A previously unrecognized plastid-to-vacuole protein trafficking pathway that is stress induced and autophagy dependent appears to involve ATI1 interaction with plastid proteins and ATG8 of the core autophagy machinery.
Abstract
Selective autophagy has been extensively studied in various organisms, but knowledge regarding its functions in plants, particularly in organelle turnover, is limited. We have recently discovered ATG8-INTERACTING PROTEIN1 (ATI1) from Arabidopsis thaliana and showed that following carbon starvation it is localized on endoplasmic reticulum (ER)-associated bodies that are subsequently transported to the vacuole. Here, we show that following carbon starvation ATI1 is also located on bodies associating with plastids, which are distinct from the ER ATI bodies and are detected mainly in senescing cells that exhibit plastid degradation. Additionally, these plastid-localized bodies contain a stroma protein marker as cargo and were observed budding and detaching from plastids. ATI1 interacts with plastid-localized proteins and was further shown to be required for the turnover of one of them, as a representative. ATI1 on the plastid bodies also interacts with ATG8f, which apparently leads to the targeting of the plastid bodies to the vacuole by a process that requires functional autophagy. Finally, we show that ATI1 is involved in Arabidopsis salt stress tolerance. Taken together, our results implicate ATI1 in autophagic plastid-to-vacuole trafficking through its ability to interact with both plastid proteins and ATG8 of the core autophagy machinery.
INTRODUCTION
Plants are exposed to multiple environmental stresses, such as limiting light levels, insufficient nitrogen in the soil, drought, and excess salt. These stresses have major physiological and metabolic effects and also lead to deprivation of energy (Baena-González and Sheen, 2008; Guiboileau et al., 2010). Hence, plants have evolved convergent stress-associated processes for protection from these negative outcomes (Baena-González and Sheen, 2008; Guiboileau et al., 2010). One of the central cellular machineries allowing plants to survive energy-depleting stresses is macroautophagy, hereafter referred to as autophagy. Starvation-induced autophagy is classically associated with bulk turnover of macromolecules and organelles in the vacuole upon exposure to stresses causing extensive energy deprivation (Han et al., 2011; Hayward and Dinesh-Kumar, 2011; Li and Vierstra, 2012; Liu and Bassham, 2012; Yoshimoto, 2012). In addition, autophagy targets specific proteins, protein aggregates, or organelles for recycling in the vacuole by a process called selective autophagy (Floyd et al., 2012; Li and Vierstra, 2012; Schreiber and Peter, 2014).
A key protein for bulk and selective autophagy is AUTOPHAGY-RELATED PROTEIN8 (ATG8). In the case of selective autophagy, ATG8 binds to specific ATG8-interacting motifs termed AIM or LIR motifs (Noda et al., 2010; Birgisdottir et al., 2013) located in proteins destined for degradation by selective autophagy, thus inducing their delivery to the lytic compartment (the vacuole in plants). ATG8 generally exists as a multiprotein family in eukaryotes, with Arabidopsis thaliana possessing nine ATG8 isoforms annotated as ATG8a up to ATG8i (Avin-Wittenberg et al., 2012). Plant ATG8 proteins, as well as other plant proteins that bind to ATG8, have been shown to be associated with multiple aspects of plant growth, such as responses to hormones and abiotic stresses (Yoshimoto et al., 2004; Slavikova et al., 2008; Zientara-Rytter et al., 2011; Zhou et al., 2013). Selective autophagy in plants has recently gained special attention through the identification of components for selective autophagy of endoplasmic reticulum (ER; Liu et al., 2012), peroxisomes (Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013; Yoshimoto et al., 2014), mitochondria (Li et al., 2014), protein aggregates (Svenning et al., 2011; Zhou et al., 2013), specific proteins (Suttangkakul et al., 2011; Derrien et al., 2012), and specific plastid components such as Rubisco (Chiba et al., 2003; Ishida et al., 2008) and starch granules (Wang et al., 2013). Further information regarding selective autophagy in plants can be found in recently published reviews (Floyd et al., 2012; Michaeli and Galili, 2014).
Our laboratory has recently identified two ATG8 binding proteins in Arabidopsis plants, ATI1 and ATI2, each of which contains two AIM motifs and a transmembrane domain (Honig et al., 2012). These two proteins defined a newly identified stress-induced compartment that moves along the ER network and is subsequently transported to the vacuole (Honig et al., 2012). Because autophagy-dependent degradation of plastid components was previously suggested (Ishida et al., 2008; Wang et al., 2013), we were interested in examining whether ATG8-INTERACTING PROTEIN1 (ATI1) might be involved in such a process. In this article, we show that in response to carbon (C) starvation stress that leads to senescence, ATI1 is associated with bodies located on the surface of plastids as well as inside their stroma. ATI1 interacts with plastid proteins and the interaction persists within the ATI-plastid bodies even following their budding off the plastid. ATI1 also interacts with ATG8f in the ATI plastid bodies to subsequently enable their delivery to the vacuole by an autophagic pathway. The pivotal role of ATI1 in this process was further demonstrated by the increased stability of PrxA, a plastid-localized ATI1-interacting protein, in ATI1-deficient mutant plants. Taken together, our results describe the involvement of ATI1 in a special stress-induced and autophagy-dependent plastid-to-vacuole protein trafficking pathway.
RESULTS
ATI1-GFP Defines Plastid-Associated Bodies, Which Are Morphologically Distinct from the Previously Described ATI1-Containing ER-Associated Bodies
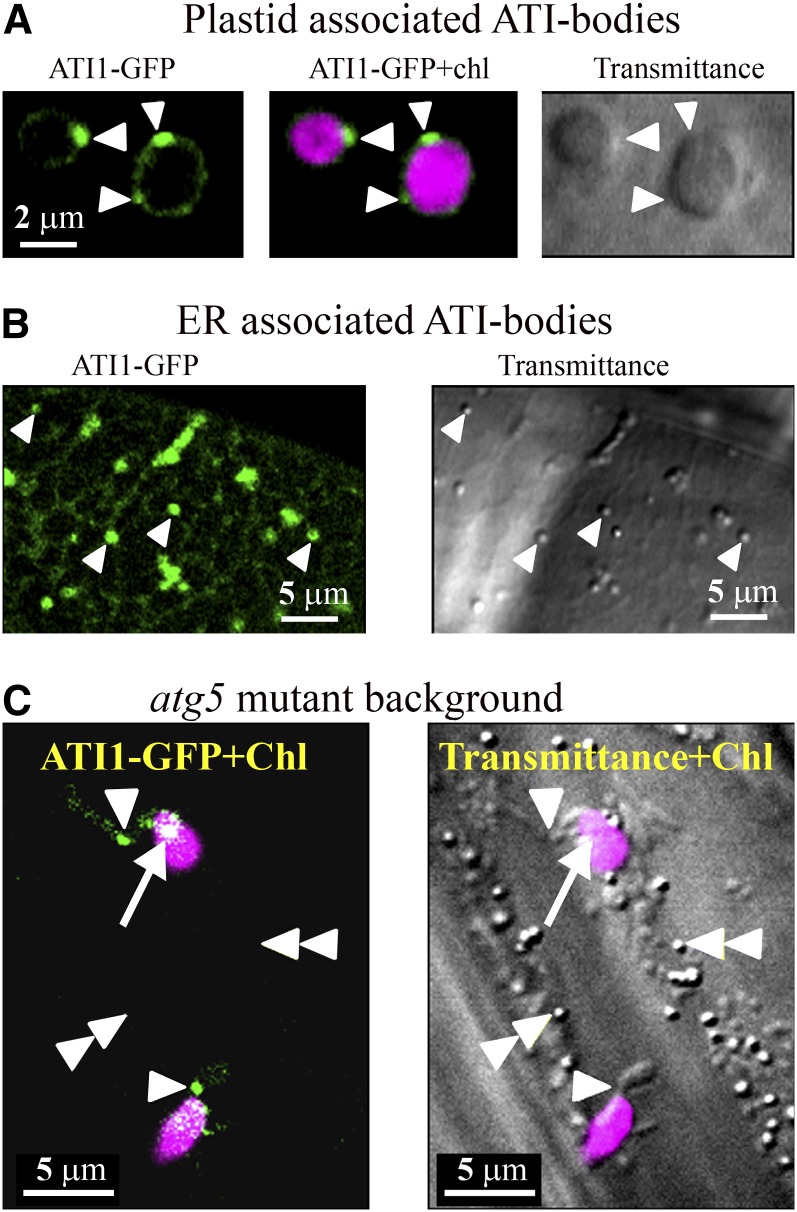
We previously showed that ATI1 is a plant-specific ATG8 binding protein defining a stress-induced ER-associated compartment that is distinct from mitochondria, Golgi, peroxisomes, and “classical” autophagosomes and is subsequently transported to the vacuole, apparently for its degradation inside this organelle (Honig et al., 2012). This ER-associated ATI1 compartment exhibits a distinct morphology that can be easily visualized in the transmittance image of a confocal microscope, thus enabling its identification without the requirement of a fluorescent ATI1-GFP (green fluorescent protein) marker. To address whether ATI1 is also associated with the plastid-localized photosynthetic apparatus, we employed confocal laser scanning microscopy with the far-red channel active (see Methods) to detect plastids via their chlorophyll autofluorescence (indicated as magenta labeling throughout this article). As shown in Figure 1A, in transgenic Arabidopsis seedlings stably expressing ATI1-GFP that were exposed to darkness (C starvation), ATI1-GFP was detected on the surface of plastids of hypocotyl epidermal cells, creating a ring-like shape that surrounds the chlorophyll autofluorescence signal (Figure 1A, left and middle). In addition, several green bodies containing ATI1-GFP (hereafter termed ATI1-PS bodies) were present on the surface of these plastids (Figure 1A, left and middle). Notably, these plastid-associated bodies were not detected in the transmission image (Figure 1A, right). These results indicated that the ATI1-PS bodies are structurally distinct from our previously reported ER-associated ATI1-GFP containing bodies (Figure 1B, left) (Honig et al., 2012), which were clearly identified in the transmission image (Figure 1B, right). For clarity, we refer to the ER-associated bodies that are visible in a transmission image as ATI1-ER bodies. Additional support for the differential structure of the ATI1-ER and ATI1-PS bodies is provided in Supplemental Figure 1.
Figure 1.
ATI1-GFP Labels Plastid-Associated Bodies (ATI-PS Bodies) That Are Distinct from the ER-Associated ATI1-GFP-Labeled Bodies (ATI-ER Bodies).
Confocal images of plastids identified by their chlorophyll autofluorescence (Chl; magenta signal) in transgenic seedlings stably expressing ATI1-GFP (green signal) in the background of the wild type ([A] and [B]) and the autophagy-deficient atg5 mutant (C) following carbon starvation (see Methods).
(A) White arrowheads indicate ATI1-GFP-labeled bodies (left and middle panels) that are not detected in the transmittance channel (right panel).
(B) ATI1-GFP-labeled bodies (white arrowheads) are seen on the ER network (left panel). These bodies are clearly detected in the transmittance image (right panel).
(C) In the left panel, both the chlorophyll and ATI1-GFP signals are shown and the white arrowheads point toward ATI-PS bodies seen in stromules, which display lower expression of ATI1-GFP compared with its expression in the ATI-PS bodies. The double white arrowheads point to ATI-ER bodies that are not fluorescently visualized but are detected in the transmittance image (right panel). The white arrow shows an ATI1-GFP labeled body that is colocalized with the magenta chlorophyll signal resulting in a body that appears white in the left panel.
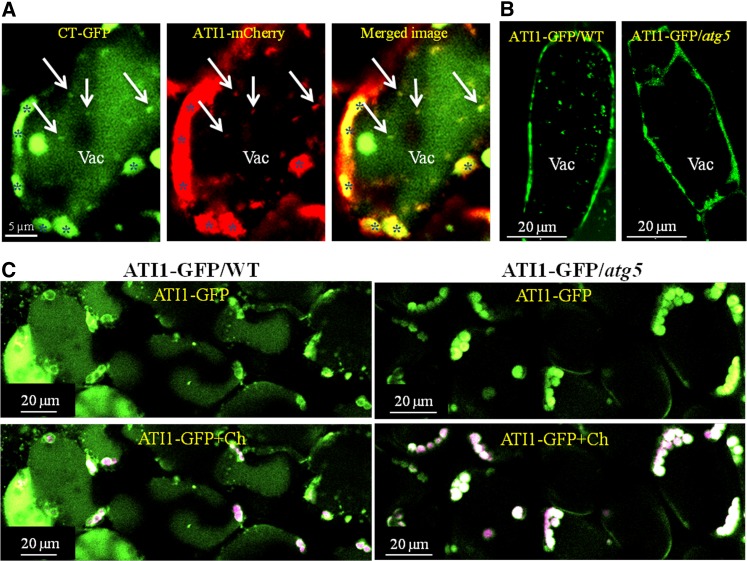
To examine the possible requirement of active autophagy for the localization of ATI1-GFP either in the ATI1-ER or in the ATI1-PS bodies, we generated a transgenic plant that expresses ATI1-GFP in the background of an Arabidopsis atg5 knockout mutant (ATI1-GFP/atg5), which lacks active autophagy (Thompson et al., 2005). As shown in Figure 1C, in a hypocotyl epidermis cell of an ATI1-GFP/atg5 plant that was exposed to C starvation, two green bodies were detected near two magenta-labeled plastids (left panel). These two bodies were located in stroma-filled tubules known as stromules (Köhler et al., 1997; Schattat et al., 2012) protruding out from these senescing plastids and displaying weak expression of ATI1-GFP compared with its expression in the ATI-PS bodies they harbored. Yet, these two bodies were not detected in the transmission image (Figure 1C, right), similarly to the ATI-PS bodies shown in Figure 1A. In this same transmission image (Figure 1C, right), several ATI-ER bodies were easily detected according to their unique “marble-like” appearance in the transmission channel of the confocal microscope (Honig et al., 2012). These same bodies were not labeled by the green ATI1-GFP signal (Figure 1C, left). This shows that while the incorporation of ATI1 to the ATI1-ER bodies requires active autophagy, the incorporation of ATI1 onto the ATI1-PS bodies does not require active autophagy. Hence, apart from the different morphology of ATI-PS and ATI-ER bodies, this is another feature by which these two types of bodies labeled by ATI1-GFP can be discriminated. Furthermore, as seen in Figure 1C, a white-labeled body was also detected inside the plastid. This body appears in white due to the mixture of the green ATI-GFP labeling and the magenta labeling of chlorophyll autofluorescence. A similar localization of a green body inside a magenta-labeled plastid (resulting in a white signal) is also shown in Supplemental Figure 2, suggesting that the ATI1-PS bodies can also be found within plastids. Additionally, a movie showing the possible penetration of ATI-PS bodies from the plastid periphery into its interior (white-labeled bodies) is provided as Supplemental Movie 1. Finally, a comparison between a stromule labeled by chloroplast-targeted GFP (CT-GFP) and ATI1-GFP is provided in Supplemental Figure 3.
Taking together the results of Figure 1, we concluded that the ATI1-PS and ATI1-ER bodies are two distinct types of bodies containing ATI1-GFP and can be located together in hypocotyl epidermis cells.
Detection of the ATI1-PS Bodies inside Plastids by Electron Microcopy
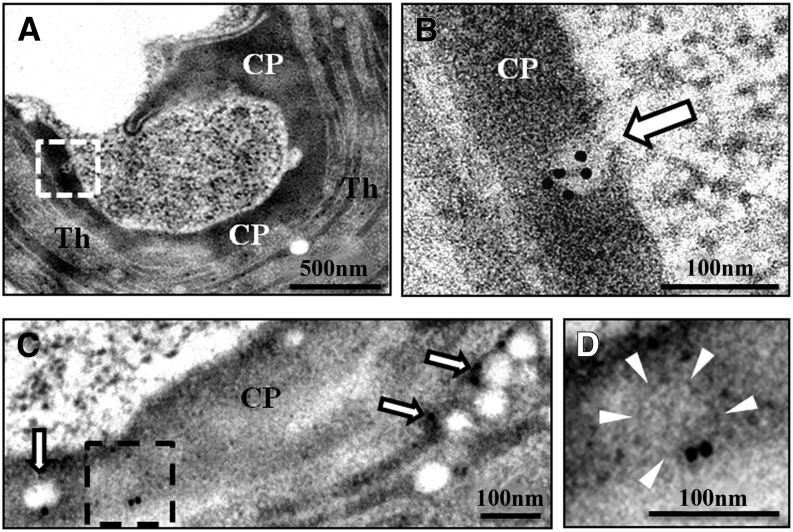
To further examine the ATI1-PS bodies, proximal regions of hypocotyl cells derived from the ATI1-GFP-expressing seedlings that were treated under C starvation were analyzed by electron microscopy (same tissue and treatment used for the confocal imaging procedure described above). Samples were subjected to a relatively mild chemical fixation and ultrathin sections were immunogold labeled with anti-GFP antibodies. We observed gold particle labeling in spherical, non-electron-dense compartments located within the plastid stroma (Figures 2B and 2C). These compartments were occasionally located in stromal areas protruding out of the grana delimited area. These chloroplast protrusions (CPs; Figures 2A and 2C) were previously shown to occur following abiotic stresses such as salt stress and temperature shifts (Holzinger et al., 2007; Yamane et al., 2012). We also observed gold particle labeling of a spherical membranous structure inside the CP (Figure 2D), which may represent one of the initial steps in the formation of ATI-PS bodies. The estimated diameter of these plastid-associated spherical bodies is ∼50 to 100 nm, which is considerably smaller than the estimated size of the ATI-ER bodies of 0.5 to 1 µm (Honig et al., 2012). Yet, the size of these ATI-PS bodies is in agreement with the size of intraplastid vesicles that are generated following low temperature stress in several dicotyledonous plant species (Morré et al., 1991; Garcia et al., 2010). These observations support our confocal imaging results describing ATI1-GFP-labeled bodies both in the periphery of plastids and within them (Figures 1 and 2; Supplemental Figures 1 and 2).
Figure 2.
Intraplastid ATI-PS Bodies Observed by Electron Microscopy.
Immunogold labeling with anti-GFP antibodies (black dots) of ultrathin sections of an ATI1-GFP transgenic plant exposed to carbon starvation (see Methods).
(A) A plastid exhibiting stress-induced CPs.
(B) Enlargement of the area designated by the white rectangle in (A). Arrow indicates an intraplastid vesicle labeled by gold particles, which report the expression and localization of ATI1-GFP.
(C) A section of a different plastid showing non-electron-dense, intraplastid compartments labeled by gold particles (arrows).
(D) Enlargement of the boxed area in (C) exhibits a membranous structure (delimited by white arrowheads) that is labeled by gold particles.
ATI-PS Bodies Bud Out and Detach from Plastids into the Cytosol by a Process That Does Not Require Active Autophagy
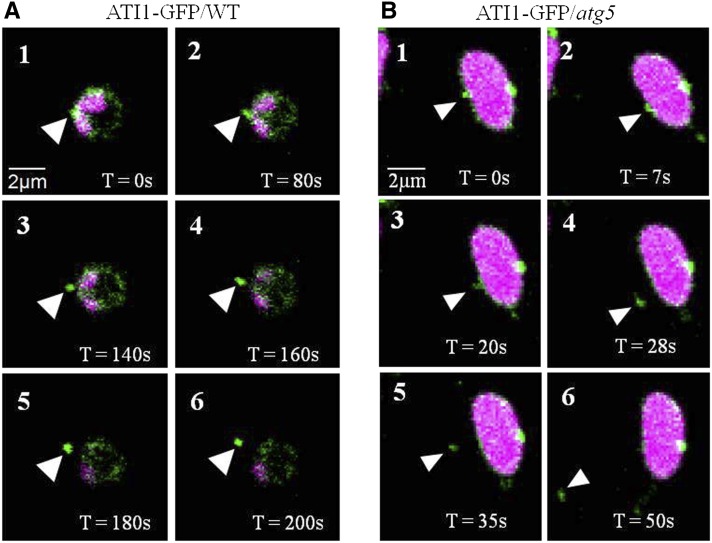
Previous studies showed that some components originating from plastids ultimately reach the vacuole, apparently for degradation (Ishida et al., 2008; Wang et al., 2013). Considering the association of ATI1 with autophagy, we hypothesized that the ATI1-GFP-labeled compartments present inside the stroma of plastids are subsequently exported to the cytosol en route to their degradation inside the vacuole. To explore this possibility, we performed live-imaging time-lapse experiments, focusing on plastids expressing ATI1-GFP. As shown in Figure 3A and Supplemental Movie 2, a plastid whose stroma was nearly depleted of chlorophyll had a small green ATI1-GFP-containing body on its left middle surface, between two small regions containing chlorophyll. Notably, as time elapsed, the green body increasingly protruded out from the plastid surface until it was completely detached (Figure 3A).
Figure 3.
The Budding and Detachment of ATI-PS Bodies from Plastids.
Confocal time-lapse imaging of plastids from hypocotyl epidermis cells of either an ATI1-GFP transgenic plant (wild-type background; [A]) or a transgenic plant expressing ATI1-GFP in the background of the atg5 autophagy-deficient mutant (B). Both plastids display chlorophyll autofluorescence (magenta signal) and ATI1-GFP expression (green signal). Numbers 1 to 6 (in both panels) represent a series of time points (T) as indicated, and the budding ATI-PS bodies are indicated by white arrowheads.
To further explore whether the budding of the ATI1-PS body out of senescing plastids requires active autophagy, we performed the same type of analysis in an Arabidopsis atg5 knockout mutant stably expressing ATI1-GFP. As shown in Figure 3B, a green body was observed budding and detaching from a chlorophyll-containing plastid into the cytosol (Supplemental Movie 3), similarly to the process visualized in the wild-type background. This suggests that active autophagy is not required for the budding of the ATI1 bodies from plastids.
ATI-PS Bodies Accumulate as Carbon Starvation Progresses in Mesophyll Cells of Seedling Cotyledons
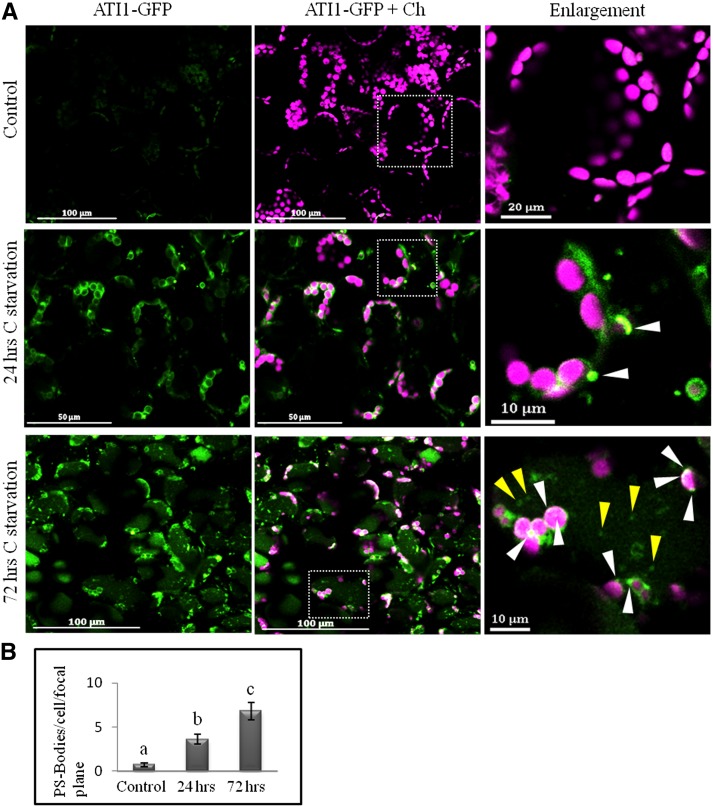
The observations described thus far were all made in hypocotyl epidermal cells known to contain a relatively low amount of plastids. In addition, epidermal cells (of both seedling cotyledons and hypocotyls) contain many ATI-ER bodies, mainly following C starvation stress (Honig et al., 2012); thus, this tissue may represent a system that is not optimal for monitoring ATI-PS bodies specifically. For these reasons, we examined the localization and expression pattern of ATI1-GFP in the plastid-rich mesophyll cells of seedling cotyledons. We examined 1-week-old ATI1-GFP transgenic seedlings that were either treated under C starvation for 24 or 72 h or grown under C-rich conditions (control). In the control mesophyll cells, the ATI1-GFP signal was almost exclusively associated with plastids and was not detected in other subcellular compartments, as demonstrated by its colocalization with the chlorophyll signal (Figure 4A, top). However, the expression of ATI1-GFP in control plastids was markedly lower (Figure 4A, top) compared with in plastids of plants under C starvation for 24 h (Figure 4A, middle). Additionally, unlike in the control cells, the ATI1-GFP signal following 24-h C starvation stress seemed to define the boundary of the chlorophyll autofluorescence (ring shapes) and plastid-associated ATI-PS bodies appeared (Figure 4, middle right). This expression pattern is similar to that observed in hypocotyl epidermal cells of seedlings exposed to 24-h C starvation (Figure 1). Following 72-h C starvation, the ATI1-GFP signal labeled increasing numbers of bodies that were either associated with plastids or within the central vacuole (Figure 4A, bottom right), which was highlighted by a diffusive green signal, most probably due to the degradation of ATI1-GFP within the vacuole.
Figure 4.
ATI-PS Bodies Accumulate with the Progression of C Starvation in Cotyledon Mesophyll Cells.
(A) Confocal imaging of cotyledon mesophyll cells of 7-d-old ATI1-GFP seedlings that were either grown under C-sufficient conditions (Control, top; see Methods) or treated under C starvation for 24 (middle) or 72 h (bottom). Right images show enlargement of the boxed area in the merged images. Parameters for the GFP channel were similar for all images. White arrowheads indicate ATI-PS bodies, whereas yellow arrowheads denote vacuole-localized ATI1-GFP-labeled bodies.
(B) Number of ATI-PS bodies per cell (in a single focal plane) in ATI1-GFP plants following the three different C availability treatments described in (A). Statistically significant differences between treatments (based on the Student’s t test, P < 0.001) are denoted as different letters above columns.
The clear association of ATI1-GFP with plastids and ATI-PS bodies in cotyledon mesophyll cells made it a good system in which to quantify ATI-PS body numbers under the various C starvation treatments. To avoid any possible confusion with ATI-ER bodies, we quantified only ATI1-GFP-labeled bodies that were associated with the chlorophyll signal, excluding independent cytosolic bodies. To that end, images of cotyledon mesophyll cells of a single focal plane were taken from ATI1-GFP seedlings grown under three different C starvation procedures. For each treatment, 24 to 50 cells, imaged from two to four different plants, were used for the analysis. The average number of ATI-PS bodies per cell (in a 2.5-µm-thick focal plane) increased significantly with the progression of the C starvation treatment (Figure 4B). Moreover, the ATI1-GFP expression pattern in hypocotyl cortex cells resembled the results in cotyledon mesophyll cells. Namely, there was relatively low expression in cortex plastids under C-rich conditions and increased plastid-associated expression following C starvation (Supplemental Figure 4). Finally, in contrast to our observations in cotyledon mesophyll cells, cotyledon epidermis cells showed labeling of the ER network, as well as labeling of the plastid periphery and ATI-PS bodies, even under C-available growth conditions (Supplemental Figure 5).
ATI1 Expression Is Intensely Enriched in the Plastids and Vacuoles of Senescing Cells of Adult Leaves
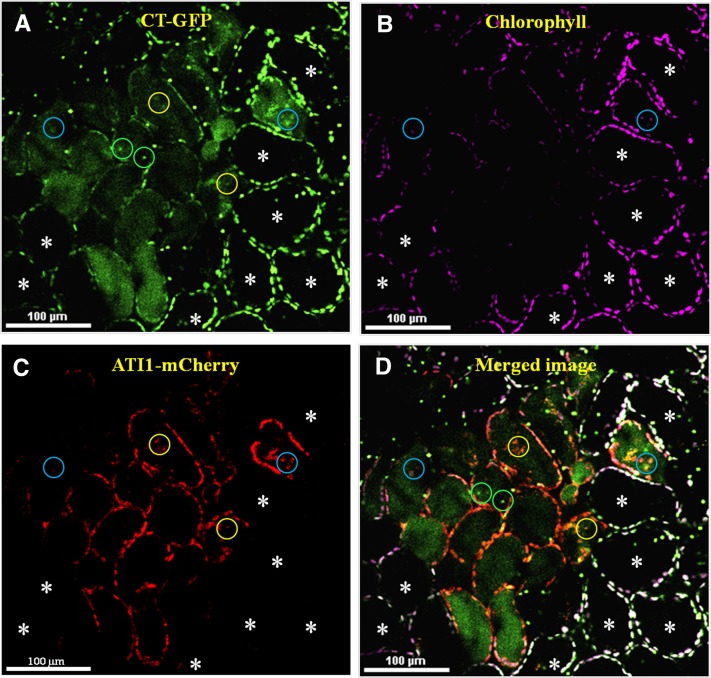
In light of the above results, we assumed that ATI-PS bodies are involved in a unique type of plastid degradation process. We hypothesized that ATI-PS bodies may not necessarily be restricted to dark-exposed seedlings but may also be observed in plastids of adult senescing leaves. Additionally, we were interested in revealing the possible cargo carried by ATI-PS bodies from the plastid. To address both of these issues, we generated transgenic Arabidopsis plants stably coexpressing (1) a chloroplast-targeted GFP (CT-GFP) containing a plastid transit peptide that directs GFP into the plastid stroma (Schattat et al., 2011), thus serving here as a representative of plastid stroma proteins; and (2) ATI1 fused to the red fluorescent protein mCherry (ATI1-mCherry). Four-week-old transgenic plants expressing both CT-GFP and ATI1-mCherry were utilized to visualize mesophyll cells of fully expanded rosette leaves that were exposed to darkness and treated with Concanamycin A (ConA). ConA prevents the acidification of the central vacuole, thus allowing improved visualization of fluorescent proteins within the vacuole lumen (Ishida et al., 2008). We identified heterogeneous mesophyll cells, some of which looked quite vital (nonsenescing), containing multiple plastids enriched with the stroma-localized CT-GFP (Figure 5A) as well as with chlorophyll (Figure 5B). The other cells contained considerably less green plastids (Figure 5A, cells not marked by asterisks) as well as less chlorophyll (Figure 5B) and accumulated considerably higher amounts of the green signal of CT-GFP in the lumen of their central vacuoles (Figure 5A). These characteristics are consistent with previous reports describing senescing cells (Wada et al., 2009; Ono et al., 2013). Notably, the ATI1-mCherry red signal (Figure 5C) was absent from the vital cells and was strictly visible in the other senescing cells, appearing in the senescing plastids. Some vacuoles seemed to contain structures with signals from all three sources (CT-GFP, ATI1-mCherry, and chlorophyll), as well as structures that were labeled with various other combinations of the fluorescent signals (Figure 5). According to the size of some of these structures, which is similar to the size of plastids, along with their emission of chlorophyll autofluorescence, we assume that the structures emitting all three signals represent the delivery of entire plastids, or plastid fractions, into the vacuole, as previously reported (Wada et al., 2009). The intravacuolar structures with CT-GFP and ATI1-mCherry signals may represent whole plastids lacking chlorophyll or ATI-PS bodies that were delivered to the vacuole (as analyzed in more detail below). This phenomenon, involving the expression of ATI1-mCherry exclusively in senescing plastids, is not restricted to adult rosette leaves and was also observed in mesophyll cells of adult stem leaves (Supplemental Figure 6). Based on evaluation of four confocal images of mesophyll cells of adult rosette leaves from four different plants, each encompassing ∼100 cells, we estimated the ratio of senescing/vital cells to be 1:7. Taken together, Figure 5 indicates that ATI1 is enriched specifically in senescing plastids and together with the results presented in Figure 4, suggests a role for ATI-PS bodies in response to C starvation and during senescence.
Figure 5.
Enriched Expression of ATI1-mCherry in Senescing Cells in Which Plastids Are Degraded.
Confocal image of a ConA-treated rosette leaf from a 4-week-old transgenic plant stably expressing chloroplast-targeted GFP (CT-GFP; [A]) and ATI1-mCherry (C). Asterisks denote nonsenescing (“vital”) mesophyll cells that display a relative high intensity of CT-GFP (A) and chlorophyll autofluorescence (B) in their plastids, along with a vacuole lumen that is devoid of any fluorescent signal. The other cells (not marked by asterisks) are senescing cells that accumulate CT-GFP or its degradation products inside their vacuoles ([A] and [D]) and exhibit relatively weak GFP signal from their chloroplasts (A) and relatively chlorophyll fluorescence (B). Expression of ATI1-mCherry (red signal) is detected solely in the senescing cells, as seen in (C) and (D). The image in (D) is the result of merging the signals of all channels (GFP, mCherry, and chlorophyll), and the white color in (D) is the result of the colocalization of the green and magenta signals in plastids of the vital cells. Green circles highlight intravacuole structures that emit only green signal while yellow and light blue circles highlight intravacuole structures that emit combinations of green + red or green + red + magenta signals, respectively.
ATI-PS Bodies That Bud from Plastids Contain a Stroma-Derived Protein Marker
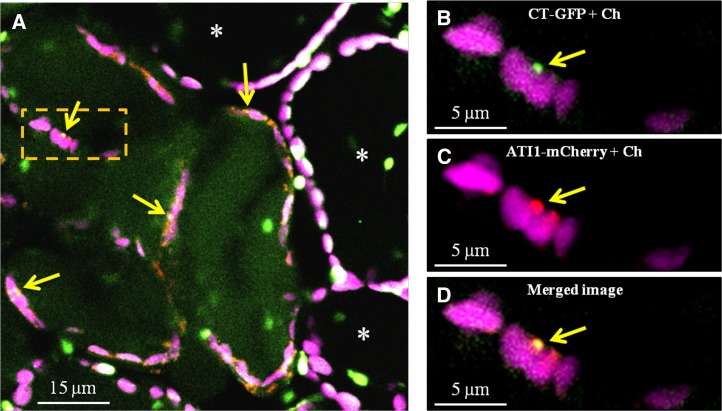
We noticed that chlorophyll-containing plastids in senescing cells apparently had relatively low amounts of stromal proteins, as these plastids were not labeled by CT-GFP (Figure 6A, see cells that are not marked by asterisks). A similar reduction in chloroplast-targeted fluorescent proteins from senescing plastids following dark treatment was previously reported (Wada et al., 2009). In accordance with the results shown in Figure 5, we observed expression of ATI1-mCherry that specifically associated with the senescing plastids, where it seemed to label bodies associating with the magenta chlorophyll signal. These bodies were also labeled by the CT-GFP signal, resulting in yellow bodies where colocalization occurred (Figure 6A). Furthermore, the appearance of the GFP signal within the vacuole lumen suggests that CT-GFP was relocated from plastids to the vacuole. CT-GFP-labeled bodies appearing on the surface of senescing plastids also contained ATI1-mCherry (Figures 6B to 6D). This indicates that ATI-PS bodies that bud from plastids contain stroma-derived proteins. It also suggests that the depletion of stromal proteins from senescing plastids occurs due to their incorporation into ATI-PS bodies as cargo.
Figure 6.
Budding ATI-PS Bodies Contain Chloroplast-Targeted GFP (CT-GFP).
Confocal imaging of an adult rosette leaf of a transgenic plant stably expressing both ATI1-mCherry (red) and CT-GFP (green) following exposure to darkness (see Methods).
(A) Plastids of vital cells (marked by asterisks) and senescing mesophyll cells (not marked by asterisks). Vital cell plastids exhibit both chlorophyll (magenta) and CT-GFP (green) signals (resulting in white color where colocalization occurs), while senescing plastids are detected by their chlorophyll autofluorescence but are devoid of the CT-GFP signal. Their CT-GFP signal is seen within the vacuoles and concentrated in bodies that are also labeled by ATI1-mCherry (red) as detected by the colocalization that results in a yellow signal (yellow arrows).
(B) to (D) Enlargements of the boxed area in (A). Panels show the magenta chlorophyll signal along with CT-GFP (B), ATI1-mCherry (C), or both (D).
ATI1 Interacts with Several Chloroplast-Localized Proteins
To learn more about the function of ATI1, which possesses a transmembrane domain, we screened for membrane-spanning proteins that might interact with it by employing the split-ubiquitin yeast two-hybrid method. Among the coding sequences recovered, 13 are known or predicted to be localized in chloroplasts (Table 1). The interaction of ATI1 with these chloroplast-associated proteins was reexamined by one-on-one yeast two-hybrid assays. Information regarding the possible intraplastid localization of nine of the 13 chloroplast-associated proteins was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/; Table 1). The other four plastid-associated proteins are considered to be localized in plastids based on computational predictions (having a predicted chloroplast targeting transit-peptide at their N terminus); thus, their localization within the plastid is indicated in Table 1 as “unknown.” Five of the chloroplast-associated ATI1-interacting proteins are involved in photosynthesis [LHCA4, NPQ4 (PsbS), PrxA (BAS1), FNR1, and APE1]. Another, ATS2, is critical for phosphatidic acid biosynthesis; thus, its loss of function results in embryo death (Kim and Huang, 2004; Yu et al., 2004). Of the nine proteins with annotated intraplastid localization, seven are localized in the plastid envelope and five in thylakoid membranes (some reside in both membranes). This is consistent with our utilization of the split ubiquitin yeast two-hybrid assay designed to screen for transmembrane spanning proteins. Also, it suggests that in addition to stroma-derived proteins (Figures 5 and 6), transmembrane proteins from the envelope and thylakoid membranes might also be incorporated into the ATI-PS bodies following their interaction with ATI1. Interestingly, NPQ4 was recently suggested to be a cargo component within plastid-localized vesicles (Khan et al., 2013). To test this in planta, we chose two candidate chloroplast-localized ATI1-interacting proteins, NPQ4 and APE1, for bimolecular fluorescence complementation (BiFC) assays. We examined whether each of them (separately) could interact with ATI1 to produce YFP fluorescence in a tobacco (Nicotiana tabacum) leaf transient assay. The results demonstrate an interaction between ATI1-YC (C-terminal fragment of YFP fused to the C terminus of ATI1) with either NPQ4-YN or APE1-YN (N-terminal fragment of YFP fused to C-terminal end of NPQ4 or APE1, respectively). The YFP fluorescence was observed within plastids, plastid stroma protrusions, and in cytosolic punctate structures (Figure 7A). This confirmed that at least APE1 and NPQ4 interact with ATI1 during their mutual localization within plastids and also within budding ATI-PS bodies.
Table 1. Chloroplast-Localized Proteins Identified in a Split Ubiquitin Yeast Two-Hybrid Assay as Potential ATI1-Interacting Proteins.
| Gene Identifier | No. of Independent Clones Identified for Each cDNA | Annotated Function or Symbol | Annotated Localization within the Plastid |
|---|---|---|---|
| At3g32930 | 1 | Uncharacterized protein | Envelope |
| At3g47470 | 1 | LIGHT HARVESTING COMPLEX A4 (LHCA4; a chlorophyll a/b binding protein). | Thylakoid, plastoglobule and envelope |
| At1g04340 | 1 | HR-like lesion inducing protein-related | Unknown |
| At1g44920 | 3 | Uncharacterized protein of chloroplast | Unknown |
| At1g44575 | 10 | NON PHOTOCHAMICAL QUENCHING4 (NPQ; a photosystem II 22-kD protein also known as PsbS) | Thylakoid |
| At4g36530 | 1 | Hydrolase, α/β fold family | Envelope |
| At1g52870 | 1 | Mpv17/PMP22 | Unknown |
| At3g11630 | 1 | 2-Cys peroxiredoxin A (PrxA; also known as BAS1) | Stroma, thylakoid and envelope |
| At5g44020 | 2 | Acid phosphatase/vegetative storage protein | Unknown |
| At5g66190 | 1 | FERREDOXIN NADP(H) OXIDOREDUCTASE1 (FNR1) | Stroma and envelope |
| At2g03140 | 1 | α/β-Hydrolase-like protein | Unknown |
| At5g38660 | 1 | ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT1 (APE1) | Thylakoid and envelope |
| At4g30580 | 1 | ATS2 (an acyltransferase) | Envelope |
Gene accession numbers, annotated function, and/or symbol and localizations within plastids (if annotated) are indicated (according to the TAIR website).
Figure 7.
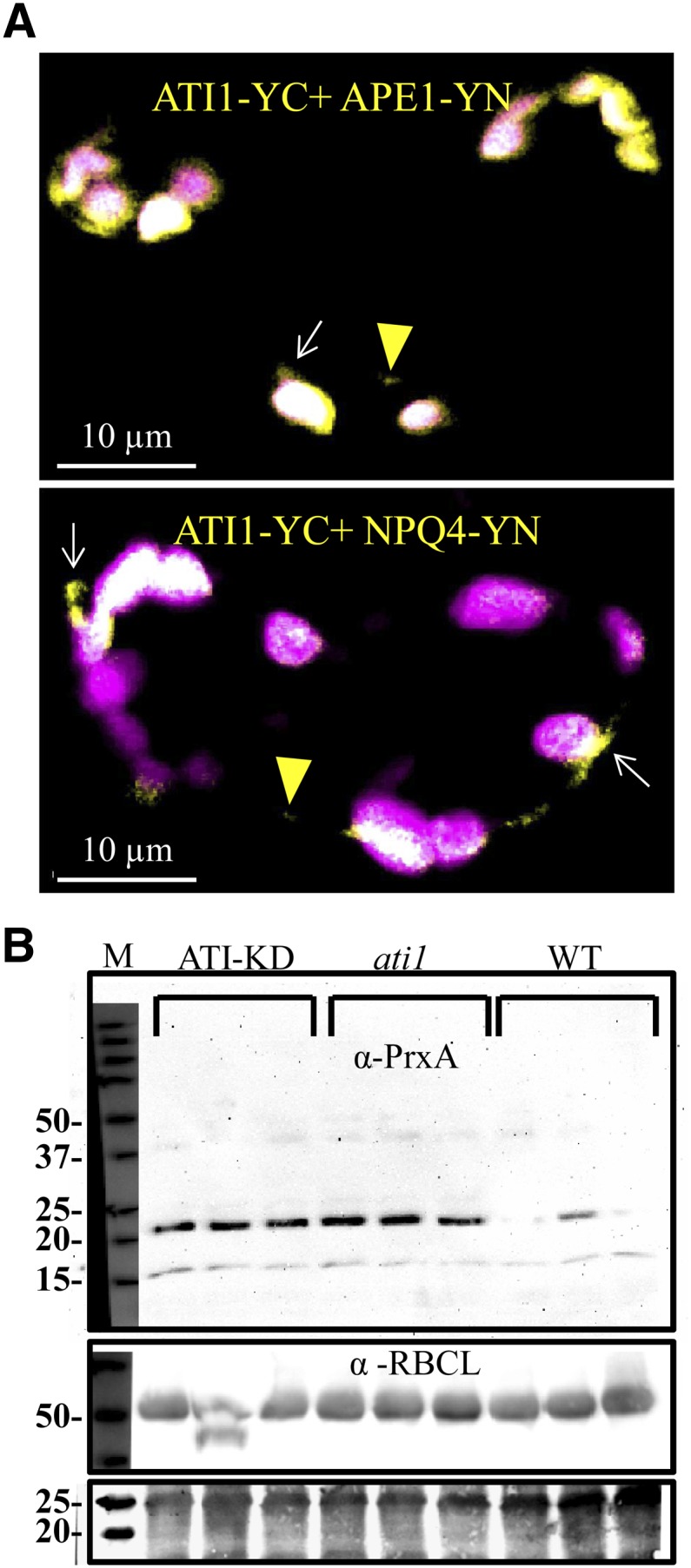
ATI1 Interacts with NPQ4 and APE1 in Planta and Is Required for the Degradation of PrxA.
(A) Confocal imaging of a BiFC assay (see Methods) involving coexpression of ATI1-YC with either APE1-YN (top panel) or NPQ4-YN (bottom panel). The yellow signal indicates an interaction between the two proteins. In both images, plastids are detected by their chlorophyll autofluorescence (magenta signal) and colocalization of the yellow and magenta signals results in white color.
(B) Immunoblot analysis on triplicate samples of rosette leaves taken from 4-week-old wild-type, ati1 knockout, and ATI-KD double knockdown plants. Immunolabeling was conducted with either an anti-PrxA antibody (labeling of the 22-kD PrxA protein, top panel) or the anti-RBCL antibody (∼55-kD large subunit of Rubisco, middle panel, as a negative control). A fraction of the total protein staining (membrane stained with amido black) is shown in the bottom panel.
ATI1 Deficiency Results in the Accumulation of the Plastidic ATI1-Interacting Protein, PrxA (BAS1), but Does Not Affect the Level of Rubisco
The above results led us to hypothesize that the plastid-localized ATI1-interacting proteins are incorporated into the ATI-PS bodies for degradation via their binding to ATI1. If so, we assumed that ATI1 deficiency might affect the level of any given ATI1-interacting protein. To examine this hypothesis, we used anti-PrxA antibodies that target the plastid-localized 2-cystein peroxiredoxin, PrxA (also known as BAS1; Dangoor et al., 2012), which was recovered following our split ubiquitin yeast two-hybrid screen (Table 1). These antibodies were utilized for immunoblot analysis on total protein extracts from 4-week-old adult rosette leaves of the wild type, ati1 knockout, and the ATI-KD double knockdown genotype (in which ATI1 is knocked out and ATI2 is knocked down; Honig et al., 2012). The amount of PrxA in ati1 and ATI-KD was significantly higher than that in the wild type (Figure 7B). These results demonstrate that a functional ATI1 is necessary for the turnover of the plastid-localized PrxA protein. As a negative control, we examined the level of Rubisco (used here as a representative of plastid proteins that do not interact with ATI1) utilizing anti-RBCL antibodies (Agrisera) that target the large subunit of Rubisco. The results showed mostly similar levels of Rubisco across the three different genotypes (except for one ATI-KD plant that displayed an apparently degraded RBCL subunit), indicating that Rubisco is probably not a target of ATI1 for degradation. As Rubisco was previously shown to be delivered to the vacuole by Rubisco-containing bodies (RCBs; Ishida et al., 2008), this result also implies that ATI-PS bodies and RBCs represent two distinct types of bodies that originate from plastids, yet deliver different plastid proteins for degradation in the vacuole.
ATI1 and ATG8f Interact in ATI-PS Bodies
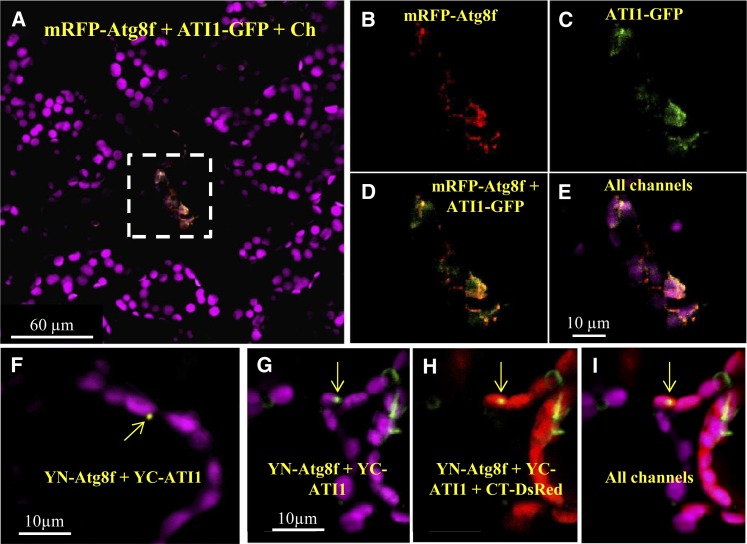
ATI1 was originally identified as an ATG8-interacting protein (Honig et al., 2012). Our observations, documenting the accumulation of ATI-PS bodies either following C starvation or in senescing leaves, implied involvement of autophagy in the function of ATI-PS bodies. To clarify this issue, we generated transgenic plants coexpressing both ATI1-GFP and mRFP-ATG8f and imaged adult rosette leaves taken from 4-week-old plants (similarly to the leaves imaged in Figures 5 and 6). We detected cells that did not express either of the fusion proteins and exhibited chlorophyll autofluorescence (Figure 8A, vital cells outside of the dashed rectangle). There were also cells with relatively low chlorophyll autofluorescence that contained both ATI1-GFP and mRFP-ATG8f (Figure 8A, a representative senescing cell is seen within the dashed rectangle). A closer look into the senescing cells revealed that mRFP-ATG8f (Figure 8B) and ATI1-GFP (Figure 8C) were colocalized in bodies (Figure 8D) that were found either surrounding or within the faint chlorophyll autofluorescence signal (Figure 8E). This result indicates that ATG8f is localized in ATI-PS bodies that are generated specifically in senescing cells of adult rosette leaves.
Figure 8.
ATG8f and ATI1 Interaction Is Associated with Plastids in Leaf Mesophyll Cells.
(A) to (E) Confocal imaging of adult rosette leaf mesophyll cells of a transgenic plant stably expressing both mRFP-ATG8f (red signal) and ATI1-GFP (green signal).
(A) A representative image showing a senescing cell (highlighted within the dashed white rectangle) exhibiting a relative low level of chlorophyll emission and expression of both the red and green signals, resulting in yellow color where colocalization occurs. This cell is surrounded by a population of “vital” cells exhibiting no detectable expression of the fluorescent proteins.
(B) to (E) Enlargements of the boxed area in (A).
(F) Confocal imaging of a BiFC assay (see Methods) involving coexpression of YC-ATI1 and YN-ATG8f. The yellow signal indicates interaction between the two proteins.
(G) to (I) A similar BiFC assay as shown in (F) except that the interaction here is presented as green signal and with the additional expression of the CT-DsRed (red) plastid stroma fluorescent marker. In (F) to (I), yellow arrows point toward bodies where the interaction between ATG8f and ATI1 occurs.
To examine whether ATI1 and ATG8f interact in ATI-PS bodies as they do in ATI-ER bodies (Honig et al., 2012), we performed a BiFC transient assay in fully expanded tobacco leaves that were deprived of light for 6 h before confocal imaging. Cotransformation of YN-ATG8f (N-terminal half of YFP fused to ATG8f) and YC-ATI1 (C-terminal half of YFP fused to ATI1; see Methods) yielded YFP emission from bodies that seemed attached to the magenta labeling of plastid chlorophyll (Figure 8F). As the chlorophyll autofluorescence is not indicative of the full volume of the plastid, we examined whether these bodies were located within the plastid or attached to its periphery. We cotransformed tobacco leaves with the BiFC constructs YC-ATI1 and YN-ATG8f, along with CT-DsRed (Ishida et al., 2008), which is used as a red fluorescent plastid stroma marker. In this case, the interaction between the proteins is presented as a green signal to better visualize colocalization with the red signal of CT-DsRed (Figures 8G to 8I). Bodies labeled by the ATI1 and ATG8f interaction did not seem to fully overlap with chlorophyll autofluorescence (Figure 8G). However, these bodies did overlap with the CT-DsRed signal (Figures 8H and 8I), indicating that the interaction of ATI1 with ATG8f apparently occurs in bodies that are localized within senescing plastids. This is consistent with the colocalization of ATI1 and ATG8f that was observed in plastids of adult rosette leaves (Figure 8E).
ATI-PS Bodies Are Transported to the Vacuole by a Process That Requires Functional Autophagy
In light of the observations of ATI1-GFP- and ATI1-mCherry-labeled bodies within vacuoles (Figures 4 and 5, respectively), we wished to get further insight into whether these are indeed ATI-PS bodies that are eventually delivered to vacuoles. For this, we examined vacuoles of senescing cells of adult rosette leaves from the CT-GFP + ATI1-mCherry line following exposure to darkness coupled with ConA treatments (see Methods; same line and treatment visualized in Figure 5). The vacuoles of senescing cells contained a number of CT-GFP-labeled bodies that were also labeled with ATI1-mCherry (Figure 9A). The fact that the ATI1-mCherry-labeled bodies within the vacuole contained a plastid stroma-derived protein (CT-GFP) indicates that ATI-PS bodies are eventually destined for the vacuole. Supplemental Movie 4 shows the random movement of the ATI-PS bodies, which is typical for bodies within the vacuole lumen. Furthermore, CT-GFP and ATI1-mCherry were also extensively colocalized in plastids situated in the cytosol surrounding the central vacuole (Figure 9A).
Figure 9.
Autophagy Is Required for the Transport of ATI-PS Bodies into the Central Vacuole.
(A) Confocal imaging of a ConA-treated adult rosette leaf of a transgenic plant stably expressing chloroplast-targeted GFP (CT-GFP; green signal) and ATI1-mCherry (red signal). Dispersed GFP signal defines the vacuole lumen (Vac) where both CT-GFP (left panel) and ATI1-mCherry (middle panel) are colocalized (right panel) in small bodies (marked by white arrows in all three images). The vacuole is surrounded by plastids that are also labeled by both CT-GFP and ATI1-mCherry, as indicated by their yellow labeling in the right panel (marked by asterisks).
(B) Confocal imaging of two different transgenic plants, both stably expressing ATI1-GFP, but one in the background of the wild-type genotype (left panel) and the second in the background of the atg5-1 mutant (right panel). Both plants were treated with ConA and incubated in the dark as described in Methods. Each image represents a stack of 10 images of 1-min live imaging (T-stack) of the vacuole focal plane to increase the probability of detecting randomly moving bodies within the vacuole lumen. Vac, vacuole lumen.
(C) Imaging of cotyledon mesophyll cells of the same lines described in (B) that were treated under C starvation for 72 h. Top panels show only ATI1-GFP, whereas bottom images show both the green ATI1-GFP signal and the magenta chlorophyll autofluorescence signal.
To explore whether the trafficking of ATI-PS bodies from plastids to the vacuole requires functional autophagy, we visualized vacuoles of hypocotyl epidermal cells of 7-d-old seedlings that were treated with ConA (see Methods). This analysis was performed in both ATI1-GFP (wild-type background) and ATI1-GFP/atg5 transgenic seedlings (Figure 9B). ATI1-GFP-labeled bodies were observed in the central vacuoles of the wild type (Figure 9B), which is consistent with the observation already reported by Honig et al. (2012). As ATI1-GFP may label both ATI-ER and ATI-PS bodies in hypocotyl epidermal cells (Figure 1; Supplemental Figure 1), we were unable to distinguish between the two types of bodies in the wild-type background. To address this, we took advantage of the fact that only ATI-PS bodies (and not ATI-ER bodies) are labeled by ATI1-GFP in the atg5 mutant background (Figure 1C). We did not detect ATI1-GFP labeled bodies in any of the vacuoles examined in the atg5 background (n = 20 vacuoles; Figure 9B). The ATI1-GFP signal surrounding the vacuole apparently represents its expression in the ER network of these cells. Time-lapse imaging of the left panel and the right panel of Figure 9B are provided as Supplemental Movies 5 and 6, respectively.
We also examined cotyledon mesophyll cells of the same two lines following 72-h C starvation stress. While the ATI1-GFP signal in the wild-type background was concentrated around the plastid chlorophyll autofluorescence (in a ring-like pattern), the ATI1-GFP signal in the atg5 background filled the entire volume of the plastids (Figure 9C). Moreover, the dispersed GFP signal filling the vacuole lumen in the wild type was absent from the vacuoles in atg5. These observations indicate that whereas ATI1-GFP is ultimately degraded in the vacuole in wild-type plants, it accumulates in the plastids in the autophagy-deficient atg5 mutants and is not degraded in vacuoles following 72 h of C starvation. Taken together, the results in Figure 9 indicate that ATI-PS bodies are transported to the vacuole through a process that requires active autophagy.
ATI1-GFP Labels ATI-PS Bodies When Driven under the Transcriptional Control of the ATI1 Endogenous Promoter
Data regarding ATI-ER (Honig et al., 2012) and ATI-PS (this report) bodies were largely obtained using ATI1 fused to GFP and driven by the constitutive 35S cauliflower mosaic virus promoter (35S:ATI1-GFP). To be sure that the labeling of these vesicles by ATI1 is not the result of its constitutive overexpression, we generated transgenic plants harboring ATI1-GFP driven by a 1552-bp DNA fragment that is naturally situated upstream of the ATI1 translation start site, which apparently includes the ATI1 endogenous promoter (proATI1:ATI1-GFP; see Methods). Confocal microscopy of proATI1:ATI1-GFP transgenic seedlings under C starvation revealed labeling of plastids in both hypocotyl cortex and cotyledon mesophyll cells (Supplemental Figure 7). Moreover, ATI-PS bodies were detected associated with plastids (Supplemental Figures 7B and 7D, indicated with white arrows), and the presence of ATI1-GFP driven by the ATI1 endogenous promoter in the plastid stroma was further confirmed by the visualization of stromules (Supplemental Figure 7D). Similarly to our observations in 35S:ATI1-GFP transgenic plants, proATI1:ATI1-GFP plants treated with the senescence-inducing “individually darkened leaves” treatment (see Methods) showed an elevated ATI1-GFP signal within plastids (Supplemental Figure 8). Finally, ATI-ER bodies, identified by their obvious appearance in the transmittance channel, were also labeled by ATI1-GFP in this transgenic line (Supplemental Figure 9). Taken together, we concluded that the ATI1-GFP accumulation pattern is similar when driven by the 35S cauliflower mosaic virus promoter and by the ATI1 endogenous promoter.
ATI1 Is Involved in Salt Stress Tolerance of Arabidopsis Plants
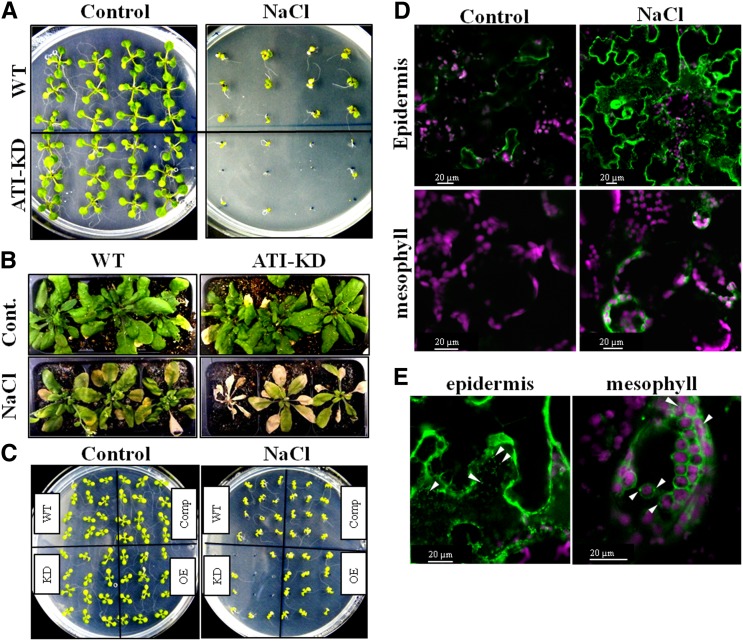
Autophagy is important for tolerance of abiotic stresses, such as salt and drought stresses (Slavikova et al., 2008; Liu et al., 2009; Zhou et al., 2013). Since salt stress was recently shown to induce the formation of stromules (Gray et al., 2012) and CPs (Yamane et al., 2012) that contain ATI-PS bodies (Figures 1C and 2), we wished to examine the possible effects of ATI1 and ATI2 deficiency on plant responses to salt stress, first focusing on the germination phase. For this, we germinated seeds of the wild type and ATI-KD on either medium containing 100 mM NaCl or medium with no added salts (control; see Methods). At least 300 seeds of each genotype from each treatment were evaluated for radicle emergence 24 h after plates were transferred to the growth chamber. This revealed 100% germination for both genotypes on control plates and 21 and 19% germination of the wild type and ATI-KD, respectively, on salt-supplemented plates. The similar germination rate of both genotypes during salt stress indicated that ATI1 and ATI2 are probably not involved in the germination of Arabidopsis seeds under salt stress. We next examined the influence of ATI1 and ATI2 deficiency on salt stress tolerance in later developmental stages. As shown in Figure 10A, at 12 d after germination, the phenotype of ATI-KD and wild-type seedlings was comparable when grown on Murashige and Skoog (MS) medium, but suppressed growth was evident for ATI-KD plants compared with the wild type in the presence of 100 mM NaCl. In addition, following our observations that ATI1 accumulates in senescing cells of mature rosette leaves (Figure 5), we exposed 3-week-old wild-type and ATI-KD plants grown in pots to salt stress for three additional weeks (see Methods). As shown in Figure 10B, ATI-KD and wild-type plants that were not exposed to salt stress (control) had comparable phenotypes. By contrast, the ATI-KD plants that were irrigated with water supplemented with 75 mM NaCl for 3 weeks displayed earlier senescence and ultimately, earlier plant death compared with the wild type. To evaluate the specific role of ATI1 in the salt-hypersensitive phenotype, we generated a transgenic line harboring 35S:ATI1-GFP in the background of the ATI-KD genotype (complementation line; see Methods). As shown in Figure 10C, seedlings of the complementation line were comparable to the wild type, indicating that ATI1-GFP is functional and sufficient to rescue the salt-hypersensitive phenotype displayed by ATI-KD. Plants overexpressing ATI1-GFP (OE) in the wild-type background were comparable to wild-type plants and did not display enhanced salt tolerance. In the control plates, all genotypes demonstrated comparable phenotypes.
Figure 10.
ATI1 Is Involved in Salt Stress Tolerance.
(A) ATI-KD and wild-type plants were grown on either half-strength MS-agar plates (left panel, control) or half-strength MS-agar plates supplemented with 100 mM NaCl (right panel, NaCl). Pictures of the seedlings were taken 12 d after germination.
(B) Six-week-old ATI-KD and wild-type plants with (NaCl) or without (Control) salt stress. Stems and inflorescences were removed to better visualize the rosette leaves.
(C) ATI-KD, wild-type, and ATI1-GFP (OE) plants were grown on MS-agar plates supplemented with 100 mM NaCl, along with plants of a transgenic complementation line that stably expresses ATI1-GFP in the background of ATI-KD genotype (Comp).
(D) Confocal imaging of leaf mesophyll and epidermis cells of ATI1-GFP transgenic plants with (NaCl) or without (Control) salt stress.
(E) A closer view of representative epidermis and mesophyll cells of an ATI1-GFP transgenic plant following exposure to salt stress. Arrowheads indicate ATI-ER bodies (left panel) or ATI-PS bodies (right panel).
Next, we wished to examine whether salt stress affects ATI1-GFP expression. For this, we grew ATI1-GFP transgenic seedlings for 5 d on standard plates (no supplements) followed by their removal to either similar plates (control plates having no supplements) or plates supplemented with 100 mM NaCl for five additional days. Confocal imaging of representative plants from both treatments is presented in Figure 10D, showing increased expression of ATI1-GFP following exposure to salt stress compared with plants transferred to control plates, both in leaf epidermis and mesophyll cells. Further examination of mesophyll and epidermis cells following exposure to salt stress revealed the appearance of ATI-PS and ATI-ER bodies, respectively (Figure 10E). Taken together, these results indicate that ATI1 is involved in the alleviation of damage caused by salt stress during plant development, probably through its involvement in plastid-to-vacuole and ER-to-vacuole trafficking.
DISCUSSION
The ATI-PS Body Is a Plastid-Associated Vesicle That Accumulates in Cells following C Starvation and Salt Stress and during Senescence
We previously reported that ATI1, a protein that binds to the autophagy-associated ATG8f protein, is partially localized on the ER membrane and upon exposure to C starvation becomes associated with a compartment that is subsequently transported from the ER to the vacuole (Honig et al., 2012). In this report, we show that upon exposure of Arabidopsis seedlings to C starvation, ATI1 is also localized in a different compartment that is associated with senescing plastids. We termed this compartment the ATI1-PS body, to distinguish it from our previously reported ER-associated compartment (Honig et al., 2012), which we now term the ATI1-ER body. While sharing a common feature of being labeled by ATI1-GFP, the two types of bodies can be distinguished by utilizing the transmission imaging of confocal microscopes wherein ATI1-ER bodies are visualized as intense circular structures (Honig et al., 2012) but ATI-PS bodies are not easily identified (Figure 1; Supplemental Figure 1). Furthermore, our results show that while active autophagy is required for the incorporation of ATI1-GFP to the ATI-ER bodies, active autophagy is not required for the incorporation of ATI1-GFP into the ATI-PS bodies (Figure 1C). This observation implies that each type of body interacts with autophagy differently. The possible association of ATI-PS bodies with autophagy was first observed by their response to dark-induced C starvation (Figure 4; Supplemental Figure 4) as well as to senescence (Figures 5 and 6). Our confocal microscopy results showed that ATI-PS bodies seem attached to the plastid periphery (Figure 1A; Supplemental Figures 1 and 3) and within the plastid stroma (Figure 1C; Supplemental Figure 2 and Supplemental Movie 1). Two additional findings led us to believe that ATI-PS bodies are located within the plastid stroma: (1) ATI-PS bodies were detected in stromules, which are stroma-filled tubules (Figure 1C; Supplemental Figures 2 and 3); and (2) our electron microscopy analysis revealed stroma-localized bodies that are labeled with ATI1-GFP (Figure 2).
Intraplastid vesicles that are derived from the plastid envelope and display similar morphology and size to ATI-PS bodies (50 to 100 nm) were previously reported following low temperature treatments in pea (Pisum sativum), soybean (Glycine max), spinach (Spinacia oleracea), and tobacco (Morré et al., 1991; Westphal et al., 2001). Morphologically similar intraplastid vesicles were shown more recently to contain a chloroplast-localized homolog of the COPII-vesicle associated Sar1 protein, termed CPSAR1 (Garcia et al., 2010). Yamane et al. (2012) also reported that similar vesicles were internalized into the plastids of rice (Oryza sativa) leaves, following exposure to salt stress. This stress, which accelerates senescence, apparently triggered the fusion of these vesicles to form larger stroma-containing bodies within the plastid (Yamane et al., 2012). Interestingly, newly formed ATI-PS bodies were observed following salt stress as well (Figure 10D), and a functional ATI1 is apparently required for salt stress tolerance of both Arabidopsis seedlings and mature plants (Figure 10).
ATI1 Interacts with Several Plastid Proteins to Enable Their Incorporation into ATI-PS Bodies and Their Subsequent Degradation in the Vacuole
As we showed in Figures 6 and 9A, ATI-PS bodies contain a plastid-derived protein marker (CT-GFP). We also showed that these ATI-PS bodies were located on the surface of plastids that were almost depleted of this plastid marker (Figure 6). This suggests that the CT-GFP marker was incorporated into ATI-PS bodies from plastids as a potential cargo destined to be sent elsewhere in the cell. A significant portion of proteins that were identified as potential binding partners with ATI1 are plastid localized (Table 1). Indeed, two of these potential ATI1-interacting proteins (NPQ4 and APE1) were further confirmed to interact with ATI1 in planta (Figure 7A). Notably, one of these potential plastid ATI1-interacting proteins, PrxA, showed significantly increased stability in transgenic plants lacking functional ATI1 (ati1 and ATI-KD; Figure 7B), compared with wild-type plants. By contrast, Rubisco, as a representative plastid-localized protein that does not interact with ATI1, was present at similar levels in plants with or without functional ATI1 (Figure 7B). Taking all these results into account, we suggest that ATI1 is required for incorporating specific plastid proteins destined for degradation into ATI-PS bodies by its ability to directly interact with them.
The ATI1-PS Compartment Buds from the Plastid and Is Transported to the Vacuole by an Autophagic Pathway That Involves ATI1
The involvement of autophagy in the turnover of plastid components was previously described. Chlorophyll degradation during senescence is delayed in the background of the autophagy-deficient atg5 and atg7 mutants (Sakuraba et al., 2014). Additionally, previous studies describe two types of bodies transporting plastid components into the vacuole by an autophagy-dependent process. The transport of small starch granule-like structures occurs during the night (Wang et al., 2013) and the transport of RCBs occurred in response to dark-induced starvation (Ishida et al., 2008). These reports showed that both RCBs and small starch granule-like structures are partially colocalized with the autophagy-associated ATG8 protein. The nature of this colocalization and how autophagy is involved in these processes are unknown. In this report, we provide evidence suggesting that the ATG8 binding protein ATI1, which associates with the plastids and the ATI-PS bodies that bud out from them, plays a regulatory role in the autophagic transport of these bodies to the central vacuole. This process apparently occurs through (1) an interaction of ATI1 located on ATI-PS bodies with ATG8f (Figure 8) and (2) the budding of ATI-PS bodies into the cytosol (Figure 3). Neither the formation of ATI-PS bodies within plastids nor their budding and release into the cytosol apparently requires the core autophagy machinery component ATG5 protein (Figure 1C; Supplemental Figure 6). However, the subsequent phase of transporting these ATI-PS bodies into the vacuole requires the functionality of ATG5 (Figures 9B and 9C). It is reasonable to suggest that the interaction of ATI1 with ATG8 eventually leads to autophagy-dependent transport, probably to enable the degradation of ATI-PS bodies and their cargo inside this lytic compartment (Figure 9A). Interestingly, the interaction of ATG8 with ATI1 colocalized with the CT-DsRed plastid stroma marker (Figures 8G to 8I). This may imply that senescing plastids that generate ATI-PS bodies might be permeable to the entry of ATG8 proteins from the cytosol. Another explanation could lie in a continuous membrane shared by both a plastid-localized ATI-PS body and the plastid envelope, thus enabling the tethering of ATG8 proteins from the cytosol to ATI-PS bodies. A continuous membrane that is shared by both the plastid envelope and RCB-like, salt-induced, and plastid-localized vesicles was recently shown (Yamane et al., 2012). The same report also showed structures with concentric membranes localized within the plastid stroma implying the activity of autophagic components already within the plastid (Yamane et al., 2012). Attachment of ATG8 to plastid-localized ATI-PS bodies could be advantageous for efficient and rapid recognition by the autophagy machinery upon their budding and release from plastids. Nevertheless, significant experimentation is required to address these hypotheses.
The budding of ATI1-PS bodies from the plastid is reminiscent of the budding of vesicles from endomembranes during vesicle trafficking, raising the possibility that similar trafficking machinery components are acting in plastids. Indeed, a recent report employing a computational approach (Khan et al., 2013) identified in Arabidopsis almost all of the plastid proteins that share sequence similarity with proteins associated with COPII vesicles that transport cargo from the ER to the Golgi apparatus. Interestingly, one of the 21 chloroplast proteins identified using this computational approach as a potential cargo protein carried by intraplastid vesicles was NPQ4, which was also recognized as an ATI1 interacting protein in our split ubiquitin Y2H assay (Table 1). NPQ4 was identified in the highest number of individual positive colonies screened (10 isolates) and was further confirmed to interact with ATI1 in vivo in plastids and cytosolic bodies (Figure 7A). This supports our hypothesis that ATI1 interacts with plastid proteins to incorporate them as cargo of the ATI-PS bodies. Taken together, we suggest that ATI1 has a dual role, one is to recruit cargo plastid proteins into ATI-PS bodies and the second is to be recognized by the autophagy machinery in order to direct the ATI-PS bodies for degradation in the vacuole.
ATI1 Is a Stress- and Senescence-Associated Multifunctional Protein Apparently Transporting Unnecessary ER and Plastid Components for Degradation in the Vacuole
This article, coupled with our previous report (Honig et al., 2012), implies that the plant-specific ATG8 binding protein ATI1 is a multifunctional protein that is associated with both ER-to-vacuole (Honig et al., 2012; Li and Vierstra, 2012) and plastid-to-vacuole (this report) trafficking by ATG8-mediated selective autophagy. The ability of a single ATG8-interacting protein to perform multiple functions is not unique to plants. In animals, the autophagy adaptor protein p62 is involved in the autophagy-dependent degradation of both pathogens (Orvedahl et al., 2010; Cemma et al., 2011) and protein aggregates (Pankiv et al., 2007) through xenophagy and aggrephagy, respectively. In addition, the Bnip3 protein induces the turnover of both mitochondria and the ER (Hanna et al., 2012). Compared with organelles known to be targets of selective autophagy, such as peroxisomes and mitochondria, the plastid and the ER are relatively larger compartments that might not be easily engulfed by an autophagosome. Thus, the processes described in our previous (Honig et al., 2012) and present reports provide a cellular method through which cargo present in these organelles is destined for degradation through incorporation into specific ATI1-containing compartments that are subsequently transported by an autophagic pathway to the vacuole.
Recently, several reports have illustrated the importance of autophagy for plants growing under regular (nonstress) conditions, especially during the nighttime (Guiboileau et al., 2012; Izumi et al., 2013; Ono et al., 2013; Wang et al., 2013). Yet, the contribution of autophagy to seedling establishment under stressful environments and the association between autophagy and leaf senescence are not completely understood (Hanaoka et al., 2002; Han et al., 2011). We have previously shown that suppression of both ATI1 and ATI2 delays seed germination on media containing the germination suppression and abiotic stress-related hormone abscisic acid (Honig et al., 2012). In this article, we show that suppression of ATI1 and ATI2 genes renders Arabidopsis plants more sensitive than wild-type plants to salt stress (Figures 10A to 10C). The apparently important role of ATI1 in salt stress tolerance was shown by its ability to complement the salt stress sensitivity of ATI-KD (Figure 10C). Furthermore, the induction of ATI1 expression and of the generation of ATI-PS and ATI-ER bodies following salt stress (Figures 10D and 10E) suggests that ATI1 defines an autophagic system that promotes seedling establishment under abiotic stress conditions.
We hypothesize that the recycling of specific plastid proteins via ATI-PS bodies following salt stress or C starvation stress may contribute to delay the destruction of the entire photosynthetic apparatus, which is the hallmark of leaf senescence in plants.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Colombia was used in this study. Both the transgenic line that stably expresses Pro35S:ATI1-GFP and the ATI-KD genotype that has no expression of ATI1 and reduced expression of ATI2 were previously described (Honig et al., 2012). The other transgenic Arabidopsis plants described in this work were generated utilizing a modification (Davis et al., 2009) of the floral dipping method (Clough and Bent, 1998), where GV3101 Agrobacterium tumefaciens strain harboring the appropriate constructs were used to deliver expression cassettes. The CT-GFP + ATI1-mCherry (see construction details below) line was generated by transforming the 35S:ATI1-mCherry cassette into a stable CT-GFP line (Marques et al., 2004; Schattat, et al., 2011) and subsequent screening for both kanamycin (CT-GFP) and BASTA (ATI1-mCherry) resistance. The mRFP-ATG8f + ATI1-GFP line was generated by introducing mRFP-ATG8f (Honig et al., 2012) into ATI1-GFP homozygous plants followed by screening for resistance to both kanamycin (ATI1-GFP) and BASTA (mRFP-ATG8f). Complementation lines (ATI-KD plants expressing 35S:ATI1-GFP) were generated by introducing ATI1-GFP into ATI-KD lines and screening plants for kanamycin resistance (ATI1-GFP). ATI1pro:ATI1-GFP (see construction details below) expressing plants were screened using kanamycin selection. Seeds were surface sterilized using bleach/HCl mixture and sown on half-strength MS medium including vitamins and 1% sucrose with the proper antibiotic selection. The seeds were further incubated in the dark at 4°C for 48 h and put in a long-day growth chamber (16 h light/8 h dark, 25°C, 40% humidity).
Plasmid Construction
To generate ATI1-mCherry fusion proteins, the coding sequence of ATI1 was amplified using the following primers: 5′-GGTACCCCATGGCTAACAATGAGGAGC-3′ and 5′-GGATCCGACCTCACTGGAGGAGCC-3′. ATI1 was cloned in frame to the coding sequence of mCherry in the vector pART7 between the KpnI and BamHI restriction sites (Gleave, 1992). The Pro35S:ATI1-mCherry cassette was further transferred into the NotI restriction site of the pBART binary vector, which is a derivative of the pART27 vector (Gleave, 1992) that possesses the BAR gene (providing plant resistance against the BASTA herbicide). To generate an expression cassette with ATI1-GFP under the transcriptional regulation of the ATI1 native promoter (ATI1pro:ATI1-GFP), a sequence of 1552 bp upstream of the start codon of ATI1 was amplified from genomic DNA of wild-type Columbia-0 plants using the following primers: ATI1-prom.F, 5′-GAGCTCACCAAAGCATCTCTATCTTCATC-3′, and ATI1-prom.R, 5′-GGTACCCCTCAACCGAAAAAAATCTC-3′. The 35S promoter sequence was cut out from the 35Spro:ATI1-GFP cassette located within pART7 (Honig et al., 2012) and replaced with the ATI1pro sequence between the SacI and KpnI restriction sites followed by its transfer to the pART27 binary vector using SacI and NotI restriction sites. To generate ATI1-YC, APE1-YN, and NPQ4-YN, we utilized the pSAT vector system for BiFC assays (Tzfira et al., 2005). Cloning was done with the In-Fusion kit (Clontech) according to the manual instructions using the following primers: APE1_pSAT-N_IF_fw, 5′-GGACTCAGATCTCGAGCATGGGATCTATAACGGTAGCTCCG-3′, APE_pSAT-N_IF_rv, 5′-GATCCCGGGCCCGCGAGAGGTAGAAGATACAGAGTTGGTGG-3′, NPQ4_pSAT-N_IF_fw, 5′-GGACTCAGATCTCGAGCATGGCTCAAACCATGCTGCTTACT-3′, NPQ4_pSAT-N_IF_rv, 5′-GATCCCGGGCCCGCGGCTTTCTTCACCATCATCGGTGATG-3′, ATI1_pSAT-C_ IF_fw, 5′-GGACTCAGATCTCGAATGGCTAACAATGAGGAGCATCC-3′, and ATI1_pSAT-C_IF_rv, 5′-GATCCCGGGCCCGCGGTCTAGACCTCACTGGAGGAGCC-3′. APE1, ATI1, and NPQ4 were cloned into pSAT1-nEYFP-N1, pSAT1-cEYFP-N1, and pSAT6-nEYFP-N1, respectively. All generated pSAT expression cassettes were transferred to the pPZP binary vector using either AscI (for pSAT1) or PI-PspI (for pSAT6) restriction enzymes (Tzfira et al., 2005).
Carbon Starvation and Salt Stress Treatments
For C starvation of seedlings, wild-type and transgenic seeds were sown on plates containing 0.5 MS medium lacking sucrose and grown for 6 d in the growth chamber. Twenty-four hours before microscopy analysis, the plates were covered by double layer aluminum foil and left in the same growth chamber. For a longer period of C starvation, 4-d-old seedlings were covered by double layer aluminum foil and left in the same growth chamber for an additional 72 h. All plants were microscopically analyzed 7 d after germination. For visualizing rosette leaves, individually darkened leaves of 4-week-old plants were gently wrapped with aluminum foil as previously described (Wada et al., 2009) for 20 h (including the 8-h dark cycle of the long-day growth chamber). Germination under salt stress was tested by sowing the different genotypes on half-strength MS-agar plates supplemented with 100 mM NaCl or with no supplements (control) and incubated at 4°C in the dark for 48 h. Twenty-four hours following their removal to the growth chamber (exposure to light), the seeds were evaluated for radicle emergence. Seedling establishment under salt stress was examined in the same manner, except that the seedlings were allowed to grow for 12 d after germination before their pictures were taken. Exposure of mature plants to salt stress was done following their growth in pots for 3 weeks while being irrigated with regular tap water. At the beginning of the fourth week, the control trays continued to be irrigated with tap water and the salt-stress trays were irrigated with tap water supplemented with 75 mM NaCl for three additional weeks. In each tray, the pots were marked according to the genotype they harbored and mixed within the tray randomly. The stem and inflorescence were removed before pictures were taken to better visualize the senescing rosette leaves. For confocal imaging of plants exposed to salt stress, ATI1-GFP seedlings were grown for 5 d on half-strength MS medium containing 1% (w/v) sucrose. On the sixth day after germination, the seedlings were gently transferred to either similar plates (harboring half-strength MS media + 1% sucrose) or similar plates supplemented with 100 mM NaCl. Under both treatments, plants were allowed to grow for an additional 5 d before they were utilized for confocal imaging.
ConA Treatment
ConA (Fluka #27689; 100 µM stock in DMSO) was used according to a previously described protocol (Ishida et al., 2008) with some modifications. Arabidopsis seedlings were grown in MS agar Petri plates without sucrose for 6 d. Then, three to five whole seedlings of each of the examined lines were transferred into wells of 24-well plates containing 1 µM ConA diluted in liquid MS medium. As a control, the same number of seedlings was subjected to the same treatment using DMSO (used to dissolve ConA). The seedlings were incubated in the liquid media for 20 h at 23°C in the dark with gentle shaking (80 rpm). Afterwards, the seedlings were directly examined by confocal microscopy. Rosette leaves of mature plants were excised and placed in the ConA-containing liquid MS media under the same conditions described for seedlings.
Quantification of ATI-PS Bodies and Statistical Analysis
For the quantification of ATI-PS bodies, plants were carbon starved for 24 or 72 h or grown under favorable conditions (1% sucrose and continuous exposure to light). Images of cotyledon mesophyll cells from different plants of each growth condition (starved and nonstarved) were acquired using confocal microscopy. Between 24 and 50 sample cells were examined from each treatment. Only ATI1-GFP-labeled bodies that were found associated with plastids (either colocalized with the chlorophyll autofluorescence or attached to it) were considered as ATI-PS bodies. The average number of ATI-PS bodies per cell was calculated for each condition, and statistical significance was determined using the Student’s t test (P < 0.01).
Protein Interaction Analysis Using the Split-Ubiquitin Yeast Two-Hybrid and BiFC Methods
The yeast two hybrid analysis was performed using the DUALhunter kit (Dualsystems Biotech), which employs split ubiquitin for detecting protein-protein interactions between transmembrane spanning proteins. The ATI1 (At2g45980) coding DNA sequence was cloned in frame between OST4 yeast ER membrane anchor and Cub coding sequence in the yeast bait vector pDHB1 (Dualsystems Biotech). This cloning was done using the primers forward, 5′-TAACAAGGCCATTACGGCCATGGCTAACAATGAGGAGCAT-3′, and reverse, 5′-CTGATTGGCCGAGGCGGCCGCGACCTCACTGGAGGA-3′. This plasmid was cointroduced with the commercial pNubG-X Arabidopsis cDNA library (P02210 Dualsystems Biotech) to the NMY51 yeast strain. This library was constructed from a mixture of 6-d-old seedlings of dark-grown (etiolated seedlings) and seedlings exposed to blue and far-red light. The library consists of 1.7 × 107 independent clones with an average insert size of 1.7 kb (ranges from 1.2 to 2.5 kb). The NMY51 yeast strain contains in its genome ADE2, HIS3, and lacZ reporter genes. The reporter genes are expressed only when in vivo two-hybrid interaction combines the Cub and Nub parts of the yeast ubiquitin protein, thus allowing the release of the LexA-VP16 transcription factor, which in turn moves to the nucleus and activates the three reporters. pDHB1 clones capable of growing on media lacking histidine, tryptophan, leucine, and adenine that were also stained blue as a result of lacZ activation were rescued and their cDNA inserts were separately sequenced by both the forward library sequencing primer, 5′-GTCGAAAATTCAAGACAAGG-3′, and the reverse library sequencing primer, 5′-GTTACTCAAGAACAAGAATTTTCG-3′. LacZ, ADE2, and HIS3 activation was rechecked once more by one-on-one yeast two-hybrid assays for all the positive clones to eliminate false positive interactions. For analyzing in planta interactions between ATG8f and ATI1, NPQ4 and ATI1, or APE1 and ATI1, different Agrobacterium strains harboring each of the following plasmids separately: pCAMBIA-YN-ATG8f (Honig et al., 2012), pCAMBIA-YC-ATI1 (Honig et al., 2012), pPZP-NPQ4-YN, pPZP-ATI1-YC, and pPZP-APE1-YN, were used to transiently cotransform the appropriate plasmid pairs in Nicotiana benthamiana leaves following their infiltration, as previously described (Sparkes et al., 2006).
Protein Extraction and Immunoblotting
For immunoblot analysis, leaves from 4-week-old plants were collected and ground to fine powder in liquid N2. Proteins were extracted in a buffer containing 100 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1% SDS, and protease inhibitor. The protein contents of the aqueous extracts were quantified spectrophotometrically using the BCA protein assay reagent (Pierce) according to the supplier’s manual. The protein extract was separated on reducing SDS-PAGE and analyzed by immunoblot using 2-Cys PrxA (BAS1) antibodies (a kind gift from A. Danon, The Weizmann Institute of Science, Israel; Dangoor et al., 2012). The same blots were used to detect the level of Rubisco with anti-RBCL antibodies (Agrisera).
Confocal Microscopy
The Olympus Fluoview 1000 IX81 and the Nikon A1 confocal microscope systems were used in this study. Generally, samples were put between two microscope glass cover slips (24 × 40 mm, No.1 thickness) in an aqueous environment. Images were taken from a single focal plane unless otherwise indicated. For image acquisition, the UPlanSApo ×60 oil objective (numerical aperture of 1.35) was generally used when using the Olympus microscope and the ×20 objective (numerical aperture of 0.75) of the Nikon microscope was used mainly to visualize leaf mesophyll cells. GFP fluorescence images were taken using 488-nm laser excitation and the emission was collected via the 525-nm filter. The mCherry images were taken using 561-nm laser excitation and emission was collected through the 595-nm filter. YFP images were taken with the same settings as for GFP, and chlorophyll autofluorescence was imaged using the 640-nm laser and collected with the 700-nm filter. Transmittance images were taken with the Olympus microscope using the differential interference contrast method. Time-lapse images were all composed from images taken using line sequence. Acquired images were analyzed using either the Olympus Fluoview 1000 viewer for images taken with the Olympus microscope or the NIS-Elements AR imaging software for images taken with the Nikon confocal microscope.
Electron Microscopy
Sample preparation for electron microscopy and immunolocalization of GFP were performed exactly as described by Honig et al. (2012).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following identifiers: At2g45980 (ATI1) and At4g16520 (At-ATG8f). The rest of the identifiers for genes mentioned in this article are listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Imaging of Both ATI-ER Bodies and ATI-PS Bodies.
Supplemental Figure 2. Confocal Imaging of ATI-PS Bodies Located within Plastids.
Supplemental Figure 3. A Comparison between Stromules Visualized by the CT-GFP and ATI1-GFP Fusion Proteins.
Supplemental Figure 4. Increasing ATI1-GFP Expression with the Progression of C Starvation Stress in Seedling Hypocotyl Cells.
Supplemental Figure 5. ATI1-GFP Expression in Cotyledon Epidermis Cells of a 7-d-Old Seedling Grown under Favorable Growth Conditions.
Supplemental Figure 6. Vital and Senescing Mesophyll Cells of a Mature Stem Leaf.
Supplemental Figure 7. ATI1-GFP Expression and Localization Driven by the ATI1 Native Promoter.
Supplemental Figure 8. C Starvation Induces the Expression of ATI1-GFP in Plastids When Driven by the ATI1 Native Promoter.
Supplemental Figure 9. ATI1-GFP Labels ATI-ER Bodies When Driven by the ATI1 Native Promoter following 24-h C Starvation Stress.
Supplemental Movie 1. The Apparently Dynamic Movement of ATI-PS Bodies in a Plastid.
Supplemental Movie 2. The Budding and Detachment of an ATI-PS Body from a Plastid into the Cytosol.
Supplemental Movie 3. The Budding and Detachment of an ATI-PS Body from a Plastid into the Cytosol in an Autophagy-Deficient atg5 Mutant.
Supplemental Movie 4. Random Movement of CT-GFP-Containing ATI-PS Bodies inside the Vacuole Lumen of a Senescing Rosette Leaf.
Supplemental Movie 5. ATI1-GFP-Labeled Bodies Randomly Moving inside the Vacuole Lumen of a Hypocotyl Epidermis Cell from a ConA-Treated ATI1-GFP Transgenic Plant.
Supplemental Movie 6. A Vacuole Lumen Depleted of any ATI1-GFP-Labeled Bodies in a Hypocotyl Epidermis Cell from a ConA-Treated ATI1-GFP/atg5 Transgenic Plant.
Supplementary Material
Acknowledgments
We thank Tamar Avin-Wittenberg for critical reading of the article and helpful comments, Vladimir Kiss for his invaluable help with confocal microscope image acquisition, Michal Habusha and Matan Levi for their assistance in some aspects of this research, Eyal Shimoni and Smadar Zaidman (Electron Microscopy Unit of the Weizmann Institute, Israel) for their invaluable help with transmission electron microscopy image acquisition, Hiroyuki Ishida (Tohoku University, Sendai, Japan) for kindly providing the CT-DsRed construct, Jaideep Mathur (Guelph University, Canada) for kindly providing the CT-GFP (FNR-GFP) construct and transgenic seeds, and Avihai Danon (Weizmann Institute of Science, Israel) for kindly providing the anti-PrxA antibodies. We also thank Niko Geldner for fruitful input following a discussion about this research. Our research was supported by grants from the Israel Science Foundation (Grant 764/07), the J & R Center for Scientific Research at the Weizmann Institute of Science and the Israeli Ministry of Agriculture. G.G. is an incumbent of the Bronfman Chair of Plant Science at the Weizmann Institute of Science.
AUTHOR CONTRIBUTIONS
S.M. and G.G. designed the research, analyzed the data, and wrote the article. S.M. performed the majority of the research. A.H., H.L., and H.P.-Z. each performed different aspects of the research.
Glossary
- ER
endoplasmic reticulum
- CP
chloroplast protrusion
- ConA
Concanamycin A
- BiFC
bimolecular fluorescence complementation
- RCB
Rubisco-containing body
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Avin-Wittenberg T., Honig A., Galili G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249: 285–299. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Sheen J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., Johansen T. (2013). The LIR motif - crucial for selective autophagy. J. Cell Sci. 126: 3237–3247. [DOI] [PubMed] [Google Scholar]
- Cemma M., Kim P.K., Brumell J.H. (2011). The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A., Ishida H., Nishizawa N.K., Makino A., Mae T. (2003). Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol. 44: 914–921. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dangoor I., Peled-Zehavi H., Wittenberg G., Danon A. (2012). A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24: 1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. (2009). Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd B.E., Morriss S.C., Macintosh G.C., Bassham D.C. (2012). What to eat: evidence for selective autophagy in plants. J. Integr. Plant Biol. 54: 907–920. [DOI] [PubMed] [Google Scholar]
- Garcia C., Khan N.Z., Nannmark U., Aronsson H. (2010). The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 63: 73–85. [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gray J.C., Hansen M.R., Shaw D.J., Graham K., Dale R., Smallman P., Natesan S.K.A., Newell C.A. (2012). Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 69: 387–398. [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Sormani R., Meyer C., Masclaux-Daubresse C. (2010). Senescence and death of plant organs: nutrient recycling and developmental regulation. C. R. Biol. 333: 382–391. [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Yoshimoto K., Soulay F., Bataillé M.P., Avice J.C., Masclaux-Daubresse C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194: 732–740. [DOI] [PubMed] [Google Scholar]
- Han S., Yu B., Wang Y., Liu Y. (2011). Role of plant autophagy in stress response. Protein Cell 2: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R.A., Quinsay M.N., Orogo A.M., Giang K., Rikka S., Gustafsson A.B. (2012). Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287: 19094–19104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576. [DOI] [PubMed] [Google Scholar]
- Holzinger A., Buchner O., Lütz C., Hanson M.R. (2007). Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma 230: 23–30. [DOI] [PubMed] [Google Scholar]
- Honig A., Avin-Wittenberg T., Ufaz S., Galili G. (2012). A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24: 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Hidema J., Makino A., Ishida H. (2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 161: 1682–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N.Z., Lindquist E., Aronsson H. (2013). New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: an Arabidopsis model. PLoS ONE 8: e59898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.U., Huang A.H.C. (2004). Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol. 134: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Lee, H., Lee, H.N., Kim, S.-H., Shin, K.D., and Chung, T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042. [DOI] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963. [DOI] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques J.P., Schattat M.H., Hause G., Dudeck I., Klösgen R.B. (2004). In vivo transport of folded EGFP by the DeltapH/TAT-dependent pathway in chloroplasts of Arabidopsis thaliana. J. Exp. Bot. 55: 1697–1706. [DOI] [PubMed] [Google Scholar]
- Michaeli S., Galili G. (2014). Degradation of organelles or specific organelle components via selective autophagy in plant cells. Int. J. Mol. Sci. 15: 7624–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré D.J., Selldén G., Sundqvist C., Sandelius A.S. (1991). Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol. 97: 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385. [DOI] [PubMed] [Google Scholar]
- Ono Y., Wada S., Izumi M., Makino A., Ishida H. (2013). Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36: 1147–1159. [DOI] [PubMed] [Google Scholar]
- Orvedahl A., MacPherson S., Sumpter R. Jr., Tallóczy Z., Zou Z., Levine B. (2010). Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Lee S.H., Kim Y.S., Park O.K., Hörtensteiner S., Paek N.C. (2014). Delayed degradation of chlorophylls and photosynthetic proteins in Arabidopsis autophagy mutants during stress-induced leaf yellowing. J. Exp. Bot. 65: 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M., Barton K., Baudisch B., Klösgen R.B., Mathur J. (2011). Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 155: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M.H., Griffiths S., Mathur N., Barton K., Wozny M.R., Dunn N., Greenwood J.S., Mathur J. (2012). Differential coloring reveals that plastids do not form networks for exchanging macromolecules. Plant Cell 24: 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A., Peter M. (2014). Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843: 163–181. [DOI] [PubMed] [Google Scholar]
- Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., Hayashi, M., Sakamoto, W., Ohsumi, Y., and Nishimura, M. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis Plant Cell 25: 4967–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59: 4029–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516. [DOI] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S., Soll J., Vothknecht U.C. (2001). A vesicle transport system inside chloroplasts. FEBS Lett. 506: 257–261. [DOI] [PubMed] [Google Scholar]
- Yamane K., Mitsuya S., Taniguchi M., Miyake H. (2012). Salt-induced chloroplast protrusion is the process of exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts into cytoplasm in leaves of rice. Plant Cell Environ. 35: 1663–1671. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53: 1355–1365. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Shibata M., Kondo M., Oikawa K., Sato M., Toyooka K., Shirasu K., Nishimura M., Ohsumi Y. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 27: 1161–1168. [DOI] [PubMed] [Google Scholar]
- Yu B., Wakao S., Fan J., Benning C. (2004). Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 45: 503–510. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.