This study analyzes how transcriptome diversity between two maize inbred lines affects gene expression patterns in individual root tissues of their F1-hybrid progeny. Tissue-specific variation in single-parent expression (SPE) illustrates the highly dynamic transcriptomic landscape of root tissues. Nonsyntenic genes predominate among SPE genes, indicating their young evolutionary age.
Abstract
Maize (Zea mays) displays an exceptional level of structural genomic diversity, which is likely unique among higher eukaryotes. In this study, we surveyed how the genetic divergence of two maize inbred lines affects the transcriptomic landscape in four different primary root tissues of their F1-hybrid progeny. An extreme instance of complementation was frequently observed: genes that were expressed in only one parent but in both reciprocal hybrids. This single-parent expression (SPE) pattern was detected for 2341 genes with up to 1287 SPE patterns per tissue. As a consequence, the number of active genes in hybrids exceeded that of their parents in each tissue by >400. SPE patterns are highly dynamic, as illustrated by their excessive degree of tissue specificity (80%). The biological significance of this type of complementation is underpinned by the observation that a disproportionally high number of SPE genes (75 to 82%) is nonsyntenic, as opposed to all expressed genes (36%). These genes likely evolved after the last whole-genome duplication and are therefore younger than the syntenic genes. In summary, SPE genes shape the remarkable gene expression plasticity between root tissues and complementation in maize hybrids, resulting in a tissue-specific increase of active genes in F1-hybrids compared with their inbred parents.
INTRODUCTION
Maize (Zea mays) is a highly polymorphic species with an extraordinary level of intraspecific genomic diversity compared with other eukaryotes (Springer et al., 2009; Swanson-Wagner et al., 2010). This diversity is mediated by single nucleotide polymorphisms (SNPs), insertion-deletion polymorphisms (INDELs), and structural variations. Major genotyping efforts determined exceptionally high frequencies of SNPs and INDELs (∼1 per 80 bp and 1 per 300 bp genomic sequence, respectively) (Bi et al., 2006; Barbazuk et al., 2007) between the commonly used inbred lines B73 and Mo17. While these polymorphisms represent small genomic alterations, structural variations such as copy number variations, presence-absence variations (PAVs), and chromosomal rearrangements refer to segmental alterations of DNA >1 kb (Lai et al., 2010; Olsen and Wendel, 2013). Between the inbred lines B73 and Mo17, thousands of structural variants have been reported (Springer et al., 2009; Swanson-Wagner et al., 2010). In addition, inbred line-specific differences in content of repetitive DNA (Kato et al., 2004) and of total genome size (Laurie and Bennett, 1985) have been detected further emphasizing the high degree of intraspecific genomic divergence. To determine how genomic variation translates to quantitative gene expression differences, transcriptome surveys of various maize inbred lines have been conducted (Hansey et al., 2012). This study reported not only extensive sequence variation in annotated gene models between 21 inbred lines but also defined the variable part of the maize transcriptome as opposed to the conserved section. Recent studies support the concept of the pan-transcriptome consisting of a core and a dispensable subtranscriptome in maize (Hirsch et al., 2014; Marroni et al., 2014), which further illustrates the extensive intraspecific variation.
As demonstrated by syntenic comparison to the genomes of other grass species, the lineage leading to maize experienced a whole genome duplication 5 to 12 million years ago (Blanc and Wolfe, 2004; Swigonová et al., 2004), which created two distinct subgenomes each of which originally contained a complete set of genes and regulatory sequences (Schnable et al., 2011). In many cases, one of the two gene copies was lost from the genome in a process known as fractionation; however 3000 to 5000 gene pairs are retained in the modern maize genome (Schnable et al., 2011). Therefore, the gene complement of maize can be divided into three categories: pairs of duplicate genes shared by both subgenomes, single-copy genes present in only one subgenome, and genes that cannot be assigned to a subgenome. The third category is characterized by a lack of syntenic orthologs in the genomes of other grass species, and most of these nonsyntenic genes are created by single gene duplication mechanisms, likely after the maize whole-genome duplication (Woodhouse et al., 2010).
The structural genomic diversity of the maize inbred lines B73 and Mo17 is also reflected on the transcriptome level. In maize roots, hundreds of genes were identified that were exclusively expressed in one parental inbred line but not in the second parent, whereas all these genes were expressed in both reciprocal hybrids, a phenomenon termed single-parent expression (SPE; Paschold et al., 2012). As a result, hybrids have hundreds more active genes than their parental inbred lines (Paschold et al., 2012).
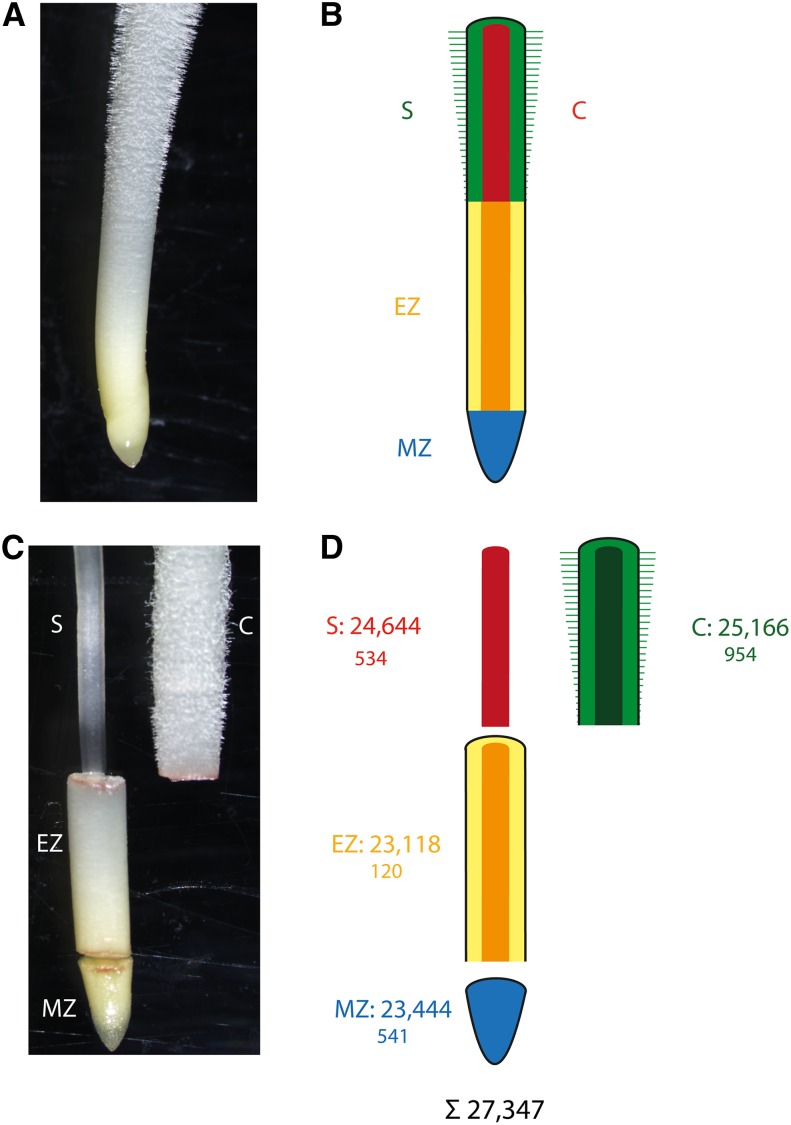
In longitudinal orientation, roots are structurally divided into the root cap at the terminal end, a subterminal meristematic zone, followed by the elongation and differentiation zones (Figure 1; Ishikawa and Evans, 1995). Cells formed by mitotic divisions in the meristematic zone are dislocated proximally into the elongation zone, where they start to elongate. The elongation zone partly overlaps with the meristematic zone because cell elongation begins in the meristematic zone (Ishikawa and Evans, 1995). Finally, root cells transit into the differentiation zone where the various cell types acquire their final functions. Hence, the cells along the longitudinal axis of a root represent a gradient of cell differentiation, with very young undifferentiated cells at the distal end near the root tip and differentiated cells toward the proximal end of the root. The differentiation zone can be spotted by the presence of epidermal root hairs (Ishikawa and Evans, 1995).
Figure 1.
Tissues of Young Maize Primary Roots Surveyed in This Study.
(A) Primary root prior to tissue sampling.
(B) Color-coded schematic of the different tissues of a maize primary root.
(C) Manually dissected primary root with separated meristematic zone, elongation zone, cortex and stele.
(D) Schematic of separated primary root tissues. Upper numbers indicate the total number of genes expressed in a tissue, while the smaller lower numbers indicate the number of genes that were exclusively expressed in a tissue (in any of the analyzed genotypes). In total, 27,347 genes were expressed in at least one tissue. Red (S), stele; green (C), cortex; yellow/orange (EZ), elongation zone; blue (MZ), meristematic zone.
In transverse orientation, the differentiation zone displays a number of functionally diverse cell types that can be distinguished by their anatomical features. The stele with the pericycle as its outermost cell layer is connected to the multilayered cortical parenchyma via a ring of endodermis cells. In maize, the stele contains differentiated xylem vessels that function in water and nutrient transport and primary phloem elements that function in the transport of photosynthates. These elements of the vasculature are embedded in parenchymal pith tissue (Hochholdinger, 2009). The cortical parenchyma that surrounds the stele consists of the endodermis, multiple layers of cortex tissue, and a single epidermal layer that connects the root to the rhizosphere (Hochholdinger, 2009). The epidermis, which is densely populated by tubular root hairs, takes up nutrients, which are then either carried via the cortex and xylem into the shoot or metabolized in the cortex of the root (Marschner, 2011). Stele and cortical parenchyma (hereafter referred to as cortex) can be separated mechanically without damaging pericycle and endodermis cells allowing subsequent analyses of these functionally diverse tissues (Saleem et al., 2009).
In this study, we globally surveyed the transcriptomes of four functionally distinct tissues of young maize primary roots in the inbred lines B73 and Mo17 and their reciprocal F1-hybrid progeny via RNA-seq. These experiments revealed dynamic tissue-specific patterns of SPE, which preferentially affected nonsyntenic genes.
RESULTS
RNA-seq Profiling of Four Root Tissues of Maize Inbred Lines and Their Hybrid Offspring
In longitudinal orientation, maize primary roots display specialized zones of development, including the meristematic zone, the elongation zone and, as parts of the differentiation zone, stele and cortex (Figures 1A and 1B), which can be mechanically separated (Figures 1C and 1D). The transcriptomes of these four distinct tissues were surveyed in 2- to 4-cm primary roots of the maize inbred lines B73 and Mo17 and their reciprocal hybrids B73xMo17 and Mo17xB73. Each tissue per genotype was analyzed in four biological replicates (Supplemental Figure 1). RNA-seq of those 64 samples resulted on average in 9 million 100-bp reads per sample (Supplemental Table 1). After quality trimming and removal of duplicate reads, 79 to 81% of all sequences mapped to the maize reference genome (ZmB73_RefGen_v2; Supplemental Table 2). For each of the four genotypes, 96% of the reads mapped to the filtered gene set of maize (ZmB73 FGS_5B_FGSv2; Supplemental Table 2), which comprises 39,656 high-confidence gene models.
In total, 27,347 of the 39,656 high-confidence genes (69%) were expressed in at least one tissue of at least one genotype (Figure 1D). Among those, 23,444 genes (86%) were expressed in the meristematic zone, 23,118 (85%) in the elongation zone, 25,166 (92%) in the cortex, and 24,644 genes (90%) in the stele (Figure 1D). Moreover, 2149 genes were exclusively expressed in only one of the four analyzed tissues (Figure 1D). While 120 genes representing 0.5% of all expressed genes in that tissue (120/23,118) were only expressed in the elongation zone, several hundred genes were exclusively expressed in cortex (954/25,166 ≙ 3.8%), stele (534/24,644 ≙ 2.2%) and meristematic zone (541/23,444 ≙ 2.3%).
Tissue-Specific Expression Complementation by SPE
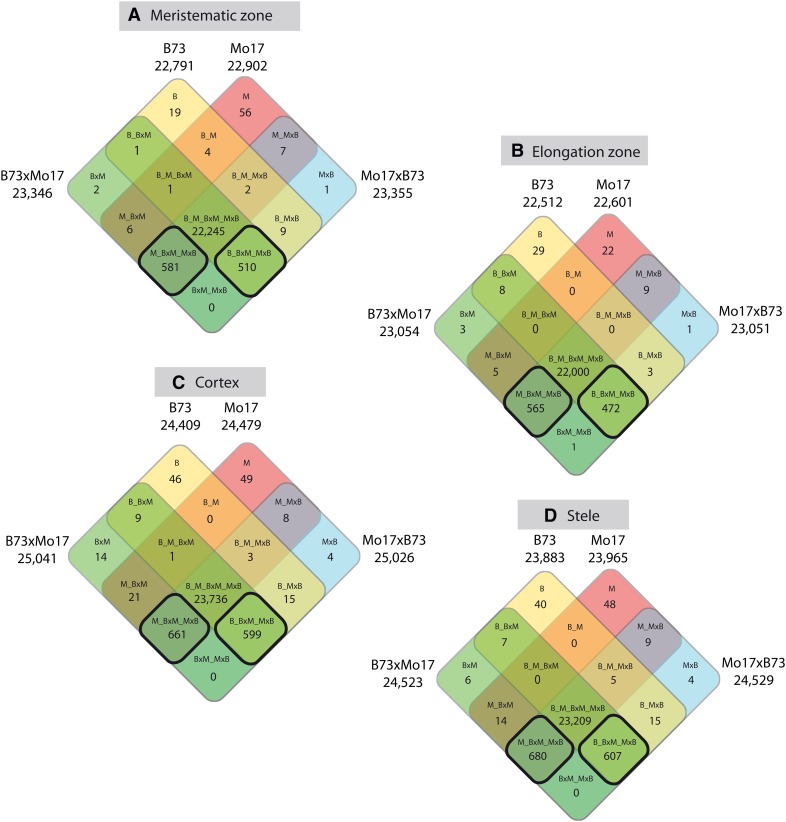
SPE is an extreme instance of expression complementation in which genes are expressed in only one parent but in both reciprocal hybrid progeny. To assay for SPE in the four root tissues, activity of all genes in all four genotypes [B73 (B), Mo17 (M), B73xMo17 (BxM), Mo17xB73 (MxB)] was analyzed by a Bayesian data-augmented Markov chain Monte Carlo approach (see Methods). The results were summarized in four-way Venn diagrams (Figure 2). In each of the four root tissues, the vast majority of genes (94 to 95%) were expressed in all four genotypes (B_M_BxM_MxB). However, most of the genes that were not expressed in all four genotypes displayed SPE (Figure 2, square fields encircled by a bold black line). SPE_B genes (B_BxM_MxB) are expressed in B73 and in the reciprocal hybrids, but not in Mo17, whereas SPE_M genes (M_BxM_MxB) are expressed in Mo17 and the reciprocal hybrids, but not in B73. SPE was detected in each of the four tissues and the number of SPE genes was similar between tissues and ranged between 1037 (elongation zone) and 1287 (stele; Figure 2). Four genes switched their SPE pattern between tissues (Supplemental Table 3) and were not considered for further calculations because it is difficult to determine if they have escaped our error rate correction or if they indeed represent rare examples where SPE has switched between different tissues. This resulted in 1096 genes showing SPE_B and 1245 genes showing SPE_M in at least one tissue. As a consequence of complementation, both hybrids displayed in all four tissues >440 active genes more than their inbred parents (Figure 2).
Figure 2.
Genotype-Specific Expression Patterns in Four Different Maize Primary Root Tissues.
Transcriptomic expression complementation by SPE is dominant among the genes that are not expressed in all four genotypes (encircled by a bold black line). SPE genes are expressed in only one parental line but both hybrids. Meristematic zone (A), elongation zone (B), cortex (C), and stele (D). The different genotypes and number of genes of the various expression classes are indicated in each square field of the four-way Venn diagrams. The total number of expressed genes per genotype and tissue are indicated at the border of each Venn diagram. Note that in each tissue, hybrids express more genes than their parental inbred lines.
To determine how many SPE_B genes are the result of PAV, i.e., these genes are not expressed in Mo17 because they are absent from that genotype, SPE_B genes were compared with a set of genes present in the B73 and absent in the Mo17 genome (Paschold et al., 2012). Overall, 4.4% (48/1096) of the SPE_B genes represent genomic PAVs (Supplemental Data Set 1).
Tissue-Specific Dynamics of SPE
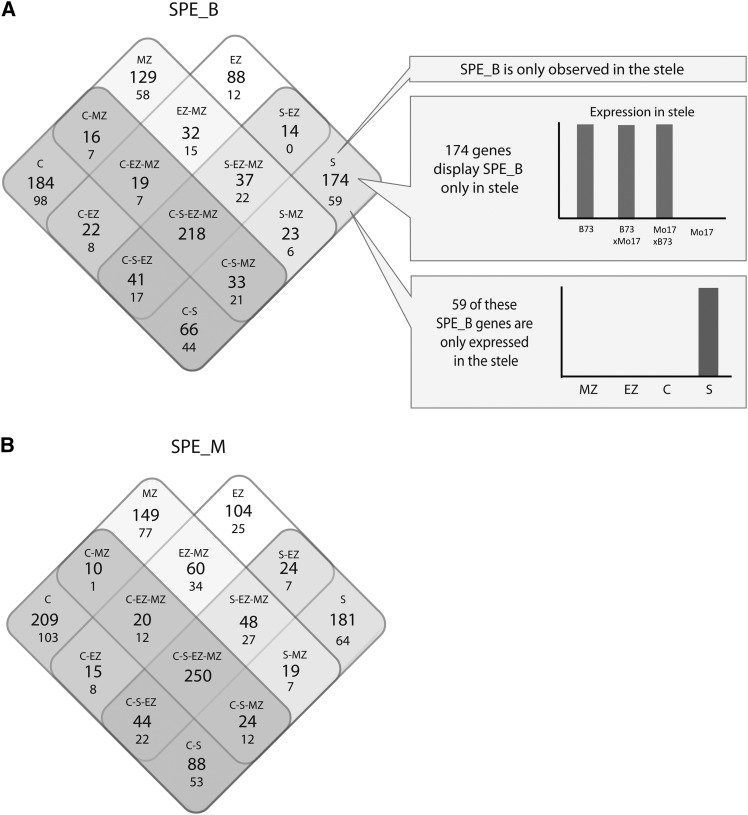
To determine their tissue-specific dynamics, SPE patterns were summarized in four Venn diagrams (Figure 3). This analysis revealed that 20% (218/1096) of the SPE_B and 20% (250/1245) of the SPE_M genes display this pattern constitutively in all four analyzed tissues (Figures 3A and 3B). By contrast, 80% (1877/2341) of all SPE patterns were tissue specific, i.e., not observed in all four tissues. Among these, 67% (1225/1877) were detected in only one of the four analyzed tissues (Figures 3A and 3B, groups C, MZ, EZ, and S). For instance, 184 SPE_B and 210 SPE_M genes showed this particular expression pattern only in the cortex of young primary roots, while in the elongation zone, meristematic zone and stele these genes did not display SPE (Figures 3A and 3B, left corners). To determine if tissue-specific SPE is the result of exclusive expression of these genes in these tissues, the number of SPE genes that are only expressed in the tissues where they display SPE was determined (Figure 3, smaller numbers in the Venn diagrams). Together with the 468 SPE genes that displayed SPE in all four tissues, a total of 55% (1294/2341) of all SPE genes displayed SPE in all tissues in which they were expressed. However, the remaining 45% (1051/2341) of SPE genes displayed non-SPE expression patterns in some or all of the remaining tissues. In summary, while 55% of all SPE genes exhibited SPE in all tissues in which they were expressed, 45% of all SPE genes displayed complex tissue-specific regulation of SPE and non-SPE patterns.
Figure 3.
Summary of SPE Genes Conserved between and Unique for Different Tissues in Four-Way Venn Diagrams.
(A) SPE_B genes expressed only in B73 and the two reciprocal hybrids.
(B) SPE_M genes expressed only in Mo17 and the two reciprocal hybrids. The larger numbers in each square field indicate all genes that display the SPE pattern, and the smaller numbers below indicate how many of the SPE genes are expressed only in the tissue(s) in which they display SPE.
Allele-Specific Contribution to SPE
To better understand the parental contribution to the SPE patterns, the allele-specific expression of SPE genes was determined in hybrids. The aim of this analysis was to determine if parental alleles that are not expressed in the inbred line are “reactivated” in the hybrid. SPE_B genes, which are known to be PAVs (Supplemental Table 3), were excluded from this analysis. Hence, for the remaining genes, both parental alleles are present in the hybrid genome. For this analysis >4 million SNPs previously called between B73 and Mo17 were used (Paschold et al., 2012). These SNPs allowed distinction between the expressed parental alleles of 2049 SPE genes. Allele-specific read mapping resulted in up to 197 tissue-specific SPE genes in which the “silent” allele was reactivated in the hybrid (Table 1). In a more conservative approach, an arbitrary threshold of 10 allele-specific reads per gene across the four replicates was introduced. Applying this cutoff reduced the number of SPE genes that expressed both alleles in the hybrids to between 0 and 4 (Table 1). This result indicates that most “silent” alleles remain inactive in hybrids if putative mapping artifacts are neglected.
Table 1. Biallelic Expression of SPE Genes in the Reciprocal Hybrids B73xMo17 and Mo17xB73.
| Group of SPE Genesa | No. of SPE Genes | No. of SPE Genes with SNPs | No. of SPE Genes with SNPs, PAVs Excludedb | Subset of SPE Genes Expressing Both Parental Alleles |
|||
|---|---|---|---|---|---|---|---|
| B73xMo17 | Mo17xB73 | ||||||
| ≥1 Read | ≥10 Reads | ≥1 Read | ≥10 Reads | ||||
| SPE_B-MZ | 510 | 399 | 374 | 73 | 0 | 74 | 0 |
| SPE_M-MZ | 581 | 525 | – | 116 | 2 | 132 | 4 |
| SPE_B-EZ | 472 | 381 | 356 | 70 | 0 | 62 | 0 |
| SPE_M-EZ | 565 | 502 | – | 134 | 2 | 124 | 2 |
| SPE_B-C | 599 | 475 | 451 | 95 | 0 | 97 | 0 |
| SPE_M-C | 661 | 611 | – | 176 | 3 | 197 | 2 |
| SPE_B-S | 607 | 488 | 457 | 107 | 1 | 117 | 1 |
| SPE_M-S | 680 | 611 | – | 183 | 2 | 169 | 3 |
An allele was considered to be expressed with either one read or 10 reads minimum. PAVs were excluded from all SPE_B genes.
SPE_B, gene expressed in B73 and hybrids but not in Mo17; SPE_M, gene expressed in Mo17 and hybrids but not in B73. B, B73; M, Mo17; MZ, meristematic zone; EZ, elongation zone; C, cortex; S, stele.
PAVs (genes present in the B73 genome but absent from the Mo17 genome).
Evolutionary Implications of Tissue-Specific SPE Patterns
To survey the evolutionary origin of the genes that display tissue-specific SPE, their assignment to maize subgenomes 1 or 2 was determined. For each of these analyses, comparisons with all 39,656 genes of the filtered gene set and with all 27,347 genes expressed in this analysis were performed. In total, 19,365 of all 39,656 high confidence FGS genes (49%) can be assigned to one of the two maize subgenomes (www.skraelingmountain.com/datasets/maize_indexed_by_subgenome.csv), which are the result of an ancient genome duplication (Table 2). Among all expressed genes 64% (17,402/ 27,347) were assigned to one of the two subgenomes pointing to their ancient origin. The extent to which this percentage exceeds the percentage of all FGS genes that can be assigned to a subgenome (19,365/39,656 [49%]; Fisher’s exact test, P < 0.001) suggests that evolutionary older (syntenic) genes are expressed more frequently than nonsyntenic genes, which likely emerged after the last whole-genome duplication of maize. By contrast, only 18 to 25% of the SPE genes identified in the individual tissues were assigned to a subgenome, which is 2.6 to 3.6 times less than expected (Table 2). This pattern was statistically significant (Fisher’s exact test, P < 0.001) for the SPE genes of each of the four tissues. Among the SPE genes that were assigned to subgenomes, the ratio of genes assigned to subgenome 1 (62%) and 2 (38%) was nearly identical to the ratio among all filtered gene set genes assigned to a subgenome and to the ratio among all expressed genes (Table 2).
Table 2. SPE Genes of the Individual Root Tissues and Their Assignment to Maize Subgenomes.
| No. of Genes | No. of Genes Assigned to Subgenomes | % | Subgenome 1 | % | Subgenome 2 | % | |
|---|---|---|---|---|---|---|---|
| FGS genesa | 39,656 | 19,365 | 49 | 12,190 | 63 | 7,175 | 37 |
| Expressed genes PRT | 27,347 | 17,402 | 64 | 10,962 | 63 | 6,440 | 37 |
| Expressed genes WPR | 28,113 | 17,996 | 64 | 11,343 | 63 | 6,653 | 37 |
| SPE class | |||||||
| SPE total | 2,341 | 696*** | 30 | 429 | 62 | 267 | 38 |
| SPE_C | 1,260 | 310*** | 25 | 197 | 64 | 113 | 36 |
| SPE_S | 1,287 | 294*** | 23 | 184 | 63 | 110 | 37 |
| SPE_EZ | 1,037 | 226*** | 22 | 156 | 68 | 71 | 31 |
| SPE_MZ | 1,091 | 199*** | 18 | 113 | 57 | 86 | 43 |
| SPE WPR | 946 | 131*** | 14 | 87 | 66 | 44 | 34 |
In addition to our tissue-specific analysis (PRT), a published WPR data set (Paschold et al., 2012) was reanalyzed using the same method as in this study. Fisher’s exact test was used to determine if more or less SPE genes than expected are assigned to subgenomes (***P < 0.001).
Filtered gene set of maize B73.
In a previous analysis, 1124 genes that displayed a SPE pattern were identified from whole primary roots (WPRs) of the same genotypes (Paschold et al., 2012). Reanalysis of this data set using the improved algorithm introduced in this study (see Methods) resulted in 28,113 expressed genes and 946 SPE genes, 17,996 (64%) and 131 (14%) of which were assigned to subgenomes, respectively (Table 2). Again, significantly fewer SPE-WPR genes than expected were assigned to the subgenomes, indicating that most SPE-WPR genes are not conserved at syntenic loci in other species (Table 2; Fisher’s exact test, P < 0.001).
Functional Classification of SPE Genes
To identify overrepresented biological functions among the four groups of tissue-specific SPE genes a Gene Ontology (GO) analysis was conducted. Only the biological processes “death” and its subcategories “cell death,” “programmed cell death,” and “apoptosis” were overrepresented in each of the four tissue-specific SPE data sets. Among the four tissue-specific sets of SPE genes, between 11 and 18 genes were assigned to these GO terms. These genes overlap between tissues, which results in a total of 29 genes assigned to the GO term apoptosis and the parent terms (Supplemental Table 3). Eighteen of the 29 genes display SPE_B, while 11/29 genes show SPE_M expression patterns, indicating that both B73 and Mo17 are deficient in the expression of cell death-related genes. The majority (24/29) of these genes cannot be assigned to subgenomes, suggesting that they emerged after the last whole-genome duplication. Many of them are suggested to encode enzymes with nucleoside triphosphatase activity, which plays a role in the degradation of nucleic acids (Supplemental Table 3).
DISCUSSION
The high degree of intraspecific genome diversity is one remarkable feature of maize that has been studied in great detail (Springer et al., 2009; Swanson-Wagner et al., 2010; Chia et al., 2012; Jiao et al., 2012; Marroni et al., 2014). Recent studies investigated how genomic diversity translates to intraspecific transcriptome diversity (Stupar and Springer, 2006; Hansey et al., 2012; Hirsch et al., 2014), which is fostered by the advent of high-throughput sequencing technologies. Using RNA-seq as one of these approaches, this study surveyed four primary root tissues of maize for gene activity in the inbred lines B73 and Mo17 and their reciprocal hybrids.
In general, between 94 and 95% of all genes active in the four tissues were expressed in all four genotypes (Figure 2). This number is similar to a recently published data set of whole primary roots of the same developmental stage in which 97% of all active genes were expressed in each of the four genotypes (Paschold et al., 2012). Remarkably, as in whole roots, the major proportion of the genes not expressed in every genotype displayed SPE, indicating that a gene is only expressed in one parental inbred line but is expressed in both reciprocal hybrids (Paschold et al., 2012). SPE represents a special instance of complementation on the level of gene expression. On the transcriptomic level, the concept of complementation implies that the combination of two distinct genomes leads to the activity of many genes in hybrids that are only active in one parental inbred line. Support for this hypothesis has been reported for whole roots where the hybrid with the lower number of active genes expressed 352 more genes than the inbred line with higher number of active genes (Paschold et al., 2012). This study went one step ahead and increased the resolution to the level of individual root tissues. The bidirectional comparison of four genotypes and four primary root tissues revealed not only hundreds of tissue-specifically expressed genes but also hundreds of SPE genes in each tissue. Similar to what has been reported in whole roots (Paschold et al., 2012) both hybrids expressed more genes in each individual tissue than either parent. Comparing the hybrid showing fewer expressed genes with the parental line displaying more expressed genes this increase ranged between 444 (meristematic zone) and 558 (stele) additional active genes.
To allow for comparison of the SPE genes of this tissue-specific data set with the results of the previously published whole root data set, we reanalyzed the whole root data (Paschold et al., 2012) with the improved SPE annotation algorithm used in this study. This approach reduced the number of whole root SPE genes from 1124 to 946 and increased the difference to the 2341 genes displaying SPE in at least one root tissue. Because both data sets were generated by the same laboratories, following the same experimental design, data analysis pipelines, and statistical procedures, the discrepancy between whole roots and individual tissues is likely the result of biological differences, not of technical artifacts or variations. Among all tissue-specific SPE genes, 55% display SPE in all tissues in which they are expressed while 45% displayed SPE in at least one tissue and general expression (i.e., the gene is expressed in each of the four genotypes) in at least one other tissue. SPE genes with such dynamic tissue-specific expression patterns are unlikely to be identified as SPE genes from complete organs because the SPE patterns are masked by the general expression of these genes in other tissues. This notion is supported by the observation that between 70% (SPE_B) and 52% (SPE_M) of all whole-root SPE patterns overlap with tissue-specific SPE patterns that are exclusively expressed in the tissues where they display SPE. This indicates that a large portion of the SPE genes identified from whole roots lack tissue-dependent gene expression dynamics. Alternatively, some of the SPE genes identified from whole roots might be expressed in each tissue, but the expression level of one particular tissue exceeds that of the others. Hence, the plasticity of SPE patterns is conditioned by the tissue-specific regulation of gene expression, indicating that a silenced allele of a SPE gene in an inbred line is not per se silenced in all tissues. This should not be confused with general tissue specificity of gene expression (Brady et al., 2007) as it refers to activity/ inactivity of genes in particular tissues. SPE, on the other hand, refers to genotype-specific gene activity and different alleles of a gene in two inbred lines.
Genotype-specific activity was detected for hundreds of expressed genes and most of these differences were complemented in the reciprocal hybrids. Only a minor proportion of these transcript variations were caused by differences in the genomic sequence (i.e., by genomic PAVs). This suggests that most of the genes are physically present in the genome but that they are inactivated. This inactivity can be caused by changes in the genomic sequence of the considered gene (cis-modification) or by alterations of proteins such as transcription factors regulating the activity of these genes (trans-modification) (Wittkopp et al., 2004). Most of the inbred line-specific differences in gene activity are complemented in the hybrids and the analysis of allele-specific gene expression levels revealed, by applying a conservative cutoff threshold, only a minor degree of reactivation of inactive alleles in the hybrids. This implies predominant cis-regulation of the SPE genes and genomic differences leading to transcriptional variation between inbred lines. Omission of any cutoff threshold resulted in biallelic expression of dozens of SPE genes in the hybrids. This observation needs to be treated with caution because the majority of these genes express the second allele at levels below 10 reads per transcript. When mapping such small read numbers to SNP positions, sequencing errors are more likely to result in mapping errors ultimately leading to false positives (Macmanes and Eisen, 2013). To account for this uncertainty, we set an arbitrary threshold cutoff of 10 reads
Phylogenetic analyses of the SPE genes identified in different root tissues were conducted to elucidate their evolutionary origin. The maize genome was duplicated several times during evolution including a whole-genome duplication (autopolyploidization) ∼5 to 12 million years ago (Blanc and Wolfe, 2004; Swigonová et al., 2004), which separated maize from its close relative, Sorghum bicolor (Paterson et al., 2009). In modern maize, the result of this most recent whole-genome duplication can be surveyed by comparing the duplicated maize and the unduplicated Sorghum genomes. Such comparisons enable the identification of recognizable homoeologs and the assignment of a considerable proportion of all current maize genes to one of the two subgenomes that resulted from this genome duplication (Schnable et al., 2011). To determine if the phylogenetic origin of a gene conditioned its contribution to intraspecific transcriptome diversity, all genes that displayed SPE patterns were analyzed for their assignment to subgenomes. In total, 49% (19,365) of all maize genes were assigned to subgenomes 1 or 2 (Schnable et al., 2011). The remaining 51% likely emerged more recently, e.g., by modular rearrangement of protein encoding domains (Kersting et al., 2012), by the transposition of existing genes (Freeling et al., 2008), or by exon shuffling from helitrons and other transposons leading to fusion genes (Barbaglia et al., 2012). Among the 27,347 genes expressed in this study, 17,402 (64%) genes were assigned to one of the two subgenomes. This implies that in general ancient genes are more frequently expressed than nonsyntenic ones and is consistent with the observation that this same population of old genes is highly enriched in genes with visible mutant phenotypes (Schnable and Freeling, 2011). Since these genes have survived more than 5 million years of natural selection they likely have crucial functions in maize development. While ancient genes are overrepresented among all expressed genes they are significantly underrepresented among the SPE genes. Only between 18 and 25% of all SPE genes in the four tissues can be assigned to subgenomes 1 or 2. The number of nonsyntenic genes (i.e., genes not assigned to any subgenome) might be inflated by false positives caused by ancestral genes present in one or both maize subgenomes but lost from the genome of Sorghum, by genes apparently present at nonsyntenic locations only as a result of assembly errors in either maize or Sorghum, or by gene prediction errors. However, these false positives, if any, are expected to be equally common among all expressed genes including both SPE and non-SPE genes, so any bias introduced would be toward proportions of nonsyntenic genes among these two categories. In fact, we observed the opposite pattern. These results imply that a disproportionate number of SPE genes emerged after the separation of maize from Sorghum and the subsequent duplication of the maize genome.
Recently emerged genes might be less likely to play essential roles during the early stages of seedling development. Maize was domesticated from its ancestor teosinte only ∼10,000 years ago (Doebley et al., 2006), resulting in the so-called landraces, which in turn have been improved during recent decades to generate elite inbred lines. However, SPE genes are likely unrelated to domestication and improvement as they were neither under nor overrepresented (data not shown) among genes found to be under selection during these processes (Hufford et al., 2012).
The observation that F1-hybrids complement gene expression patterns of contrasting parental inbred lines and express more genes could have implications for the manifestation of heterosis, which describes the increased vigor of hybrid plants compared with the average of their parental inbred lines (Shull, 1908) and is of enormous economic relevance in maize breeding (Duvick, 2005). There is consensus that heterosis requires genetic variation between the parents leading to highly heterozygotic hybrids and that heterosis is conditioned by the action of many loci (reviewed in Schnable and Springer, 2013).
It has been suggested that genotypic differences, such as polymorphisms, the number and distribution of repetitive sequences, gene copy number variation, or PAV contribute to heterosis (Zhang et al., 2008; Lai et al., 2010; Swanson-Wagner et al., 2010). Moreover, it has been demonstrated that despite the consensus that all organs have the same complement of the nuclear genome, the degree of heterosis largely varies for different tissues or phenotypic traits (Flint-Garcia et al., 2009). This observation suggests that heterosis is associated with tissue-specific gene activity, which in turn is conditioned by genetic variation. Tissue-specific SPE patterns as they are reported from this study support this hypothesis; however, it is likely that several other processes contribute to heterosis (Schnable and Springer, 2013).
In line with this concept, one could hypothesize that the ability to express ∼2% more genes than their parental inbred lines provides a phenotypic advantage to hybrid plants. Their evolutionary young age argues against SPE genes encoding essential proteins and rather suggests that these genes encode proteins having redundant functions thus making the respective biological processes more robust, e.g., in stress situations.
The finding that apoptosis-related genes, which are involved in several developmental processes in roots (Fukuda, 1996; Buckner et al., 1998; Drew et al., 2000), are overrepresented among the SPE genes supports this hypothesis. Hybrids might benefit from the complementation of these deficiencies, e.g., by improved regulation of programmed cell death.
In summary, SPE is observed for hundreds of genes in very dynamic tissue-specific patterns. Many genes display SPE only in certain tissues while they show general expression in other tissues, which underpins the subtle regulation of these expression patterns and tightly controlled transcriptomic plasticity in root tissues. Finally, most of the SPE genes are not conserved at syntenic loci in the genomes of other grasses such as Sorghum, suggesting that in the maize genome, the average SPE gene is evolutionarily younger than the average expressed gene. These findings imply a role for SPE genes in the manifestation of heterosis in maize, which needs to be further analyzed in the future.
METHODS
Plant Material and Growth Conditions
Seeds of the maize (Zea mays) inbred lines B73 and Mo17 and the two reciprocal hybrids B73xMo17 and Mo17xB73 were germinated in paper rolls in distilled water as previously described (Hoecker et al., 2006). For each genotype, 18 paper rolls each containing 10 kernels were prepared. All material was germinated in a single 10-liter bucket in which the 72 paper rolls were randomly arranged. Primary roots of 2 to 4 cm length were harvested 4 d after germination. Even at this very early developmental stage, many hybrid primary roots were longer than those of their parental inbred lines. To avoid differential gene expression patterns based simply on root length, pools of roots having the same lengths (2 to 4 cm) were selected for all genotypes and replicates. From each root, four different tissues were sampled (Figure 1). First, 3 mm of the root tip was collected, which corresponded to the terminal root cap and the subterminal meristematic zone (Dembinsky et al., 2007). Second, the zone adjacent to the root tip up to the part of the root where root hairs became visible was separated, which basically corresponds to the elongation zone. Finally, we separated the beginning of the root hair zone up to the coleorhiza, which is connected to the differentiation zone. In the differentiation zone, cortex and stele were mechanically separated without damaging pericycle and endodermis cells (Saleem et al., 2009). For each tissue, 14 roots were pooled for each of the four biological replicates per genotype.
RNA Isolation and Sequencing Library Construction
The pooled primary root tissues were ground under liquid nitrogen and RNA was isolated as previously described (Winz and Baldwin, 2001). RNA quality was assayed via agarose gel electrophoresis and a Bioanalyzer (Agilent Technologies). Only samples with an RNA integrity number (Schroeder et al., 2006) ≥ 7.2 were used for downstream analyses. The cDNA libraries for sequencing-by-synthesis were constructed according to the protocol of the manufacturer (Illumina). The 64 samples were loaded on two flow cells in a predefined order (Supplemental Table 1). The experiment was randomized according to a split plot design. Four contiguous lanes of a flow cell were regarded as a complete block, so there were four complete blocks (biological replicates), distributed across two flow cells with eight lanes each. The four tissues were randomized within each block (set of four lanes). A lane was considered as a main plot. For the same tissue, the four genotypes were allocated to each lane, so the genotypes correspond to subplots of the design. The different tissues for the same genotype in a block originated from the same plants. Cluster generation and sequencing (100 bp, single read) was performed as per the instructions of the manufacturer (Genome Analyzer IIx; Illumina).
Analysis of Raw Sequencing Data, Read Mapping, and Allele-Specific Read Calling
Sequencing was performed on the GAIIx genome analyzer equipped with on-instrument sequencing control software SCS Version 2.8 and real-time analysis RTA1.8.7. The genome analyzer data analysis pipeline OLB version 1.8.0 was used for base calling and run statistics on all 16 lanes. On average, a raw cluster density of 387,145 clusters per tile was achieved, yielding between ∼3.25 and ∼3.76 Gb per lane after sample post filtering (see Supplemental Figure 1 and Supplemental Table 1 for sample allocation and sequencing performance per lane).
The maize B73 reference genome (Schnable et al., 2009) was indexed using GMAP_BUILD (k-mer size of 15 and step size 3) and all high-quality reads were aligned to the whole reference genome using the short reads aligner GSNAP (http://research-pub.gene.com/gmap/, version 2012-01-11). Only reads uniquely mapping to the B73 reference genome (Schnable et al., 2009) with a maximum of two mismatches out of 36 bp and with at most 5-bp tails for every 76 bp were extracted and subsequently analyzed. In addition, redundant reads mapping at the same starting coordinate and mapping orientation (stacked reads) were removed from the set of uniquely mapping RNA-seq reads. The remaining reads of all samples were projected to the filtered gene set (http://ftp.maizesequence.org/release-5b/filtered-set/, release 5b.60) of the B73 reference genome derived from maize genome sequencing project using a Perl script. Allele-specific gene expression was analyzed with a set of 4,034,683 SNPs (identified between the B73 and Mo17 genotypes via 123SNP software, which is available for download from http://schnablelab.plantgenomics.iastate.edu/software. B73-Mo17 SNPs were called by aligning B73 RefGen_v2 sequences and Mo17 sequences retrieved from GenBank Sequence Read Archive records (accession numbers SRA009756, SRA049859, SRA048055, SRA053592, and SRA050451) and maizegdb (http://ftp.maizegdb.org/MaizeGDB/FTP/Mo17/).
The allele-specific reads were extracted from the reads uniquely projected onto the FGSv2 and associated with any B73-Mo17 SNP using a customized Perl script. Since the mapping procedure presented here did not allow for quantifying sequence reads mapping to repeats throughout the genome, only uniquely mapping genes were considered. In addition, transcript isoforms of a given gene locus could not be distinguished.
Detection of Expression Complementation and Functional Analysis
Classification of the gene-wise transcriptional activity (presence/absence) for each genotype was determined by a hierarchical Bayesian latent class model using Markov Chain Monte Carlo methods, similar to Paschold et al. (2012). We again modeled read counts via the mean-dispersion parameterization of the negative binomial distribution, using the median read count multiplied by the exon length (in kilobases) of the specific transcript as an offset value for normalization. Expression was modeled log-linearly, with overall gene and gene-specific genotype effects, which we assumed were both normally distributed. To account for the split-plot design, random normally distributed plot effects were also included in the model. Each gene was assumed to have its own unique dispersion parameter, which was treated as random from a common log-normal distribution. To account for falsely mapped reads, we included a fixed background error rate estimated from the data. The model was defined using MCMC software JAGS (Plummer, 2003) and fit in R using the R2jags library package (Su and Yajima, 2011). Due to the size of such a model under the full data, we initially trained the model on a representative subset (4000 total) of the genes, each selected at random. We ran two Markov chains at a total of 15,000 iterations each, treating the first 10,000 as a burn-in and then thinning the remaining iterations by saving every 5th iteration, resulting in posterior sample parameter traces of length 1000. We analyzed the posterior sample to verify that the model had converged by visually inspecting the mixture of the chains and using posterior checking procedures from the coda output diagnostics package (Plummer et al., 2006) in R. We then applied Monte Carlo integration under the learned posterior distributions of the hyperparameters to obtain gene-wise posterior probabilities of each possible state vector over the entire set of genes, conditional on the observed read counts. Classification of each gene was determined by the maximum posterior predictive probability of a given state vector. Separate model fits were run for each analysis. PAVs were called as previously described (Paschold et al., 2012).
The GO of specific genes and the overrepresentation of GO terms were conducted using the single enrichment analysis of the AgriGo platform (http://bioinfo.cau.edu.cn/agriGO/analysis.php).
Accession Numbers
Raw sequencing data are stored at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP029742.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of 64 Samples across 16 Lanes on Two Flow Cells for RNA-seq.
Supplemental Table 1. Summary of the Sequencing Output of the 64 Primary Root Tissue Samples.
Supplemental Table 2. Summary of the Sequencing Output after Trimming of the Reads, Mapping Them to the Maize Genomic Sequence, Removal of Redundant Reads, and Determination of Reads in the Gene Space of the Maize Genome.
Supplemental Table 3. Summary of the SPE Genes Assigned to the GO Term Apoptosis.
Supplemental Data Set 1. Summary of All Expressed Genes and Their Distribution across Tissues, Presence-Absence Variations, and Their Assignment to Subgenomes.
Supplementary Material
Acknowledgments
This article is dedicated to Guenter Feix on the occasion of his 80th birthday. We thank Nahal Ahmadinejad (INRES, Crop Bioinformatics, University of Bonn) for assistance with the allele-specific expression analyses and Jens Riexinger (Max Planck Institute for Developmental Biology, Tuebingen, Germany) for technical assistance during library production and sequencing. This work was funded by Deutsche Forschungsgemeinschaft Grants HO2249-9/1 to F.H. and PI 377/12-1 to H.-P.P.
AUTHOR CONTRIBUTIONS
A.P. carried out the experiment and tissue sampling, conducted downstream analyses, drafted the article, and designed figures. N.B.L. carried out the complementation analysis. C.M. isolated RNAs, constructed RNA-seq libraries, and participated in writing the article. J.C.S. participated in the subgenome analysis and in article writing. C.-T.Y. carried out the mapping and analysis of raw sequencing data. C.L. carried out the RNA sequencing and participated in library construction. D.N. and H.-P.P. provided the sequencing design and performed the statistical analysis. P.S.S. participated in data interpretation and helped to draft the article. F.H. conceived and coordinated the study and participated in data interpretation and drafting the article. All authors approved the final article.
Glossary
- SNP
single nucleotide polymorphism
- PAV
presence-absence variation
- SPE
single-parent expression
- WPR
whole primary root
- GO
Gene Ontology
Footnotes
Online version contains Web-only data.
References
- Barbaglia A.M., Klusman K.M., Higgins J., Shaw J.R., Hannah L.C., Lal S.K. (2012). Gene capture by Helitron transposons reshuffles the transcriptome of maize. Genetics 190: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W.B., Emrich S.J., Chen H.D., Li L., Schnable P.S. (2007). SNP discovery via 454 transcriptome sequencing. Plant J. 51: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi I.V., McMullen M.D., Sanchez-Villeda H., Schroeder S., Gardiner J., Polacco M., Soderlund C., Wing R., Fang Z., Coe E.H. (2006). Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci. 46: 12–21. [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Buckner B., Janick-Buckner D., Gray J. (1998). Cell-death mechanisms in maize. Trends Plant Sci. 3: 218–223. [Google Scholar]
- Chia J.M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Dembinsky D., et al. (2007). Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 145: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J.F., Gaut B.S., Smith B.D. (2006). The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Drew M.C., He C.J., Morgan P.W. (2000). Programmed cell death and aerenchyma formation in roots. Trends Plant Sci. 5: 123–127. [DOI] [PubMed] [Google Scholar]
- Duvick D.N. (2005). The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 86: 83–145. [Google Scholar]
- Flint-Garcia S.A., Buckler E.S., Tiffin P., Ersoz E., Springer N.M. (2009). Heterosis is prevalent for multiple traits in diverse maize germplasm. PLoS ONE 4: e7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Lyons E., Pedersen B., Alam M., Ming R., Lisch D. (2008). Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18: 1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. (1996). Xylogenesis: Initiation, progression, and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 299–325. [DOI] [PubMed] [Google Scholar]
- Hansey C.N., Vaillancourt B., Sekhon R.S., de Leon N., Kaeppler S.M., Buell C.R. (2012). Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS ONE 7: e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger, F. (2009). The maize root system: morphology, anatomy, and genetics. In Handbook of Maize: Its Biology, J.L. Bennetzen and S.C. Hake, eds (New York: Springer), pp. 145–160. [Google Scholar]
- Hoecker N., Keller B., Piepho H.P., Hochholdinger F. (2006). Manifestation of heterosis during early maize (Zea mays L.) root development. Theor. Appl. Genet. 112: 421–429. [DOI] [PubMed] [Google Scholar]
- Hufford M.B., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Evans M.L. (1995). Specialized zones of development in roots. Plant Physiol. 109: 725–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44: 812–815. [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J.C., Birchler J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting A.R., Bornberg-Bauer E., Moore A.D., Grath S. (2012). Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biol. Evol. 4: 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., et al. (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42: 1027–1030. [DOI] [PubMed] [Google Scholar]
- Laurie D.A., Bennett M.D. (1985). Nuclear-DNA content in the genera Zea and Sorghum - intergeneric, interspecific and intraspecific variation. Heredity 55: 307–313. [Google Scholar]
- Macmanes M.D., Eisen M.B. (2013). Improving transcriptome assembly through error correction of high-throughput sequence reads. PeerJ 1: e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F., Pinosio S., Morgante M. (2014). Structural variation and genome complexity: is dispensable really dispensable? Curr. Opin. Plant Biol. 18: 31–36. [DOI] [PubMed] [Google Scholar]
- Marschner, P. (2011). Mineral Nutrition of Higher Plants. (London: Academic Press). [Google Scholar]
- Olsen K.M., Wendel J.F. (2013). A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64: 47–70. [DOI] [PubMed] [Google Scholar]
- Paschold A., Jia Y., Marcon C., Lund S., Larson N.B., Yeh C.T., Ossowski S., Lanz C., Nettleton D., Schnable P.S., Hochholdinger F. (2012). Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res. 22: 2445–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Plummer, M. (2003). JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. In 3rd International Workshop on Distributed Statistical Computing, K. Hornik, F. Leisch, and A. Zeileis, eds (Vienna, Austria: Technical University Vienna). [Google Scholar]
- Plummer, M., Best, N., Cowles, K., Vines, K. (2006). Convergence diagnosis and output analysis for MCMC. R News 6: 7–11. [Google Scholar]
- Saleem M., Lamkemeyer T., Schützenmeister A., Fladerer C., Piepho H.P., Nordheim A., Hochholdinger F. (2009). Tissue specific control of the maize (Zea mays L.) embryo, cortical parenchyma, and stele proteomes by RUM1 which regulates seminal and lateral root initiation. J. Proteome Res. 8: 2285–2297. [DOI] [PubMed] [Google Scholar]
- Schnable J.C., Freeling M. (2011). Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., Springer N.M. (2013). Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64: 71–88. [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull G.F. (1908). The composition of a field of maize. American Breeders’ Association. Annual Report 4: 296–301. [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R.M., Springer N.M. (2006). Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., and Yajima, M. (2011). R2jags: A package for running JAGS from R. In R Package Version 0.02-17, http://cran.r-project.org/web/packages/R2jags/. [Google Scholar]
- Swanson-Wagner R.A., Eichten S.R., Kumari S., Tiffin P., Stein J.C., Ware D., Springer N.M. (2010). Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winz R.A., Baldwin I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-Induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125: 2189–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P.J., Haerum B.K., Clark A.G. (2004). Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.Y., He H., Chen L.B., Li L., Liang M.Z., Wang X.F., Liu X.G., He G.M., Chen R.S., Ma L.G., Deng X.W. (2008). A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol. Plant 1: 720–731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.