Abstract
Reverse transcription-quantitative PCR (RT-qPCR) has been widely adopted to measure differences in mRNA levels; however, biological and technical variation strongly affects the accuracy of the reported differences. RT-qPCR specialists have warned that, unless researchers minimize this variability, they may report inaccurate differences and draw incorrect biological conclusions. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines describe procedures for conducting and reporting RT-qPCR experiments. The MIQE guidelines enable others to judge the reliability of reported results; however, a recent literature survey found low adherence to these guidelines. Additionally, even experiments that use appropriate procedures remain subject to individual variation that statistical methods cannot correct. For example, since ideal reference genes do not exist, the widely used method of normalizing RT-qPCR data to reference genes generates background noise that affects the accuracy of measured changes in mRNA levels. However, current RT-qPCR data reporting styles ignore this source of variation. In this commentary, we direct researchers to appropriate procedures, outline a method to present the remaining uncertainty in data accuracy, and propose an intuitive way to select reference genes to minimize uncertainty. Reporting the uncertainty in data accuracy also serves for quality assessment, enabling researchers and peer reviewers to confidently evaluate the reliability of gene expression data.
Reverse transcription followed by real-time (or quantitative) PCR (RT-qPCR) has been widely adopted for quantification of gene expression by estimating steady state mRNA levels (Taylor et al., 2010). Alternatives such as RNA gel blotting or other RT-PCR methods reveal only relatively large differences in mRNA levels. However, the higher sensitivity of RT-qPCR necessitates accurate and precise pipetting, high-quality RNA, accurate estimation of RNA concentration, and efficient reverse transcription. Any variation in these technical parameters can influence the accuracy and precision of the results (Nolan et al., 2006; Udvardi et al., 2008; Bustin et al., 2009; Baker, 2011). Normalization to internal controls, such as spiked foreign RNA or internal reference genes, can control for technical variation (Huggett et al., 2005; Taylor et al., 2010; Baker, 2011). Unfortunately, both of these controls have weaknesses. Spiked RNA cannot correct for differences in extraction efficiency or overall transcriptional activity, and spiking itself can introduce bias if the small quantities of spiked control RNA are inaccurately pipetted.
The use of reference genes to normalize RT-qPCR data has been preferred by researchers, since variation in the experimental workflow affects all genes similarly (Huggett et al., 2005; Vandesompele et al., 2009; Bustin et al., 2013). A reference gene must show stable expression under the conditions of the experiment. Historically, and similarly to RNA gel blot experiments, PCR experiments have used a single housekeeping gene, assumed to be stably expressed, as a reference. However, the highly sensitive nature of RT-qPCR requires more stability of expression for reference genes; therefore, the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines call for the use of multiple reference genes with validated expression stability in each experiment (Bustin et al., 2009).
To ensure accurate RT-qPCR results, researchers must limit technical variability and select appropriate, stably expressed reference genes. To evaluate reports using RT-qPCR data, reviewers must determine if the technical quality of the experiment and the normalization strategy justify the reported RT-qPCR data. However, this judgment may be difficult, since many variables can affect the accuracy of RT-qPCR results, and most reviewers are not RT-qPCR specialists. In this commentary, we review essential guidelines that ensure reliable RT-qPCR results, direct researchers to useful resources, and provide a new way to represent RT-qPCR data that allows the nonspecialist reader to assess the quality and accuracy of the data.
USE OF MIQE GUIDELINES TO ENSURE HIGH-QUALITY EXPERIMENTAL DATA
RT-qPCR experiments lack a standard method; rather, they may include different steps, multiple protocols, and diverse commercial kits. Nonetheless, following basic guidelines can help researchers make appropriate experimental choices. Bustin et al. (2009) described the MIQE guidelines and provided a template (Bustin et al., 2010) to help researchers adopt the guidelines and report their adherence (as supplemental data in publications). A number of other publications can help with the design, analysis, and reporting of RT-qPCR experiments. Nolan et al. (2006) provide detailed experimental procedures for real-time PCR quantification and discuss some critical experimental and data analysis issues. Udvardi et al. (2008) summarize RT-qPCR essentials for plant scientists. Further guidelines for reliable data analysis using multiple reference genes can be found in Hellemans et al. (2007) and D’haene and Hellemans (2010), and Taylor et al. (2010) present a practical approach to producing RT-qPCR data that conform to MIQE guidelines.
Although RT-qPCR data are considered to be reliable when best practices are followed as described in the MIQE guidelines, Bustin et al. (2013) documented a widespread lack of adherence to these guidelines in the published literature. As the MIQE guidelines address the principal criteria that determine the quality of RT-qPCR data, assessment of these data relies on transparent reporting of those variables. Therefore, we urge the research community to adopt MIQE reporting and journal editors to consider MIQE reporting as a required addition to articles with RT-qPCR data (Bustin et al., 2013). To further aid researchers in this endeavor, we provide some critical discussion and useful tools below for selecting reference genes, assessing qPCR efficiency, and reporting RT-qPCR data.
SELECTING REFERENCE GENES FOR RT-qPCR IN PLANTS
Producing accurate RT-qPCR results requires the selection of reference genes with adequately stable expression under the chosen experimental conditions; using inappropriate reference genes may lead to inaccurate and misleading results (Gutierrez et al., 2008; Bustin et al., 2013). The use of multiple reference genes with validated minimal expression variation has been the standard for RT-qPCR data normalization for more than a decade (Vandesompele et al., 2002; Bustin et al., 2009). Nevertheless, the survey by Bustin et al. (2013) found numerous examples of inadequate reference gene selection. Selection of appropriate reference genes involves identifying candidates, validating the candidates under the specific experimental conditions, and then revalidating the selected reference genes in each subsequent experiment.
Candidate reference genes can be selected from recent publications that provide validation of many reference genes for diverse plant species, in different organs and developmental stages, and under different stress conditions (Zhang et al., 2013; Luo et al., 2014), from self-produced transcriptome data, or by using Refgenes (Hruz et al., 2011) or PlantRGS (Patel and Jain, 2011) to mine public databases for closely related experimental conditions. If data are not available for the species under study, then orthologs of known reference genes may be considered as candidates. Candidate reference genes of different functional classes should be selected to avoid coregulated genes.
The candidate reference genes must be validated using the same set of cDNAs used to quantify the gene of interest to ensure that the selected reference genes show stable expression across the conditions of the experiment. Indeed, differences in the experimental setup can influence the suitability of different reference genes when used in different laboratories or even in slightly modified experimental setups, such as alternative growth systems or conditions, other genotypes (mutants) or ecotypes, or different environmental conditions. Various mathematical and statistical algorithms such as geNorm (Vandesompele et al., 2002), Normfinder (Andersen et al., 2004), Bestkeeper (Pfaffl et al., 2004), and others (reviewed in Vandesompele et al., 2009) can be used to identify the combination of reference genes that is minimally affected by the experimental conditions.
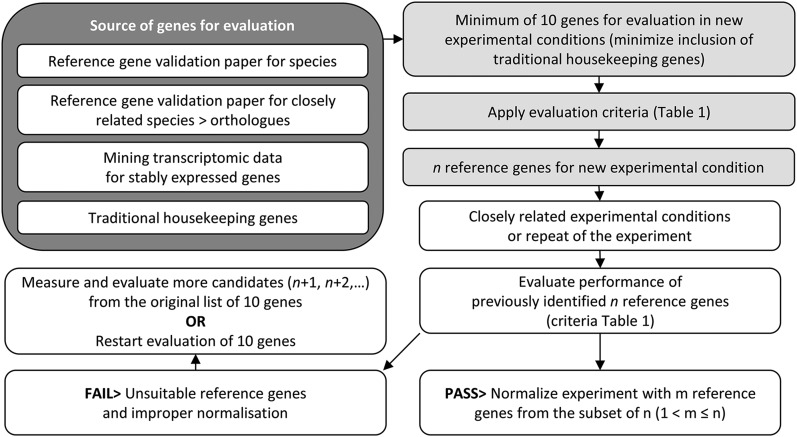
Selection of about 10 candidate genes provides a good starting point for validation (Remans et al., 2008). After identifying the subset that constitutes the best combination of reference genes, a normalization factor is calculated as the geometric mean of the relative expression of multiple reference genes and used to normalize the expression values for the genes of interest (GOIs; Vandesompele et al., 2002; Hellemans et al., 2007). Additionally, the selected reference genes should be revalidated in each subsequent experiment, even in repeated experiments, by reassessing their performance in each new sample set, and evaluation of additional candidates is necessary if some of these genes fail the revalidation (Hellemans et al., 2007). To facilitate the use of these procedures, we provide a flowchart for reference gene selection (Figure 1) and useful criteria for (re)validation (Table 1).
Figure 1.
Flow Chart for the Identification of Suitable Reference Genes in New Experimental Conditions and Subsequent Related Experiments.
A starting pool of minimum 10 reference genes, which may or not include a minimum number of traditional housekeeping genes, can be selected from sources such as reference gene papers or from transcriptome data. This pool should be evaluated by a chosen algorithm and evaluation criteria to identify appropriate reference genes. For subsequent related experiments (closely related experimental conditions or repeated experiments), it may be sufficient to start with the evaluation of the small number of genes that were used for normalization in previous experiments, but when these genes fail the criteria, additional ones need to be incorporated. Strongly deviating experimental conditions may require reevaluation of the original starting pool to identify the best genes for normalization, or may even need the incorporation of additional candidate reference genes.
Table 1. Checkpoints and criteria for validation of reference gene performance.
| Checkpoint | Criteria | Notes | Reference |
|---|---|---|---|
| qPCR efficiency | E ≥ 1.8 with se ≤ 5%; use E−ΔCq for most accurate quantification | Cq versus log(dilution) calibration curve for a dilution series of a pooled sample containing an equal fraction of all cDNA samples in the experiment. Determine slope and its se by linear regression analysis. E = (10−1/slope); se measures deviation of dilution points from the fitted slope. Determine the linear quantification range and analytical sensitivity: Samples with low Cq should fall within calibration curve without suffering from background signal due to primer dimer formation. qPCR efficiency also applies to GOIs. | 1, 2 |
| geNorm analysis | Minimum two genes with M < 1 and V < 0.15 | Advisable to use at least three genes with M < 1 and V < 0.15. Alternatively use y genes when Vx/y shows minimal value (that is <0.15). | 3, 4, 5 |
| Coefficient of variation of NRQs (CV) | Homogenous sample panels CV < 0.25 Heterogenous sample panels CV < 0.5 | Heterogeneity of sample panel depends on complexity of experimental comparisons, e.g., multiple conditions to be compared (mutants, treatments). | 1, 3 |
| Visualize NFs for each sample | NF should be > 0.3 when highest NF = 1 | NF < 0.3 indicates potential problem with sample. | 1, 3 |
| If RQ of all reference genes are low for a sample or group of samples, then technical problem, e.g., (RT-)PCR inhibition. | |||
| If RQ of single reference gene is low for a sample or group of samples, then technical problem, e.g., accidental lower cDNA input. | |||
| If RQ consistently low for all genes in all samples of the same condition, then possible treatment influence on expression of this reference gene. | |||
| NRQs, check for patterns in expression and possible coregulations | Check NRQs for each sample within a reference gene; should be close together | Outlying values for samples within a gene may point to inappropriate normalization. | 3 |
A number of criteria for evaluating the expression stability of reference genes have been published. Here, we compile checkpoints that we have chosen for evaluating reference genes before implementation in our experimental conditions. It should be noted that this is our experimental choice; a number of other suitable algorithms besides geNorm exist that can be used. RQ, relative quantity (2−ΔCq or E−ΔCq); NF, normalization factor = the geometric mean of the RQs of the chosen reference genes; NRQ, normalized relative quantity (2−ΔCq/NF or E−ΔCq/NF); CV, coefficient of variation (sd divided by mean); M and V, geNorm parameters of expression stability and pairwise variation, respectively. 1, Hellemans et al. (2007); 2, Pfaffl (2004); 3, D’haene and Hellemans (2010); 4, Vandesompele et al. (2002); 5, Ling and Salvaterra (2011).
THE IMPORTANCE OF ASSESSING qPCR EFFICIENCY
PCR efficiency refers to the actual increase in PCR amplicon quantity in each cycle of amplification; at 100% efficiency, the quantity of amplicons doubles in each PCR cycle during the exponential phase. However, different sequences amplify with different efficiencies, and differences in PCR efficiency between reference genes and GOIs can affect the accuracy of the results. PCR efficiency can be measured for the actual assay (also referred to as primer efficiency), as well as for each sample within that assay. Determination of sample-specific efficiency is possible (e.g., LinRegPCR; Ramakers et al., 2003), although it lacks precision (Hellemans et al., 2007). Nevertheless, estimation of sample-specific PCR efficiency can help detect outliers, i.e., samples whose amplification efficiencies clearly deviate from the assay efficiency (Hellemans et al., 2007). Such samples may contain a substantial amount of inhibitors, which can also produce amplification plots that have a low slope and low plateau (D’haene and Hellemans, 2010; Huggett and Bustin, 2011).
By contrast, assay-specific efficiency provides an essential parameter for normalization to reference gene expression. When the assays for the reference gene and the GOI assay differ in efficiency, the difference between target gene and reference gene relative quantities will vary with varying template amounts. Thus, assuming an efficiency of 100% can produce false expression ratios, resulting in over- or underestimation of normalized expression of the GOI (Pfaffl, 2004). It is also more accurate to use efficiency-corrected relative quantities to calculate the normalization factor from multiple reference genes.
The assay efficiency is measured using a dilution series of a pooled sample containing an equal fraction of all cDNAs in the experiment. The linear calibration curves are set up by linear regression analysis of Cq versus log(dilution), and the efficiency relative to 2 is derived as E = 10−1/slope (Pfaffl, 2004). Relative quantities (RQs) are then calculated as E−ΔCq or derived from the calibration curve equation. Calibration curves also provide a linear quantification range and analytical sensitivity (dilution at which cDNA amplification stays below background or where primer dimer formation becomes more important than specific amplicons, hindering accurate quantification). The linear range of the calibration curve ideally includes the interval for the targets to be quantified (Bustin et al., 2009). The se on the slope of the calibration curve is a reflection of how close the dilutions points match the fitted regression line (Hellemans et al., 2007). An efficiency of at least 1.80 with a se of <5% is appropriate (Table 1; Supplemental Data Set 1).
A NEW WAY OF REPORTING RT-qPCR DATA INCLUDING UNCERTAINTY IN ACCURACY
The goal of RT-qPCR experiments is to estimate the true in vivo GOI mRNA levels. Selection of appropriate reference genes and assessing qPCR efficiency as described above are critical procedures to minimize both biological and technical variation and ensure the reliability of RT-qPCR data. However, it is not possible to remove all variability from an RT-qPCR experiment, even when applying appropriate procedures, as reference genes always show small or large differences in expression between tissues or treatments. Thus, ideal reference genes do not exist (Huggett et al., 2005), and the normalization factor includes technical and biological variation in expression of the reference genes. Although a number of publications have sought to address these problems, no practical guidelines for data representation or quantitative measures for data accuracy have been adopted. Here, we provide a method to represent RT-qPCR data that allows for assessment of the accuracy of the data. Importantly, this data representation allows visualization of improper procedures and accuracy assessment even by nonspecialists.
The accuracy of RT-qPCR data is related to how close the obtained values are to the true in vivo GOI mRNA levels, whereas the precision is related to how close repeated measurements are clustered together. These repeated measurements should be derived from at least three biological replicates (Udvardi et al., 2008). Precision of measurements can be increased by minimizing technical variation and selection of appropriate reference genes to normalize the remaining technical variation. However, while using reference genes to increase precision, the accuracy can be affected, as explained below. In practice, RQs are derived from Cq values by the 2−ΔCq or preferably the E−ΔCq method, and for a single gene these RQs vary between samples due to both real biological differences in gene expression and technical variation. The reference genes and GOIs in the same sample experience the same technical variation; hence, the RQs of selected reference genes can be used to normalize technical GOI variation. However, this calculation assumes that the variation in the reference gene RQ includes only technical variation, whereas it also comprises biological variation. Thus, while normalizing to reduce technical variation, the biological variation in reference gene expression introduces a systematic error that affects the accuracy of the measurement. Unfortunately, technical and biological variation cannot be disconnected and the degree of technical variation relative to the biological variation erroneously imposed on GOI data remains unclear.
Note that RT-qPCR data normalized by reference gene expression is accurate only if all observed variation in the normalization factor originates from technical variation. Conversely, the alternate possibility—that all observed variation in the normalization factor originates from biological variation—must also be considered. In this case, normalization would not be necessary and in fact would be detrimental, such that GOI levels would be most accurately represented by the non-normalized data. In reality, experiments use imperfect reference genes and also have technical variation, and the true in vivo GOI expression most likely is situated between the normalized and non-normalized values. Thus, the normalized and non-normalized data set the boundaries of the uncertainty in the accuracy. In conclusion, as we cannot separate biological and technical variation, we also cannot accurately represent GOI levels with one data set. Rather, we must report both the normalized and non-normalized data as an interval that most likely includes the true gene expression levels. As examples, we use two experimental data sets to demonstrate that uncertainty in the accuracy of the data exists, even when the selected reference genes pass current evaluation algorithms, and that the choice of reference genes influences this level of uncertainty. We show how the uncertainty in data accuracy of RT-qPCR experiments can be calculated and visualized for fold changes in gene expression between sample groups.
Example 1
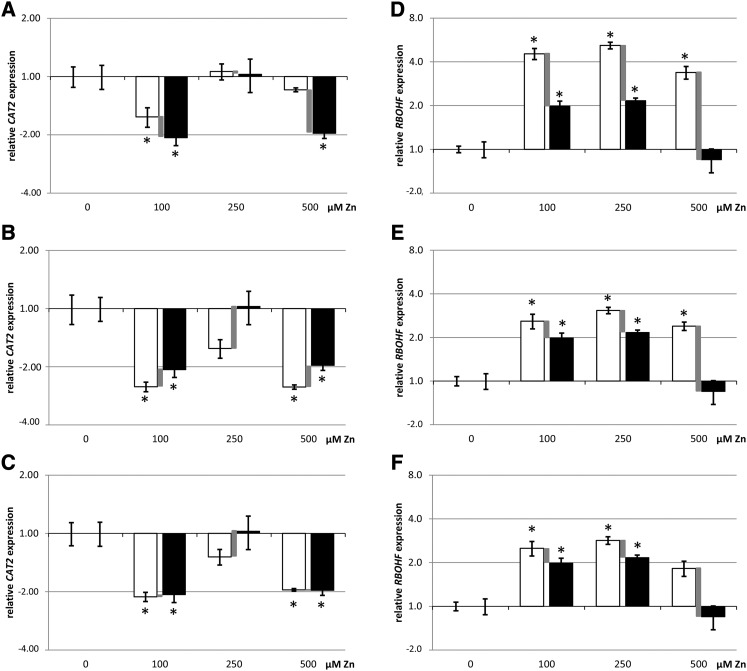
Using data from Arabidopsis thaliana exposed to excess zinc (Zn) (Remans et al., 2012), we revalidated four reference genes using the geNorm algorithm and additional criteria (Vandesompele et al., 2002; D’haene and Hellemans, 2010; criteria summarized in Table 1), using the geometric mean as a sample-specific factor to normalize GOI data. The evaluation of reference genes is provided in Supplemental Data Set 1. Using four reference genes selected according to appropriate standards produced a lower uncertainty (Figures 2B and 2E), compared with using the single, nonvalidated housekeeping gene ACT2 (Figures 2A and 2D; Supplemental Data Set 2). A one-way ANOVA with Dunnett posthoc testing to correct for the multiple comparison of Zn treatments with controls was performed separately on both the non-normalized and the normalized data. When both the normalized and non-normalized data showed a statistical difference, we considered the GOI to be up- or downregulated by a factor between the values for the normalized and the non-normalized data. When only the normalized or non-normalized data but not both showed a statistical difference, we considered the up- or downregulation to be uncertain, as it fell within the experimental noise and the accuracy was not high enough to allow a confident conclusion. Statistically, this is considered a sensitivity analysis: The primary analysis (e.g., on normalized data) is repeated, substituting the data set with another set (non-normalized data) to assess the impact of the data input on the statistical outcome. We thus explore if the same conclusion can be drawn for both data sets that define the gene expression level. The requirement for both analyses to yield the same conclusion is more stringent than analyzing only one data set. For example, when using only ACT2 for normalization, downregulation of CATALASE2 (CAT2) expression at 500 µM Zn exposure in the leaves was uncertain (Figure 2A), but using four appropriately selected reference genes produced a lower level of uncertainty, allowing a more confident conclusion about the downregulation (Figure 2B).
Figure 2.
Representation of RT-qPCR Data in Column Graphs Including the Uncertainty on the Accuracy.
Relative expression data (RT-qPCR; average ± se, n = 4 biological replicates from one experiment) of CAT2 in the leaves ([A] to [C]) and RBOHF in the roots ([D] to [F]) of Arabidopsis exposed to excess Zn (100, 250, and 500 μM) and relative to the control (0 μM = 1.0). Fold up- or downregulations are represented on a log2 scale y axis. (The control condition equals 1.0; hence, bars are not visible but standard errors are.) The normalized data are represented in white and the non-normalized data are in black. The difference between these is indicated by a gray bar and visualizes the uncertainty on the accuracy of the estimated GOI up- or downregulation, which varies according to the reference genes used for normalization. Normalization of data was performed with ACT2 only ([A] and [D]), four previously validated reference genes ([B] and [D]), or the gene(s) proposed by the GrayNorm algorithm yielding the lowest level of uncertainty ([C] and [F]). Statistics (one-way ANOVA and Dunnett comparison after testing normality with Shapiro-Wilk test and homoscedasticity with Bartlett test; *P < 0.05) were performed on both normalized and non-normalized data, and both data sets should yield significance to conclude a treatment effect. Uncertainty remains no matter the set of reference genes chosen, but the uncertainty is smaller in (C) and (F), allowing more accurate estimation of true GOI levels. This revealed, for example, that RBOHF is not upregulated in the roots at 500 μM Zn.
Example 2
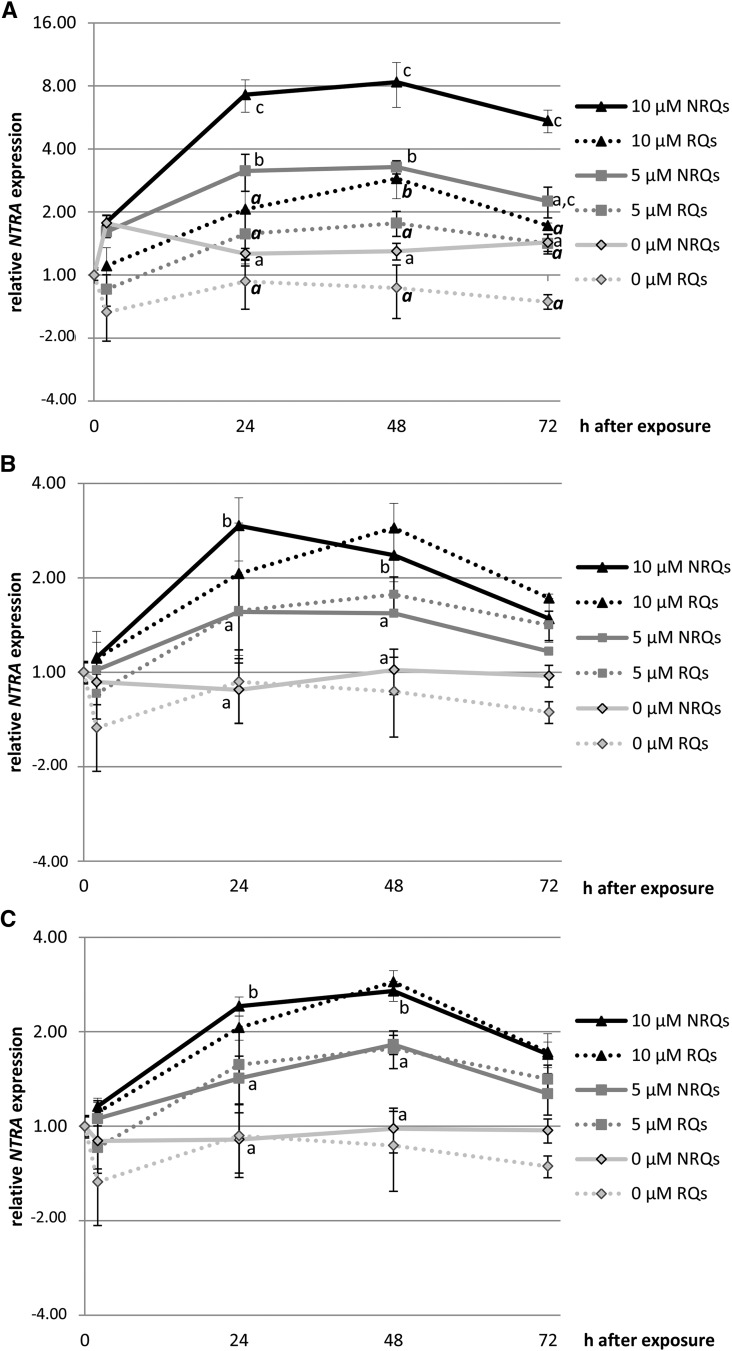
In a second example of data representation including uncertainty, changes over a 72-h period in expression of NADPH-DEPENDENT THIOREDOXIN REDUCTASE A (NTRA) were quantified in the leaves of Arabidopsis plants exposed to 5 or 10 μM CdSO4 (data in Supplemental Data Set 3). Again, true gene expression levels would be contained between the normalized and non-normalized data. Both data sets should indicate significant concentration dependent effects within a time point after one-way ANOVA and Tukey-Kramer posthoc test. Normalization with ACT2 indicated an 8-fold upregulation after 48 h exposure to 10 μM Cd, but included a very large interval of uncertainty between 3- and 8-fold (Figure 3A). Using three previously validated reference genes showed that a 3-fold upregulation at that time point is more accurate (Figure 3B). Hence, normalization influences the uncertainty interval.
Figure 3.
Representation of RT-qPCR Data in Line Graphs Including the Uncertainty on the Accuracy.
Representation of log2 relative expression levels (RT-qPCR; average ± se, n = 3 to 8 biological replicates from one experiment) of NTRA in leaves of Arabidopsis exposed to Cd (5 and 10 μM) over a time period of 0, 2, 24, 48, and 72 h and relative to the control (0 μM, 0 h = 1.0). The normalized data are represented by full lines and the non-normalized data by dotted lines, which visualizes the uncertainty on the accuracy of estimated GOI up- or downregulation. The uncertainty level varies with the (set of) reference genes used for normalization. Normalization of data was performed with ACT2 (A), three previously validated reference genes (B), or the gene proposed by the GrayNorm algorithm yielding the lowest level of uncertainty (C). If statistical differences (one-way ANOVA and Tukey-Kramer adjustment; P < 0.05) were observed between treatments within a time point, they are indicated with different lowercase letters for normalized data and bold italic for non-normalized data (only indicated in [A], as they are the same for [B] and [C]). Both data sets should yield significance to conclude a concentration dependent effect. Uncertainty remains no matter which normalization chosen, but the uncertainty is smaller in (C), allowing more accurate estimation of true GOI expression levels.
Such representation of the uncertainty interval, delimited by the normalized and non-normalized data, and statistical analysis of both data sets compared with their control conditions, should allow more confident assessment of the reliability of observed fold changes in GOI expression within the limitations of variation of biological and technical origin and help peer reviewers to assess the quality of RT-qPCR experiments and the validity of the conclusions.
In experiments in which it is difficult to obtain good quality RNA from certain tissues or conditions, the uncertainty interval on the data would remain large irrespective of the reference genes selected. Given the probability of high technical variation, the normalized data likely should be trusted more than the non-normalized data. This conclusion could be strengthened by presenting additional data showing that the use of different sets of reference genes does not significantly lower the uncertainty interval. Guénin et al. (2009) argued that if the differences between the patterns obtained using the candidate reference genes separately are small, then the choice of candidate will not greatly affect the gene expression profiles, thus providing reassurance regarding the reliability of normalization. In Example 1 described above, there was a large uncertainty interval at 500 μM Zn exposure in the roots (Figures 2D and 2E). These samples had low expression of all reference genes (Supplemental Data Set 1), which may point to condition-specific (RT-)PCR inhibition (e.g., due to residual Zn ions in the extract) or an overall lowered transcriptional activity due to the severe stress imposed by exposure to 500 μM Zn. Although the uncertainty level remains, it can be argued that the normalized data provide better accuracy.
GRAYNORM: A NEW ALGORITHM TO IDENTIFY REFERENCE GENES THAT YIELD THE LOWEST LEVEL OF UNCERTAINTY
As is clear from the above examples, a small uncertainty interval increases the reliability of the data. The non-normalized GOI data contain both genuine expression differences and technical variation and remain fixed once an experiment has been measured by RT-qPCR. The size of the uncertainty interval can vary only with the choice of reference genes. What combination of measured reference genes would yield the lowest level of uncertainty? Reference genes are used to calculate the normalization factor (NF), and the GOI data are divided by the NF during normalization. Thus, 1/NF is the imposed deviation from the non-normalized data, creating the uncertainty interval. The closer the average 1/NF per sample group is to 1.0, the smaller the potential erroneous influence of reference genes and the higher the resolution of the experiment, allowing more accurate estimation of fold-changes in GOI expression. The calculation of the deviation of the average 1/NF from 1.0 is as follows: (1) calculate the 1/NF for each sample, (2) calculate the average 1/NF per condition and relative to the control condition (=1.0), and (3) calculate the coefficient of variation of these 1/NF per condition, which measures the general deviation from 1.0 over all the conditions. This calculation is relatively simple for one set of reference genes; to perform all the calculations for each possible combination of measured reference genes, we developed the freely available algorithm GrayNorm, which minimizes the “gray zone” of uncertainty. The algorithm then ranks the combinations of genes by lowest coefficient of variation of the 1/NFs over the conditions (full details and explanation in Supplemental Methods). GrayNorm produces a table that can be sorted for other parameters, such as cumulative deviation of 1/NF from 1 over all conditions, or lowest deviation from 1 for a certain condition, to provide better accuracy for a particular condition.
We tested GrayNorm on Example 1 (Zn exposure) and identified an optimal combination of three reference genes for the leaves (ACT2, AT5G15710, and MSD1; Supplemental Data Set 4) and a single reference gene for the roots (AT5G08290; Supplemental Data Set 4). For experiment 2 (kinetics of NTRA expression after Cd exposure) GrayNorm returned a single reference gene (AT5G25760; Supplemental Data Set 4). In these particular data sets, using the reference genes identified by GrayNorm produced a higher experimental resolution and a lower level of uncertainty, even when using a single validated gene for normalization, compared with the current standard of using multiple reference genes (Figures 2C, 2F, and 3C; data in Supplemental Data Sets 2 and 3). Remarkably, a gene discarded for its relatively low expression stability in the leaves by the criteria of Table 1 (ACT2; evaluated in Supplemental Data Set 1) produced the most accurate normalization when used in combination with MSD1 and AT5G15710 as suggested by GrayNorm. This could be due to selection of two genes showing opposite variation, resulting in decreased NF variation (Andersen et al., 2004).
Assessment of the uncertainty in data accuracy and a high experimental resolution increase the confidence in the results. This is especially relevant for unexpected effects, such as the lack of downregulation of CAT2 at 250 μM Zn exposure or the lack of NADPH OXIDASE ISOFORM F (RBOHF) upregulation at 500 μM (Figure 2). Responses to metals like Zn and Cd depend on exposure time and concentration, which influence metal uptake. Furthermore, responses may peak in time and disappear (Figure 3) and may be biphasic or provoked by different mechanisms at different exposure concentrations. A confident analysis of an appropriate number of biological replicates, and repeated experiments with similar conclusions, would be needed before further investigating the molecular basis or consequences of changes in gene expression.
Conclusions
We present a method of reporting the uncertainty in data accuracy that allows for visual assessment of experimental variation and more confident estimation of fold changes in gene expression in RT-qPCR analysis. Additionally, the GrayNorm algorithm can be used to select the combination of reference genes measured in the experiment that yield the highest possible accuracy. The choice of reference genes for normalization influences experimental variation, and we showed that this influences the uncertainty level in data accuracy. The strength of the data presentation proposed here is that, irrespective of the individual experimental choices regarding technical procedures and reference genes, the effect of these choices on GOI data becomes more obvious for researchers and reviewers. The consequences of inappropriate practices become assessable, such as using a single reference gene without validation, as well as variations of technical origin that yield a larger uncertainty interval. The issue of biological significance also deserves attention. The biological consequences of small changes in expression of single genes should not be overinterpreted, and it is important to remember that changes in steady state levels of mRNA are not necessarily correlated with changes in protein abundance or activity. By establishing an accuracy interval, false positive up- or downregulations can be uncovered with high confidence, so that researchers can focus on using other tools to pursue the biological consequences of the changes in gene expression that are true, repeatable, and interesting.
METHODS
A description of the GrayNorm algorithm is outlined in Supplemental Methods, and GrayNorm input and output data sets for the experiments of this article are in Supplemental Data Set 4. Plant materials, growth conditions, and RT-qPCR procedures were according to Remans et al. (2012) and Keunen et al. (2013) and are included in Supplemental Methods. Adherence to MIQE guidelines is described in Supplemental Table 1. The GrayNorm algorithm is also written as a python script and is included as Supplemental File 1, along with a License and a README file on how to use it on your data (Supplemental Methods).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. MIQE Reporting for Examples 1 and 2.
Supplemental Methods. Methodologies for Examples 1 and 2.
Supplemental Data Set 1. Data Set and Reference Gene Evaluation for Example 1 (Zn Exposure).
Supplemental Data Set 2. GOI and Reference Gene Data Set for Example 1 (Zn Exposure).
Supplemental Data Set 3. GOI and Reference Gene Data Set for Example 2 (Kinetics Cd Exposure).
Supplemental Data Set 4. GrayNorm Input and Output Data for Examples 1 and 2.
Supplemental Data Set 5. MIQE Reporting for Examples 1 and 2 Using Template Table.
The following materials are freely available at the GitHub repository: https://github.com/gjbex/GrayNorm.
Supplemental File 1. graynorm.txt (Python Script for GrayNorm Algorithm).
Supplementary Material
Acknowledgments
This work was supported by Research Foundation-Flanders (FWO) projects (G.0436.06, G.0807.09, and G.0606.10). T.R. was funded by a postdoctoral fellowship of the FWO and E.K. by a doctoral fellowship of the FWO. We thank Michal Kicinski (Centre for Environmental Sciences) and Christel Faes (Center for Statistics, Hasselt University) for their aid with statistical analysis.
AUTHOR CONTRIBUTIONS
T.R., K.S., A.C., and J.V. designed experiments and analyzed data. T.R. and E.K. performed experiment and analyzed data. G.J.B. contributed new computational tools and analyzed data. T.R. wrote the article.
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Andersen C.L., Jensen J.L., Ørntoft T.F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- Baker M. (2011). qPCR: quicker and easier but don’t be sloppy. Nat. Methods 8: 207–212. [Google Scholar]
- Bustin S.A., Beaulieu J.F., Huggett J., Jaggi R., Kibenge F.S.B., Olsvik P.A., Penning L.C., Toegel S. (2010). MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., et al. (2013). The need for transparency and good practices in the qPCR literature. Nat. Methods 10: 1063–1067. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. [DOI] [PubMed] [Google Scholar]
- D’haene B., Hellemans J. (2010). The importance of quality control during qPCR data analyis. Int. Drug Discov., 18–24. [Google Scholar]
- Gutierrez L., Mauriat M., Guénin S., Pelloux J., Lefebvre J.F., Louvet R., Rusterucci C., Moritz T., Guerineau F., Bellini C., Van Wuytswinkel O. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6: 609–618. [DOI] [PubMed] [Google Scholar]
- Guénin S., Mauriat M., Pelloux J., Van Wuytswinkel O., Bellini C., Gutierrez L. (2009). Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 60: 487–493. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., et al. (2011). RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Bustin S.A. (2011). Standardisation and reporting for nucleic acid quantitation. Accredit. Qual. Assur. 16: 399–405. [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6: 279–284. [DOI] [PubMed] [Google Scholar]
- Keunen E., Jozefczak M., Remans T., Vangronsveld J., Cuypers A. (2013). Alternative respiration as a primary defence during cadmium-induced mitochondrial oxidative challenge in Arabidopsis thaliana. Environ. Exp. Bot. 91: 63–73. [Google Scholar]
- Ling D., Salvaterra P.M. (2011). Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One 6: e17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H.L., Luo L.P., Guan B.C., Li E.X., Xiong D.J., Sun B.T., Peng K., Yang B.Y. (2014). Evaluation of candidate reference genes for RT-qPCR in lily (Lilium brownii). J. Hortic. Sci. Biotechnol. 89: 345–351. [Google Scholar]
- Nolan T., Hands R.E., Bustin S.A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1: 1559–1582. [DOI] [PubMed] [Google Scholar]
- Patel R.K., Jain M. (2011). PlantRGS: a web server for the identification of most suitable candidate reference genes for quantitative gene expression studies in plants. DNA Res. 18: 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2004). Quantification strategies in real-time PCR. In A-Z of Quantitative PCR, S.A. Bustin, ed (La Jolla, CA: International University Line; ), pp. 87–112. [Google Scholar]
- Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509–515. [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Remans T., Opdenakker K., Guisez Y., Carleer R., Schat H., Vangronsveld J., Cuypers A. (2012). Exposure of Arabidopsis thaliana to excess Zn reveals a Zn-specific oxidative stress signature. Environ. Exp. Bot. 84: 61–71. [Google Scholar]
- Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227: 1343–1349. [DOI] [PubMed] [Google Scholar]
- Taylor S., Wakem M., Dijkman G., Alsarraj M., Nguyen M. (2010). A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50: S1–S5. [DOI] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034.1–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., Kubista M., Pfaffl M.W. (2009). Reference gene validation software for improved normalization. In Real-Time PCR: Current Technology and Applications, J. Logan, K. Edwards, and N. Saunders, eds (Poole, UK: Caister Academic Press), pp. 47–64. [Google Scholar]
- Zhang L., He L.L., Fu Q.T., Xu Z.F. (2013). Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int. J. Mol. Sci. 14: 24338–24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.