This work shows that the guard cell master regulator FAMA has a role in idioblast myrosin cell differentiation in Arabidopsis. Brassicales plants evolved a common regulatory pathway for specification of two distinct types of leaf cells: epidermal guard cells for gas exchange and myrosin cells in leaf inner tissue for defense.
Abstract
Brassicales plants, including Arabidopsis thaliana, have an ingenious two-compartment defense system, which sequesters myrosinase from the substrate glucosinolate and produces a toxic compound when cells are damaged by herbivores. Myrosinase is stored in vacuoles of idioblast myrosin cells. The molecular mechanism that regulates myrosin cell development remains elusive. Here, we identify the basic helix-loop-helix transcription factor FAMA as an essential component for myrosin cell development along Arabidopsis leaf veins. FAMA is known as a regulator of stomatal development. We detected FAMA expression in myrosin cell precursors in leaf primordia in addition to stomatal lineage cells. FAMA deficiency caused defects in myrosin cell development and in the biosynthesis of myrosinases THIOGLUCOSIDE GLUCOHYDROLASE1 (TGG1) and TGG2. Conversely, ectopic FAMA expression conferred myrosin cell characteristics to hypocotyl and root cells, both of which normally lack myrosin cells. The FAMA interactors ICE1/SCREAM and its closest paralog SCREAM2/ICE2 were essential for myrosin cell development. DNA microarray analysis identified 32 candidate genes involved in myrosin cell development under the control of FAMA. This study provides a common regulatory pathway that determines two distinct cell types in leaves: epidermal guard cells and inner-tissue myrosin cells.
INTRODUCTION
Plants have evolved various strategies for herbivore defense, including the release of toxic compounds. The myrosinase (thioglucoside glucohydrolase [TGG])-glucosinolate defense system is characteristic of Brassicales. When herbivores damage tissues, myrosinase is released from its subcellular compartment to interact with its substrate glucosinolate, and the reaction products are toxic to herbivores (Rask et al., 2000; Wittstock and Halkier, 2002; Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Hopkins et al., 2009; Kissen et al., 2009). Large amounts of myrosinase are stored in myrosin cell vacuoles (Rask et al., 2000; Andréasson et al., 2001; Husebye et al., 2002; Ueda et al., 2006), whereas the glucosinolate substrates are stored in different cells at the leaf periphery and along veins (Koroleva et al., 2000; Shroff et al., 2008). Myrosin cells were first discovered as idioblasts by Heinricher in 1884 (Heinricher, 1884). They were designated as myrosin cells by Guignard in 1890 (Guignard, 1890). Arabidopsis thaliana myrosin cells specifically develop along leaf veins (Xue et al., 1995; Andréasson et al., 2001; Husebye et al., 2002; Thangstad et al., 2004; Barth and Jander, 2006; Ueda et al., 2006). Several mutants with defective myrosin cell distribution have been identified (Ueda et al., 2006; Shirakawa et al., 2010, 2014). However, the molecular mechanism regulating myrosin cell development is largely unknown.
Stomatal guard cells function as specialized valves that mediate vapor and gas exchange in plants. Guard cell differentiation proceeds through a series of steps originating from meristemoid mother cells (Nadeau and Sack, 2002; Lau and Bergmann, 2012; Pillitteri and Torii, 2012; Pillitteri and Dong, 2013) and is positively regulated by two distinct basic helix-loop-helix (bHLH) transcription factor subfamilies. One subfamily contains three paralogs, SPEECHLESS (SPCH), MUTE, and FAMA, which regulate distinct developmental steps (Bergmann et al., 2004; Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). These three paralogs are not functionally exchangeable (MacAlister et al., 2007; MacAlister and Bergmann 2011). The other subfamily contains two paralogs, ICE1/SCREAM (SCRM) and SCRM2/ICE2, which redundantly regulate all steps of stomatal development (Kanaoka et al., 2008). Three different bHLH heterodimers, SPCH-ICEs, MUTE-ICEs, and FAMA-ICEs, are proposed to specifically promote the three distinct differentiation steps of stomatal lineages (Kanaoka et al., 2008). ICE1 and SCRM2 also function in freezing tolerance regulation (Chinnusamy et al., 2003; Fursova et al., 2009), but no other biological functions are reported for SPCH, MUTE, and FAMA.
We performed in silico analysis to identify transcription factors that were coexpressed with myrosinase-glucosinolate system genes and identified FAMA as an essential component for myrosin cell differentiation. Before differentiation of stomatal lineages in leaf primordia, a subset of ground meristem cells transiently expresses FAMA; these cells subsequently differentiate into idioblasts (myrosin cells) expressing myrosinase. Differentiation of myrosin and guard cells requires ICE1 and SCRM2. By contrast, guard cell differentiation requires SPCH and MUTE, but these are not required for myrosin cell differentiation. Our study elucidates the molecular mechanism underlying myrosin cell development. The data indicate that regulatory mechanisms for cell differentiation can be shared by two different developmental pathways that generate different cell types.
RESULTS
FAMA Expression in Corniculate-Shaped Cells of the Leaf Inner Layer and Stomatal Lineage Cells
To identify a key regulator of myrosin cell development, we analyzed transcription factor coexpression with genes involved in the myrosinase-glucosinolate system. We performed in silico screening with the Arabidopsis ATTED-II transcriptome database (Obayashi et al., 2009). We identified FAMA as a gene coexpressed with EPITHIOSPECIFIER MODIFIER1 (Supplemental Figure 1), which encodes a protein in the myrosinase-glucosinolate pathway (Zhang et al., 2006). FAMA is a bHLH transcription factor that acts as a master regulator of stomatal development (Bergmann et al., 2004; Ohashi-Ito and Bergmann, 2006).
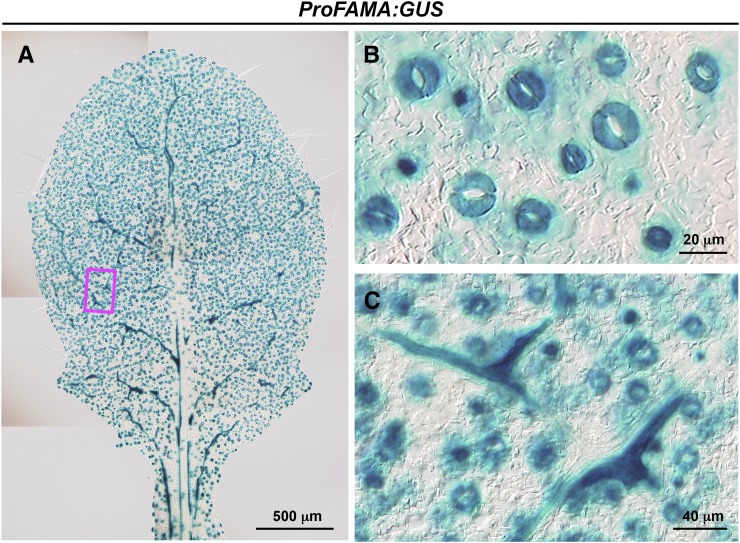
We investigated the spatial expression pattern of FAMA in greater detail by generating transgenic plants expressing β-glucuronidase (GUS) under control of the 3.1-kb FAMA promoter (ProFAMA:GUS). GUS activity was detected in stomatal lineage cells of the leaf epidermis (Figures 1A and 1B), consistent with previous results (Ohashi-Ito and Bergmann, 2006), and in corniculate-shaped cells with horn-like extensions that were distributed along veins in inner layer leaf tissues (Figures 1A and 1C). The characteristic shape and distribution of GUS-positive cells in inner layer tissues was similar to that of myrosin cells. Myrosin cells are localized in aerial parts but excluded from the hypocotyl in Arabidopsis (Husebye et al., 2002; Barth and Jander, 2006). GUS-positive corniculate-shaped cells were not observed in roots or hypocotyls (Supplemental Figure 2). These observations suggest that FAMA-expressing cells of the inner leaf tissues correspond to myrosin cells.
Figure 1.
FAMA Expression in Leaf Inner Tissue Layer.
GUS staining of a rosette leaf of wild-type Arabidopsis (Col-0) expressing ProFAMA:GUS.
(A) An image of the whole leaf.
(B) Enlarged image of epidermal stomatal lineage cells.
(C) Enlarged image of the boxed area in (A). Note that GUS activity is detected in the corniculate cells of the inner tissue layer.
FAMA Expression in Leaf Primordia Identifies Myrosin Cells and Stomatal Cells
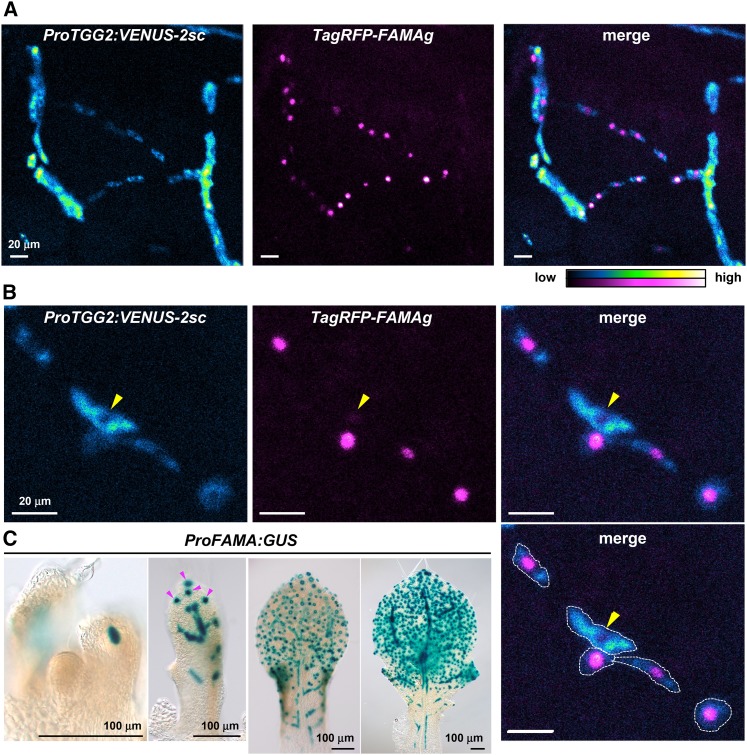
To determine whether FAMA-expressing cells of inner leaf tissues are myrosin cells and/or their precursors, we generated transgenic plants coexpressing the mature myrosin cell reporter ProTGG2:VENUS-2sc (Shirakawa et al., 2014) and the FAMA reporter ProFAMA:TagRFP-FAMAg. The FAMA reporter contained a translational fusion of TagRFP and a full genomic FAMA sequence; this reporter was functional because expressing ProFAMA:TagRFP-FAMAg rescued growth defects of fama-1 mutants (Supplemental Figure 3). The Venus signals of mature myrosin cell reporters were detected in cells with TagRFP-FAMA-positive nuclei in leaf inner tissues (Figure 2A). The maturing and/or mature myrosin cells with high Venus fluorescence had low TagRFP-FAMA expression levels, whereas immature myrosin cells with low Venus fluorescence had high TagRFP-FAMA expression levels (Figure 2B). Typically, mature myrosin cells had almost no TagRFP-FAMA signals (Figure 2B, arrowhead). These results suggest that FAMA is expressed in myrosin cell precursors and promotes myrosin cell development.
Figure 2.
FAMA Is Expressed before a Mature Myrosin Cell Marker in Myrosin Cells.
(A) and (B) Confocal images of the inner tissue of leaf primordia coexpressing both ProTGG2:VENUS-2sc (blue-to-yellow) and ProFAMA:TagRFP-FAMAg (magenta-to-white). Images are maximum intensity projections of a series of images in the Z-plane. Signal intensities are shown in blue-to-yellow or magenta-to-white according to increasing intensity levels. VENUS-2sc localizes to the endoplasmic reticulum and vacuoles, whereas TagRFP-FAMAg localizes to the nucleus. Note that very low expression of ProFAMA:TagRFP-FAMAg was found in mature myrosin cells (arrowheads in [B]). The approximate border of the myrosin cell is indicated by a white outline (lower right panel in [B]).
(C) Developmental change in ProFAMA:GUS expression pattern. Arrowheads indicate stomatal lineage cells.
We examined changes in ProFAMA:GUS expression patterns during development of inner leaf tissues. A GUS-positive cell first emerged at the middle point of a future primary vein in leaf primordia with 85 to 140 μm length (Figure 2C; Supplemental Figure 4). At this stage, the GUS-positive cell was morphologically indistinguishable from neighboring ground meristem cells, which start to differentiate into several cell types including vascular and mesophyll cells (Kang and Dengler, 2004; Scarpella et al., 2004; Sawchuk et al., 2008). At a subsequent stage, leaf primordia with a length of 240 μm expressed the mature myrosin cell marker ProTGG2:VENUS-2sc at the middle point of a future primary vein axis (Supplemental Figure 5). These results suggest that FAMA is first expressed in a subset of ground cells and that these FAMA-positive cells begin to express TGG2 in later developmental stages. At much later stages, FAMA was expressed in stomatal-lineage cells in leaves of ∼340 μm in length (Figure 2C).
Loss-of-Function fama Mutants Fail to Develop Myrosin Cells
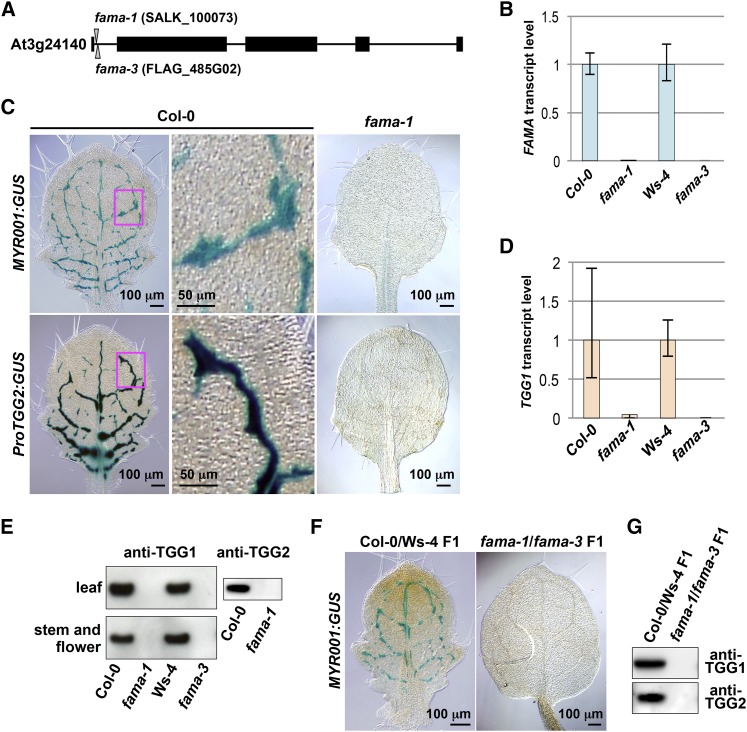
We examined the effect of the FAMA null mutation on myrosin cell development by generating two T-DNA insertion mutants (fama-1 and fama-3) that lacked detectable FAMA transcripts (Figures 3A and 3B). The myrosin cell reporters MYR001:GUS (Shirakawa et al., 2014) and ProTGG2:GUS (Barth and Jander, 2006) distributed along leaf veins and formed network patterns in wild-type plants (Figure 3C, Columbia-0 [Col-0]). When these myrosin cell markers were introduced into fama-1, GUS activity was not detected in leaves (Figure 3C, fama-1). This result was supported by the undetectable level of the TGG1 transcript in fama-1 and fama-3 (Figure 3D). TGG1 and TGG2 are endogenous myrosin cell markers because they accumulate to high levels in myrosin cells. Immunoblot analysis showed that leaves, stems, and flowers of both fama mutants lacked detectable TGG1 and TGG2 (Figure 3E; Supplemental Figure 6). The F1 progeny of fama-1 × fama-3 was defective in myrosin cell reporter MYR001:GUS expression (Figure 3F) and accumulation of both TGG1 and TGG2 (Figure 3G), indicating that fama-1 and fama-3 are allelic to each other. Collectively, these results suggest that FAMA is essential for myrosin cell development in Arabidopsis.
Figure 3.
Myrosin Cell Development in Leaf Primordia of fama Loss-of-Function Mutants.
(A) Exon-intron organization of FAMA. T-DNA insertions are shown for fama-1 (SALK_100073) and fama-3 (FLAG_485G02). Closed boxes, exons; solid lines, introns.
(B) Quantitative RT-PCR of FAMA in 28 d after germination plants of fama-1, fama-3, and their respective wild-type lines (Col-0 and Ws-4) using Actin2 as a control. Error bars indicate 95% confidence intervals (n = 3).
(C) GUS staining of the rosette leaves of Col-0 and fama-1 plants expressing the myrosin cell markers MYR001:GUS (upper panels) and ProTGG2:GUS (lower panels). The boxed areas in the left panels are enlarged (middle panels).
(D) Quantitative RT-PCR of TGG1 in 28 d after germination plants of fama-1, fama-3, and their respective wild-type lines (Col-0 and Ws-4) using Actin2 as a control. Error bars indicate 95% confidence intervals (n = 3).
(E) Immunoblot analysis of rosette leaves (upper panel) and stems and flowers (lower panel) of fama-1, fama-3, and their respective wild-type lines (Col-0 and Ws-4) with anti-TGG1 antibody (left panels) and anti-TGG2 antibody (right panel).
(F) GUS staining of the rosette leaves of F1 progenies of Col-0 × Ws-4 (left panel) and fama-1 × fama-3 (right panel), both of which expressed MYR001:GUS.
(G) Immunoblot analysis of rosette leaves of the indicated F1 progenies with anti-TGG1 antibody (upper) and anti-TGG2 antibody (lower).
The enhancer trap line E1728 was originally identified as a guard-cell-specific green fluorescent protein (GFP) line (Gardner et al., 2009). E1728 also had GFP fluorescence in corniculate-shaped cells along veins in leaf inner tissue and in epidermal guard cells (Supplemental Figure 7A). GFP fluorescence along leaf veins was not detected in fama-1 (Supplemental Figure 7B). These characteristics suggest that GFP-positive corniculate-shaped cells in the E1728 line are myrosin cells. The fluorescent activity of E1728 in the leaf inner tissues may be overlooked previously because of difficulty to detect fluorescence in inner tissues. E1728 can be used as a line for the analysis of both myrosin cells and guard cells.
FAMA Is Required for Its Own Expression in the Myrosin Cell Lineage but Not in the Stomatal Cell Lineage
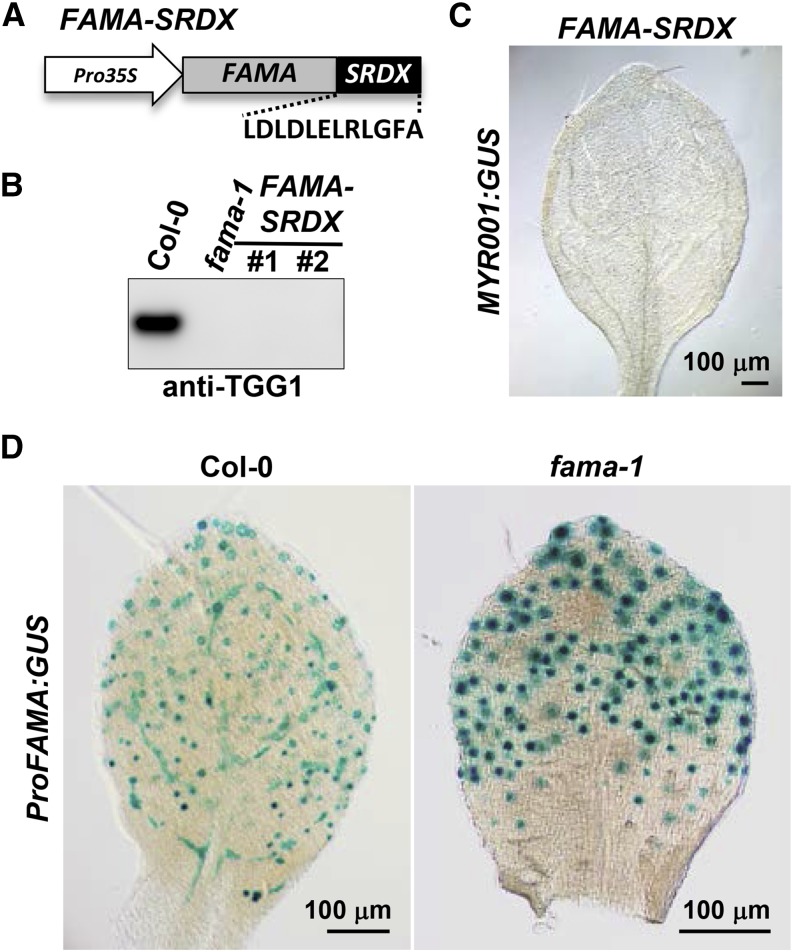
To examine the mode of action of FAMA in myrosin cell development, we generated transgenic plants expressing FAMA fused to the ethylene response factor-associated amphiphilic repression (EAR) domain (FAMA-SRDX), under the control of the 35S promoter (Figure 4A). Transgenic plants neither accumulated endogenous TGG1 (Figure 4B) nor expressed MYR001:GUS (Figure 4C). These results indicate that FAMA-SRDX has dominant-negative activity. FAMA might act primarily as a transcriptional activator in myrosin cell development.
Figure 4.
FAMA Is Required for Its Own Expression in Myrosin Lineage Cells.
(A) Structural organization of the FAMA-SRDX construct. FAMA-SRDX expresses FAMA fused to the EAR repression domain under the control of the cauliflower mosaic virus 35S promoter.
(B) Immunoblot analysis of rosette leaves of Col-0, fama-1, and transgenic plants expressing FAMA-SRDX (independent lines #1 and #2) with anti-TGG1 antibody.
(C) GUS staining of the rosette leaves of FAMA-SRDX transgenic plants expressing MYR001:GUS.
(D) GUS staining of rosette leaves of Col-0 and fama-1 plants expressing ProFAMA:GUS.
Positive feedback regulation is a common mechanism that enables many transcription factors to stabilize their own expression. A previous report indicated that positive autoregulation is not absolutely required to promote FAMA expression in stomatal lineage cells (Ohashi-Ito and Bergmann, 2006), although FAMA binds its own promoter (Hachez et al., 2011). In agreement, we detected GUS activity in stomatal lineage cells of transgenic plants expressing ProFAMA:GUS in the fama background (Figure 4D). However, GUS activity was not detected in the myrosin-lineage cells of transgenic plants expressing ProFAMA:GUS in the fama background (Figure 4D; Supplemental Figure 8). These results suggest that FAMA is required for its own expression in the myrosin lineage cells. Positive feedback regulation of FAMA expression might function in the myrosin lineage cells, but not in the stomatal lineage cells.
FAMA Expression Potentially Confers Myrosin Cell Identity to Various Cell Types
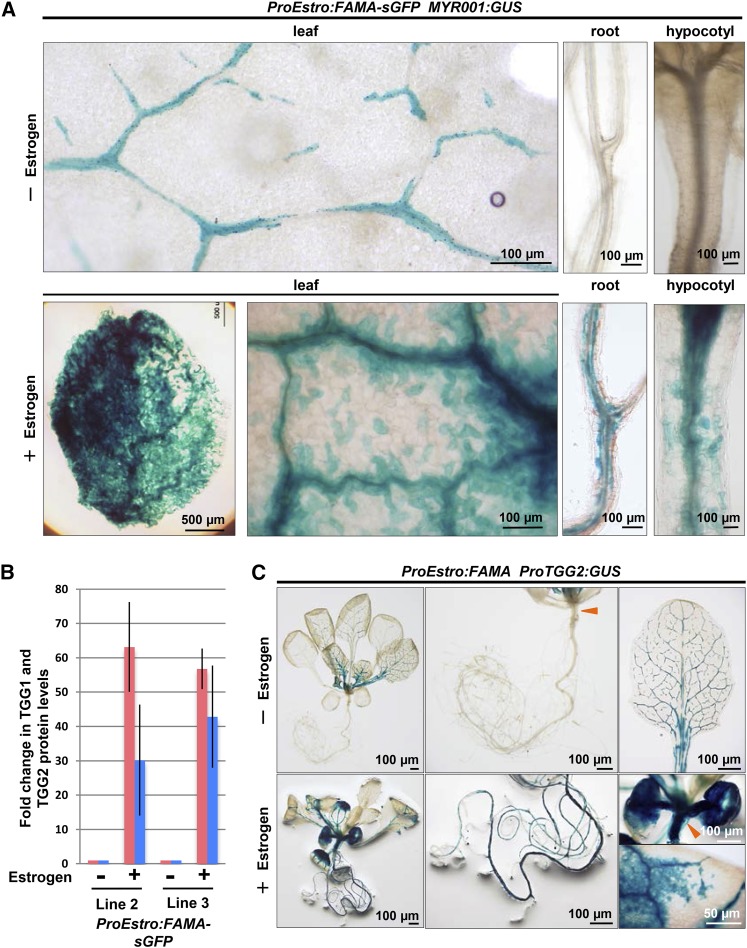
To investigate whether FAMA is sufficient for myrosin cell development, we expressed FAMA-sGFP under the estrogen-inducible promoter in MYR001:GUS-expressing transgenic plants. Estrogen treatment strongly induced GUS expression in cells throughout the leaf in two independent lines (Figure 5A; Supplemental Figure 9). Consequently, the TGG1 and TGG2 protein levels in estrogen-treated leaves were more than 30-fold higher in two independent lines compared with those in untreated leaves (Figure 5B; Supplemental Figure 10). The estrogen-treated leaves became pale green during seedling growth, which is probably due to the overproliferation of myrosin cells in mesophyll tissue (Supplemental Figure 11). Estrogen treatment induced the ectopic development of myrosin cells in hypocotyls and roots, both of which lack myrosin cells in wild-type plants (Figure 5A; Supplemental Figure 9). Similarly, transgenic plants expressing both ProEstro:FAMA and ProTGG2:GUS exhibited the ectopic development of myrosin cells under the estrogen-treated condition (Figure 5C). These results suggest that FAMA expression potentially confers the myrosin cell identity to various cell types.
Figure 5.
FAMA Expression Confers Myrosin Cell Identity to Various Cell Types.
(A) GUS staining of transgenic ProEstro:FAMA-sGFP plants expressing MYR001:GUS. The ProEstro:FAMA-sGFP plants express FAMA-sGFP under the control of the estrogen-inducible promoter. The 8-d-old plants were transplanted onto inductive medium containing 10 μM estrogen (+Estrogen) or no estrogen (−Estrogen) and incubated for 2 weeks.
(B) Immunoblot analysis of rosette leaves of ProEstro:FAMA-sGFP plants (independent lines 2 and 3) with anti-TGG1 (pink) and anti-TGG2 (blue) antibodies. The immunoblot signal intensities were quantified by densitometry. Error bars indicate standard deviations (n = 3). See also Supplemental Figure 10.
(C) GUS staining of transgenic plants expressing ProTGG2:GUS and ProEstro:FAMA. This plant expressed FAMA under the control of the estrogen-inducible promoter. The 8-d-old plants were transplanted onto inductive medium containing 10 μM estrogen (+Estrogen) or no estrogen (−Estrogen) and then incubated for 2 weeks. Arrowheads indicate hypocotyl.
FAMA Expression in the Myrosin Cell Lineage Is Independent of SPCH and MUTE
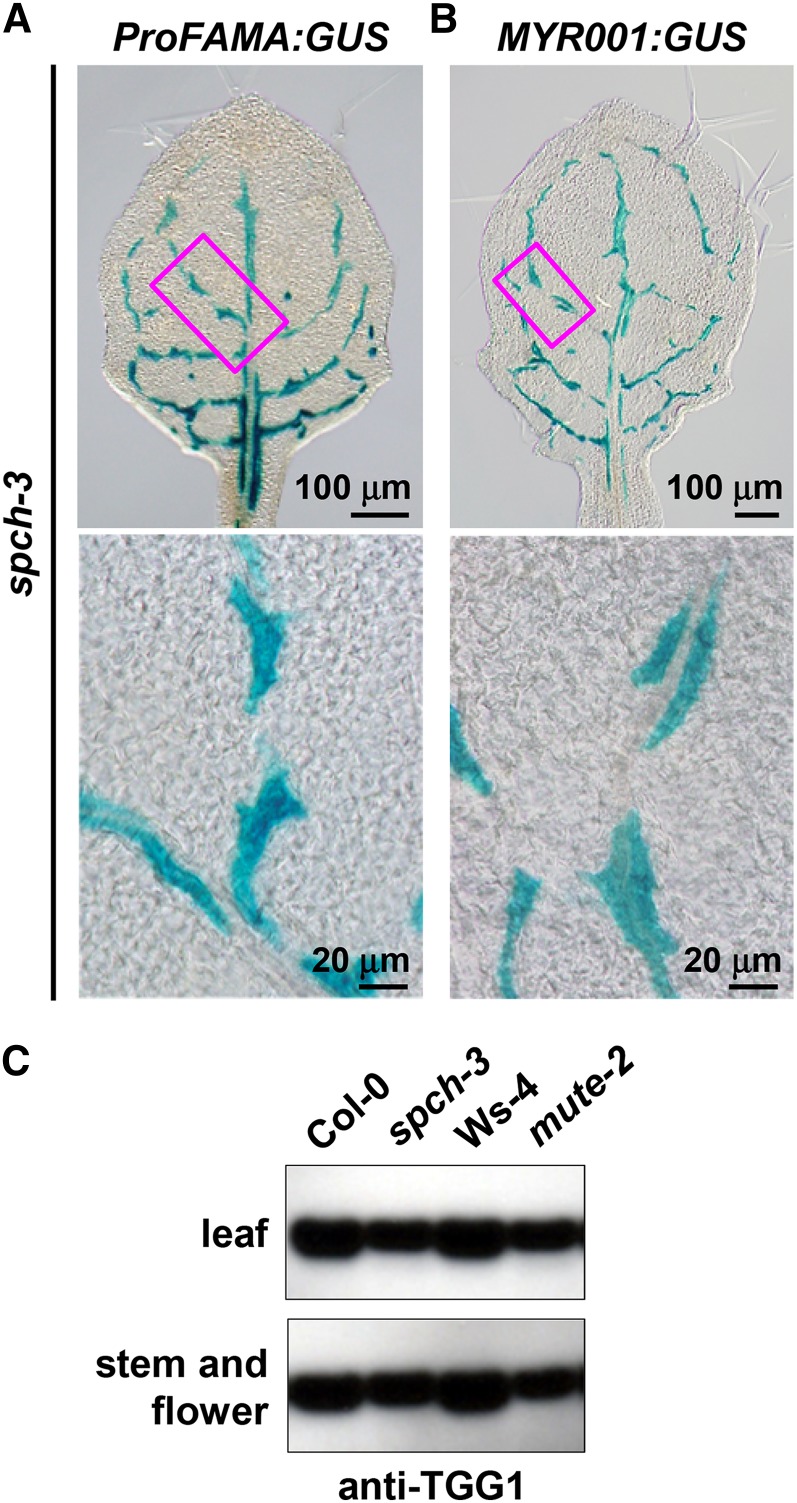
FAMA functions downstream of the bHLH transcription factors SPCH and MUTE in the stomatal cell lineage (MacAlister et al., 2007; Pillitteri et al., 2011). To investigate the epistatic effect of FAMA on SPCH and MUTE during the generation of FAMA-expressing cells in the leaf inner tissues, ProFAMA:GUS was expressed in spch-3 and mute-2 mutants, both of which lack stomata. In these lines, GUS signal was detected in cells along leaf veins (Figure 6A; Supplemental Figure 12). This distribution pattern of the GUS-positive cells in spch-3 expressing ProFAMA:GUS was very similar to that in spch-3 expressing MYR001:GUS (Figure 6B). By contrast, no GUS signals were detected in stomatal lineage cells in the epidermis of spch-3 and mute-2 expressing ProFAMA:GUS (Figure 6A; Supplemental Figure 12). RT-PCR analysis revealed that both FAMA and TGG1 were expressed in spch-3 at almost the same levels as in wild-type plants (Supplemental Figure 13). Consistent with this, TGG1 accumulation levels in spch-3 and mute-2 were similar to those in the wild type (Figure 6C). Notably, the TGG1 levels in both mutants were slightly less than in the wild type, which is consistent with the report that TGG1 is also expressed in guard cells (Husebye et al., 2002; Barth and Jander, 2006). These results suggest that FAMA expression in myrosin cells is independent of SPCH and MUTE.
Figure 6.
FAMA Expression in Myrosin Cells Is Independent of SPCH and MUTE.
(A) and (B) GUS staining of the rosette leaves of spch-3 expressing ProFAMA:GUS (A) or MYR001:GUS (B). Each boxed area in the upper panel is enlarged in the corresponding lower panel.
(C) Immunoblot analysis of rosette leaves (upper panel) and stems and flowers (lower panel) of wild-type lines (Col-0 and Ws-4), spch-3, and mute-2 with anti-TGG1 antibody.
FAMA-Interacting Partners ICE1 and SCRM2 Are Required for Myrosin Cell Development
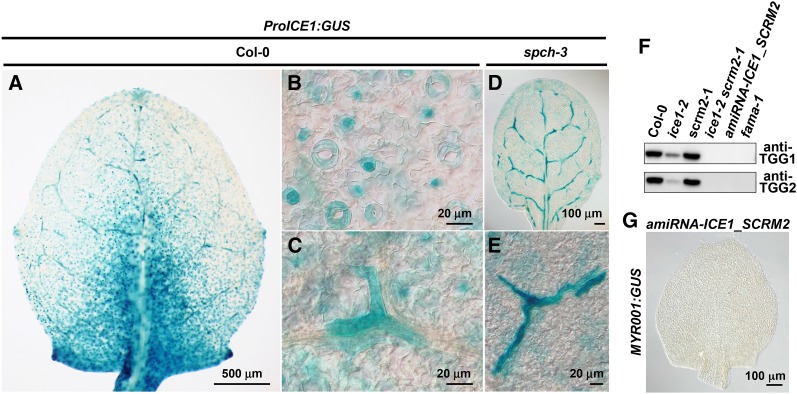
ICE1 and its closest paralog SCRM2 encode bHLH transcription factors that form heterodimers with FAMA and act together in guard cell differentiation although they are expressed throughout leaves (Chinnusamy et al., 2003; Kanaoka et al., 2008; Fursova et al., 2009). We examined their involvement in myrosin cell development using the GUS reporter gene of ICE1 (ProICE1:GUS). GUS activity was predominantly detected in corniculate-shaped cells along leaf veins (Figures 7A and 7C) and in stomatal lineage cells (Figure 7B), in addition to weak signals in mesophyll (Figures 7A and 7C). The spch-3 mutant exhibited GUS-positive corniculate-shaped cells along leaf veins (Figures 7D and 7E) and GUS-positive mesophyll cells (Figures 7D and 7E). This expression pattern of ProICE1:GUS except for mesophyll cells was similar to the ProFAMA:GUS pattern in spch-3 (Figure 6A). These results suggest that ICE1 is expressed in myrosin lineage cells.
Figure 7.
ICE1 and SCRM2 Are Required for Myrosin Cell Development.
(A) to (E) GUS staining of rosette leaves of Col-0 ([A] to [C]) and spch-3 ([D] and [E]) expressing ProICE1:GUS. In the Col-0 background, GUS activity is detected in the corniculate cells of leaf inner tissue (C) as well as in epidermal stomatal lineage cells (B). In spch-3, GUS activity is detected only in the corniculate cells of leaf inner tissue (E).
(F) Immunoblot analysis of rosette leaves of the wild type and the indicated mutant or transgenic lines with anti-TGG1 antibody (upper) and anti-TGG2 antibody (lower).
(G) GUS staining of a rosette leaf of transgenic amiRNA-ICE1_SCRM2 plants expressing MYR001:GUS.
We questioned whether ICE1 and SCRM2 were required for myrosin cell development. The TGG1 protein level was markedly reduced in ice1-2, but not in scrm2-1 (Figure 7F; Supplemental Figure 14). No TGG1 was detected in ice1-2 scrm2-1, similar to that observed for fama mutants (Figure 7F; Supplemental Figure 14). Similar defects in both TGG1 and TGG2 accumulations were detected in transgenic plants expressing an artificial microRNA targeted to both ICE1 and SCRM2 (amiRNA-ICE1_SCRM2) (Figure 7F). In addition, no GUS activity conferred by MYR001:GUS was detected in amiRNA-ICE1_SCRM2 (Figure 7G). Taken together, these results suggest that ICE1 and SCRM2 act redundantly during myrosin cell development.
Identification of Novel Components of FAMA-Regulated Myrosin Cell Development
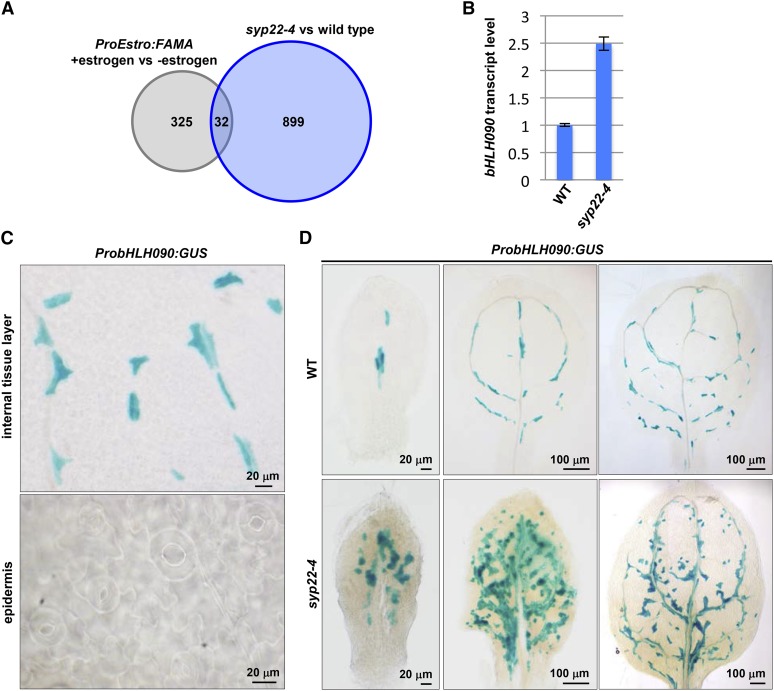
We identified the following novel components of myrosin cell development under the control of FAMA. First, we performed comparative microarray experiments using the wild type and the myrosin cell overproliferation mutant syp22-4 (Ueda et al., 2006), which exhibited normal distribution and density of stomata (Supplemental Figure 15). In syp22-4, 931 genes were significantly upregulated relative to the wild type. Next, by comparing these microarray data with those of the 4-h estrogen-inducible FAMA overexpression lines (Hachez et al., 2011), we found that 32 genes were upregulated commonly in both lines (Figure 8A, Table 1). These genes were candidate components in the myrosin cell development regulated directly by FAMA. They did not include the myrosin cell marker genes TGG1 and TGG2 because both TGG1 and TGG2 were significantly upregulated in syp22-4 but were not included in the 357 genes of the microarray data (Hachez et al., 2011).
Figure 8.
bHLH090 Specifically Expressed at the Myrosin Lineage Cells in Leaves.
(A) Summary of comparative microarray results showing 32 candidate genes involved in myrosin cell development. See Table 1 for details of these genes. The genes upregulated in syp22-4 are listed in Supplemental Data Set 1. They were compared with the genes upregulated in 4-h estrogen-treated plants expressing ProEstro:FAMA (Hachez et al., 2011).
(B) Quantitative RT-PCR of bHLH090 in 21 d after germination plants of wild-type lines (Col-0) and syp22-4 using Actin2 as a control. Error bars indicate sd (n = 3).
(C) GUS staining of the rosette leaves of ProbHLH090:GUS.
(D) Developmental change in ProbHLH090:GUS expression pattern in the first pair of rosette leaves of the wild-type lines (Col-0) and syp22-4.
Table 1. Candidate Genes Involved in Myrosin Cell Development.
| AGI Code | Gene Name | Gene Description |
|---|---|---|
| Transcription factors | ||
| AT1G10610 | bHLH090 | bHLH transcription factor |
| AT1G52890 | NAC019 | NAC transcription factor |
| AT2G40340 | DREB2C | DREB subfamily A-2 of ERF/AP2 transcription factor |
| AT2G40350 | DREB subfamily A-2 of ERF/AP2 transcription factor | |
| AT2G47260 | WRKY23 | WRKY transcription factor |
| AT3G57600 | DREB subfamily A-2 of ERF/AP2 transcription factor | |
| AT5G65590 | SCAP1 | Dof-type transcription factor |
| Others | ||
| AT1G03440 | Leucine-rich repeat (LRR) family protein | |
| AT1G19160 | F-box family protein | |
| AT1G27680 | APL2 | ADPGLC-PPase large subunit |
| AT1G30040 | GA2OX2 | Gibberellin 2-oxidase |
| AT2G11810 | MGDC | Monogalactosyldiacylglycerol synthase type C |
| AT2G25780 | Protein of unknown function (DUF1677) | |
| AT2G28420 | GLYI8 | Lactoylglutathione lyase/glyoxalase I family protein |
| AT3G05640 | Protein phosphatase 2C family protein | |
| AT3G46230 | HSP17.4 | Heat shock protein 17.4 |
| AT3G55840 | Hs1pro-1 protein | |
| AT3G56620 | Nodulin MtN21 /EamA-like transporter family protein | |
| AT4G09740 | GH9B14 | Glycosyl hydrolase 9B14 |
| AT4G15120 | VQ motif-containing protein | |
| AT4G21730 | Pseudogene of N-ethylmaleimide sensitive factor (NSF) | |
| AT4G23560 | GH9B15 | Glycosyl hydrolase 9B15 |
| AT4G24480 | Protein kinase superfamily protein | |
| AT4G29570 | Cytidine/deoxycytidylate deaminase family protein | |
| AT4G35070 | SBP (S-ribonuclease binding protein) family protein | |
| AT5G17830 | Plasma membrane choline transporter family protein | |
| AT5G43860 | CLH2 | Chlorophyllase 2 |
| AT5G66400 | RAB18 | Dehydrin family protein |
| Unknown proteins | ||
| AT2G21560 | ||
| AT3G09730 | ||
| AT3G26390 | ||
| AT4G28330 |
Top priority genes were transcription factors, especially of the bHLH class, given the fact that this family plays key roles in many cell fate decisions in animals and plants. We found only one bHLH transcription factor, bHLH090 (At1g10610), in the candidate gene list (Table 1). Real-Time PCR analysis confirmed that the expression level of bHLH090 in syp22-4 was 2.5-fold higher than in the wild type (Figure 8B). To investigate the spatiotemporal expression pattern of bHLH090 in leaf primordia, we generated transgenic plants expressing GUS under the control of the 2-kb bHLH090 promoter (ProbHLH090:GUS). GUS activity was detected in corniculate-shaped cells with horn-like extensions that were distributed along veins in ProbHLH090:GUS lines as in ProFAMA:GUS lines and was not detected in stomatal lineage cells of the leaf epidermis (Figure 8C). In syp22-4 ProbHLH090:GUS, GUS-positive cells were more abundant than in the wild type and formed a drastically denser network than observed in the wild type (Figure 8D). These results suggest that bHLH090 is a novel component for the myrosin cell development, and our comparative microarray experiments may identify new components in the myrosin cell development.
DISCUSSION
FAMA Is an Essential Transcription Factor for Myrosin Cell Differentiation
No essential component for idioblast myrosin cell development has been identified, although several factors required for the development of Arabidopsis myrosin cells have been identified (Ueda et al., 2006; Shirakawa et al., 2010, 2014). In this study, we show that the bHLH transcription factor FAMA is essential for myrosin cell differentiation in Arabidopsis. FAMA has a dual functional role: it regulates differentiation of myrosin cells in leaf inner tissues, and it regulates differentiation of guard cells in epidermal tissues (Ohashi-Ito and Bergmann, 2006). We propose that FAMA is a master regulator of myrosin cell differentiation based on three results. First, FAMA was expressed in the ground meristem cell at very early developmental stages of leaf primordia (Figures 1 and 2). Second, no mature myrosin cells developed in fama loss-of-function mutants (Figure 3). Third, ectopic FAMA expression was sufficient to confer idioblast myrosin cell identity to various cell types such as root cells, which do not differentiate into myrosin cells in wild-type Arabidopsis (Figure 5). It was reported that TGG1 was also detected in a subset of guard cells, called myrosin guard cells (Husebye et al., 2002; Barth and Jander, 2006). In fama, differentiations of both idioblast myrosin cells and myrosin guard cells were blocked (Figure 3) (Bergmann et al., 2004; Ohashi-Ito and Bergmann, 2006). Taken together, these results indicate that FAMA plays a crucial role in myrosin cell differentiation in Arabidopsis.
How FAMA Generates Two Different Cell Types in Plants
Myrosin cells are a recent innovation that is restricted to Brassicales (Rask et al., 2000). By contrast, stomata composed of a pair of guard cells represent a very old plant innovation found in all land plants except liverwort (Bowman, 2011). Arabidopsis has two FAMA paralogs, SPCH and MUTE. These three paralogs are involved in the development of stomatal lineage cells at different steps (MacAlister et al., 2007; Pillitteri et al., 2007). Phylogenetic analysis indicates that the FAMA amino acid sequence is better conserved among land plants than is SPCH or MUTE (Ran et al., 2013). The question arises how FAMA exerts its dual function for guard cell and myrosin cell development, without dynamic changes in its sequence.
One possible explanation is as follows: Expression of FAMA in epidermal guard mother cells results in guard cell differentiation, whereas expression of FAMA in ground meristem cells of leaf inner tissue results in myrosin cell differentiation. FAMA might have different downstream targets in different cell types due to differences in chromatin structure in these cells. Another possible explanation could be spatiotemporal differences in the dynamics of FAMA expression in the two cell types. FAMA expression is regulated by a positive feedback loop in myrosin lineage cells (Figure 4), but not in stomatal lineage cells (Ohashi-Ito and Bergmann, 2006). FAMA expression is transient in myrosin lineage cells (Figure 2), whereas FAMA expression is prolonged in stomatal lineage cells (Ohashi-Ito and Bergmann, 2006). Regulatory mechanisms governing FAMA expression dynamics might differ in the two cell types. Further analysis is necessary to identify the downstream targets and regulatory mechanism of FAMA.
Upstream Factors of FAMA at the Start of Myrosin Lineage Cell Specification
FAMA expression in stomatal lineage cells depends on SPCH and MUTE, indicating that FAMA is a downstream target (MacAlister et al., 2007; Pillitteri et al., 2011). By contrast, this study demonstrates that FAMA is required for myrosin cell development, but SPCH and MUTE are not. We show that myrosin lineage cells are derived from ground meristem cells. Vascular precursor cells (preprocambium/procambium) are also derived from ground meristem cells (Kang and Dengler, 2004; Scarpella et al., 2004). These two cell lineages independently arise from ground meristem cells because both ProFAMA:GUS and ProAtHB8:GUS (AtHB8 is a master transcription factor of vascular precursor cells) are initially expressed simultaneously in a subset of the ground meristem cells with different spatial patterns (Figure 2) (Baima et al., 1995, 2001; Kang and Dengler, 2004; Scarpella et al., 2004). AtHB8 expression in ground meristem cells is determined by the phytohormone auxin (Mattsson et al., 2003; Donner et al., 2009; Ohashi-Ito and Fukuda, 2010; Krogan et al., 2012). It is possible that FAMA expression in ground meristem cells also is determined by auxin. Auxin involvement in the onset of FAMA expression and synchronous division in stomatal lineage cells was recently reported (Le et al., 2014). The involvement of auxin in myrosin lineage cell specification is supported further by our previous observation that abnormal myrosin cell development is observed in syp22/vam3 mutants, which exhibit an abnormal distribution of auxin (Ueda et al., 2006; Shirakawa et al., 2009). An investigation of FAMA in syp22/vam3 would help reveal the fate determination mechanism of ground meristem cells. Thus, it is necessary to examine the relationship between auxin and FAMA expression during myrosin lineage cell specification.
bHLH090 Is a Novel Component in Myrosin Cell Development
Comparative DNA microarray experiments identified 32 candidates involved in myrosin cell development (Table 1). Indeed, one of these candidates, bHLH090, was expressed specifically in myrosin lineage cells in leaves (Figure 8). The expression of bHLH090 was quickly triggered by FAMA (Hachez et al., 2011) and the 2-kb promoter region of bHLH090 contains 11 G-box sequences (CANNTG) that are typical binding motifs of bHLH transcription factors. These results suggest that bHLH090 might be a direct target of FAMA. Although bHLH090 is classified as an orphan bHLH protein (Pires and Dolan, 2010), bHLH090 shows high levels of sequence similarities with bHLH093, ICE1, and SCRM2, all of which interact with FAMA (Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008), suggesting that bHLH090 might bind FAMA to modulate the activity of FAMA. Because bHLH090 homologs seem to be specific to Brassicales (Supplemental Figure 16), bHLH090 may be an evolved type of bHLH transcription factors for the production of myrosin lineage cells.
How Did Brassicales Plants Acquire Myrosin Cells during Evolution?
It is intriguing that the different cell lineages are regulated by the same FAMA transcription factor in Arabidopsis. FAMA is also expressed around leaf veins in rice (Oryza sativa) (Liu et al., 2009), but rice lacks myrosin cells. Although the nature of these FAMA-expressing cells is unknown, FAMA might function as an idioblast regulator in leaf inner tissue. It is possible that the FAMA-regulating idioblasts were specified to accumulate myrosinase and differentiate into myrosin cells when the Brassicales acquired myrosinase during evolution. It would be interesting to study the nature of FAMA-expressing cells near leaf veins in rice and other plants. In an alternative scenario, transcription networks including FAMA and myrosinases might first have been established in myrosin guard cells. Subsequently, these transcription networks might have been co-opted from guard cells to idioblasts in leaf inner tissue during the evolution of Brassicales.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana Columbia (Col-0) ecotype was used for all lines except for fama-3 and mute-2 (Ws-4). The T-DNA insertion mutants and an enhancer trap line were obtained from the following sources: SALK_100073 (fama-1), SAIL_36_B06 (spch-3), SALK_003155 (ice1-2), and SAIL_808_B10 (scrm2-1) from the ABRC at Ohio State University; FLAG_485G02 (fama-3) and FLAG_225D03 (mute-2) from INRA; and E1728 from The European Arabidopsis Stock Center. The ProTGG2:VENUS-2sc and MYR001:GUS constructs were reported previously (Shirakawa et al., 2014). ProTGG2:GUS (Barth and Jander, 2006), fama-1 E1728 (Ohashi-Ito and Bergmann, 2006), and ice1-2 scrm2-1 (Kanaoka et al., 2008) were provided by G. Jander (Boyce Thompson Institute for Plant Research), D.C. Bergmann (Stanford University), and K.U. Torii (University of Washington), respectively. syp22-4 was previously described (Ohtomo et al., 2005). Seeds were surface-sterilized with 70% ethanol and then sown onto 0.5% w/v gellan gum (Wako) containing 1% w/v sucrose and Murashige and Skoog medium (Wako). The seeds were incubated at 4°C for 3 to 5 d to break seed dormancy and were grown at 22°C for 20 d under continuous light. Plants were transferred onto vermiculite for subsequent growth.
Plasmid Construction and Transgenic Plants
The Gateway Cloning System (Life Technologies) was used for plasmid constructions. For transcriptional GUS fusion constructs, the 3.1-kb promoter of FAMA, the 2.6-kb promoter of ICE1, and the 2-kb promoter of bHLH090 were cloned into pENTR D-TOPO. They were introduced into the binary vector pBGWFS7 (BASTA selection for plants) or pHGWFS7 (hygromycin B selection for plants) using LR reactions. For translational fusion constructs, the cDNA encoding TagRFP was inserted in front of the start codon of the 5.8-kb FAMA genomic fragment (including 3.1 kb of the 5′-flanking sequence and 0.5 kb of the 3′-flanking sequence). After cloning into pENTR D-TOPO, the construct was introduced into the binary vector pBGW (BASTA selection for plants) using LR reactions. For the ProEstro:FAMA and the ProEstro:FAMA-sGFP constructs, FAMA coding sequence and FAMA coding sequence fused to the cDNA encoding sGFP were cloned into pENTR D-TOPO and introduced into pMDC7 (Curtis and Grossniklaus, 2003) using LR reactions, respectively. An artificial microRNA (amiRNA) against both ICE1/SCRM and ICE2/SCRM2 was designed using the WMD2-Web MicroRNA designer (http://wmd3.weigelworld.org) and was amplified using the following primers from the pRS300 vector: amiRNA-F, amiRNA-R, ICE1/2_amiRNA_1, ICE1/2_amiRNA_2, ICE1/2_amiRNA_3, and ICE1/2_amiRNA_4. The primer sets are presented in Supplemental Table 1. The amplified amiRNA-ICE1_SCRM2 DNA fragment was cloned into the pENTR D-TOPO plasmid. The plasmid was introduced into binary vector pFAST-G02 (Shimada et al., 2010) using the LR reaction to generate amiRNA-ICE1_SCRM2. For the FAMA-SRDX construct, FAMA coding sequence fused to the EAR motif (Hiratsu et al., 2003) was cloned into pENTR D-TOPO. The plasmid was introduced into the binary vector pFAST-G02 (Shimada et al., 2010) using the LR reaction. Agrobacterium tumefaciens (strain GV3101) was transformed with these constructs. Plants were transformed with Agrobacteria using the floral dip method (Clough and Bent, 1998). T1 seeds were selected using medium containing 10 mg L−1 BASTA or 25 to 50 mg L−1 hygromycin B.
GUS Staining
Samples were first placed into ice-cold acetone for 15 min and then into GUS staining solution containing 0.5 mg/mL X-Gluc, 0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.5 to 5 mM potassium ferricyanide, 0.5 to 5 mM potassium ferrocyanide, and 0.1% Triton X-100. Samples in the GUS staining solution were placed under a vacuum and incubated at room temperature for 4 to 24 h.
SDS-PAGE and Immunoblot Analysis
SDS-PAGE and immunoblot analysis were performed as described previously (Shimada et al., 2003). The antibodies used in this analysis were anti-TGG1 (diluted 5000-fold) (Ueda et al., 2006) and anti-TGG2 (diluted 5000-fold) (Ueda et al., 2006).
Confocal Laser Scanning Microscopy
Fluorescence micrographs were obtained with a confocal laser scanning microscope (LSM780; Carl Zeiss) using a water immersion objective (63× 1.20 numerical aperture [NA]) and dry objectives (40× 0.95 NA, 20× 0.80 NA, and 10× 0.50 NA). The laser wavelengths used include 488 nm (GFP and Venus) and 543 nm (TagRFP). The images were analyzed using LSM image software (Carl Zeiss) and were processed using ImageJ (NIH) and Photoshop (Adobe Systems) software.
RT-PCR
Total RNA was prepared from wild-type and mutant plants at 14, 21, and 28 d after germination using an RNeasy Plant Mini Kit (Qiagen). After DNase I (Invitrogen) treatment, reverse transcription was performed using a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with an oligo(dT)12-18 primer (Invitrogen). We performed PCR using 30 and 35 cycles with each primer set. Quantitative RT-PCR was performed using a gene-specific primer set (FAMA, At02279294_g1; TGG1, At02185835_g1; Actin2, At02335270_gH; Applied Biosystems) and a TaqMan Gene Expression Assay Kit (Applied Biosystems) or SYBR Premix Ex Taq II (Takara Bio) in a 7500 Real-Time PCR system (Applied Biosystems). The relative quantity of target mRNA was calculated using Actin2 as a control. Primer sets except TaqMan Probe are presented in Supplemental Table 1.
Microarray Experiments
Total RNA was isolated from each developing leaf of both the wild type and syp22-4 using the RNeasy Plant Mini Kit. The extracted RNA was quantified using a NanoDrop ND-1000 UV-VIS spectrophotometer (NanoDrop), and the quality of the RNA samples was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA (200 ng) was subjected to fluorescent labeling. Labeling was performed using an Agilent Low Input Quick Amp Labeling Kit 1-Color (Agilent Technologies) according to the manufacturer’s protocol. The labeled cRNA was fragmented and hybridized on a slide of the Arabidopsis V4 Gene Expression Microarray 4 × 44K (Agilent Technologies; G2519F-021169) at 65°C for 17 h. Hybridization and washing of the hybridized slides were performed according to the manufacturer’s instructions. Slides were scanned using a G2505B DNA microarray scanner (Agilent Technologies), and background correction of the raw signals was performed using the Agilent Feature Extraction software. All microarray data were transformed into a log2 scale and normalized using the qspline normalization method (Workman et al., 2002). The microarray data collected in this study are available in Supplemental Data Set 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FAMA (At3g24140), ICE1/SCREAM (At3g26744), ICE2/SCREAM2 (At1g12860), SPEECHLESS (At5g53210), MUTE (At3g06120), TGG1 (At5g26000), TGG2 (At5g25980), VSR1 (At3g52850), EPITHIOSPECIFIER MODIFIER1 (At3g14210), SYP22/VAM3 (At5g46860), bHLH090 (At1g10610), and ACT2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FAMA Coexpression Network Predicted by ATTED-II.
Supplemental Figure 2. FAMA Expression Pattern in Wild-Type Plants.
Supplemental Figure 3. ProFAMA:TagRFP-FAMAg Expression Rescues fama-1 Dwarfism.
Supplemental Figure 4. ProFAMA:GUS Expression Pattern in Early Developmental Stages of Leaves.
Supplemental Figure 5. ProTGG2:VENUS-2sc Expression Pattern in Early Developmental Stages of Leaves.
Supplemental Figure 6. The fama-Deficient Mutant Lacks Endogenous TGG1 Protein.
Supplemental Figure 7. Myrosin Cell Development in Leaf Primordia of fama Loss-of-Function Mutants.
Supplemental Figure 8. FAMA Expression Pattern in the fama-Deficient Mutant.
Supplemental Figure 9. Development of Myrosin Cells in FAMA-sGFP Overexpression Lines.
Supplemental Figure 10. Accumulation Levels of TGG1 and TGG2 in FAMA-sGFP Overexpression Lines.
Supplemental Figure 11. Plant Morphology of FAMA-sGFP Overexpression Lines.
Supplemental Figure 12. FAMA Expression in Myrosin Cells Is Independent of MUTE.
Supplemental Figure 13. TGG1 and FAMA Transcript Levels in spch.
Supplemental Figure 14. Accumulation Levels of TGG1 in ICE1 and SCRM2 Mutants.
Supplemental Figure 15. Distribution of Stomata in the Wild Type and syp22-4.
Supplemental Figure 16. The bHLH Domain and Alignment of bHLH090 Homologs.
Supplemental Table 1. Primer Sets Used in This Study.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.m9160.
Supplemental Data Set 1. Microarray Data of Col_vs_syp22.
Supplementary Material
Acknowledgments
We thank Yoichi Ogawa (Chiba University) for discussions. We thank Georg Jander (Boyce Thompson Institute for Plant Research), Dominique Bergmann (Stanford University), and Keiko Torii (University of Washington) for sharing materials. We also thank ABRC, INRA, and NASC for providing seeds of Arabidopsis lines. This work was supported by Specially Promoted Research of Grant-in-Aid for Scientific Research to I.H-.N. (no. 22000014) and Grants-in-Aid for Scientific Research to H.U. (nos. 21200065 and 25440132) from the Japan Society for the Promotion of Science (JSPS). M.S. was supported by a research fellowship from JSPS (nos. 20002057 and 24005453) and by a Grant-in-Aid for Plant Graduate Students from the Nara Institute of Science and Technology.
AUTHOR CONTRIBUTIONS
M.S., H.U., T.S., and I.H.-N. designed research. M.S. performed all experiments. A.J.N. analyzed DNA microarray experiments. M.S., H.U., T.S., and I.H.-N. wrote the article. T.K. and I.H.-N. supervised the study.
Glossary
- bHLH
basic helix-loop-helix
- NA
numerical aperture
- Col-0
Columbia-0
Footnotes
Online version contains Web-only data.
References
- Andréasson E., Bolt Jørgensen L., Höglund A.S., Rask L., Meijer J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127: 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima S., Possenti M., Matteucci A., Wisman E., Altamura M.M., Ruberti I., Morelli G. (2001). The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46: 549–562. [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bowman J.L. (2011). Stomata: active portals for flourishing on land. Curr. Biol. 21: R540–R541. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner T.J., Sherr I., Scarpella E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246. [DOI] [PubMed] [Google Scholar]
- Fursova O.V., Pogorelko G.V., Tarasov V.A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429: 98–103. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Baker A.J., Assie J.M., Poethig R.S., Haseloff J.P., Webb A.A. (2009). GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J. Exp. Bot. 60: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11: 89–100. [DOI] [PubMed] [Google Scholar]
- Guignard L. (1890). Recherches sur la localization des principes actifs des Cruciferes. J. Bot. 4: 385–395. [Google Scholar]
- Hachez C., Ohashi-Ito K., Dong J., Bergmann D.C. (2011). Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 155: 1458–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Heinricher E. (1884). Uber Eiweisstoffe fuhrennde Idioblasten bei einigen Cruceren. Ber. Dtsch. Bot. Ges. 2: 463–467. [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Hopkins R.J., van Dam N.M., van Loon J.J. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54: 57–83. [DOI] [PubMed] [Google Scholar]
- Husebye H., Chadchawan S., Winge P., Thangstad O.P., Bones A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128: 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Dengler N. (2004). Vein pattern development in adult leaves of Arabidopsis thaliana. Int. J. Plant Sci. 165: 231–242. [Google Scholar]
- Kissen R., Rossiter J.T., Bones A.M. (2009). The ‘mustard oil bomb’: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 8: 69–86. [Google Scholar]
- Koroleva O.A., Davies A., Deeken R., Thorpe M.R., Tomos A.D., Hedrich R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.T., Ckurshumova W., Marcos D., Caragea A.E., Berleth T. (2012). Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 194: 391–401. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Bergmann D.C. (2012). Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139: 3683–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., et al. (2014). Auxin transport and activity regulate stomatal patterning and development. Nat. Commun. 5: 3090. [DOI] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K., Bergmann D.C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276. [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Bergmann D.C. (2011). Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 13: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- Mattsson J., Ckurshumova W., Berleth T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Stomatal development in Arabidopsis. The Arabidopsis Book 1: e0066, doi/10.1199/tab.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Fukuda H. (2010). Transcriptional regulation of vascular cell fates. Curr. Opin. Plant Biol. 13: 670–676. [DOI] [PubMed] [Google Scholar]
- Ohtomo I., Ueda H., Shimada T., Nishiyama C., Komoto Y., Hara-Nishimura I., Takahashi T. (2005). Identification of an allele of VAM3/SYP22 that confers a semi-dwarf phenotype in Arabidopsis thaliana. Plant Cell Physiol. 46: 1358–1365. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Dong J. (2013). Stomatal development in Arabidopsis. The Arabidopsis Book 11: e0162, doi/10.1199/tab.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Peterson K.M., Horst R.J., Torii K.U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23: 3260–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J.H., Shen T.T., Liu W.J., Wang X.Q. (2013). Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants. PLoS ONE 8: e78997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42: 93–113. [PubMed] [Google Scholar]
- Sawchuk M.G., Donner T.J., Head P., Scarpella E. (2008). Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol. 148: 1908–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131: 3445–3455. [DOI] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Koumoto Y., Fuji K., Nishiyama C., Kohchi T., Hara-Nishimura I., Shimada T. (2014). CONTINUOUS VASCULAR RING (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol. 55: 764–772. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Shimada T., Koumoto Y., Shimada T.L., Kondo M., Takahashi T., Okuyama Y., Nishimura M., Hara-Nishimura I. (2010). Arabidopsis Qa-SNARE SYP2 proteins localized to different subcellular regions function redundantly in vacuolar protein sorting and plant development. Plant J. 64: 924–935. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Ueda H., Shimada T., Nishiyama C., Hara-Nishimura I. (2009). Vacuolar SNAREs function in the formation of the leaf vascular network by regulating auxin distribution. Plant Cell Physiol. 50: 1319–1328. [DOI] [PubMed] [Google Scholar]
- Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105: 6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangstad O.P., Gilde B., Chadchawan S., Seem M., Husebye H., Bradley D., Bones A.M. (2004). Cell specific, cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum. Plant Mol. Biol. 54: 597–611. [DOI] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., Takahashi T., Ohtomo I., Nishimura M., Hara-Nishimura I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47: 164–175. [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7: 263–270. [DOI] [PubMed] [Google Scholar]
- Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Saxild H.H., Nielsen C., Brunak S., Knudsen S. (2002). A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3: h0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Jørgensen M., Pihlgren U., Rask L. (1995). The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol. Biol. 27: 911–922. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ober J.A., Kliebenstein D.J. (2006). The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18: 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.