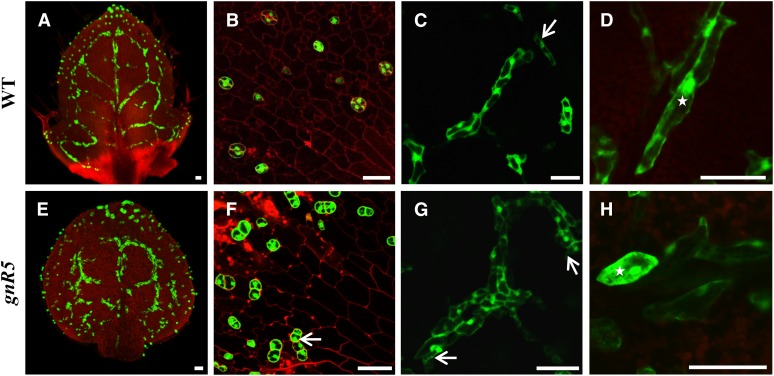
The gnomR5 allele mutant harbors a mutation in a single nucleotide and impairs GNOM function (Geldner et al., 2003). In the gnomR5 mutant, GCs formed abnormally in contact (Le et al., 2014). While wild-type plants frequently display MIs in contact (Figures 7A and 7C), this number increased in a gnomR5 background (Figures 7E and 7G). The gnomR5 allele also induced abnormal swelling of MIs (as well as of GCs) (Figures 7F and 7B), a phenotype also observed and quantified following BFA treatment (Figures 6O and 6T). These mutants, as well as chemically treated plants, showed decreased seedling growth and disrupted rosette leaf shape, as well as many short vascular bundles (Supplemental Figures 12D to 12F). Moreover, quantification of MI numbers in gnomR5 (visualized using a FAMA-GFP transcriptional-reporter fusion) confirmed that a significantly higher percentage of MIs formed abnormally in direct contact (Supplemental Figure 13C). However, the total number and density of MIs in gnomR5 mutant leaves were similar to that of the wild type (Supplemental Figures 13A and 13B).
Figure 7.
Wild-type and gnomR5 Phenotypes Visualized with ProFAMA:GFP Fluorescence in GCs and MIs.
(A) Wild-type rosette leaf showing MI and GC distribution using ProFAMA:GFP.
(B) Higher magnification of wild-type GC patterning.
(C) Normal patterning includes multiple MIs linearly arranged in contact (vertically arranged cluster), as well as side-by-side (lower right). Arrow indicates a single MI.
(D) Wild type showing higher magnification of groups of MIs with normal, elongated shape. Star indicates an irregularly elongated MI.
(E) Low magnification of MIs in the gnomR5 mutant showing many abnormally in contact.
(F) Aberrantly swollen GCs in gnomR5. Arrow denotes two stomata in direct contact.
(G) Loss of GNOM function increases the number of MIs in contact. Arrows indicate a large MI cluster. Higher magnification of the same leaf in (E).
(H) Abnormally swollen MIs in gnomR5 mutant. Star indicates a single MI.
All samples were from 10 DAG first rosette leaves visualized using confocal microscopy. Red fluorescence derives from PI staining as well as from chloroplast autofluorescence. Bars = 50 μm.
VAM3 encodes Syntaxin of Plants 22 that normally contributes to the regulation of vesicular trafficking (Shirakawa et al., 2009). The loss of VAM3 function induces the formation of excessive numbers of MIs (Ueda et al., 2006), as well as the elevated expression of both TGG1 and TGG2 (Supplemental Figures 12G and 12H). In contrast to vam3-1, the gnomR5 mutant did not harbor any significant changes in the transcription levels of TGG1 and TGG2 (Supplemental Figures 12G and 12H). Collectively, these data show that the normal distribution and shape of myrosin idioblasts depend upon GNOM-mediated vesicular trafficking as well as polar auxin efflux.
DISCUSSION
The bHLH Transcription Factor FAMA Regulates the Fate of Both Types of Myrosin Cells
This work reveals that MI development is marked by the expression of FAMA as well as the enhancer trap E1728. Previously both markers had only been shown to exhibit expression in guard cells (Figure 1; Hachez et al., 2011). These findings reveal that FAMA displays additional functions in promoting both MI identity and development (Supplemental Figure 14). Moreover, FAMA confers the cell fate of both types of myrosin cells (Figure 4), as well as regulates the expression of two key myrosinase genes, TGG1 and TGG2 (Supplemental Figures 7 to 9). Thus, this study provides possible entrée into analyzing the regulation of myrosin cell formation and the events upstream of myrosinase expression.
FAMA regulatory networks activate stomatal differentiation genes and directly target and regulate cell cycle genes such as CDKB1;1, which is essential for GMC symmetric division (Hachez et al., 2011). Thus, FAMA likely acts as both a transcriptional activator and a repressor in the stomatal pathway. These findings might help identify genes that directly regulate MI differentiation and number, such as those related to cell cycling and MI cell growth. In addition, the transformation of a native FAMA gene into wild-type or fama mutant plants can induce a gain-of-function phenotype, that of a stoma forming inside a preexisting guard cell (Lee et al., 2014). Such transformants also disrupt epigenetic marks on stem cell genes in the stomatal lineage (Lee et al., 2014), raising the possibility that FAMA-conferred MI fate is also epigenetically maintained.
It remains to be seen whether FAMA regulates the identity of MIs and GCs in other crucifers, but since putative orthologs of FAMA genes also regulate stomatal development in grasses (Liu et al., 2009), exploring this possibility might be fruitful.
Comparison of the Origins of MIs, Stomata, and the Vasculature
MI initiates as a square-shaped ground meristem cell, then undergoes cell elongation and expansion of nucleus and vacuole and finally forms an irregularly shaped idioblast (Supplemental Figure 14). Although GCs as well as MIs are myrosin cells, their development differs significantly. Stomata arise via successive cell types that include stem cells (MMCs and meristemoids) as well as GMCs (Pillitteri and Torii, 2012). The loss of FAMA function leads to two different phenotypes in the stomatal lineage, an absence of GC fate, as well as clusters of cells that form in place of guard cells and that arise from extra symmetric divisions. GMCs have a specific cell fate and are strictly separated from each other (Pillitteri and Torii, 2012). By contrast, MI precursor cells arise from adjacent as well as single ground meristem cells (Figures 3A and 3G) sometimes resulting in MIs forming in direct contact in the wild type. Since adjacent MIs are often offset positionally from each other, it is unlikely that both arise from the division of the same parent cell.
The spch-1, mute, and fama-1 alleles result in plants that exhibit poor growth as well as reduced vascular formation, and infertility (Supplemental Figures 6 and 8). However, only fama-1 eliminates MI formation (Figure 4; Supplemental Figures 7 and 8). Clearly, the lack of stomata in these three mutants prevents gas exchange and thus greatly reduces plant productivity, which might in turn lead to a secondary defect in vascular development. Moreover, the loss of myrosin cells and of selected TGG functions presumably makes fama plants more susceptible to predation, since half of the glucosinolate-myrosinase defense system is absent. Although MIs do not derive from the procambium, their proximity to veins normally deters a certain degree of phloem feeding, such as by insects. Seedling growth is severely inhibited in fama plants presumably, in part, because vein networks are so poorly developed.
MIs originate from young ground meristem cells located under elongating procambial strands (Supplemental Movies 2 to 9 and Supplemental Figures 16 and 17). Young MIs as well as procambial strands exhibit high auxin transcriptional output activity as shown by the DR5 marker (Figure 5; Supplemental Movie 14 and Supplemental Figure 19A). Auxin maxima and transport are required for procambial patterning (Scarpella et al., 2010). Very high auxin levels are found in hydathodes and leaf tips where IAA is transported from the leaf margins to inner tissues via the polar localization of PIN proteins (Scarpella et al., 2006). The auxin efflux inhibitor NPA, that blocks PIN function, reduces auxin transport, and leads to the formation of excess vascular tissue in leaf margins as well as in the mid-region of the leaf (Scarpella et al., 2006). In addition, the chemical inhibitor BFA has been shown to block GNOM-mediated PIN endosomal recycling resulting in the formation of excessive numbers of veins of which many are discontinuous (Drdová et al., 2013; Tanaka et al., 2014). Our results show that NPA radializes the distribution of MIs much like the effect of this chemical on the vasculature (Figures 6E, 6F, and 6N); by contrast, BFA application increases the number of MIs in direct contact with each other (Figure 6I, 6J, and 6O). Thus, the spacing of the procambium and MIs in leaves is regulated by local auxin activity, and is altered by chemical inhibitors of auxin efflux. It is also possible that PIN proteins act during MI development (Supplemental Movies 15 and 16 and Supplemental Figures 19B and 19C). Collectively, these data are consistent with the auxin pathway coregulating the number and spacing of MIs in addition to its well known effect on the vasculature.
MI Morphogenesis and Patterning Require AtVAM3, COV1, and GNOM Endosomal Trafficking
In addition to the Arabidopsis Q-SNARE protein, VAM3, MI number is also regulated by the trans-Golgi network-localized membrane protein COV1 that contributes to the morphogenesis of the Golgi apparatus (Ueda et al., 2006; Shirakawa et al., 2014). The vam3 mutant shows a slight reduction in the number of veins, as well as in their junctions (Ueda et al., 2006). VAM3 likely induces excess MI formation, and these cells become abnormally arranged in contact (Ueda et al., 2006). Similarly, the loss of COV1 function disrupts and increases vein junctions as well as elevating the number of MI cells (Shirakawa et al., 2014). VAM3 is constitutively expressed in meristematic and developing cells and controls the docking and fusion of vesicles with the tonoplast membrane (Shirakawa et al., 2009). Although the gnom mutants do not elevate total MI number as well as the transcription levels of TGG1 and TGG2 (Supplemental Figures 12 and 13), we found that the number of Idioblasts in contact increased when GNOM function is abrogated (Figure 7; Supplemental Figure 13). The loss of GNOM function also induces gaps in the vasculature yielding some discontinuities (Geldner et al., 2003). This defect arises when auxin transport is disrupted and instead flows ectopically through inner tissues (Naramoto et al., 2009; Friml, 2010). Directional auxin efflux depends upon the polar localization of PIN proteins in the cell membrane that involves PIN recycling between the cell membrane and the endosomal compartment (Drdová et al., 2013). Previous reports showed that both GNOM and VAM3 are required for the polar localization and cycling of PIN proteins, a process that regulates vectorial auxin transport as well as IAA accumulation in the procambial/provascular cell lineages (Shirakawa et al., 2009; Ohashi-Ito and Fukuda, 2010). These data are consistent with GNOM, COV1, and VAM3 collectively regulating MI and vascular distribution, e.g., by disrupting PIN localization during endosomal trafficking that in turn leads to abnormal auxin transport. However, it cannot be excluded that an auxin-independent pathway operates, e.g., by VAM3 affecting TGG enzymatic activity (Ueda et al., 2006).
Evolutionary Significance of Myrosin Cells and Glucosinolate Cells in the Mustard Oil Bomb System
Myrosinase genes have been characterized in B. napus and S. alba as well as in Arabidopsis (Morant et al., 2008). The functions of these enzymes, as well as those in the glucosinolate biosynthesis pathway, are well described (Wang et al., 2011), as are the spatial distributions of the cells that synthesize and store myrosinases (Kissen et al., 2009). We found two types of myrosin cells present in Arabidopsis, in the epidermis and outside of the phloem (Figure 2). The rupture of these cells located in or relatively close to the epidermis, by fungi and insects, activates glucosinolate breakdown, thus deterring deeper penetration, e.g., into the stem cortex and the phloem (Chen et al., 2010). Thus, myrosin cells provide two lines of defense that reduce attacks to stomata and that discourage predator access to sugars in the phloem.
Similar to myrosinases in MIs, glucosinolates are especially abundant in sulfur-rich idioblasts termed S-cells (Bones et al., 1991). Although recent studies suggest that both myrosinases and glucosinolates are stored in separate subcellular compartments in the same cells (Koroleva and Cramer, 2011), S-cells can form groups or clusters close to MIs, and S-cells can also be located between the endodermis and phloem of each vascular bundle (Supplemental Figure 15; Kissen et al., 2009). Glucosinolate biosynthesis is costly metabolically and diverts energy from plant growth (Kliebenstein, 2013). The accumulation of highly concentrated glucosinolates can lead to organelle degradation and even programmed cell death (Koroleva et al., 2010). While S-cells are limited in number, they are located strategically. The proximity of S-cells and MIs means that the contents of both cell types can be simultaneously disrupted by predation. This in turn leads to interactions between myrosinases and glucosinolates that foster the enzymatic reactions that characterize the “mustard oil bomb” and thus deter predation. Thus, the proximity of myrosin cells and S-cells discourages predation while minimizing metabolic costs.
The MI/S-cell system has only been described in crucifers. Myrosinases and glucosinolates appear to be unique β-glucosidases and glucosides that are present in most members of the mustard family. However, some insect herbivores have parallel adaptations representing coevolution with their Brassica host plants. For example, some specialist aphids in the Brassicaceae store myrosinases in crystalline microbodies in their digestive system and defend against predators by ingesting plant glucosinolates (Bridges et al., 2002). Parallel or comparable defense mechanisms have also been identified in other angiosperm families, such as flavonoids and corresponding glucosides in legumes (Morant et al., 2008). This raises the possibility that cellular antipredation systems comparable to that of MIs/GCs might also have evolved in other angiosperm species. Thus, our identification of a gene and a hormone that together regulate MI development might help in defining novel genes that regulate glucosinolate-rich cells and their pathway, as well in recognizing novel antipredation cell types in crucifers and other angiosperms.
METHODS
Plant Materials and Growth Conditions
Wild type Arabidopsis thaliana Columbia (Col-0) plants were used, including as the background for transgenes and all mutants. The ProTGG1:GUS reporter line and tgg1 tgg2 double mutant were obtained from Barth and Jander (2006). Seeds of ProDR5:3XVENUS-N7 were as described (Heisler et al., 2005), and the 35S:DII-3XVENUS-N7 seeds as described by Vernoux et al. (2011). Seeds harboring ProPIN3:PIN3-GFP were obtained from Le et al. (2014). Two procambial markers, ProAt-HB8:HTA6-YFP and Q0990 (an enhancer trap), were used as described by Scarpella et al. (2010). The E1728 enhancer trap line, as well as other stomatal reporter lines including ProFAMA:GUS, ProFAMA:GFP, and ProFAMA:FAMA-GFP, were as described (Lee et al., 2014). Seeds of the vam3-1 mutant allele, i.e., SALK_060946, an allele previously reported by Saito et al. (2011), were obtained from Masao Tasaka (Nara Institute, Japan). The ProSPCH:SPCH-GFP reporter line and the spch-1 null mutant allele were obtained from MacAlister et al. (2007), while the ProMUTE:MUTE-GFP reporter line and mute mutants were as described by Pillitteri et al. (2007). Seeds of the fama Salk 049126 allele were obtained from the ABRC. The fama-1 (Salk_100073) and the gnomR5 mutant alleles were described by Ohashi-Ito and Bergmann (2006) and Geldner et al. (2003), respectively, and were confirmed by genotyping and mutant phenotypes. Primers used for genotyping are in Supplemental Table 1.
Chemical Inhibition
Seeds were germinated on media containing 10 μM NPA or 20 μM BFA (Sigma-Aldrich). These chemicals were dissolved in DMSO (Sigma-Aldrich) and then added to half-strength Murashige and Skoog plant growth medium. The final DMSO concentration was <0.1% (v/v).
GUS Activity
To check GUS activity, all samples were dissected and vacuum infiltrated in a 5-bromo-4-chloro-3-indolyl-d-glucuronide (X-Gluc) solution for 15 min at room temperature and then incubated at 37°C in the dark for 16 to 24 h as described (Malamy and Benfey, 1997). X-Gluc-treated samples were rinsed with water and then fixed in ethanol:acetic acid (6:1 [v/v]) for 2 h at room temperature. Samples were rinsed with 95% (v/v) ethanol and placed successively in 70, 50, and 30% (v/v) ethanol each for 10 min and then mounted in chloral-hydrate:glycerol:water (8:1:2 [g/v/v]) as described (Berleth and Jurgens, 1993). Tissue samples were mounted on microscope slides and observed using differential interference contrast optics.
Microscopy
Selected low-magnification images of the leaf vasculature were acquired using dark-field optics and an Olympus SZX10 stereomicroscope. Samples stained for GUS were visualized using oil immersion differential interference contrast objectives on an Olympus AX-70 microscope or a Nikon ECLIPSE 80i confocal microscope. For two-channel fluorescence imaging using GFP/YFP and propidium iodide (PI) fluorescence filter sets, fresh tissue was immersed in 1 mg/mL PI solution for 5 min and then rinsed briefly with water before visualization with the Nikon microscope. Confocal fluorescence images including quantitative data (e.g., leaf area) were processed using ImageJ and Adobe Photoshop 5.0 software. The expression patterns shown in the figures were representative and were based upon at least 20 separate images from different plants.
Generation of Transgenic Plants
Plants harboring mutations (fama-1 or gnomR5) or reporters (ProTGG1:GUS, ProFAMA:GUS, ProFAMA:GFP, and E1728:YFPer) were obtained via introgression and then by selecting F1 and F2 plants by phenotyping or reporter visualization. Individual F2 lines were selected on plates using kanamycin (50 μg/mL) or hygromycin (25 μg/mL) and then confirmed using PCR-based genotype analysis with primers described above.
RNA Extraction, RT-PCR, and Real-Time PCR
Total RNA was extracted from 3-week-old whole shoot tissues using an RNeasy Plant Mini Kit (Qiagen) and then treated with DNase (Invitrogen) to remove genomic DNA. First-strand cDNAs were generated by 1 mg RNA using Thermo Script Reverse Transcriptase and an oligo(dT) 20 primer (Invitrogen). Real-time PCR was performed with 1 μL of the first-strand template at 98°C for 30 s, 56°C for 30 s, and 72°C for 30 s, with 25 cycles. The primers TGG1-F(ex7) and TGG1-R(ex8) were designed to amplify TGG1 transcripts, TGG2-F(ex7) and TGG2-R(ex8) to amplify TGG2 transcripts, and ACT2-F and ACT2-R to amplify ACTIN transcripts (see Supplemental Table 2 for primer details). PCR products for RT-PCR underwent electrophoresis using 1% agarose gels and were visualized by staining with Safe View Nucleic Acid Stain (NBS). Real-time quantitative PCR based on SYBR-Green chemistry was performed using the same conditions and gene-specific primers described above. Real-time quantitative PCR was monitored over 40 cycles using a Bio-Rad iCycler (iQ5) Multicolor Real-Time PCR detection system. Gene expression levels are shown in supplemental figures as the ratio relative to the ACT2 gene control and were calculated using the 2^ (2Δ Δ Ct) method. Two biological replicates were performed for each real-time PCR experiment.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: TGG1, AT5G26000; TGG2, AT5G25980; TGG3, AT5G48375; TGG4, AT1G47600; TGG5, AT1G51470; TGG6, AT1G51490; FAMA, AT3G24140; COV1, AT2G20120; VAM3, AT5G46860; At-HB8, AT4G32880; SPCH, AT5G53210; MUTE, AT3G06120; FLP, AT1G14350; MYB88, AT2G02820; GNOM, AT1G13980; PIN1, AT1G73590; and PIN3, AT1G70940.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A ProFAMA:GUS Transcriptional Fusion Is Expressed throughout the Shoot in GCs and MIs.
Supplemental Figure 2. Coexpression of FAMA and E1728 with TGG1 in MIs.
Supplemental Figure 3. A FAMA-GFP Translational Fusion (ProFAMA:FAMA-GFP) Is Expressed in Young but Not Mature GCs and MIs.
Supplemental Figure 4. GFP Expression Patterns of Procambial and MI Markers.
Supplemental Figure 5. Visualization of MIs with Three Different Fluorescent Markers Showing Positional Relationships between MIs and Surrounding Tissues.
Supplemental Figure 6. Plant and Vasculature Phenotypes of Three Genotypes Compared: the Wild Type, fama-1, and FAMA trans fama-1 Plants.
Supplemental Figure 7. MIs Are Still Present in spch-1, mute, and Double Mutants tgg1-3 tgg2-1 and flp-7 myb88.
Supplemental Figure 8. Test for Complementation as well as qRT-PCR Analysis of Wild-type, fama-1, and FAMA trans fama-1 Plants.
Supplemental Figure 9. Plant Phenotypes in fama Mutant Allele SALK_049126.
Supplemental Figure 10. Gain-of-Function Phenotypes in FAMA trans Plants.
Supplemental Figure 11. Myrosin Idioblasts Are Located between the Procambium and the Mesophyll as Shown in One of Four Different Focal Planes Using Ubiquitous 35S:GFP Expression.
Supplemental Figure 12. Plant Phenotypes of Wild-Type, Chemical-Treated, and gnomR5 Seedlings.
Supplemental Figure 13. Quantification of MI Number in Wild-Type and gnomR5 Mutant Backgrounds.
Supplemental Figure 14. Diagram Showing Developmental Model of a Single Myrosin Idioblast.
Supplemental Figure 15. Diagram Reproduced from Kissen et al. (2009) Showing the Distribution of Myrosin Cells and Glucosinolate-Rich/S-Cells in a Partial Cross Section of an Arabidopsis Stem.
Supplemental Figure 16. Each Row Shows Three Frames from Z-Stacks of GCs and MIs from Supplemental Movies, with Expression from FAMA andE1728.
Supplemental Figure 17. Each Shows Three Frames from Supplemental Movies from Z-Stacks of Procambial Expression Markers At-HB8 and Q0990.
Supplemental Figure 18. Three Frames (First, Middle, and Last) from Supplemental Movies of spch-1 and mute Mutants as well as SPCH and MUTE Expression.
Supplemental Figure 19. Three Frames (First, Middle, and Last) from Supplemental Movies Showing Expression from the Auxin Activity Markers DR5, PIN1, and PIN3.
Supplemental Table 1. List of Primers and DNA Sequences Used for Genotype Analysis.
Supplemental Table 2. List of Primers and DNA Sequences Used for Real-Time PCR.
Supplemental Movie 1. ProFAMA:GFP Expression in Mature Seed Cotyledon.
Supplemental Movie 2. ProFAMA:cFAMA-GFP Expression in Differentiating Hydathode Region.
Supplemental Movie 3. High Magnification of ProFAMA:cFAMA-GFP Expression in Young and Elongating MI.
Supplemental Movie 4. ProFAMA:GFP Expression in a Differentiating Hydathode Region.
Supplemental Movie 5. High Magnification of ProFAMA:GFP Expression in Differentiating Hydathode Region.
Supplemental Movie 6. E1728 Expression in Differentiating Hydathode Region.
Supplemental Movie 7. ProATHB8:HTA6-YFP Expression in Differentiating Hydathode Region.
Supplemental Movie 8. Q0990:GFPer Expression in Differentiating Hydathode Region.
Supplemental Movie 9. High Magnification of Q0990:GFPer Expression in Differentiating Hydathode Region.
Supplemental Movie 10. spch-1 Mutant Leaf.
Supplemental Movie 11. mute Mutant Leaf.
Supplemental Movie 12. ProSPCH:SPCH-GFP Expression in Differentiating Hydathode Region.
Supplemental Movie 13. ProMUTE:MUTE-GFP Expression in Differentiating Hydathode Region.
Supplemental Movie 14. ProDR5:3XVENUS-N7 Expression Near Differentiating Procambium Region.
Supplemental Movie 15. ProDR5:3XVENUS-N7 and ProPIN3:PIN3-GFP Expression Near Differentiating Hydathode Region.
Supplemental Movie 16. ProPIN1:PIN1-GFP Expression Near Differentiating Hydathode Region.
Supplemental Movie Legends.
Supplementary Material
Acknowledgments
Seeds of fama-1 and spch-1 as well as those harboring ProFAMA:FAMA-GFP and ProSPCH:SPCH-GFP constructs were obtained from Dominique Bergmann and EunKyoung Lee, respectively. Seeds of the ProMUTE:MUTE-GFP reporter line and the mute mutant were a gift from Keiko Torii’s lab. We thank Jie Le for valuable discussions, especially about the role of auxin in MI development. George Barth provided seeds of ProTGG1:GUS and of the tgg1 tgg2 double mutant. Enrico Scarpella gave us seeds harboring ProATHB:HTA6-YFP as well as Q0990 constructs. Markus Heisler graciously provided seeds harboring ProDR5:3XVENUS-N7, and Teva Vernoux and Masao Tasaka provided seeds harboring a 35S:DII-3XVENUS-N7 construct as well as vam3-1 seeds.
AUTHOR CONTRIBUTIONS
M.L. and F.D.S. conceived and designed the research and wrote the article. M.L. performed the research. Both authors read and approved the article.
Glossary
- GC
guard cell
- bHLH
basic helix-loop-helix
- GMC
guard mother cell
- MI
myrosin idioblast
- IAA
indole-3-acetic acid
- NPA
1-N-naphthylphthalamic acid
- BFA
Brefeldin A
- DAG
days after germination
- PI
propidium iodide
Footnotes
Online version contains Web-only data.
References
- Ahuja I., Rohloff J., Bones A.M. (2010). Defense mechanisms of Brassicaceae: implications for plant–insect interactions and potential for integrated pest management. Agron. Sustain. Devel. 30: 311–348. [Google Scholar]
- Andréasson E., Bolt Jørgensen L., Höglund A.S., Rask L., Meijer J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127: 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46: 549–562. [DOI] [PubMed] [Google Scholar]
- Borgen B.H., Thangstad O.P., Ahuja I., Rossiter J.T., Bones A.M. (2010). Removing the mustard oil bomb from seeds: transgenic ablation of myrosin cells in oilseed rape (Brassica napus) produces MINELESS seeds. J. Exp. Bot. 61: 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Jurgens G. (1993). The role of the MONOPTEROS gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587. [Google Scholar]
- Bridges M., Jones A.M., Bones A.M., Hodgson C., Cole R., Bartlet E., Wallsgrove R., Karapapa V.K., Watts N., Rossiter J.T. (2002). Spatial organization of the glucosinolate-myrosinase system in brassica specialist aphids is similar to that of the host plant. Proc. Biol. Sci. 269: 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Bones A.M., Thangstad O.P., Haugen O., Espevik T. (1991). Fate of myrosin cells - Characterization of monoclonal antibodies against myrosinase. J. Exp. Bot. 42: 1541–1549. [Google Scholar]
- Chadchawan S., Bishop J., Thangstad O.P., Bones A.M., Mitchell-Olds T., Bradley D. (1993). Arabidopsis cDNA sequence encoding myrosinase. Plant Physiol. 103: 671–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., Jackson C.L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12: 475–482. [DOI] [PubMed] [Google Scholar]
- Donner T.J., Sherr I., Scarpella E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246. [DOI] [PubMed] [Google Scholar]
- Drdová E.J., Synek L., Pečenková T., Hála M., Kulich I., Fowler J.E., Murphy A.S., Zárský V. (2013). The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 73: 709–719. [DOI] [PubMed] [Google Scholar]
- Eriksson S., Andréasson E., Ekbom B., Granér G., Pontoppidan B., Taipalensuu J., Zhang J., Rask L., Meijer J. (2002). Complex formation of myrosinase isoenzymes in oilseed rape seeds are dependent on the presence of myrosinase-binding proteins. Plant Physiol. 129: 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, K. (1965). Plant Anatomy, 2nd ed. (New York: McGraw-Hill). [Google Scholar]
- Friml J. (2010). Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235. [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Hachez C., Ohashi-Ito K., Dong J., Bergmann D.C. (2011). Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 155: 1458–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Ann. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Husebye H., Chadchawan S., Winge P., Thangstad O.P., Bones A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128: 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O.A., Gibson T.M., Cramer R., Stain C. (2010). Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J. 64: 456–469. [DOI] [PubMed] [Google Scholar]
- Koroleva O.A., Cramer R. (2011). Single-cell proteomic analysis of glucosinolate-rich S-cells in Arabidopsis thaliana. Methods 54: 413–423. [DOI] [PubMed] [Google Scholar]
- Kissen R., Rossiter J.T., Bones A.M. (2009). The “mustard oil bomb”: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 8: 69–86. [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J. (2013). Making new molecules—evolution of structures for novel metabolites in plants. Curr. Opin. Plant Biol. 16: 112–117. [DOI] [PubMed] [Google Scholar]
- Lai L.B., Nadeau J.A., Lucas J., Lee E.K., Nakagawa T., Zhao L., Geisler M., Sack F.D. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17: 2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., et al. (2014). Auxin transport and activity regulate stomatal patterning and development. Nat. Commun. 5: 3090. [DOI] [PubMed] [Google Scholar]
- Lee E., Lucas J.R., Goodrich J., Sack F.D. (2014). Arabidopsis guard cell integrity involves the epigenetic stabilization of the FLP and FAMA transcription factor genes. Plant J. 78: 566–577. [DOI] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K., Bergmann D.C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276. [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Morant A.V., Bjarnholt N., Kragh M.E., Kjaergaard C.H., Jørgensen K., Paquette S.M., Piotrowski M., Imberty A., Olsen C.E., Møller B.L., Bak S. (2008). The β-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus. Plant Physiol. 147: 1072–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.A., Friml J., Zažímalová E. (2010). The transport of auxins. In Plant Hormones: Biosynthesis, Signal Transduction, Action!, P.J. Davies, ed (Dordrecht, The Netherlands: Springer), pp. 451–484. [Google Scholar]
- Naramoto S., Sawa S., Koizumi K., Uemura T., Ueda T., Friml J., Nakano A., Fukuda H. (2009). Phosphoinositide-dependent regulation of VAN3 ARF-GAP localization and activity essential for vascular tissue continuity in plants. Development 136: 1529–1538. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Fukuda H. (2010). Transcriptional regulation of vascular cell fates. Curr. Opin. Plant Biol. 13: 670–676. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42: 93–113. [PubMed] [Google Scholar]
- Saito C., Uemura T., Awai C., Tominaga M., Ebine K., Ito J., Ueda T., Abe H., Morita M.T., Tasaka M., Nakano A. (2011). The occurrence of ‘bulbs’, a complex configuration of the vacuolar membrane, is affected by mutations of vacuolar SNARE and phospholipase in Arabidopsis. Plant J. 68: 64–73. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Barkoulas M., Tsiantis M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Koumoto Y., Fuji K., Nishiyama C., Kohchi T., Hara-Nishimura I., Shimada T. (2014). CONTINUOUS VASCULAR RING (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol. 55: 764–772. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Shimada T., Nishiyama C., Hara-Nishimura I. (2009). Vacuolar SNAREs function in the formation of the leaf vascular network by regulating auxin distribution. Plant Cell Physiol. 50: 1319–1328. [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C.L., Paris S., Gälweiler L., Palme K., Jürgens G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Nodzyłski T., Kitakura S., Feraru M.I., Sasabe M., Ishikawa T., Kleine-Vehn J., Kakimoto T., Friml J. (2014). BEX1/ARF1A1C is required for BFA-sensitive recycling of PIN Auxin transporters and auxin-mediated development in Arabidopsis. Plant Cell Physiol. 55: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., Takahashi T., Ohtomo I., Nishimura M., Hara-Nishimura I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47: 164–175. [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu J., Sun S., Liu B., Cheng F., Sun R., Wang X. (2011). Glucosinolate biosynthetic genes in Brassica rapa. Gene 487: 135–142. [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7: 263–270. [DOI] [PubMed] [Google Scholar]
- Xue J.P., Lenman M., Falk A., Rask L. (1992). The glucosinolate-degrading enzyme myrosinase in Brassicaceae is encoded by a gene family. Plant Mol. Biol. 18: 387–398. [DOI] [PubMed] [Google Scholar]
- Zhang J., Pontoppidan B., Xue J., Rask L., Meijer J. (2002). The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant. 115: 25–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.