CAPE1, a conserved peptide elicitor derived from tomato PR-1, was induced by wounding and found to regulate immune responses against biological threats. As PR-1 is highly conserved across many organisms and the putative peptide from AtPR1 was also found to be bioactive in Arabidopsis, the results suggest that this peptide may be useful for enhancing resistance to stress in other plant species.
Abstract
Many important cell-to-cell communication events in multicellular organisms are mediated by peptides, but only a few peptides have been identified in plants. In an attempt to address the difficulties in identifying plant signaling peptides, we developed a novel peptidomics approach and used this approach to discover defense signaling peptides in plants. In addition to the canonical peptide systemin, several novel peptides were confidently identified in tomato (Solanum lycopersicum) and quantified to be induced by both wounding and methyl jasmonate (MeJA). A wounding or wounding plus MeJA-induced peptide derived from the pathogenesis-related protein 1 (PR-1) family was found to induce significant antipathogen and minor antiherbivore responses in tomato. This study highlights a role for PR-1 in immune signaling and suggests the potential application of plant endogenous peptides in efforts to defeat biological threats in crop production. As PR-1 is highly conserved across many organisms and the putative peptide from At-PR1 was also found to be bioactive in Arabidopsis thaliana, our results suggest that this peptide may be useful for enhancing resistance to stress in other plant species.
INTRODUCTION
All multicellular organisms have evolved mechanisms to perceive and respond to extracellular chemical signals. External signals include endogenous hormones and cues from the environment, pathogens, and symbiotic organisms. In animal systems, intercellular communications are mostly mediated by steroids, peptides, and other small compounds (Ryan et al., 2002). Among them, peptides are the most common mediators of cell-to-cell interactions in animals because they provide great variety in their sequences, lengths, and/or posttranslational modifications to represent different physiological responses (Boller, 2005). In contrast to peptide discovery in animals, only a few signaling peptides have been identified in plants (Farrokhi et al., 2008a; Butenko et al., 2009). The completion of the Arabidopsis thaliana genome has revealed that plants have up to 10 times as many predicted peptide transporters (Initiative, 2000) and receptors (Shiu and Bleecker, 2003) as animals. Therefore, it is expected that most of the endogenous plant signaling peptides that play prominent roles in cell-to-cell communication are still undiscovered. Among the currently identified peptides in plants, only relatively few have been found to function in defense signaling. This may be because defense signaling peptides are mostly derived from the selective action of proteases on larger precursor proteins, are expressed at low levels, and are highly dynamic. The identification of defense signaling peptides in plants has the potential not only to advance plant stress biology, but also to aid in the development of alternative ways to improve stress tolerance or resistance for better crop productivity and minimization of the use of agrochemicals (Pearce et al., 1991, 2001a; Huffaker et al., 2006). So far, defense signaling peptides have been discovered mainly by bioassay-guided screening. However, this approach requires large amounts of plant tissue and complicated purification of bioactive peptides (Pearce et al., 1991, 2001a). Furthermore, the detection of the signaling peptides can be limited by the selection of bioassay. One type of stress may produce several defense and physiological responses, which may be regulated by different signaling peptides. Therefore, the use of a single bioassay may not be sufficient to identify all the defense signaling peptides induced by a stress.
Peptidomics approaches can provide comprehensive and unbiased detection of transient amounts of signaling peptide and even identify their modification status (Fricker et al., 2006). In animal tissues, peptidomics approaches have been used to elucidate novel bioactive peptides and their functions (Svensson et al., 2003; Che et al., 2005; Sasaki et al., 2009; Tinoco and Saghatelian, 2011). However, very few successful cases of discovery of plant signaling peptides have been reported. This may be due to the high complexity and low abundance of plant signaling peptides. More sensitive and reliable peptidomics approaches are required (Murphy et al., 2012). Recent developments in mass spectrometry (MS) have significantly improved the accuracy and sensitivity of peptide detection. MS is routinely applied to proteomics analysis that analyzes the total peptides generated from the digestion of complex protein mixtures using tandem mass spectrometry (MS/MS) (Mallick and Kuster, 2010). A reliable protein identification result can be obtained by matching peptide MS/MS spectra to theoretical fragment spectra (Aebersold and Mann, 2003). Unfortunately, proteomics approaches cannot be directly applied to the detection of signaling peptides because it is difficult to create an endogenous peptide library for MS/MS spectra matching, as how plants utilize proteases to produce peptides from proproteins is still unclear (Yamaguchi and Huffaker, 2011). Identification of endogenous peptides using a protein database and consideration of all the possible peptide cleavages during MS/MS spectra matching would generate large quantities of false positive hits (Kapp et al., 2005). Therefore, more stringent matching criteria are required to accept significant peptide hits to reduce the detection sensitivity (Ding et al., 2008).
In this study, an MS-based peptidomics approach using a hypothetical peptide database combining a target-decoy search strategy and differential database match scoring was developed to discover defense signaling peptides. This platform was demonstrated by the identification of defense peptides induced by wounding plus methyl jasmonate (MeJA) treatment in tomato (Solanum lycopersicum). It is already known that tomato wounding can induce an antiherbivore response, which is regulated by the peptide hormone systemin, and the small molecule hormone jasmonic acid (JA) and its methyl ester, MeJA (Pearce et al., 1991; Orozco-Cardenas et al., 2001). Antipathogen responses are activated in plants through signals known as damage-associated molecular patterns (DAMPs) (Huffaker et al., 2006; Boller and Felix, 2009a; Malinovsky et al., 2014). DAMPs are endogenous molecules produced by the wounding or damaging of the tissue (which can occur through pathogen or herbivore attack). Systemin was the first identified signaling peptide and also the first confirmed peptide elicitor of DAMPs in plants. It is expected that several signaling peptides are involved in combating herbivore and pathogen attack (Cheong et al., 2002; Francia et al., 2007; Chassot et al., 2008), but the details of the regulation of antiherbivore and antipathogen responses by peptides during wounding stress still await elucidation. Several DAMP peptides have been discovered in other plant species and suggested to be bioactive in tomato (Boller and Felix, 2009b; Campos et al., 2014); these include HypSys (Pearce et al., 2001a; Narvaez-Vasquez et al., 2007), RALF (Pearce et al., 2001b), and Pep1 (Huffaker et al., 2006; Trivilin et al., 2014). Pep1 was clearly identified to be pathogen related in Arabidopsis, and its putative precursor in tomato was recently found to involve in the antipathogen response (Trivilin et al., 2014). However, its endogenous level in tomato has not yet been proved to be induced by tissue damage or MeJA, a potent inducer of systemic wound signaling and response in tomato (Scheer and Ryan, 1999). To our knowledge, no study to date has quantitatively profiled the global change in cellular peptide expression in plants before and/or after the induction of stress responses. Using this platform, several peptides including systemin were identified and quantified to be wounding plus MeJA induced. One of the peptides induced by wounding only or wounding plus MeJA was found to activate immune signals for defense against biological threats. The setup of this platform and functional studies of this peptide are described here.
RESULTS
Platform for the Discovery of Defense-Related Peptides
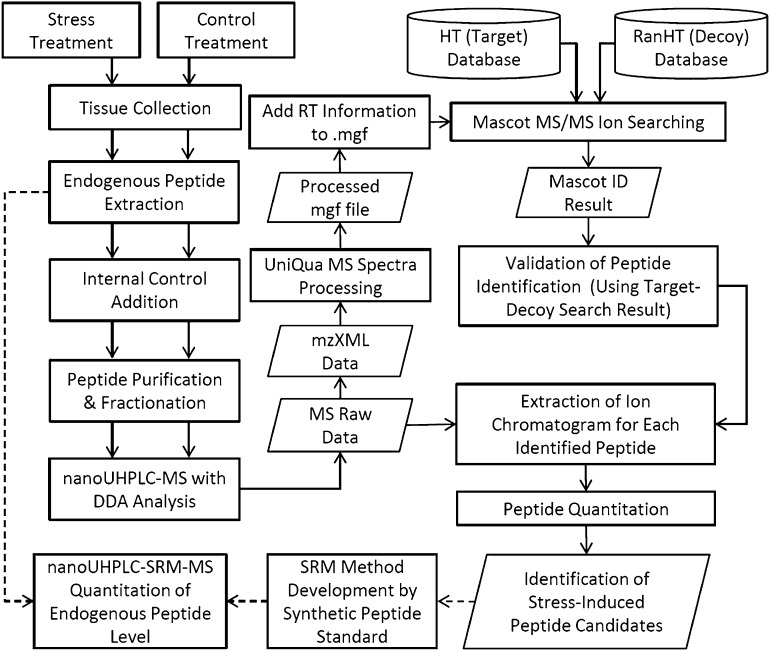
A MS-based platform using a hypothetical database was developed to discover peptides involved in defense that are induced by stress, as outlined in Figure 1. Global endogenous peptides were profiled and compared before and after stress induction. The endogenous peptide mixtures from unstressed and stressed plants were extracted and fractionated. Each of the fractions was analyzed by nanoflow ultrahigh performance liquid chromatography mass spectrometry (nanoUHPLC-MS) using the data-dependent acquisition (DDA) mode. The acquired MS/MS spectra were processed by UniQua (Chang et al., 2013) and searched against a peptide database using Mascot MS/MS ion search. To identify the endogenous peptides with higher sensitivity, it is important to reduce the false positive rate in MS/MS ion searching. This is because there is no specific prediction method for the generation of endogenous peptides from the precursor proteins by plants. Therefore, the identification of endogenous peptide signals using MS/MS ion search requires considering all possible cleavage events. Although the protein database of tomato is not large, considering that all subsequences can be degraded from the proteins without any specificity would generate several orders of magnitude more candidate sequences to match the MS/MS spectra, in comparison with identification of peptides with specific protease cleavage sites (Zhou et al., 1999; Svensson et al., 2003; Farrokhi et al., 2008a). The large number of candidate sequences for peptide MS/MS ion matching would generate more false positive hits and increase the matching score to accept confident peptide hits. Therefore, in this study, a hypothetical peptide database (HT database) using partial protein sequences was used as the target database for the peptide identification. To date, most defense-related peptides have been identified to be derived from the C-terminal end of proproteins (Farrokhi et al., 2008b). Therefore, the HT database was composed of 50 amino acid sequences from the C-terminal end of protein sequences in the protein database. To evaluate the criteria for positive peptide identification, the target-decoy database search strategy was applied (Elias and Gygi, 2007). The randomized hypothetical peptide database (RanHT database) was used as the decoy database.
Figure 1.
Hypothetical Database-Assisted Peptidomics Platform for the Discovery of Stress-Induced Peptides.
Arrows show experimental priority for endogenous peptide extraction and analysis, and dashed arrows show the SRM method for target peptide quantitation.
The changes in abundance level of each identified peptide before and after induction of stress were quantified by the extracted ion chromatogram (XIC) peak area of the peptide precursor ion. If one peptide was detected in several gel filtration fractions, all of the XIC peak areas were summarized and compared. To normalize the sample recovery during the sample preparation and column trapping efficiency, the relative peptide abundances in stressed and unstressed samples were normalized by the signal ratio of doped internal control peptides from stressed and unstressed samples. In addition, when peptides are fractionated before performing nanoUHPLC-MS, quantitation accuracy can be impaired by the retention reproducibility during peptide fractionation. To confirm that the observed peptide was responding to stress, a nanoUHPLC-MS operated in selected reaction monitoring (SRM) was used to quantify the peptide of interest in a more sensitive and reliable way without prefractionation of the extracted peptides.
Identification of Wounding- plus MeJA-Induced Peptides in Tomato Leaves
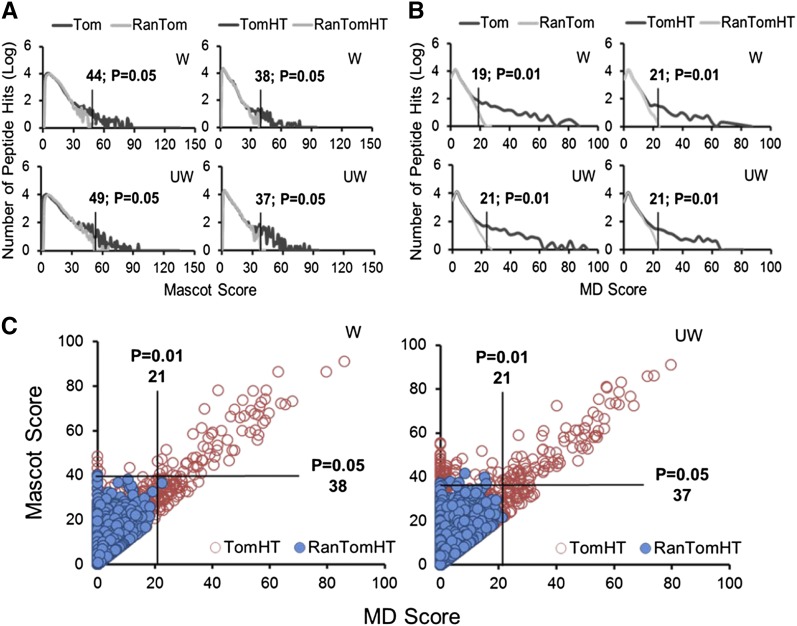
To discover the defense peptides in tomato that are induced by wounding, the endogenous peptides extracted from tomato plants with or without wounding plus MeJA treatment were profiled and compared. To assign significant peptide hits, the distribution of Mascot and Mascot Delta (MD) scores using randomized tomato protein (RanTom) and hypothetical (RanTomHT) databases were used as the model for the null hypothesis. As shown in Figure 2A, the Mascot scores for P value < 0.05 using RanTom and RanTomHT databases were >49 and >38, respectively. As shown in Figure 2B, the MD score was >21 with P value < 0.01 using RanTom and RanTomHT databases. To obtain confident peptide identification results, in this study, Mascot score > 38 and MD score > 21 were considered as true positive hits using the TomHT database. The Mascot and MD score distribution for all of the identified peptides in unwounded and wounding plus MeJA-treated tomato is illustrated in Figure 2C. In this analysis, a total of 46 unique peptides derived from 25 proproteins were identified and quantified in tomato leaves using the TomHT database (Supplemental Table 1). However, only 25 unique peptides were identified with the use of the Tom database because the Mascot score for P < 0.05 and MD score for P < 0.01 were >49 and >21, respectively.
Figure 2.
Score Distribution of the Peptides Identified in Unwounded and Wounded plus MeJA-Treated Tomato.
(A) and (B) The Mascot (A) and MD (B) score distribution using a target-decoy search based on tomato (Tom) and randomized tomato (RanTom) protein database or tomato hypothetical (TomHT) and randomized tomato hypothetical (RanTomHT) peptide database, respectively. W, wounded; UW, unwounded.
(C) Mascot versus MD score distribution using a target-decoy search based on TomHT and RanTomHT database, respectively. The red circles and blue dots indicated the peptide hits from TomHT and RanTomHT, respectively.
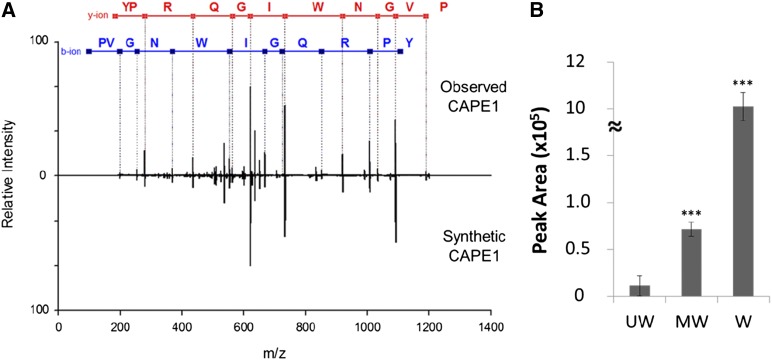
Quantitative analysis revealed 14 novel peptides and a known peptide (systemin) with expression levels more than 2-fold upregulated after wounding plus MeJA treatment (Table 1). Systemin showed no significant expression in the unwounded plant but was highly expressed after wounding plus MeJA treatment. One novel peptide derived from PATHOGENESIS-RELATED PROTEIN1b (PR-1b) showed a similar expression response to systemin. Since PR-1b is classified as a member of the cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins (CAP) superfamily (Gibbs et al., 2008), this peptide was designated as CAP-derived peptide 1 (CAPE1). In CAPE1 identification, with the exception of the y1 and b1 ions, most of the y and b fragment ions were matched to the theoretical fragments of the CAPE1 sequence, as shown in Figure 3A. Using this approach, the tissue quantity used for global peptide identification was <150 g. To confirm the matched sequences, synthetic CAPE1 was analyzed by MS/MS and the resulting spectrum was totally matched to the endogenous CAPE1 (Figure 3A). Without peptide prefractionation, total peptides extracted from the unwounded, mechanically wounded, and wounded plus MeJA-treated tomato plants were directly analyzed by nanoUHPLC-SRM-MS targeted on the specific CAPE1 collisional induced dissociation reaction (Supplemental Figure 1). The quantitation result showed that CAPE1 was expressed in low level in unwounded plants but significantly induced after wounding or wounding plus MeJA treatments (Figure 3B).
Table 1. Endogenous Peptides Induced by Wounding plus MeJA Treatment.
| Proprotein Information |
Peptide Identification |
Peptide Ratio (W/UW)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession Number | Description | Peptide Sequence | MD Score | MASCOT Score | Observed m/z | Z | Observed MW | Theor. MW | Mass Error | Scoring Ions | RT (min) | Ratio | Norm. Ratio |
| Internal Standard | β-Casein | F.LLYQEPVLGPVR.G | 36.84∼61.66 | 51.47∼78.69 | 692.42 | 2 | 1382.83 | 1382.79 | ±0.08 | 6∼10 | 38.75∼40.20 | 1.63 | 1.00 |
| Solyc05g051750.2.1 | Prosystemin | L.AVQSKPPSKRDPPKMQTD.N (systemin) | 21.35 | 38.18 | 1005.54 | 2 | 2009.07 | 2009.04 | 0.04 | 18 | 21.52 | 9999b | 9999b |
| Solyc05g025600.1.1 | Chloroplast photosystem II subunit X | G.VSNFDPVKR.T | 21.71 | 40.69 | 531.28 | 2 | 1060.55 | 1060.57 | −0.01 | 5 | 26.16 | 9999b | 9999b |
| Solyc00g174340.1.1 | Pathogenesis-related protein 1b | D.PVGNWIGQRPY.- (CAPE1) | 41.91 | 53.37 | 643.86 | 2 | 1285.7 | 1285.66 | 0.05 | 8 | 36.84 | 9999b | 9999b |
| Solyc07g044860.2.1 | Oxygen-evolving enhancer protein 2, chloroplastic | K.KFVENAATSFSI.A | 39.23 | 49.38 | 657.35 | 2 | 1312.69 | 1312.67 | 0.03 | 8 | 40.20 | 111.62 | 68.48 |
| Solyc01g007350.2.1 | Photosystem I reaction center subunit VIII | M.ASLFLHVQKNK.I | 38.52 | 56.85 | 642.88 | 2 | 1283.75 | 1283.73 | 0.01 | 7 | 29.04 | 19.08 | 11.71 |
| Solyc12g035280.1.1 | Photosystem II CP47 chlorophyll apoprotein | K.LGDPTTKRQAA.- | 26.31 | 45.66 | 579.33 | 2 | 1156.65 | 1156.62 | 0.03 | 5 | 21.47 | 7.70 | 4.72 |
| Solyc02g038690.1.1 | Histone H2B | E.IQTAVRLVLPGE.L | 29.20 | 38.23 | 648.41 | 2 | 1294.8 | 1294.76 | 0.04 | 6 | 41.81 | 5.35 | 3.28 |
| Solyc11g071640.1.1 | β-d-Glucosidase | D.SHNDPLFHFGFGLTTKPVK.A | 45.86 | 46.63 | 714.73 | 3 | 2141.16 | 2141.11 | 0.05 | 14 | 41.75 | 4.43 | 2.72 |
| Solyc01g104170.2.1 | Ankyrin repeat domain-containing protein 2 | Q.DVLKLLEKDAFL.- | 21.47∼32.00 | 38.68∼42.18 | 702.42 | 2 | 1402.82 | 1402.81 | 0.01 | 8 | 50.47 | 4.04 | 2.48 |
| Solyc06g063370.2.1 | Chlorophyll a/b binding protein 1A, chloroplastic | L.LTVIGGASERVPT.L | 22.63∼62.90 | 41.92∼86.53 | 650.39 | 2 | 1298.77 | 1298.72 | ±0.05 | 6∼10 | 33.38∼35.15 | 3.73 | 2.29 |
| Solyc01g087520.2.1 | Ferredoxin-thioredoxin reductase variable chain | L.EGRSTPVKFSAHLKED.E | 27.28 | 39.74 | 600.97 | 3 | 1799.9 | 1799.92 | −0.02 | 8 | 26.58 | 29.71 | 18.23 |
| L.EGRSTPVKFSAHLKEDE.F | 24.72∼59.75 | 37.34∼67.73 | 644.01 | 3 | 1929.01 | 1928.96 | ±0.05 | 8∼9 | 26.67∼27.01 | 6.23 | 3.82 | ||
| Solyc09g063130.2.1 | Photosystem I reaction center subunit IV A | R.FNKVNYANVSTNNYALDEVEEVK.- | 31.18∼85.88 | 40.12∼91.04 | 887.46 | 3 | 2659.37 | 2659.28 | ±0.09 | 8∼20 | 38.14∼38.75 | 20.97 | 12.87 |
| D.PKTRYPVVVR.F | 20.95∼27.72 | 37.64∼46.28 | 607.86 | 2 | 1213.71 | 1213.73 | −0.02 | 6 | 25.38 | 9.90 | 6.07 | ||
| F.NKVNYANVSTNNYALDEVE.E | 56.12∼79.63 | 57.96∼86.34 | 1079.02 | 2 | 2156.03 | 2156 | ±0.02 | 12∼14 | 36.34∼36.41 | 5.70 | 3.50 | ||
Figure 3.
Validation of the Endogenous CAPE1 Identity and the Expression Level Triggered by Wounding and Wounding plus MeJA.
(A) The MS/MS spectra matching the endogenous and synthetic CAPE1 peptide. Red fonts labeled y ions and blue fonts labeled b ions, respectively.
(B) The direct analysis of endogenous CAPE1 expression level in unwounded (UW), mechanically wounded (MW), and wounded plus MeJA (W) treated tomato using LC-SRM-MS represented the means and sd of three biological samples. The statistically significant differences between the unwounded, mechanically wounded, and wounded plus MeJA samples are indicated with three asterisks (P < 0.001) based on Student’s t test.
Bioactivity of CAPE1
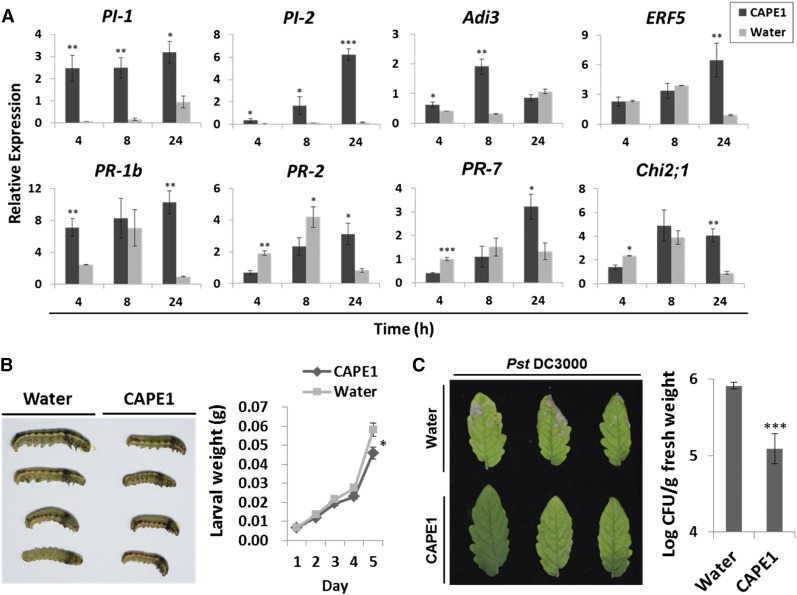
CAPE1 treatment induces H2O2 formation in tomato leaves as detected by 3,3-diaminobenzidine (DAB) staining (Supplemental Figure 2). The profiles of induced genes obtained by microarray analysis (Supplemental Data Set 1) suggest that CAPE1 elevates the expression of several genes known to be involved in the antiherbivore and antipathogen defense response. CAPE1 mainly induced genes involved in the stress response, defense response, innate immune response, bacterial defense, and systemic acquired resistance (Supplemental Figure 3). Quantitative RT-PCR (qRT-PCR) analysis further confirmed that the antiherbivore genes PROTEINASE INHIBITOR1 (PI-1) and PI-2 and pathogen-related genes PR-1b (CAPE1 precursor gene), BETA-1,3-GLUCANASE (PR-2), CYS PROTEASE (PR-7), CLASS II CHITINASE (Chi2;1), ETHYLENE RESPONSE FACTOR5 (ERF5), and AvrPto-DEPENDENT Pto-INTERACTING PROTEIN3 (Adi3) were activated after CAPE1 treatment (Figure 4A).
Figure 4.
Antipathogen and Antiherbivore Defense Genes and Responses of Tomato Plants Induced by Spraying with CAPE1.
(A) Relative expression of antiherbivore and antipathogen defense genes were quantified by comparing the expression levels in untreated plants with plants treated with water or 250 nM CAPE1 for 4, 8, and 24 h (n = 5 for each time point). The internal control EF-1α was used for normalization.
(B) Changes of the larval sizes after feeding with water or 250 nM CAPE1-treated tomato leaves. The photographs of S. litura larvae were taken after 5 d of feeding. The larval weights of 30 larvae were recorded each day.
(C) The Pst DC3000 infection phenotypes of the plants presprayed with water or 100 nM CAPE1 peptide for 2 h (n = 3) prior to pathogen inoculation. The infection symptoms were observed after inoculation for 7 d. The bacterial numbers, presented as log colony-forming units (Log CFU) per gram of leaf tissue, represent the means and sd of three biological samples. All statistically significant differences between the CAPE1- and water-treated samples are indicated with asterisks, *P < 0.05, **P < 0.01, or ***P < 0.001, based on Student’s t test.
The antiherbivore response was evaluated by average larval weights of 30 Spodoptera litura larvae fed with tomato leaves pretreated with water or CAPE1. Tomato plants presprayed with CAPE1 suppressed larval growth and reduced larval weight by ∼20% (Figure 4B). To demonstrate that plant resistance can also be enhanced by CAPE1, two groups of tomato plants were presprayed with water or synthetic CAPE1 for 2 h. After the treatment, the two plant groups underwent challenge with the pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000). As shown in Figure 4C, the water pretreated tomato plants showed severe pathogen infection symptoms. However, the plants pretreated with CAPE1 showed no significant symptoms of infection or hypersensitive response (HR) (Heath, 2000) after the pathogen challenge. Although the HR was not induced to limit pathogen spread, CAPE1 was still able to reduce the bacterial population in the plant through activating a defense response (Figure 4C).
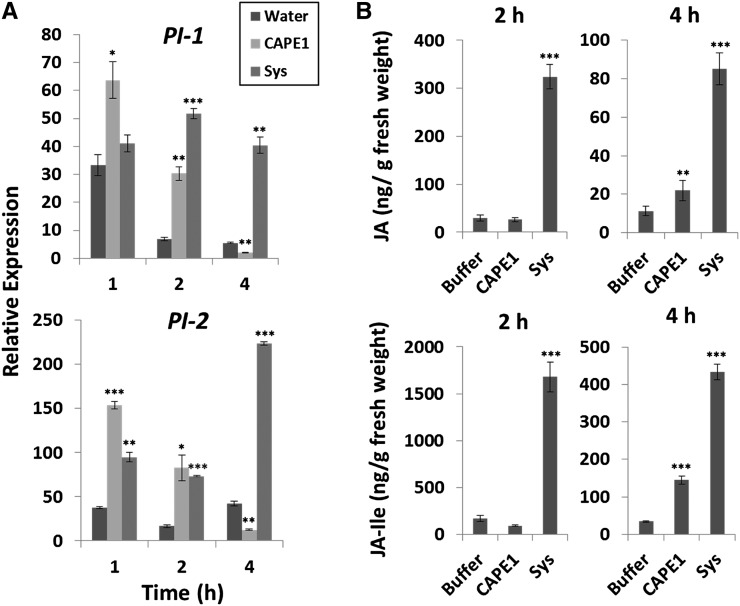
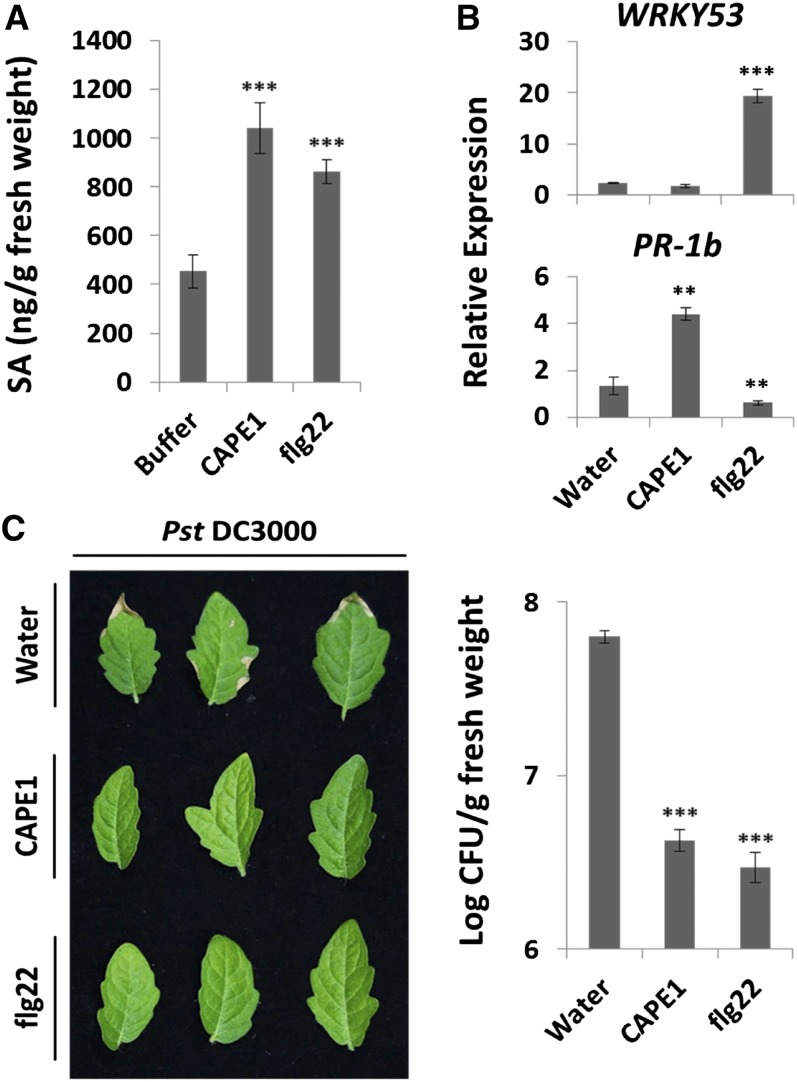
To compare the antiherbivore response of systemin with CAPE1, excised tomato plants were used, since the excised plant treated with peptide solution has been used previously to test the bioactivity of systemin (Schaller et al., 1995; Howe et al., 1996). A comparison of systemin and CAPE1 showed that with CAPE1, a higher PI-1 and PI-2 expression level was induced after 1 h of treatment, but expression decreased rapidly after 2 h of treatment (Figure 5A). With systemin, PI-1 was induced after 2 h and PI-2 was induced after 1 h. Both PI-1 and PI-2 were observed to be induced by systemin at 2 h and the effect lasted for more than 4 h (Figure 5A). JA and JA-IIe were observed to be significantly induced by treatment for 2 h with systemin but was induced after 4 h treatment with CAPE1, and the expression level was ∼4-fold lower than that of systemin (Figure 5B). As the results suggested that CAPE1 may be a novel DAMP signal for the induction of immunity to pathogenesis, next, CAPE1 was compared with the canonical pathogen/microbe-associated molecular pattern peptide flg22 (Hayashi et al., 2001). As shown in Figure 6A, both CAPE1 and flg22 significantly induced salicylic acid (SA) when supplied to excised plants. In Figure 6B, the flg22 highly induced WRKY TRANSCRIPTION FACTOR53 (WRKY53) expression but not PR-1b in tomato. This result was consistent with the public RNA-seq data in the Tomato Functional Genomics Database (Fei et al., 2011), which are based on the experiment “transcriptome sequencing of tomato leaves treated with different bacteria and PAMPs” (Rosli et al., 2013). The RNA-seq data showed that WRKY53 could be induced but PR-1b, Adi3, and ERF5, but could not be significantly induced using 30 min or 6 h treatment of flg22 on the tomato. However, the CAPE1 did not induce WRKY53 but highly induced its precursor gene PR-1b. Spraying of plants for 2 h with either CAPE1 or flg22 resulted in plant resistance to Pst DC3000 infection (Figure 6C).
Figure 5.
Induction of PI Genes and Jasmonate Accumulation by CAPE1 and Systemin in Excised Plants.
(A) The relative expression of PI-1 and PI-2 was quantified by comparing the expression levels in untreated detached leaves with detached leaves treated with water, 250 nM CAPE1, or 250 nM systemin (Sys) for 1, 2, and 4 h (n = 4 for each time point). The internal control Ubi3 was used for normalization.
(B) The level of JA and JA-Ile induced by CAPE1 and systemin. The plants treated with 10 mM phosphate buffer (buffer), 365 nM systemin (Sys), or 365 nM CAPE1 in buffer through the cut stem for 2 and 4 h (n = 3). The quantities of JA and JA-Ile were quantified by LC-SRM-MS and calculated by the abundance of spiked standard H2JA. Data represent the means and sd of three independent biological replicates. A statistically significant difference compared with the correspondingly treated water (or buffer) samples is indicated with asterisks, *P < 0.05, **P < 0.01, or ***P < 0.001, based on Student’s t test.
Figure 6.
The SA Accumulation, Antipathogen Genes, and Responses Induced by CAPE1 and flg22 in Tomato.
(A) The level of SA induced by CAPE1 and flg22. The excised plants were pretreated with 10 mM phosphate buffer (buffer) or 365 nM CAPE1 or 365 nM flg22 in buffer through the cut stem for 8 h (n = 3). The quantity of SA was quantified by LC-SRM-MS and calculated based on the abundance of spiked SA isotopic standard (d6-SA).
(B) The relative expression of WRKY53 and PR-1b were quantified by comparing the expression levels in untreated detached leaves with the detached leaves treated with water, 250 nM CAPE1, or 250 nM flg22 for 2 h (n = 4). The internal control Ubi3 was used for normalization.
(C) The Pst DC3000 infection phenotypes for plants presprayed with water, 100 nM CAPE1, or 100 nM flg22 peptide for 2 h (n = 3) prior to the pathogen inoculation. The infection symptoms were observed 4 d after inoculation. The bacterial numbers were calculated 4 d after inoculation and represented as log colony-forming units (Log CFU) per gram leaf tissue. Data represent the means and sd of three biological samples. A statistically significant difference compared with the correspondingly treated water (or buffer) samples is indicated with asterisks, **P < 0.01 or ***P < 0.001, based on Student’s t test.
CAPE1 Proprotein
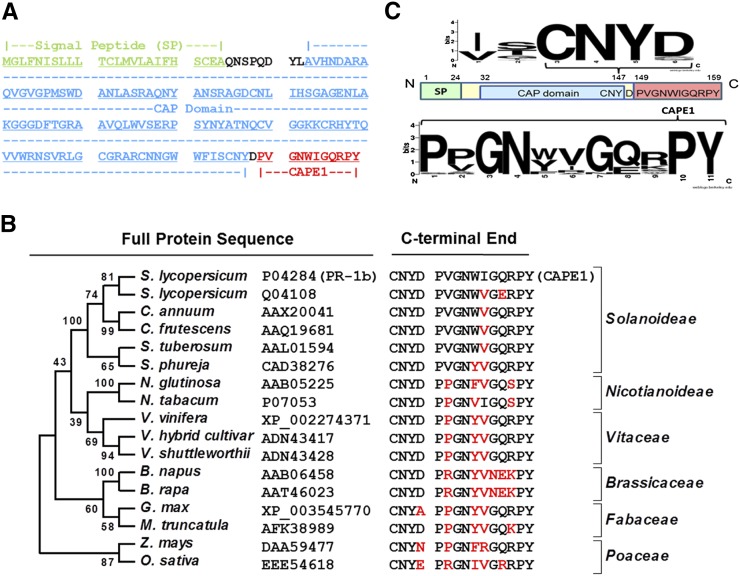
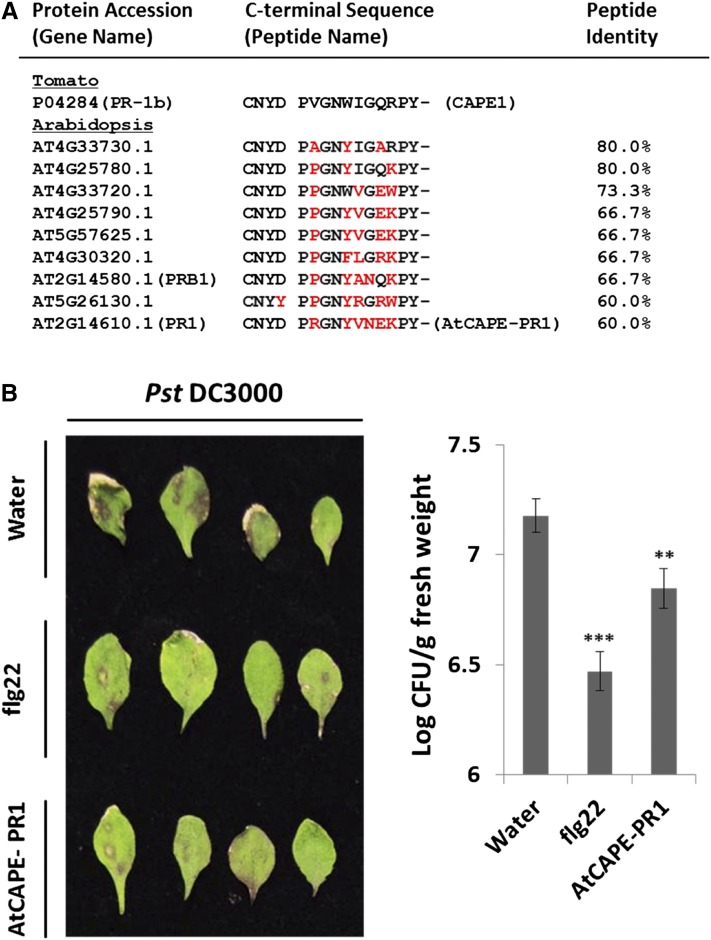
The mature CAPE1 peptide is derived from the C-terminal end of tomato PR-1b. This proprotein consists of an N-terminal signal peptide, a CAP domain, and an extended C-terminal end (Figure 7A). The phylogenetic analysis using Molecular Evolutionary Genetics Analysis version 5.2 (MEGA5.2)(Tamura et al., 2011) and C-terminal alignment of the PR-1b protein demonstrated that the full protein and extended C-terminal end are highly conserved across different flowering plants ranging from monocots to dicots (Figure 7B; Supplemental Data Set 2). It is interesting that the PxGNxxxxxPY- motif was conserved in the CAPE1 sequence and that the three residue sequences before the cleavage site had a conserved CNYx motif (Figure 7C). This suggests that CNYx.PxGNxxxxxPY- could be a functional motif that may mark bioactive peptides in other species. To demonstrate that a peptide derived from the CNYx.PxGNxxxxxPY- motif could be bioactive, the Arabidopsis pathogenesis-related protein 1 (At-PR1), which contains this motif but shows the lowest level of sequence identity with CAPE1 in tomato, was selected (Figure 8A). This putative CAPE peptide from At-PR1 (designated as AtCAPE-PR1) was shown to increase immunity against Pst DC3000 infection (Figure 8B).
Figure 7.
Identification of Conserved CAPE Sequences and Proproteins in Diverse Species Based on Tomato CAPE1.
(A) The sequence and the classified motifs of CAPE1 proprotein (Tomato PR-1b).
(B) Phylogenetic analysis of 17 selected CAPE proproteins generated by MEGA5.2 using the Maximum Likelihood method based on the Whelan and Goldman model. Bootstrap values set to 1000 replicates.
(C) The sequence identities and logo illustration of 30 CAPE1 homologs generated by Weblogo (http://weblogo.berkeley.edu/logo.cgi). SP, signal peptide; CAP domain, cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 protein domain.
Figure 8.
Identification of CAPE Homologs in Arabidopsis and the Antipathogen Activity of AtCAPE-PR1.
(A) The putative AtCAPE peptides derived from the CAP proteins in Arabidopsis containing a conserved cleavage (CNYx) and a signaling peptide (PxGNxxxxxPY-) motif. One of the putative AtCAPEs is derived from At-PR1, designated AtCAPE-PR1. Red characters indicated different amino acids compared with SolCAPE1.
(B) The Pst DC3000 infection phenotypes for the plants presprayed with water, 100 nM AtCAPE-PR1, or 100 nM flg22 peptide for 2 h (n = 3) prior to pathogen inoculation. The infection symptoms were observed 4 d after inoculation. The bacterial numbers were calculated 4 d after inoculation and are represented as log colony-forming units (Log CFU) per gram of leaf tissue. Data represent the means and sd of three biological samples. A statistically significant difference compared with the corresponding water-treated samples is indicated with asterisks, **P < 0.01 or ***P < 0.001, based on Student’s t test.
DISCUSSION
In this study, a novel peptidomics platform was successfully applied to the analysis of low abundance plant peptides. The sensitivity for the identification of endogenous peptide is determined by the score distribution of false positive hits. The use of the Tom or TomHT database had a significant effect on the low score hits but only a mild effect on the high score hits. With the use of RanTom and RanTomHT to evaluate the score distribution of false positive hits for using Tom and TomHT, respectively, the use of RanTomHT showed a lower Mascot score threshold to accept significant hits. This implies that the use of the TomHT database can enhance the detection sensitivity of endogenous peptide because the score threshold required to exclude most random peptide hits can be reduced. The combination of Mascot and MD scoring can identify the plant peptides with higher confidence without significantly compromising peptide ID sensitivity. Quantitative peptidomics analysis revealed three peptides, including systemin, that were not significantly expressed in the unwounded plant but were expressed at high levels after wounding plus MeJA treatment. This peptide is a signaling molecule for the systemic activation of the antiherbivore response (Pearce et al., 1991). Systemin is the upstream component of the antiherbivore signaling cascade and systemic signal transmission is mediated by JA (Li et al., 2003; Stratmann, 2003). We show here the concentration change of endogenous systemin, a well-known wound-induced peptide, before and after the induction of wounding plus MeJA treatment. The detection of systemin also proved that the platform proposed in this study is able to detect defense signaling peptides. The second peptide found to be upregulated upon treatment was derived from the chloroplast photosystem II subunit X. This peptide may be associated with the induction of reactive oxygen species (ROS) in chloroplasts. The third peptide (designated as CAPE1) was derived from PR-1b, a protein of unclear function.
The H2O2 and defense gene responses induced by CAPE1 indicated that this peptide regulates plant defense responses. H2O2 is a ROS involved in several defense responses during wounding, insect attacks, and pathogen infections (Doke et al., 1996; Lamb and Dixon, 1997; Orozco-Cardenas and Ryan, 1999). CAPE1 was shown to be a DAMP elicitor in this study as it was induced by wounding and activated defense responses. Although several peptides in tomato are proposed to be DAMPs, the evidence for a peptide DAMP is mainly based on the consideration of the precursor gene induced by the damage or the bioactivity of synthesized putative peptides (Pearce et al., 1991; Huffaker et al., 2006; Trivilin et al., 2014). Microarray and qRT-PCR analysis showed that CAPE1 can induce defense genes to produce immune responses against herbivores and pathogens. Both JA and SA hormones can be induced by CAPE1, which explains why antiherbivore and antipathogen genes were induced by the peptide treatment. The JA and SA biosynthesis pathways are known to be antagonistic (Robert-Seilaniantz et al., 2011; Thaler et al., 2012), but they may also function synergistically in the SA-JA-ethylene backbone of the plant immune signaling network, thereby redirecting defense output (Verhage et al., 2010). In comparison with the activation of antiherbivore and antipathogen responses by systemin and flg22, respectively, CAPE1 showed a mild antiherbivore response but activated a comparable antipathogen response. The mild antiherbivore response induced by CAPE1 can be explained by a lower induction level of PI genes and JA hormones than that seen with systemin treatment. CAPE1 significantly induced several pathogen-related marker genes, including PR-2, PR-7, Chi2;1, and the precursor of CAPE1 (PR-1b). Unlike flg22, which induces WRKY53 (Xiao et al., 2007), the CAPE1-triggered immunity did not induce the PTI-responsive gene WRKY53 but induced PR-1b. This implies that flg22 and CAPE1 act as an elicitor for pathogen/microbe-associated molecular pattern and DAMP, respectively, and thereby regulate different mechanisms in the antipathogen response. In addition, ERF5, a GCC box (AGCCGCC) binding protein, was induced by CAPE1. We suggest that ERF5 is a mediator of CAPE1 defense responses because of the GCC box, a cis-acting element found in the promoter of many JA/ethylene-inducible and PR genes. ERF5 was also demonstrated to positively regulate SA signaling and plant immunities against the bacterial pathogen Pst DC3000 and improve plant resistance to pathogens by activating several PR genes (Moffat et al., 2012; Son et al., 2012). In tomato, the overexpression of ERF5 was observed to induce PR genes and conferred tolerance to Ralstonia solanacearum (Li et al., 2011). This study suggested an alternative approach to enhance plant resistance through ERF5, which can be regulated by a low concentration of peptide without the use of transgenes. Furthermore, Adi3, encoding a component of the effector-triggered immunity response, which negatively regulates programmed cell death (Devarenne et al., 2006), was induced after CAPE1 treatment. Adi3 is a cell death suppressor (CDS) and its localization is dictated by a nuclear localization signal found in the Adi3 T-loop extension, which is phosphorylated for kinase activation (Ek-Ramos et al., 2010b). The deactivation of Adi3 CDS function is initiated by the interaction of Pto only when Pto interacts with the Pst effector protein AvrPto. This deactivation of CDS activity can lead to HR, which functions to limit pathogen spread. The HR through deactivation of Adi3 function was demonstrated to be compensated for by overexpression of Adi3 (Devarenne et al., 2006; Ek-Ramos et al., 2010a). In this study, CAPE1 was found to activate a “defense-no-death” phenotype to enhance plant resistance against the bacterial pathogen Pst DC3000 without induction of the HR (Yu et al., 1998). This phenotype could be explained by the elevated level of transcription of antipathogen and cell death suppressor genes as well as the level of SA. It also suggests that systemically induced immune responses can be activated by CAPE1, since SA and JA are essential hormones for the induction of systemic acquired resistance and induced systemic resistance, respectively (Pieterse et al., 2009). Plant insects and pathogens are responsible for substantial crop losses worldwide every year, and amid increasing environmental concerns, the use of agrochemicals to defeat the biological stress is more and more restricted. CAPE1 may potentially be used to activate resistance against biological threats in tomato. Furthermore, the highly conserved sequence of CAPE1 and its proprotein suggests that CAPE1 may also exist and be biologically active in other species. This study demonstrated the role of PR-1b in tomato defense signaling and demonstrated that a putative CAPE peptide with a PxGNxxxxxPY- motif derived from At-PR1 induces resistance against Pst DC3000 in Arabidopsis. Although At-PR1 is considered to be a common marker gene for the antipathogen response, its function was unclear previously. This study highlights the biological role of PR1 and CAP proteins in defense signaling.
METHODS
Chemicals, Enzymes, and Materials
Tris (2-carboxyethyl) phosphine hydrochloride, methyl methanethiosulfonate, DAB, KOH, NaOH, HCl, 10× Murashige and Skoog basal salt micronutrient mixture, King Agar B medium, isopropanol, ethanol, chloroform, MeJA (95% solution), Triton X-100, β-casein, SA, 2-hydroxybenzoic acid-[2H6] (d6-SA), and JA were purchased from Sigma-Aldrich. Dihydrojasmonic acid (H2JA) was purchased from OlChemim. Analytical grade methanol, acetonitrile (ACN), and trifluoroacetic acid (TFA) were purchased from Merck. Liquid chromatography-mass spectrometry (LC-MS) grade ACN with 0.1% formic acid was from J.T. Baker. Deionized water (18.1 MΩ·cm resistivity) from Milli-Q system (Millipore) was used throughout this work. C18 Zip Tip and Millex HA 0.45-μm filters were purchased from Millipore. The TriPure RNA Isolation Reagent and FastStart Universal SYBR Green Master (ROX) kit were purchased from Roche. The RNA purification reagent RNAmate was from BioChain. The SuperScript III Reverse Transcriptase Kit was purchased from Invitrogen. Fast-Run HotStart PCR Mix was from Postech. Miracloth was purchased from Calbiochem. The customized Sep-Pak C18 Cartridge 60 cc (20 g) was purchased from Waters. Gel filtration XK 16/40 column and packing gel (Sephadex G-25 Fine) were purchased from GE Healthcare Bio-Sciences. Tryptic enolase and [Glu1]-Fibrinopeptide (GFP) was purchased from Waters. Trypsin (modified, sequencing grade) was purchased from Promega. Systemin (AVQSKPPSKRDPPKMQTD), CAPE1 (PVGNWIGQRPY), and AtCAPE-PR1 (PRGNYVNEKPY) were synthesized and purified to >95% purity by Yao-Hong Biotechnology. The flg22 (QRLSTGSRINSAKDDAAGLQIA) with purity >95% purity was purchased from KareBay Biochem. The purity of synthetic CAPE1 peptide was further checked to be 97.68% using nanoUHPLC-MS (Supplemental Figure 4).
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum cv CL5915) seeds were provided by AVRDC-The World Vegetable Center (Tainan, Taiwan). The tomato plants were kept at 25°C d/20°C night temperature under a 12-h-light/12-h-dark photoperiod. Tomato seeds were germinated in soil and grown in a growth chamber for 2 weeks. For detection of endogenous peptides, the 2-week-old plants were transferred and maintained in a phytotron for 6 weeks. The tomato plants for peptide treatments were continuously grown in a growth chamber for 5 weeks. To examine the peptide activity in Arabidopsis thaliana, Arabidopsis (ecotype Columbia) seeds were germinated in soil and grown in a growth chamber at 22°C d/20°C night temperature under a 8-h-light/16-h-dark photoperiod for 4 weeks.
Plant Treatments
To extract the wound-induced peptides, the tomato plants were mechanically wounded by cutting across the surface of the mesophyll with a pair of scissors and spraying with 1.25 mM MeJA in 0.1% Triton X-100 solution for 15 h (Pearce et al., 2001a). For direct quantitation of CAPE1 in tomato, unwounded, wounded, or wounded plus MeJA-treated plants for 15 h were used to study peptide induction. To examine the possible function of peptide using cDNA microarray analysis, detached tomato leaves were immersed in water or 250 nM CAPE1 in aqueous solution for 1, 2, 4, and 8 h, respectively. To confirm the gene expression induced by the peptide, the tomato plants were collected after spraying with 250 nM CAPE1 or water for 0, 4, 8, and 24 h. To compare the ROS induced by different treatments, detached tomato leaves were treated with water (control), mechanical wounding, 1.25 mM MeJA, 250 nM systemin, or 250 nM CAPE1 for 4 h. To test the antiherbivore activity induced by the peptide, the tomato plants were collected after spraying with 250 nM CAPE1 or water for 24 h before feeding with insects. To compare the PI genes induced by the peptides, detached tomato leaves were immersed in 250 nM CAPE1, 250 nM systemin, or water for 1, 2, and 4 h. To compare the jasmonates induced by the peptides, the excised tomato plants were treated with 10 mM phosphate buffer, 365 nM systemin, or 365 nM CAPE1 in buffer through the cut stem for 2 and 4 h (Schaller et al., 1995; Howe et al., 1996). To compare the salicylic acid induced by peptides, the tomato plants were treated with 10 mM phosphate buffer, 365 nM flg22, or 365 nM CAPE1 in buffer through the cut stems of excised plants for 8 h. To compare the WRKY53 and PR-1b genes induced by the peptides, detached tomato leaves were immersed in 250 nM CAPE1, 250 nM flg22, or water for 2 h. To test the antipathogen activity induced by peptides, three groups of the tomato plants were sprayed with 100 nM CAPE1, 100 nM flg22, or water, respectively, for 2 h prior to pathogen challenge. To test the antipathogen activity induced by peptides in Arabidopsis, the plants were sprayed with 100 nM AtCAPE-PR1, 100 nM flg22, or water, respectively, for 2 h prior to pathogen challenge.
Endogenous Peptide Extraction
The unwounded and wounding plus MeJA-treated tomato leaves were collected and individually ground into powder under liquid nitrogen by a blender (Waring Commercial). Frozen leaf powder (150 g) was dissolved in 200 mL of 1% TFA and homogenized to leaf juice by a blender for 2 min. The leaf juice was filtered through four layers of cheesecloth and one layer of Miracloth. The filtrated leaf juice was then centrifuged at 10,000g for 20 min at 4°C. The supernatant was adjusted to pH 4.5 with 10 n NaOH and centrifuged at 10,000g for 20 min at 4°C. Then the supernatant was readjusted to pH 2.5 using TFA and 150 μg tryptic β-casein peptides were added to the supernatant as an internal control for peptide quantity before purification. To avoid the trypsin residue reacting with the endogenous proteins or peptides, the tryptic β-casein peptides were acidified by TFA and purified using C18 Zip Tip. Before purifying the supernatant using a Sep-Pak cartridge, the stationary phase was first equilibrated by 60 mL of 0.1% TFA. The supernatant was loaded into the Sep-Pak cartridge, washed with 120 mL 0.1% TFA, and eluted by 200 mL of 60% methanol in 0.1% TFA. The eluted solution was vacuum-evaporated to remove methanol using a vacuum centrifugation concentrator (miVac Duo Concentrator; Genevac) first and then using a lyophilizer (EYELA) to dryness (Pearce et al., 1991). To profile total endogenous peptides using LC-MS operated in DDA mode, the lyophilized crude extract was dissolved in 1 mL of 0.1% TFA, centrifuged at 10,000g for 10 min at 4°C, and filtered through a Millex HA 0.45-μm filter before peptide fractionation. For the peptide fractionation, the filtrated peptide extract was injected into a Sephadex G-25 column and eluted by 1 mL/min of 0.1% TFA with 1 fraction/min collection. Ten fractions from elution times of 22 to 31 min were collected and evaporated to dryness using a vacuum centrifugation concentrator. Each fraction was purified by C18 Zip Tip for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. For targeted peptide analysis using LC-MS operated in SRM mode, the endogenous peptides were extracted from the unwounded, wounding only, and wounding plus MeJA-treated tomato leaves using the same procedure but without gel filtration fractionation.
Endogenous Peptide Profiling Using LC-MS/MS
For endogenous peptide profiling, LC-MS/MS analysis was performed with a nanoUHPLC system (nanoACQUITY UPLC; Waters) coupled online to the nanoelectrospray source of a hybrid quadruple time-of-flight mass spectrometer (SYNAPT HDMS G1; Waters). The SYNAPT HDMS G1 instrument was operated in the positive ion mode and DDA methods for detection of endogenous peptides. The sample was loaded into a 180 μm × 50-mm tunnel frit trap column packed with 20 mm of 5-μm Symmetry C18 particles (Waters) and separated online with a 75 μm × 250-mm tunnel frit analytical column packed with 250 mm of 1.7-μm BEH C18 particles (Waters) using a 95 min gradient flow with 300 nL/min and 5 to 90% ACN/0.1% formic acid ratio (Chen et al., 2012). The DDA acquisition parameters were set to one full MS scan (m/z 400 to 1600) with a scan time of 0.6 s and switched to three product ion scans (m/z 100 to 1990) with a scan time of 1.2 s when a precursor ion charge was 2+, 3+, and 4+ and an intensity greater than 20 counts was detected. The data generated from SYNAPT HDMS G1 were first converted into mzXML format (Pedrioli et al., 2004) using massWolf (version 4.3.1) and then processed by UniQua with default parameters for SYNAPT HDMS G1 (Chang et al., 2013). The UniQua processed spectra were converted into Mascot generic format (.mgf) using mzXML2Search from Trans Proteomics Pipeline (TPP) version 4.4 rev. 1 (Pedrioli, 2010).
Hypothetical and Decoy Database
The tomato hypothetical peptide database (TomHT database) was composed by extracting 50 residues of all protein C-terminal sequences from the International Tomato Annotation Group protein database (Tom Database) (release version 2.3, total protein entries = 34,728) with the addition of the bovine β-casein sequence. The randomized databases (Ran Databases) were generated by shuffling sequences in the target databases using Perl script (decoy.pl) provided by Matrix Science.
Endogenous Peptide Identification and Quantitation
The processed mgf files were searched against the TomHT or Tom database without specifying enzyme cleavage rules using a Mascot MS/MS ion search (Matrix Science, server version 2.3). The mass tolerance in the MS/MS ion search for peptide precursors and fragments was ± 0.1 D. The Mascot search results from the randomized database were used to evaluate the score to cutoff the random matched peptides. The identified peptides were further quantified by TargetLynx (MassLynx version 4.1; Waters). For the peptide quantitation using TargetLynx, the observed m/z and retention time of each identified peptide was imported into the software to obtain the peptide abundance according to the peak area of the XIC. The XIC peak areas were summarized if one peptide was detected across several gel filtration fractions. The abundance of tryptic β-casein peptides was used as the internal control to normalize the expression level of the endogenous peptides.
Phytohormone Extraction
After peptide treatment, the metabolites were extracted from leaf tissues for phytohormone quantitation. The extraction procedure was modified from a previously published protocol (Pan et al., 2010). The leaf tissues (∼0.6 g fresh weight) were ground into powder under liquid nitrogen and transferred to a 50-mL screw-cap tube. The frozen leaf powder was dissolved in 6 mL extraction solvent and d6-SA (3 ng to 0.6 g leaf tissue) and H2JA (15 ng to 0.6 g leaf tissue) were added as internal standards. The samples were extracted by shaking at a speed of 100 rpm at 4°C for 30 min and then 12 mL dichloromethane was added to each sample and shaken at 100 rpm at 4°C for 30 min. The samples were centrifuged at 13,000g at 4°C for 5 min, and two phases were formed. The lower phase was transferred carefully into a new tube and evaporated to dryness by a vacuum centrifugal concentrator for ∼1 h. The dried samples were dissolved in 300 μL methanol, mixed well, and centrifuged at 10,000g at 4°C for 5 min and then the supernatant was transferred to the sample vial for targeted quantitation analysis using LC-MS/MS.
Targeted Peptide and Phytohormone Quantitation Using LC-MS/MS
For targeted peptide quantitation, the nanoUHPLC method was the same as for endogenous peptide profiling and the mass spectrometer (LTQ Velos Pro; Thermo Fisher Scientific) was set to one full MS scan (m/z 400 to 1600) with enhanced scan speed and switched to one SRM scan with normal scan speed. For SRM targeted on CAPE1, the doubly charged CAPE1 precursor ion m/z was selected (m/z 643.84) for fragmentation and product ions m/z of 620.34, 733.37, and 1090.57 were monitored. The relative abundances of CAPE1 in wounded and unwounded samples were estimated by combining SRM peak areas of product ions.
For phytohormone quantitation, a linear ion trap-orbitrap mass spectrometer (Orbitrap Elite; Thermo Fisher Scientific) coupled online with a UHPLC system (ACQUITY UPLC; Waters) was used. The phytohormones were separated by a HSS T3 column (Waters) using gradients of 0.5 to 25% ACN at 0 to 2 min, 25 to 75% ACN at 2 to 7 min, and 75 to 9.5% ACN at 7 to 7.5 min. The mass spectrometer was operated in the negative ion mode and set to one full FT-MS scan (m/z 100 to 600) with 60,000 resolution and switched to five FT-MS product ion scans (in 30,000 resolution) for five precursors: m/z of 137.02 (for SA), 209.12 (for JA), 322.20 (for JA-Ile), 141.05 (for d6-SA dissociated to d4-SA), and 211.13 (for H2JA). The fragmentation reactions of m/z 137.02 to 93.03 for SA, 209.12 to 59.01 for JA, 322.20 to 130.09 for JA-Ile, 141.05 to 97.06 for d6-SA, and 211.13 to 59.01 for H2JA were selected for quantitation. The absolute abundances of JA, JA-Ile, and SA were calculated by the abundance of d6-SA and H2JA.
cDNA Microarray and qRT-PCR
The transcriptomes of the tomato leaf tissues after treatment with 250 nM CAPE1 for 1, 2, 4, and 8 h were compared with the transcriptomes from four independent water-treated tomato leaf tissues using two-color cDNA microarray. The two-color gene expression microarray chip for tomato was purchased from Agilent (Agilent-022270). The hybridization of Cy3- and Cy5-labeled cDNA for each of CAPE1/water sample pair and slide washing were performed according to the manufacturer’s instructions and reagent kits (Agilent Technologies). The array was scanned with a microarray scanner (Agilent Technologies) using 535 nm for Cy3 and 635 nm for Cy5. The images were analyzed by Feature Extraction Software version 10.7.1.1 (Agilent Technologies). The microarray data were further interpreted by GeneSpring GX11.5.1 (Agilent Technologies). A gene with raw intensity >100 was considered as identified and its expression ratio for CAPE1/water >1.5-fold and P < 0.05 was considered as induced by CAPE1. There were 95, 204, 179, and 105 genes found to be induced by 1-, 2-, 4-, and 8-h CAPE1 treatments, respectively. A total of 485 nonredundant genes were determined to be CAPE1 induced. Those CAPE1-induced genes were analyzed by BLAST2GO (Conesa et al., 2005) (http://www.blast2go.de/) to obtain the functional annotation and classification. Two hundred and twenty four genes out of 485 genes were annotated and used for the functional categorization based on Gene Ontology (http://www.geneontology.org). Functional enrichment analysis was then used to assign the biological relevance of these annotated genes using an integrated Web-based GO toolkit, agriGO (Du et al., 2010).
qRT-PCR was used to validate the expression of some specific genes. Three biological replicates were used for qRT-PCR analyses. The qRT-PCR was performed using SYBR Green reagent and ABI 7500 Real Time PCR systems. The PCR cycling steps were 50°C for 2 min and 94°C for 10 min for initial steps and followed by 95°C for 15 s and 60°C for 1 min for 40 cycles. The gene expressions across different samples were normalized with internal control EF-1α or Ubi3. The primers used are listed in Supplemental Table 2. The melting curve was used to verify the specificity of the PCR product.
In Vivo Detection of H2O2
DAB was dissolved with 1 n HCl and adjusted to pH 3.8 with NaOH to a final concentration of 1 mg/mL. After plant treatments, the detached leaves were continuously supplied with DAB solutions for 8 h in the dark and then decolorized by boiling ethanol (96%) for 10 min. The leaves were cooled to room temperature and preserved in fresh ethanol.
Herbivory Treatments
The Spodoptera litura larval eggs were originally obtained from Taiwan Agricultural Chemicals and Toxic Substances Research Institute (Taichung County, Taiwan). Thirty uniformly sized larvae of the first instar stage were used for the anti-insect bioassay study. The larvae were continuously fed with tomato leaves harvested from water or CAPE1 presprayed plants every 24 h for 5 d. All larval weights were recorded for each day and the averages of larval weights were calculated. The larval sizes were observed after 5 d of feeding.
Pathogen Growth and Challenge
The bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000) was grown on King’s B agar medium containing 100 mg/L rifampicin for 2 d at 28°C. Before the challenge, the bacteria were cultured in King’s B liquid medium at 28°C with 230 rpm shaking overnight. The bacteria were pelleted by centrifugation and resuspended in 10 mM MgSO4 at A600 = 0.25 (∼108 colony-forming units/mL). The plants were dipped into a diluted suspension of 105 colony-forming units/mL Pst DC3000 in 10 mM MgSO4 containing 0.005% Silwet L-77 under vacuum for 30 s. Pst DC3000 were grown in water- or peptide-treated plants for several days to observe the symptoms and then the bacteria were collected from the leaves and evaluated by bacterial titers according to a method outlined previously (Zimmerli et al., 2000).
Accession Numbers
Sequence data from this article can be found in the International Tomato Annotation Group protein database or GenBank/EMBL databases under the accession numbers listed in Table 1, Supplemental Tables 1 and 2, and Supplemental Data Sets 1 to 3. Sequence data of putative CAPE peptides in Arabidopsis from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: AT4G33730.1, AT4G25780.1, AT4G33720.1, AT4G25790.1, AT5G57625.1, AT4G30320.1, AT2G14580.1 (PRB1), AT5G26130.1, and AT2G14610.1 (PR1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Abundances of Selected CAPE1 Transitions in Unwounded, Mechanically Wounded, and Wounded plus MeJA-Treated Tomato.
Supplemental Figure 2. Study of CAPE1 Bioactivities in the Triggering of Leaf Tissue H2O2 Production.
Supplemental Figure 3. Functional Categorization of the Defense Genes Induced by CAPE1 Using Hierarchy of GO Categorization Analyzed by Blast2GO and agriGO.
Supplemental Figure 4. Purity Evaluation of CAPE1 Peptide Using NanoUHPLC-MS.
Supplemental Table 1. Total Identified and Quantified Endogenous Peptides Observed in Unwounded and Wounded plus MeJA-Treated Tomato.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Time-Course Analysis of the Tomato Transcriptome Regulated by CAPE1 Using Microarray.
Supplemental Data Set 2. Alignments of 17 CAPE Proproteins Selected from Different Species.
Supplemental Data Set 3. Mascot MS/MS Ion Matching Result of Total Identified Peptides.
Supplementary Material
Acknowledgments
This work was financially supported by Academia Sinica and the National Science Council of Taiwan. The MS analysis was supported by the Metabolomics Facilities of the Scientific Instrument Center at Academia Sinica. The microarray analysis was supported by the DNA Microarray Core Laboratory of the Institute of Plant and Microbial Biology at Academia Sinica. The larval eggs were obtained from Taiwan Agricultural Chemicals and Toxic Substances Research Institute, Taichung County, Taiwan.
AUTHOR CONTRIBUTIONS
Y.-L.C. contributed ideas, performed experiments, and analyzed data. C.-Y.L. analyzed peptide analysis result. K.-T.C. performed parts of experiments. W.-H.C. processed mass spectrometry spectra. R.-N.H. provided larvae and supported insect experiments. H.G.N. commented on the biological result prior to submission. Y.-R.C. initiated project, contributed ideas, and wrote the article.
Glossary
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- DAMP
damage-associated molecular pattern
- MeJA
methyl jasmonate
- JA
jasmonic acid
- nanoUHPLC-MS
nanoflow ultrahigh performance liquid chromatography mass spectrometry
- DDA
data-dependent acquisition
- XIC
extracted ion chromatogram
- SRM
selected reaction monitoring
- MD
Mascot Delta
- qRT-PCR
quantitative RT-PCR
- HR
hypersensitive response
- ROS
reactive oxygen species
- CDS
cell death suppressor
- DAB
3,3-diaminobenzidine
- SA
salicylic acid
- H2JA
dihydrojasmonic acid
- ACN
acetonitrile
- TFA
trifluoroacetic acid
- LC-MS
liquid chromatography-mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aebersold R., Mann M. (2003). Mass spectrometry-based proteomics. Nature 422: 198–207. [DOI] [PubMed] [Google Scholar]
- Boller T. (2005). Peptide signalling in plant development and self/non-self perception. Curr. Opin. Cell Biol. 17: 116–122. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009a). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009b). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M. (2009). Plant peptides in signalling: looking for new partners. Trends Plant Sci. 14: 255–263. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Kang J.H., Howe G.A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40: 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.H., Lee C.Y., Lin C.Y., Chen W.Y., Chen M.C., Tzou W.S., Chen Y.R. (2013). UniQua: A universal signal processor for MS-based qualitative and quantitative proteomics applications. Anal. Chem. 85: 890–897. [DOI] [PubMed] [Google Scholar]
- Chassot C., Buchala A., Schoonbeek H.J., Metraux J.P., Lamotte O. (2008). Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 55: 555–567. [DOI] [PubMed] [Google Scholar]
- Che F.Y., Lim J., Pan H., Biswas R., Fricker L.D. (2005). Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol. Cell. Proteomics 4: 1391–1405. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Chen W.Y., Tseng M.C., Chen Y.R. (2012). Tunnel Frit: A nonmetallic in-capillary frit for nanoflow ultra high-performance liquid chromatography-mass spectrometry applications. Anal. Chem. 84: 297–303. [DOI] [PubMed] [Google Scholar]
- Cheong Y.H., Chang H.S., Gupta R., Wang X., Zhu T., Luan S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129: 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Devarenne T.P., Ekengren S.K., Pedley K.F., Martin G.B. (2006). Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Choi H., Nesvizhskii A.I. (2008). Adaptive discriminant function analysis and reranking of MS/MS database search results for improved peptide identification in shotgun proteomics. J. Proteome Res. 7: 4878–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke N., Miura Y., Sanchez L.M., Park H.J., Noritake T., Yoshioka H., Kawakita K. (1996). The oxidative burst protects plants against pathogen attack: mechanism and role as an emergency signal for plant bio-defence–a review. Gene 179: 45–51. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos M.J., Avila J., Cheng C., Martin G.B., Devarenne T.P. (2010a). The T-loop extension of the tomato protein kinase AvrPto-dependent Pto-interacting Protein 3 (Adi3) directs nuclear localization for suppression of plant cell death. J. Biol. Chem. 285: 17584–17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos M.J., Avila J., Cheng C., Martin G.B., Devarenne T.P. (2010b). The T-loop extension of the tomato protein kinase AvrPto-dependent Pto-interacting protein 3 (Adi3) directs nuclear localization for suppression of plant cell death. J. Biol. Chem. 285: 17584–17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J.E., Gygi S.P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- Farrokhi N., Whitelegge J.P., Brusslan J.A. (2008a). Plant peptides and peptidomics. Plant Biotechnol. J. 6: 105–134. [DOI] [PubMed] [Google Scholar]
- Farrokhi N., Whitelegge J.P., Brusslan J.A. (2008b). Plant peptides and peptidomics. Plant Biotechnol. J. 6: 105–134. [DOI] [PubMed] [Google Scholar]
- Fei Z., Joung J.G., Tang X., Zheng Y., Huang M., Lee J.M., McQuinn R., Tieman D.M., Alba R., Klee H.J., Giovannoni J.J. (2011). Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 39: D1156–D1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia D., Demaria D., Calderini O., Ferraris L., Valentino D., Arcioni S., Tamietti G., Cardinale F. (2007). Wounding induces resistance to pathogens with different lifestyles in tomato: role of ethylene in cross-protection. Plant Cell Environ. 30: 1357–1365. [DOI] [PubMed] [Google Scholar]
- Fricker L.D., Lim J., Pan H., Che F.Y. (2006). Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom. Rev. 25: 327–344. [DOI] [PubMed] [Google Scholar]
- Gibbs G.M., Roelants K., O'Bryan M.K. (2008). The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr. Rev. 29: 865–897. [DOI] [PubMed] [Google Scholar]
- Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., Aderem A. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Heath M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44: 321–334. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Lightner J., Browse J., Ryan C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative T.A.G. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Kapp E.A., Schutz F., Connolly L.M., Chakel J.A., Meza J.E., Miller C.A., Fenyo D., Eng J.K., Adkins J.N., Omenn G.S., Simpson R.J. (2005). An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5: 3475–3490. [DOI] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. 48: 251–275. [DOI] [PubMed] [Google Scholar]
- Li C., Liu G., Xu C., Lee G.I., Bauer P., Ling H.Q., Ganal M.W., Howe G.A. (2003). The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15: 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.W., Su R.C., Cheng C.P., Sanjaya, You S.J., Hsieh T.H., Chao T.C. Chan M.T. (2011). Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 156: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky F.G., Fangel J.U., Willats W.G. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick P., Kuster B. (2010). Proteomics: a pragmatic perspective. Nat. Biotechnol. 28: 695–709. [DOI] [PubMed] [Google Scholar]
- Moffat C.S., Ingle R.A., Wathugala D.L., Saunders N.J., Knight H., Knight M.R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24: 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez-Vasquez J., Orozco-Cardenas M.L., Ryan C.A. (2007). Systemic wound signaling in tomato leaves is cooperatively regulated by systemin and hydroxyproline-rich glycopeptide signals. Plant Mol. Biol. 65: 711–718. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M., Ryan C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96: 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas M.L., Narvaez-Vasquez J., Ryan C.A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13: 179–191. [PMC free article] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. (2001a). Production of multiple plant hormones from a single polyprotein precursor. Nature 411: 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. Jr. (2001b). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98: 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrioli P.G. (2010). Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol. Biol. 604: 213–238. [DOI] [PubMed] [Google Scholar]
- Pedrioli P.G., et al. (2004). A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22: 1459–1466. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Leon-Reyes A., Van der Ent S., Van Wees S.C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rosli H.G., Zheng Y., Pombo M.A., Zhong S., Bombarely A., Fei Z., Collmer A., Martin G.B. (2013). Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 14: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A., Pearce G., Scheer J., Moura D.S. (2002). Polypeptide hormones. Plant Cell 14 (suppl.): S251–S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Satomi Y., Takao T., Minamino N. (2009). Snapshot peptidomics of the regulated secretory pathway. Mol. Cell. Proteomics 8: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A., Bergey D.R., Ryan C.A. (1995). Induction of wound response genes in tomato leaves by bestatin, an inhibitor of aminopeptidases. Plant Cell 7: 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Ryan C.A. (1999). A 160-kD systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 11: 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son G.H., Wan J., Kim H.J., Nguyen X.C., Chung W.S., Hong J.C., Stacey G. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25: 48–60. [DOI] [PubMed] [Google Scholar]
- Stratmann J.W. (2003). Long distance run in the wound response–jasmonic acid is pulling ahead. Trends Plant Sci. 8: 247–250. [DOI] [PubMed] [Google Scholar]
- Svensson M., Skold K., Svenningsson P., Andren P.E. (2003). Peptidomics-based discovery of novel neuropeptides. J. Proteome Res. 2: 213–219. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T., Whiteman N.K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270. [DOI] [PubMed] [Google Scholar]
- Tinoco A.D., Saghatelian A. (2011). Investigating endogenous peptides and peptidases using peptidomics. Biochemistry 50: 7447–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivilin A.P., Hartke S., Moraes M.G. (2014). Components of different signalling pathways regulated by a new orthologue of AtPROPEP1 in tomato following infection by pathogens. Plant Pathol. 63: 1110–1118. [Google Scholar]
- Verhage A., van Wees S.C., Pieterse C.M. (2010). Plant immunity: it's the hormones talking, but what do they say? Plant Physiol. 154: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F.M., He P., Abramovitch R.B., Dawson J.E., Nicholson L.K., Sheen J., Martin G.B. (2007). The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 52: 595–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A. (2011). Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14: 351–357. [DOI] [PubMed] [Google Scholar]
- Yu I.C., Parker J., Bent A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95: 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Webb G., Zhu X., Steiner D.F. (1999). Proteolytic processing in the secretory pathway. J. Biol. Chem. 274: 20745–20748. [DOI] [PubMed] [Google Scholar]
- Zimmerli L., Jakab G., Metraux J.P., Mauch-Mani B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc. Natl. Acad. Sci. USA 97: 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.