This work presents a technique to identify specific regions of maize chromatin that are hypersensitive to digestion by micrococcal nuclease, which preferentially cleaves DNA not bound into nucleosomes. This chromatin profiling technique helps elucidate how global changes in gene expression relate to changes in nucleosome position and should be broadly applicable to other species.
Abstract
The eukaryotic genome is organized into nucleosomes, the fundamental units of chromatin. The positions of nucleosomes on DNA regulate protein-DNA interactions and in turn influence DNA-templated events. Despite the increasing number of genome-wide maps of nucleosome position, how global changes in gene expression relate to changes in nucleosome position is poorly understood. We show that in nucleosome occupancy mapping experiments in maize (Zea mays), particular genomic regions are highly susceptible to variation introduced by differences in the extent to which chromatin is digested with micrococcal nuclease (MNase). We exploited this digestion-linked variation to identify protein footprints that are hypersensitive to MNase digestion, an approach we term differential nuclease sensitivity profiling (DNS-chip). Hypersensitive footprints were enriched at the 5′ and 3′ ends of genes, associated with gene expression levels, and significantly overlapped with conserved noncoding sequences and the binding sites of the transcription factor KNOTTED1. We also found that the tissue-specific regulation of gene expression was linked to tissue-specific hypersensitive footprints. These results reveal biochemical features of nucleosome organization that correlate with gene expression levels and colocalize with functional DNA elements. This approach to chromatin profiling should be broadly applicable to other species and should shed light on the relationships among chromatin organization, protein-DNA interactions, and genome regulation.
INTRODUCTION
The eukaryotic nuclear genome is organized into nucleosomes, 147-bp segments of DNA that are wrapped 1.65 times around histone octamers (Luger et al., 1997). Nucleosomes regulate the access of proteins to DNA and thus can regulate and respond to various nuclear processes through their position, modifications, and remodeling to influence DNA-templated events (Venter et al., 1994; Stünkel et al., 1997; Lieb and Clarke, 2005; Lomvardas and Thanos, 2002; Lam et al., 2008). The positions of nucleosomes can be determined with micrococcal nuclease (MNase), an enzyme that preferentially cleaves internucleosomal DNA (Noll, 1974). Consequently, MNase has been used to generate genome-wide nuclease protection footprint maps using DNA microarrays or next-generation sequencing, in order to characterize the chromatin landscapes in a variety of model organisms (Yuan et al., 2005; Johnson et al., 2006; Dennis et al., 2007; Mavrich et al., 2008, Schones et al., 2008; Chodavarapu et al., 2010; Kent et al., 2011; Labonne et al., 2013).
Despite advances in chromatin profiling of genomes in animals and fungi, information on the chromatin landscapes in plants is relatively limited. This is particularly true for maize (Zea mays), an important crop species with a large and complex genome (Schnable et al., 2009). Nucleosome positioning in Arabidopsis thaliana has established that plants exhibit many aspects of chromatin structure that are considered canonical for eukaryotes, including the rotational phasing of nucleosomes with regard to AT-containing dinucleotides, and high nucleosome occupancy in exons, heterochromatic regions, and methylated DNA (Chodavarapu et al., 2010). In maize, nucleosomes containing the histone variant CENH3 appear to be rotationally phased with respect to AT-containing dinucleotides at the CentC (but not CRM1 and CRM2) repeats (Gent et al., 2011). In maize mutants of mediator of paramutation1, which are impaired in the RNA-directed DNA methylation pathway, nucleosome positioning differences compared with wild-type plants have been identified at several loci, although these differences were not associated with differences in gene expression (Labonne et al., 2013). While there has been a general expectation that large-scale changes in gene expression are accompanied by large-scale changes in nucleosome repositioning, this relationship has only recently begun to be characterized (Cairns, 2009; Druliner et al., 2013; Sexton et al., 2014).
A modification of nucleosome-mapping assays, the digestion of chromatin to different degrees with MNase, has recently been shown to result in variability of nucleosome occupancy profiles in yeast (Weiner et al., 2010; Henikoff et al., 2011; Xi et al., 2011). Specifically, numerous specific regions of the yeast genome contain footprints that show significantly higher signal under light-digest conditions than under heavy-digest conditions, complicating interpretation of genome-wide nucleosome occupancy mapping studies. These differences have been attributed to an altered chromatin conformation that renders these nucleosomes hypersensitive to MNase cleavage (Henikoff et al., 2011). These so-called “fragile nucleosomes” partially overlap with regions enriched in histone H2A.Z-containing nucleosomes (Xi et al., 2011) and are associated with highly transcribed genes in yeast (Weiner et al., 2010). Fragile nucleosomes have been identified at individual loci in other organisms (Lombraña et al., 2013). While thousands of fragile nucleosomes have been identified in yeast, the widespread occurrence and significance of such fragile nucleosomes remain largely uncharacterized in multicellular organisms, particularly plants.
Here, we show that technical variability in preparing nucleosomal DNA, particularly with respect to the degree of digestion with MNase, leads to variability in nucleosome occupancy signals at specific regions in the maize genome. Exploiting the degree of digestion as an experimental variable in nucleosome occupancy mapping, an approach we term differential nuclease sensitivity profiling (DNS-chip), we identified the widespread presence of hypersensitive and hyperresistant nucleosomes and characterized them in relation to functional DNA elements, gene structure, and tissue-specific mRNA levels. We demonstrate and discuss the utility of DNS-chip to identify biochemically distinct and biologically functional regions within the epigenomic background of nucleosome occupancy information.
RESULTS
Buffer Effects on Nuclear Morphology Preservation Are Not Reflected in Nucleosome Occupancy Profiles
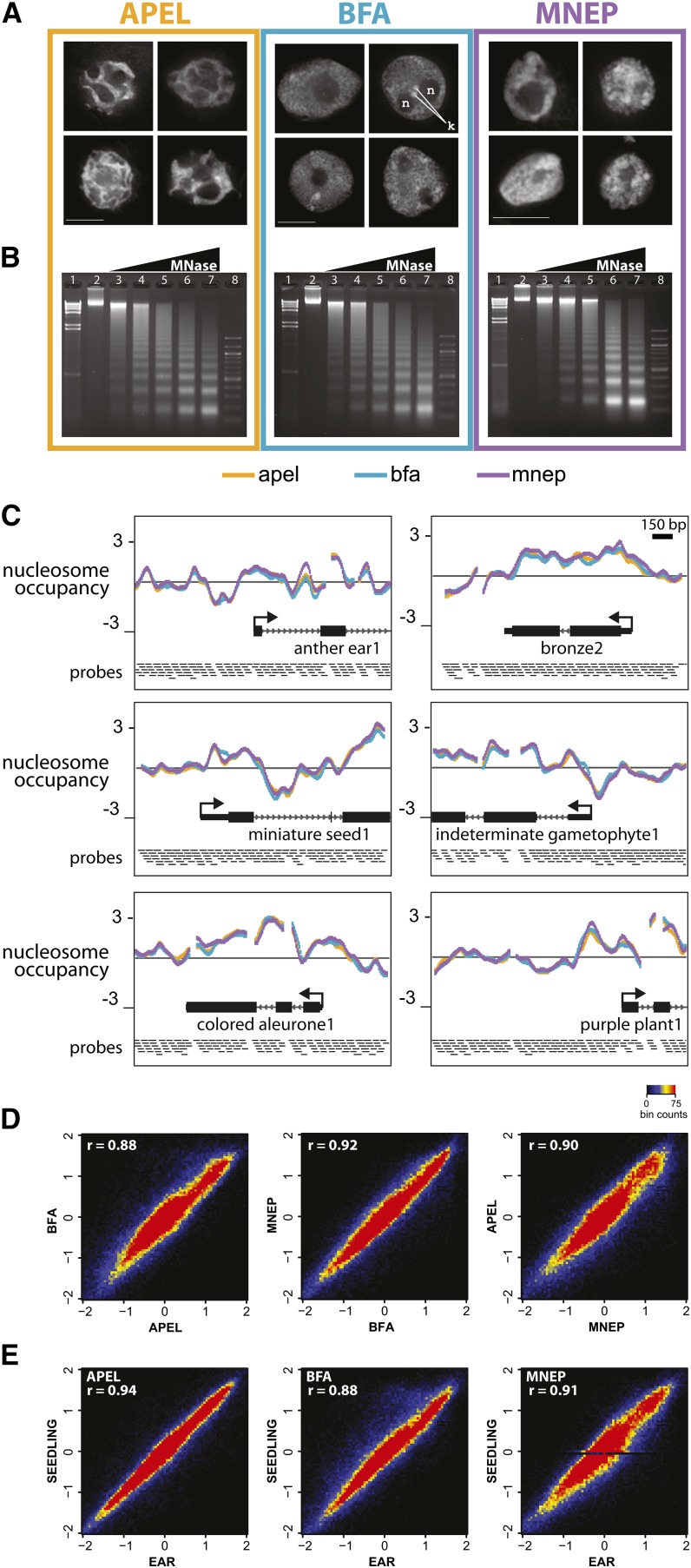
To develop a nucleosome occupancy mapping protocol in maize, we first examined the effects of different nucleus isolation buffers on the preservation of chromatin structure at several levels. Formaldehyde was used to cross-link chromatin in its native state in the presence of one of three different nucleus isolation buffers: a plant nucleus isolation buffer that was developed for examining plant chromatin and transcription (APEL; Steinmüller and Apel, 1986), a buffer that was developed to preserve chromatin structure for cytological and ultrastructural studies (BFA; Belmont et al., 1987; Howe et al., 2012), and a nucleus isolation buffer that was developed for nucleosome occupancy mapping assays in human cell lines (MNEP; Dennis et al., 2007). Frozen, ground tissue from developing maize ears (3 to 5 cm) was suspended and cross-linked in one of the three different formaldehyde-containing buffers and nuclei were subsequently isolated.
To compare the preservation of chromatin structure among the three different buffers at the cytological level, nuclei were stained with 4′,6-diamidino-2-phenylindole and imaged by three-dimensional deconvolution microscopy, as shown in Figure 1. We examined nuclei for the presence of features associated with well-preserved nuclei, including a spherical shape, distinguishable chromatin fibers within the nucleoplasm, one or two conspicuous nucleoli (n, Figure 1A) per nucleus, and distinct heterochromatic knobs (k, Figure 1A), several of which are present in the genome of the maize inbred line used for this study, B73. BFA best preserved these cytological features, followed by MNEP and then APEL (Figure 1A). Despite the conspicuous cytological differences among buffers, the canonical nucleosomal ladders following partial digest with MNase were remarkably similar, as assayed by agarose gel electrophoresis (Figure 1B).
Figure 1.
Different Fixation Buffers Preserve Nuclear Morphologies to Different Degrees, but Yield Comparable Nucleosome Occupancy Profiles.
Developing ear tissue was formaldehyde-fixed and isolated in three different buffers (APEL, BFA, and MNEP), and nuclei were analyzed by three-dimensional deconvolution microscopy (A), MNase digestion products (B), and microarray-based nucleosome occupancy profiles ([C] to [E]).
(A) Single-optical middle sections of deconvolved three-dimensional data sets showing the differences in preservation of nuclear shape and chromatin appearance of immature ear nuclei among the three buffers examined. ; n, nucleolus; k, knobs. Bar = 10 μm.
(B) Developing ear nuclei were treated with titrated amounts of MNase and DNA was subjected to 1% agarose gel electrophoresis.
(C) Nucleosome occupancy plots at six example genes. Nucleosome occupancy is measured as the log2 ratio of nucleosomal/genomic fluorescence signal on a high-density microarray that includes 400 transcription start sites.
(D) and (E) Probe-by-probe scatterplots of nucleosome occupancy scores for each pairwise buffer comparison (D) and comparison of seedling against ear for each buffer (E).
To examine the buffers’ effects on microarray-based nucleosome occupancy profiles, nuclei extracted in the three different buffers were digested with MNase to near completion, in which the majority of DNA was present in the mononucleosomal (100 to 200 bp) fraction. The 100- to 200-bp DNA fragments were gel-extracted, fluorescently labeled with Cy3, and cohybridized with Cy5-labeled genomic DNA to a custom high-density NimbleGen microarray that was designed to cover the transcription start site (TSS) regions of 400 maize genes primarily derived from the list of Classical Maize Genes (Schnable and Freeling, 2011). Nucleosome occupancy profiles were generated by plotting the log2 ratio (nucleosomal/genomic) of hybridization signals from the microarray (Figure 1C). Nucleosome occupancy scores were very similar for representative genes regardless of which buffer was used. This similarity extended across the majority of the data set in pairwise buffer comparisons (r between 0.88 and 0.92; Figure 1D). Nucleosome occupancy profiles were also generated from 9-d-old seedling shoots and were also highly correlated to immature ear nucleosome occupancy profiles (Figure 1E). The uniformity of nucleosome occupancy profiles from probe-by-probe global correlation analysis revealed remarkably uniform nucleosome occupancy signals across the 3-kb windows surveyed.
Different Degrees of Digestion with MNase Reveal Site-Specific Variability in Nucleosome Occupancy Profiles
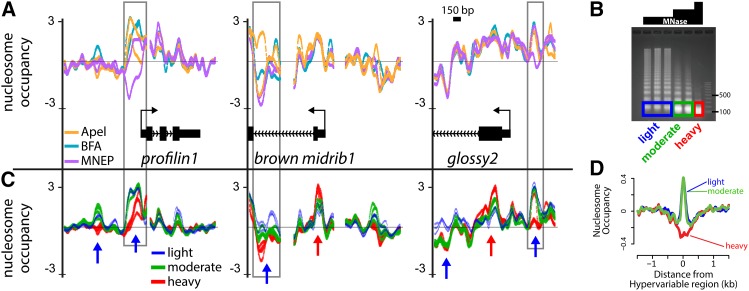
Despite the overall uniformity of nucleosome occupancy profiles from nuclei isolated with different buffers, we found that some genomic regions displayed prominent, localized signal variation (Figure 2A). This localized hypervariability (boxed in gray, Figure 2A) was not associated with a particular buffer and was not consistent between replicates. However, recent studies in yeast have shown that the degree of nuclease digestion used to isolate nucleosomal DNA can have an influence on nucleosome occupancy profiles in particular genomic regions (Weiner et al., 2010; Henikoff et al., 2011; Xi et al., 2011). In order to determine whether the hypervariable regions we observed might have resulted in part from variation in the extent of MNase digestion, we mapped nucleosome occupancy in earshoots using three different concentrations (hereafter referred to as light, moderate, and heavy; Figure 2B) of MNase as the controlled variable. Plotting of nucleosome occupancy profiles as a function of digestion levels (Figure 2C) exhibited many of the same localized regions of signal variability, but this variability typically was associated with the degree of digestion and was consistently observed across the four replicate digestions.
Figure 2.
The Degree of MNase Digestion Is a Significant Source of Variability of Nucleosome Occupancy Profiles.
(A) Nucleosome occupancy profiles for three representative loci. Profiles are color coded according to buffer, for which two replicates exist for each. Regions of notable signal variability are boxed in gray. Bar = 150 bp.
(B) Nucleosomal DNA was isolated from ear nuclei after digestion with three different concentrations of MNase, corresponding to light, moderate, and heavy digestions.
(C) Nucleosome occupancy profiles of differentially digested nucleosomal DNA at the same genes as in (A), which include four profiles for each digestion level corresponding to replicate digestions. Regions displaying higher signal under light-digest conditions relative to heavy digestion conditions are marked with blue arrows; regions with the reciprocal pattern are marked with red arrows.
(D) Hypervariable regions, defined as regions with an sd > 1, were aligned by their midpoint and used to calculate average nucleosome occupancy profiles (n = 174).
A commonly observed pattern of variation was one in which a nucleosome occupancy signal from the lightly and moderately digested samples was greater than that of the heavily digested samples (blue arrows, Figure 2C). These regions, which were relatively more enriched in mononucleosomal DNA under the light-digest conditions than under the heavy-digest conditions, are interpreted as biochemically distinct footprints that are highly susceptible to MNase cleavage (Weiner et al., 2010; Henikoff et al., 2011; Xi et al., 2011) and will hereafter be called MNase-sensitive footprints (MSFs). We also observed the inverse pattern, in which nucleosome occupancy signals were greater under heavy-digest conditions than light-digestion conditions (red arrows, Figure 2C), indicative of footprints relatively resistant to being digested to nucleosomal monomers and therefore not detectable in the 100- to 200-bp fraction except under heavy-digest conditions. These regions exhibit persistent footprints even after extensive nuclease digestion, similar to those described in yeast (Xi et al., 2011), and are hereafter called MNase-resistant footprint (MRFs). The majority of regions that appeared hypervariable in the buffer comparison experiment frequently overlapped specifically with MSFs. This relationship was also observed when we averaged light, moderate, and heavy-digested nucleosome occupancy profiles at regions we identified as hypervariable in the buffer comparison experiment (sd > 1, n = 174; Figure 2D). These features were not a result of normalization artifacts because they are detectable in raw data and log-transformed data prior to normalization (Supplemental Figure 1). Thus, nucleosome occupancy mapping is susceptible to technical variability in the form of digestion-linked variation. The approach of mapping the nucleosome landscape with different degrees of digestion, or DNS-chip, was used to further explore the chromatin landscape in maize.
MSFs and MRFs Are Nonrandomly Distributed within and Around Genes
To expand the DNS-chip profiling beyond the 400 TSS regions to a larger and less biased sample of the maize genome, we designed a high-density microarray spanning three large blocks of the maize genome, each ∼20 Mb. This “big-block” microarray includes 1688 of the 39,570 high-confidence genes (the maize “filtered gene set”) in addition to single-copy intergenic regions. We performed DNS-chip with immature ears using the big-block microarray as shown in Figure 3. To quantify hypersensitivity and hyperresistance from DNS-chip, we subtracted the heavy-digest profile from the light-digest profile to create a “difference” profile, where positive values in the indicate MNase sensitivity and negative values indicate MNase resistance (Figure 3A).
Figure 3.
MNase-Hypersensitive and Hyperresistant Footprints Are Nonrandomly Distributed in and Around Genes.
(A) Four example genes with nucleosome occupancy profiles (“nucleosome occupancy”), difference profiles (light-heavy “difference”), and the segmentation of the difference profiles (“segmentation”). Bars = 200 bp.
(B) to (D) Genes were scaled so that each was 2 kb in size and subsequently aligned by their boundaries (“metagene”). Scaled genes were used to calculate average of nucleosome occupancy scores (B), the difference profile (light-heavy; [C]), and predicted nucleosome occupancy (NOL; [D]).
We computationally defined MSF and MRF segments at several statistical stringencies by applying a segmentation algorithm (iSeg; see Methods) to the difference profile data (example in segmentation track, Figure 3A, tabulated in Table 1). We found that MSFs and MRFs were not limited to TSS-proximal regions of known genes but were also observed within and between genes (Figure 3A). In particular, MSFs were frequently observed at the 5′ and 3′ boundaries of genes, whereas MRFs were frequently observed within the coding regions of genes (Figure 3A). Using the iSeg biological cutoff of 1.5 (see Methods and Table 1), we defined 3290 MSFs and 4908 MRFs within the 11 Mb of unique genomic space surveyed by the big-block microarray. A total of 58% of MSFs and 43% of MRFs were located in or near (within 500 bp) known protein-coding genes. Over half (58%) of all genes contained one or more MSFs. Similarly, most (67%) of all genes contained one or more MRFs. The MSFs had similar median segment size, 167 to 168 bp, in overall (n = 3290) and within-gene (n = 1602) regions. The MRFs had a median segment size of 115 bp in the overall region (n = 4908) region and 151 bp for those (n = 1815) found within genes. These findings show that MSFs and MRFs are widespread and abundant, occurring at most genes and representing previously unrecognized but major parts of the maize epigenomic landscape.
Table 1. Summary Features of MSF and MRF Segments Defined at Different Statistical Stringencies by iSeg.
| Segmenta | sd Cutoffb | Countsc | Total Array (%)d | Ave. No. of Segments per 10 kb | Segments Neare Genes (%)f | Segments within Genes (%) | Genes Neare Segments (%) | Median Segment Size (bp) | Median Size of Segments Neare Genes (bp) | Median Size of Segmentse within Genes (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| MSF | 1.0 | 4267 | 6.4 | 2.96 | 59 | 45 | 64 | 172 | 172 | 172 |
| 1.5 | 3290 | 4.4 | 2.28 | 58 | 47 | 58 | 167 | 168 | 168 | |
| 2.0 | 2597 | 3.2 | 1.80 | 57 | 44 | 52 | 161 | 160 | 160 | |
| 3.0 | 1753 | 2.0 | 1.22 | 55 | 42 | 40 | 152 | 151 | 152 | |
| MRF | 1.0 | 6783 | 9.3 | 4.71 | 41 | 36 | 72 | 117 | 127 | 151 |
| 1.5 | 4908 | 6.3 | 3.41 | 43 | 32 | 67 | 115 | 131 | 151 | |
| 2.0 | 3582 | 4.4 | 2.49 | 47 | 41 | 59 | 118 | 130 | 142 | |
| 3.0 | 2104 | 2.1 | 1.46 | 48 | 44 | 41 | 112 | 115 | 116 |
sd cutoff: a parameter in the iSeg segmentation algorithm related to the statistical stringency of segmentation.
Counts: the total number of segments within the 11 Mb of unique genomic space surveyed by the big-block microarray.
Proportion: the sum of all segments divided by total array size (bp).
Near: within 500 bp.
Genes: The 1688 genes of the 39,570 canonical gene models (Maize Filtered Gene Set version 5b) represented on the microarray.
Aggregate DNS-chip profiles at the 1688 genes aligned by their boundaries revealed the presence of a prominent signal directly upstream of TSSs under light-digest conditions, at a location commonly referred to as the “nucleosome-depleted region” (arrow, Figure 3B). This signal is notably absent from both the moderate- and heavy-digest profiles. This finding is particularly important because the moderate- and heavy-digest conditions used here are typical of those used in published nucleosome mapping studies (Yuan et al., 2005; Johnson et al., 2006; Schones et al., 2008). Interestingly, this footprint signal is not predicted by a computational model of nucleosome occupancy likelihood trained on human chromatin (compared with Figures 3B to 3D; Fincher et al.., 2013). Single gene examples (Figure 3A) and composite metagene analysis (Figure 3B) reveal a pattern in which many genes exhibit a tripartite chromatin structure in which MSFs are present at both ends of genes and MRFs reside within the open reading frames of genes. Thus, MSFs and MRFs show a nonrandom distribution in and around genes, which may indicate a function in gene regulation.
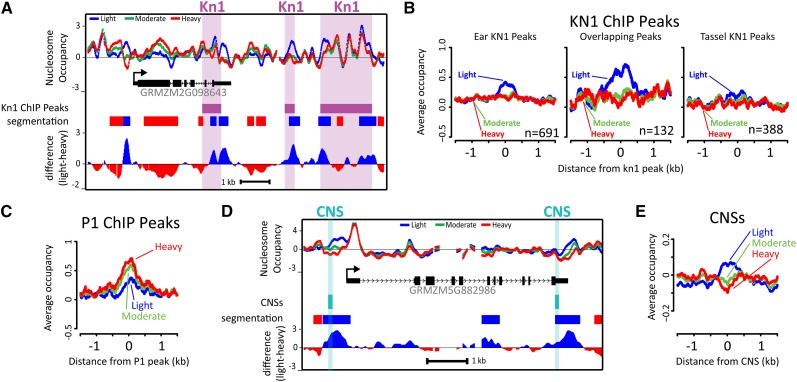
MSFs Are Enriched at KNOTTED1 Binding Sites and Conserved Noncoding Sequences
The prevalence and locations of MSFs around genes prompted us to ask whether these regions represent functional elements such as transcription factor binding sites. The binding sites for KNOTTED1 (KN1), a homeobox transcription factor expressed in meristematic cells (Vollbrecht et al., 1991), have been mapped via chromatin immunoprecipitation sequencing (ChIP-seq) in several tissues including immature ear (5 to 10 mm) and immature tassel (Bolduc et al., 2012). Although the immature ears we used for our study were harvested at a later stage of development (30 to 50 mm), we observed frequent overlaps between MSFs and KN1 binding sites (Figure 4). To examine the possible tissue specificity of MSF signatures, we classified kn1 ChIP-seq peaks into three exclusive groups: ear-specific (absent in tassel), tassel-specific (absent in ear), or common (present in both). On average, ear-specific kn1 ChIP-seq peaks displayed a conspicuous MSF signature, whereas tassel-specific kn1 ChIP-seq peaks showed only a slight MSF signature (Figure 4B). Interestingly, kn1 ChIP-seq peaks shared between ear and tassel displayed a very prominent MSF signature. We also examined the relationship between MSFs we identified in immature ears and ChIP-seq peaks identified in maize pericarps of the MYB-domain-containing transcription factor PERICARP COLOR1 (P1) (Morohashi et al., 2012). P1, which shows little expression in maize ears compared with seeds (pericarp included) (Supplemental Figure 2), displayed average P1 ChIP-seq peaks that resembled an MRF-like signature (Figure 4C).
Figure 4.
MSFs Overlap with Regulatory Elements.
(A) Nucleosome occupancy, Kn1 ChIP-seq peaks (Bolduc et al., 2012), the difference profile (light-heavy “difference”), and the segmentation of the difference profile (“segmentation”) at a representative locus.
(B) Knotted1 ChIP-seq peaks were classified as tassel-specific, ear-specific, or shared between tissues, and each were aligned by their midpoints and used to calculate average nucleosome occupancy scores.
(C) Average nucleosome occupancy scores from developing ears at P1 ChIP-seq peaks identified in pericarps.
(D) Representative gene with data tracks for nucleosome occupancy, conserved noncoding sequences (Turco et al., 2013), the difference profile (light-heavy), and the segmentation of the difference profile.
(E) Average nucleosome occupancy scores at CNSs aligned by their midpoints.
We next examined DNS-chip profiles at other potential regulatory elements defined as conserved noncoding sequences (CNSs; Turco et al., 2013). Like kn1 ChIP-seq peaks, CNSs also frequently overlapped with MSFs (Figure 4D). When we computed average DNS-chip profiles at CNSs aligned by their midpoint, we also found an MSF signature associated with CNSs (Figure 4E). Thus, CNS elements appear to exhibit a unique chromatin structural feature of an MSF. Together, these data show that many KN1 binding sites and CNSs in the maize genome can adopt a chromatin state that can be detected by DNS-chip, confirming the expectation that MSFs reflect functionally important genomic regions.
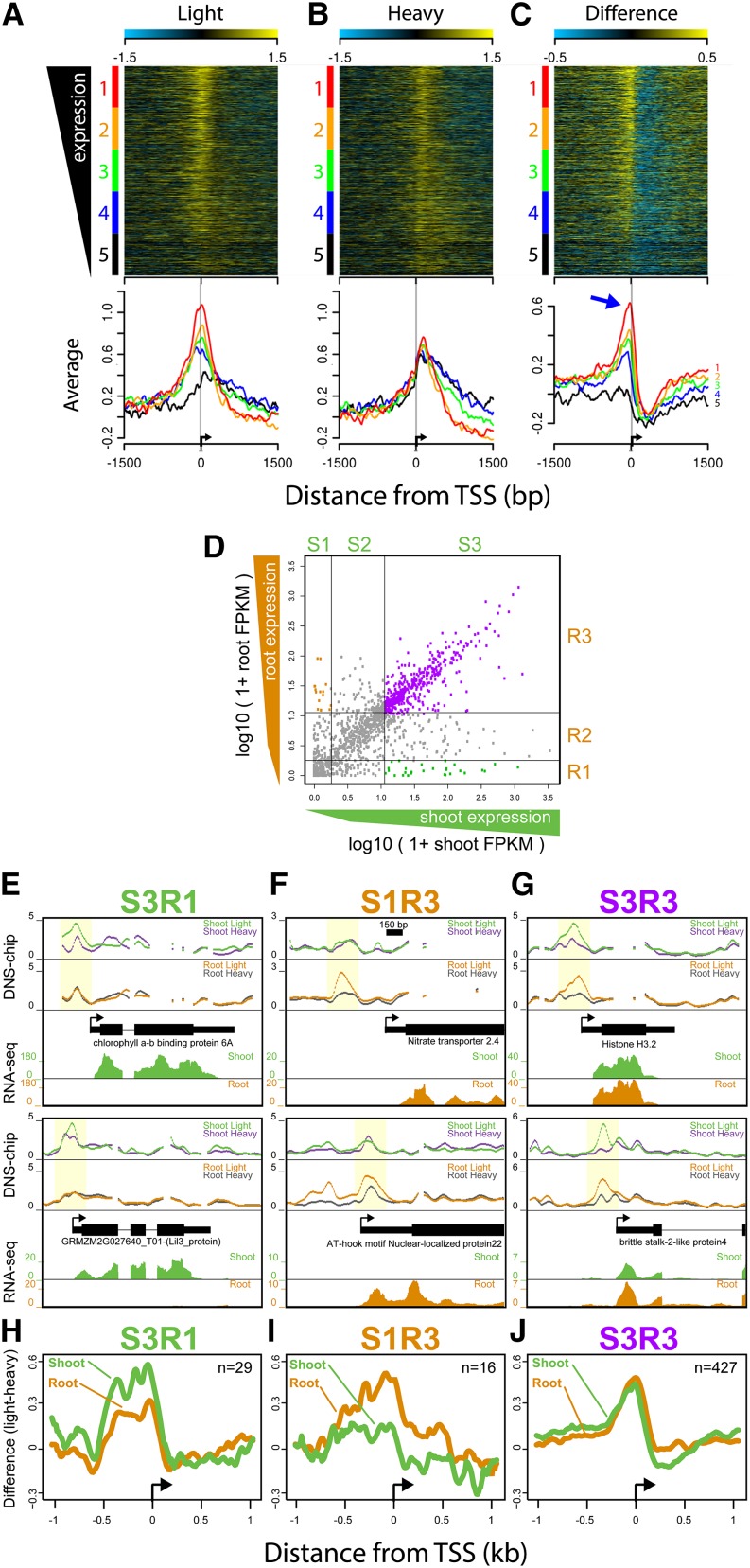
Promoter MSF Signals Are Associated with mRNA Levels and the Regulation of Gene Expression
To examine the relationship between MSFs and gene expression levels, we performed DNS profiling and RNA-seq on 9-d-old shoots. We then sorted the 1688 genes by their steady state mRNA levels and generated heat maps of DNS-chip profiles at the TSSs (Figures 5A and 5B). Gene expression levels were proportional to nucleosome occupancy signals under light-digest but not heavy-digest conditions. This trend was also observed when we grouped the 1688 genes into five equally sized groups sorted by expression levels and plotted average DNS-chip signals for each group (bottom plots, Figures 5A and 5B). When a heat map of the difference values (light-heavy) is generated, an expression-associated MSF signature (arrow, Figure 5C) is observed in all but the lowest quintile, a group with mostly nonexpressed genes (black line, Figure 5C). This analysis establishes that in general, the MSFs just upstream of TSSs are dynamic chromatin features, quantitatively captured by the DNS profiling method. Within gene bodies, expression levels were inversely proportional to nucleosome occupancy signals under both digestion conditions, a relationship consistent with that observed in other organisms (Valouev et al., 2011).
Figure 5.
MSFs Are Associated with Gene Expression Levels.
(A) to (C) Heat maps of nucleosome occupancy scores (light-digest and heavy-digest) and their difference (light-heavy) at 3 kb surrounding the 1688 TSSs included on the microarray. Gene TSSs were sorted based on their steady state gene expression levels (FPKM) and grouped into five equally sized groups (color coded at left of heat maps, from highest [red] to lowest [black]). Average scores for each of the five groups are plotted below each heat map. Blue arrow marks the −1 nucleosome region within which the difference profile covaries with gene expression levels.
(D) A scatterplot of root and shoot gene expression scores (log10(1+FPKM)) for each of the 1688 genes. Genes were grouped into three equally sized tertiles based on their gene expression level for both root and shoot, demarcated by the gray lines, which generated nine groups of genes (S1-3, R1-3).
(E) to (G) Examples DNS-chip profiles for genes highly expressed in shoots but not in roots (S3R1), genes highly expressed in roots but not shoots (S1R3), and genes highly expressed in both tissues (S3R3). Examples include nucleosome occupancy profiles for each tissue along with RNA-seq read densities for each tissue.
(H) to (J) Average difference profiles (light-heavy) at all genes within the three groups aligned by their TSS, with the number of genes in each group indicated by “n=.”
The correlation between gene expression and MSFs at TSSs suggests that differentially expressed genes between two given tissues may be associated with differences in the presence of MSFs within those genes. To further explore this relationship, we performed DNS-chip on root tissue from the same 9-d-old seedlings. Gene expression levels of each of the 1688 genes for the two organs were used to generate a shoot-versus-root scatterplot of gene expression values (Figure 5D). Each gene was grouped into expression tertiles for each tissue (Figure 5D, gray lines distinguishing groups S1-S3 and R1-R3). The root-versus-shoot expression-level scatterplot revealed that the majority of the 1688 genes represented on our microarray showed similar expression levels in the two organs. However, these groupings allowed us to examine a small subset of genes that were highly expressed in one tissue but showed little to no expression in the other. Examination of representative genes whose RNA levels were high in shoot but low in root (S3R1) include those encoding chlorophyll A-B binding proteins, which contained an MSF in shoots but not roots (highlighted in yellow, Figure 5E). Conversely, genes whose RNA levels were low in shoot but high in root (S1R3) include those encoding an AT-hook motif Nuclear-localized protein22 and Nitrate transporter 2.4 (Figure 5F). These genes showed an MSF in roots but not shoots. Finally, genes whose RNA levels were high in both organs, such as Histone H3.2 and brittle stalk-2-like protein4, had pronounced MSF signals in both organs (Figure 5G). Aggregate difference profiles confirmed that the patterns observed at the individual genes were representative of the overall trend (Figures 5H to 5J; Supplemental Figure 3). As expected, nucleosome occupancy profiles based on heavy-digest data were highly correlated between the two tissues (r = 0.92). Interestingly, genes that were differentially expressed in root versus shoot did not on average display different levels of nucleosome occupancy based on heavy-digest data alone. Thus, the negative correlation between gene expression and nucleosome occupancy in the gene body is not reflected in genes that are differentially expressed between the two tissues (Supplemental Figure 3). Our findings show that MSFs can be tissue specific and that their presence, especially at the 5′ end of the genes, is coupled to mRNA expression levels. These observations, together with the colocalization of MSFs with CNSs and KN1 binding sites, demonstrate that DNS profiling is a powerful experimental approach for comprehensive mapping of functional chromatin landscapes in a plant genome.
DISCUSSION
In the process of developing a nucleosome occupancy mapping technique for maize, we found localized variability in nucleosome occupancy mapping that was likely caused by technical differences in the preparation of nucleosome-protected DNA. These variable regions were easily digested, or nuclease hypersensitive, resembling the “fragile nucleosomes” recently described for yeast (Weiner et al., 2010; Henikoff et al., 2011; Xi et al., 2011). We demonstrated the ability of DNS-chip to detect thousands of regions of fragile chromatin, which we termed MSFs, and identified their widespread presence and distribution in plants.
MSFs, experimentally defined as footprint signals evident under light-digest conditions that are less detectable under heavy-digest conditions, could have different causes (Figure 6). One possible biochemical basis for MSFs is a stable nucleosomal footprint that is preferentially released under light-digest conditions, which could result from a chromatin conformation that renders the surrounding linker DNA more susceptible to cleavage than that of most nucleosomes (Figure 6A). Nucleosomes that contain the histone variants H2A.Z and H3.3, which are more readily solubilized under moderate salt concentrations than bulk nucleosomes, are enriched at the “nucleosome-depleted” region in humans (Jin and Felsenfeld, 2007; Jin et al., 2009). Thus, the presence of MSFs in maize promoters may be a result of histone-variant-containing nucleosomes, which are conserved in maize. Given the association of MSFs with KN1 binding sites, another possible basis for MSFs could be non-nucleosomal footprints. Non-nucleosomal footprints such as those resulting from transcription factors could potentially yield a 100- to 200-bp fragment that may not persist under higher nuclease digest levels (Figure 6B). In addition, it is possible that some MSFs may be caused by the simultaneous binding of transcription factors or chromatin remodelers and nucleosomes, which have been previously described (Adams and Workman, 1995; Floer et al., 2010; Mirny, 2010) (Figure 6C). In addition to nucleosome footprints that show different enrichment under different digestion conditions, there are also genomic regions that display a nucleosomal footprint under light-digestion conditions but display virtually no footprint under heavy-digest conditions (“Fragile,” Figure 6). These regions may represent a nucleosome conformation that renders the DNA on the nucleosomal core susceptible to cleavage with high concentrations of MNase, giving rise to <75-bp fragments that were not isolated when we gel-extracted 100- to 200-bp fragments.
Figure 6.
Potential Causes of MSFs and MRFs.
A hypothetical set of nucleosome occupancy profiles using light and moderate digestion conditions, with “Open” MSF, “Fragile” MSF, and MRF signatures (top). Footprints that could potentially give rise to the observed nucleosome occupancy profiles are shown. Regions that are preferentially cut under light‐digestion conditions are indicated by blue lightning bolts, whereas regions that are preferentially cut under heavy‐digestion conditions are indicated by red lightning bolts. An “Open” MSF signature could potentially be caused by a nucleosomal footprint (A), a transcription factor footprint (B), or the simultaneous binding of both (C) that render the surrounding DNA hypersensitive to MNase digestion. A “Fragile” MSF signature could potentially be caused by a nucleosomal footprint (A) or transcription factor footprint (B), or the simultaneous binding of both (C) that are susceptible to over digestion by MNase. MRFs could potentially be caused by arrays of tightly packed nucleosomes ([A] to [C]).
Though MSFs may represent a variety of chromatin structures and footprints, we believe that most of the MSFs observed in this study result from biochemical properties of nucleosomes and not transcription factors. This expectation is largely based on the fact that these features are detected by the use of nucleosome-sized fragments (100 to 200 bp) on our microarrays and that transcription factor footprints are typically much smaller (Gidoni et al., 1985). This interpretation is also consistent with the observation that the majority of hypersensitive footprints identified in yeast overlap with signals of histone H3 (Xi et al., 2011). Though we do not have a maize DNase-hypersensitive site (DHS) data set to compare with, we believe that MSFs are distinct from DHSs. MSFs result from a pair of cleavages between 100 and 200 bp apart, in contrast to DHSs, which are defined by single cleavage events and are interpreted as nucleosome-depleted regions. It is conceivable that MSFs and DHSs may overlap at regions that possess appropriately spaced DHSs in individual cells, though the relationship between fragile nucleosomes and DHSs has not been examined.
Nucleosome mapping studies in a variety of model systems have reported the pervasive presence of nucleosome-depleted regions directly upstream of TSSs (Lee et al., 2007; Schones et al., 2008; Valouev et al., 2008, 2011). In humans, many so-called nucleosome-depleted regions in promoters overlap with H2A.Z/H3.3-containing nucleosomes located just upstream of TSSs, and these nucleosomes are more readily solubilized in moderate-salt-containing buffers (Jin and Felsenfeld, 2007; Jin et al., 2009). This suggests that some regions that have been referred to as nucleosome-depleted may in fact be nucleosomal footprints that are not efficiently recovered under conditions commonly used to prepare nucleosomal DNA. In this study, we showed that occupancy footprints are readily detected in the nucleosome-depleted region of many genes under light digest conditions, supporting the idea that some regions classified as “nucleosome-free” may in fact represent fragile nucleosomes.
Another pattern revealed in our study is the presence of genomic regions whose relative abundance increased with higher levels of nuclease digestion (MRFs, Figure 6). This pattern could result from tightly packed arrays of nucleosomes with shorter or protected linker DNA, rendering the nucleosomes relatively resistant to MNase cleavage. Whatever the cause, this pattern is typical of open reading frames (e.g., the Bronze1 open reading frame region [Figure 3A] and the trend plot in Figure 3B). The localization of MRFs within gene bodies, particularly directly downstream of the TSS, indicates that they may be involved in transcriptional elongation, but their importance remains to be characterized.
DNS profiling is an approach to chromatin profiling that reveals multiple dimensions of chromatin structure information, including nucleosome occupancy, nuclease sensitivity, and chromatin compaction. In this study, we identified the presence of thousands of hypersensitive footprints in a plant genome that are associated with multiple different features, including transcription factor binding sites, conserved noncoding sequences, gene expression levels, and tissue-specific gene expression. This approach has great potential as a framework to identify functionally important regions of the genome and to integrate different types of genomic annotations.
METHODS
Plant Material
Maize (Zea mays) B73 seeds were obtained from the Maize Genetics Cooperation Stock Center. To obtain seedling tissue, seeds were germinated in Fafard Seedling Mix in the greenhouse (Department of Biological Science, Florida State University, Tallahassee, FL). At midday, 9 d after germination, aboveground tissue was cut at the soil line, flash frozen in liquid nitrogen, and stored at −80°C until use. Immature ears 3 to 5 cm in size were obtained from field-grown plants at the Florida State University Mission Road Research Facility (Tallahassee, FL).
Nucleus Isolation Buffer Compositions
APEL (20 mM Tris HCl, pH 7.8, 250 mM sucrose, 5 mM MgCl2, 5 mM KCl, 40% glycerol, 0.25% Triton X-100, and 0.1% BME; Steinmüller and Apel, 1986), BFA (15 mM PIPES-NaOH, pH 6.8, 0.32 mM sorbitol, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 1 mM DTT, 0.15 mM spermine, and 0.5 mM spermidine; Belmont et al., 1987), and MNEP (15 mM Tris-HCl, pH 7.5, 300 mM sucrose, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM EGTA, 0.5 mM DTT, and 1% Triton X-100; Dennis et al., 2007) were used for buffer comparison experiments. All other experiments used BFA for nucleus isolation. Immediately before use, 10 mM o-phenanthroine and 0.1 mM phenylmethylsulfonyl fluoride were added to nuclear fixation buffers.
Nucleus Isolation
Ten grams of seedling tissue, or 1 g of immature ear tissue, were ground under liquid nitrogen with a mortar and pestle and cross-linked by stirring for 10 min in 10 mL ice-cold nuclear isolation buffer with 1% formaldehyde. Fixation was stopped with 125 mM glycine for 5 min. Tissue was pelleted at 2000g for 10 min at 4°C in a swinging-bucket centrifuge, then the tissue pellet was resuspended in 25 mL nucleus isolation buffer with 1% hexylene glycol and 1% Triton X-100. After 5 min of gentle rotary shaking, nuclear suspensions were filtered through two layers of Miracloth (Calbiochem) and nuclei were pelleted at 2000g for 10 min at 4°C. Nuclei were washed once with 10 mL nuclear isolation buffer and resuspended in 10 mL MNase digestion buffer (50 mM Tris-HCl, pH 7.5, 320 mM sucrose, 4 mM MgCl2, and 1 mM CaCl2). Nuclei were sequentially filtered through 150, 100, 50, and 30 µm CellTric filters (Partec), pelleted at 2000g for 10 min at 4°C, resuspended in 1 mL of MNase digestion buffer, flash frozen in liquid nitrogen, and stored at −80C until use.
Cytology and Quantitation of Purified Nuclei
To assess nuclear preservation, nuclei were placed on standard glass microscopy slides, stained with 3 μg/mL 4′,6-diamidino-2-phenylindole in 1× PBS, and mounted in Vectashield antifading reagent with a nail-polish-sealed 22 × 30 × 1.5-mm cover slip. Images were collected on an Olympus IX-70 epifluorescence microscope and deconvolved and analyzed with the SoftWorx computerized workstation.
Isolation of Nucleosomal DNA
To compare the effect of nuclear isolation buffer on nucleosome occupancy, 100 μL nuclei were digested with 2 units/mL MNase at 37 C for 5 min and stopped with 20 mM EGTA. Deionized H2O (100 μL) was added to each sample, and nuclei were de-cross-linked with 1% SDS and 100 μg/mL proteinase K overnight at 65°C. DNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in deionized water. Digested DNA were treated with 40 μg/mL RNase A and run in a 1% agarose gel. DNA fragments (100 to 200 bp) were excised and gel extracted with the Qiaex II gel extraction kit (Qiagen) following the manufacturer’s instructions.
To examine the effect of digestion level on nucleosome occupancy in immature ears, nuclei isolated with the three buffers (APEL, HNIB, and BFA) were pooled, and 500 μL nuclear suspensions was digested with 2 units/mL MNase (light), 30 units/mL MNase (moderate), or 300 units/mL MNase (heavy). A total of four replicates were performed for each digestion level; however, replicate 4 of the light digest was discarded because of an anomalous microarray probe score distribution.
RNA-seq Library Preparation and Data Analysis
RNA was prepared from the same pools of seedling shoot and root tissue used for DNS-seq experiments. One hundred milligrams of frozen, ground tissue was resuspended in 1.5 mL Trizol (Invitrogen) and prepared according to the manufacturer’s instructions. RNA-seq data were kindly provided by Cornell University Genomics Center (courtesy of E.S. Buckler, E. Rodgers-Melnick, and P.A. Schweitzer) using RNA from our samples. The RNA-seq libraries were prepared using the Illumina TruSeq RNA sample preparation kit (v2) according to the manufacturer’s instructions. RNA-seq libraries were subjected to 100-cycle, single-end sequencing with an Illumina HiSequation 2500. Single-end reads were aligned to the AGPv2 genome assembly with TopHat (Kim et al., 2013) using default parameters. Fragments per kilobase per million (FPKM) values for each gene were obtained with Cuffdiff (Trapnell et al., 2013) using default parameters.
Microarray Design and Hybridization
The NimbleGen microarrays contain isothermal 49- to 74-bp probes at ∼15-bp spacing, representing both strands of genomic sequence. The 12-plex TSS-based microarray covered 3000 bp of genomic sequence surrounding 400 transcription start sites primarily derived from the Classical Maize Genes (Schnable et al., 2009). The 3-plex “big-block” NimbleGen microarrays represent unique regions within three ∼20-Mb gene-rich chromosome termini of chromosomes 4L, 5S, and 9S. Purified nucleosomal DNA and B73 bare genomic DNA were labeled and hybridized to the microarray according to manufacturer’s instructions for Dual-color CGH microarrays (Nimblegen).
Data Analysis and Sources
Slides were scanned with a Nimblegen MS 200 Scanner, and probe-by-probe fluorescence intensity ratios (nucleosomal/genomic) were produced with Nimblescan 2.6. Subsequent data analysis and plotting were performed primarily using BEDtools (Quinlan and Hall, 2010) and the R statistical programming language (Ihaka and Gentleman, 1996). Nucleosome occupancy profiles at each base pair were calculated as the average score of all overlapping probes at each base pair. Profiles were then smoothed using a 60-bp sliding-mean window at 10-bp step sizes. Data for each sample were quantile normalized. Segmentation of nucleosome occupancy and difference profiles was performed by first discarding regions with probes covering fewer than 500 bp of contiguous sequence, allowing for small (<60 bp) gaps. The resulting data were then used for segmentation with iSeg (http://cloud.stat.fsu.edu/iSeg). Segments exceeding a sd cutoff of 1.5 were used for “segmentation” tracks and overlap quantifications. Segmentation tracks for each profile will be available on the Greenome browser, a new UCSC genome browser (Kent et al., 2002) for plant genomes (http://greenomebrowser.com/). ChIP-seq peaks were obtained from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) for kn1 (GSE39161; Bolduc et al., 2012) and p1 (GSE38587; Morohashi et al., 2012). Canonical gene models were obtained from the B73_RefGen_v2 5b version of Filtered Gene Set (ftp://ftp.ensemblgenomes.org/pub/plants/release-15/gtf/zea_mays/). p1 gene expression profiles were obtained from qTeller (qTeller.org) using published RNA-seq data (Jia et al., 2009; Wang et al., 2009; Li et al., 2010; Davidson et al., 2011; Waters et al., 2011).
Accession Numbers
Microarray data have been deposited in the National Center for Biotechnology Information GEO database under accession number GSE60092. ChIP-seq peaks were obtained from GEO for kn1 (GSE39161) and p1 (GSE38587).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MNase-Sensitive and MNase-Resistant Footprints Are Observed in Raw and Un-Normalized Data.
Supplemental Figure 2. PERICARP COLOR1 Gene Expression Levels in Various Maize Tissues.
Supplemental Figure 3. Average Nucleosome Occupancy and Difference Profiles at Genes Grouped by Their Expression Levels in Seedling Shoot and Roots.
Supplementary Material
Acknowledgments
We thank E.S. Buckler, E. Rodgers-Melnick, and the Cornell Genomics Facility (P.A. Scheitzer) for the RNA-seq library preparation and sequencing. We also thank the FSU Research Computing Center (http://rcc.fsu.edu/) and especially D. Shrum and J. Wilgenbusch for computational support throughout this project. This work was supported by the National Science Foundation (Award NSF-ACI-1245758 to H.W.B. and Plant Genome Research Program Award NSF-IOS-1025954 to H.W.B., K.M.M., and J.H.D.) and the National Institutes of Health (Award NIH-R01-DA033775 to J.H.D. and J.Z.).
AUTHOR CONTRIBUTIONS
D.L.V., T.F.M., J.D.L., M.P.A., and G.G.H. performed experiments. All authors contributed to analysis and interpretation of the results. D.L.V., S.B.G., and J.Z. analyzed data and contributed computational tools. D.L.V., T.F.M., J.D.L., M.P.A., K.M.M., J.H.D., and H.W.B. designed experiments. The article was written primarily by D.L.V. and H.W.B.
Glossary
- MNase
micrococcal nuclease
- DNS-chip
differential nuclease sensitivity profiling
- TSS
transcription start site
- MSF
MNase-sensitive footprint
- MRF
MNase-resistant footprint
- ChIP-seq
chromatin immunoprecipitation sequencing
- CNS
conserved noncoding sequence
- DHS
DNase-hypersensitive site
- GEO
Gene Expression Omnibus
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adams C.C., Workman J.L. (1995). Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol. Cell. Biol. 15: 1405–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S., Sedat J.W., Agard D.A. (1987). A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J. Cell Biol. 105: 77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R. (2009). The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198. [DOI] [PubMed] [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R.M., et al. (2011). Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203. [Google Scholar]
- Dennis J.H., Fan H.Y., Reynolds S.M., Yuan G., Meldrim J.C., Richter D.J., Peterson D.G., Rando O.J., Noble W.S., Kingston R.E. (2007). Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome Res. 17: 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druliner B.R., Fincher J.A., Sexton B.S., Vera D.L., Roche M., Lyle S., Dennis J.H. (2013). Chromatin patterns associated with lung adenocarcinoma progression. Cell Cycle 12: 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher J.A., Vera D.L., Hughes D.D., McGinnis K.M., Dennis J.H., Bass H.W. (2013). Genome-wide prediction of nucleosome occupancy in maize reveals plant chromatin structural features at genes and other elements at multiple scales. Plant Physiol. 162: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., et al. (2010). A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Schneider K.L., Topp C.N., Rodriguez C., Presting G.G., Dawe R.K. (2011). Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning. Epigenetics Chromatin 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J.T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. (1985). Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science 230: 511–517. [DOI] [PubMed] [Google Scholar]
- Henikoff J.G., Belsky J.A., Krassovsky K., MacAlpine D.M., Henikoff S. (2011). Epigenome characterization at single base-pair resolution. Proc. Natl. Acad. Sci. USA 108: 18318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe E.S., Clemente T.E., Bass H.W. (2012). Maize histone H2B-mCherry: a new fluorescent chromatin marker for somatic and meiotic chromosome research. DNA Cell Biol. 31: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R. (1996). R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Jia Y., Lisch D.R., Ohtsu K., Scanlon M.J., Nettleton D., Schnable P.S. (2009). Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G. (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.M., Tan F.J., McCullough H.L., Riordan D.P., Fire A.Z. (2006). Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 16: 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N.A., Adams S., Moorhouse A., Paszkiewicz K. (2011). Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 39: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonne J.D., Dorweiler J.E., McGinnis K.M. (2013). Changes in nucleosome position at transcriptional start sites of specific genes in Zea mays mediator of paramutation1 mutants. Epigenetics 8: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam F.H., Steger D.J., O’Shea E.K. (2008). Chromatin decouples promoter threshold from dynamic range. Nature 453: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. (2007). A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39: 1235–1244. [DOI] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lieb J.D., Clarke N.D. (2005). Control of transcription through intragenic patterns of nucleosome composition. Cell 123: 1187–1190. [DOI] [PubMed] [Google Scholar]
- Lombraña R., et al. (2013). High-resolution analysis of DNA synthesis start sites and nucleosome architecture at efficient mammalian replication origins. EMBO J. 32: 2631–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S., Thanos D. (2002). Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110: 261–271. [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Mavrich T.N., et al. (2008). Nucleosome organization in the Drosophila genome. Nature 453: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny L.A. (2010). Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. USA 107: 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize Pericarp Color1 controlled genes. Plant Cell 24: 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. (1974). Subunit structure of chromatin. Nature 251: 249–251. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Freeling M. (2011). Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. (2008). Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton B.S., et al. (2014). The spring-loaded genome: nucleosome redistributions are widespread, transient, and DNA-directed. Genome Res. 24: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmüller K., Apel K. (1986). A simple and efficient procedure for isolating plant chromatin which is suitable for studies of DNase I-sensitive domains and hypersensitive sites. Plant Mol. Biol. 7: 87–94. [DOI] [PubMed] [Google Scholar]
- Stünkel W., Kober I., Seifart K.H. (1997). A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol. Cell. Biol. 17: 4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco G., Schnable J.C., Pedersen B., Freeling M. (2013). Automated conserved non-coding sequence (CNS) discovery reveals differences in gene content and promoter evolution among grasses. Front. Plant Sci. 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A., et al. (2008). A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A., Johnson S.M., Boyd S.D., Smith C.L., Fire A.Z., Sidow A. (2011). Determinants of nucleosome organization in primary human cells. Nature 474: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243. [DOI] [PubMed] [Google Scholar]
- Venter U., Svaren J., Schmitz J., Schmid A., Hörz W. (1994). A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13: 4848–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.J., Makarevitch I., Eichten S.R., Swanson-Wagner R.A., Yeh C.T., Xu W., Schnable P.S., Vaughn M.W., Gehring M., Springer N.M. (2011). Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. (2010). High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Yao J., Chen R., Li W., He X. (2011). Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 21: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. (2005). Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.