Abstract
Background
Clostridium difficile infection (CDI) remains a major health problem worldwide. Antibiotic use, in general, and clindamycin and ciprofloxacin, in particular, have been implicated in the pathogenesis of CDI. Here, we hypothesized that antibiotics that are highly active in vitro against C. difficile are less frequently associated with CDI than others. The primary goals of our study were to determine if antibiotic susceptibility and CDI are associated and whether the antimicrobial susceptibility of C. difficile changed over the years.
Methods and results
We examined a large panel of C. difficile strains collected in 2006-2008 at the University Hospital of Zurich. We found that the antimicrobial susceptibilities to amoxicillin/clavulanate, piperacillin/tazobactam, meropenem, clindamycin, ciprofloxacin, ceftriaxone, metronidazole and vancomycin were similar to those reported in the literature and that they are similar to those reported in other populations over the last two decades. Antibiotic activity did not prevent CDI. For example, thre use of meropenem, which is highly active against all strains tested, was a clear risk factor for CDI. Most of the antibiotics tested also showed a higher minimum inhibitory concentration distribution than that of EUCAST. All strains were susceptible to metronidazole. One strain was resistant to vancomycin.
Conclusions
Antibiotic susceptibilities of the collection of C. difficile from the University Hospital of Zurich are similar to those reported by others since the 1980. Patients treated with carbapenems and cephalosporins had the highest risk of developing CDI irrespective of the antimicrobial activity of carbapenems.
Electronic supplementary material
The online version of this article (doi:10.1186/s12879-014-0607-z) contains supplementary material, which is available to authorized users.
Keywords: Clostridium difficile, Antibiotic susceptibility, Diarrhea, EUCAST
Background
Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia was first recognized by Bartlett et al. [1]. Today this manifestation is best known as Clostridium difficile infection (CDI). CDI is the major cause of nosocomial diarrhea. However, incidence rates for community- vs. hospital-acquired CDI nowadays are similar with 11.16 and 12.1 per 100,000 person years, respectively [2].
C. difficile is a Gram-positive spore-forming anaerobic rod that replicates exclusively in the lumen of the gastrointestinal (GI) tract. Disease manifestations are due to enterotoxin A, which affects barrier integrity, and cytotoxin B, which induces cell death [3],[4]. The clinical manifestations of CDI include mild-to-profuse diarrhea with abdominal pain and, in the most severe form, toxic megacolon and perforation. The latter presentation has a mortality rate of 25-40% [5]. The overall mortality rate of CDI increased significantly since 2000 to approximately 6% today [6]. That increase is usually attributed to the emergence of the ribotype 027 strain [6],[7], but its cause is most likely multifactorial [6]. Mutations in the ribotype 027 strain are thought to increase production of toxins A and B [8]. A binary toxin is also produced [9]-[11]. Fatality rates of CDI caused by strains with genes for the binary toxin are twice those of CDI caused by strains that lack the toxin. However, it is unknown if the binary toxin directly contributes to the increased fatality rate or if it is an epiphenomenon of strains with higher virulence [9].
The main risk factor for developing CDI is prior or ongoing antibiotic use [12] that disrupts the flora of the GI tract. C. difficile exists in a vegetative form or as a spore. Disruption of the flora of the GI tract enables C. difficile to sporulate and the vegetative forms to proliferate, and if they contain the genetic information for toxin formation, to induce CDI.
Various antibiotics have antimicrobial activity against C. difficile. However, it is largely unknown whether the inherent activity of those antibiotics protects against CDI. In other words, some antibiotics with activity against C. difficile (e.g., metronidazole) may prevent or delay CDI despite disrupting the microbial flora [13],[14]. Loss of activity of fluoroquinolones against C. difficile and the associated increase of CDI suggest that antibiotic activity against C. difficile may, at least partially, hinder proliferation and toxin production despite disrupting the GI flora [15].
We hypothesized that antibiotics, which are highly active in vitro against C. difficile, are less frequently associated with CDI than others. Our primary goal was to determine if antibiotic susceptibility and CDI are associated, and thus, we determined the in vitro activity of antibiotics against C. difficile recovered in 2006-2008 at a tertiary university hospital. Furthermore, the minimum inhibitory concentrations (MICs) were compared with published MICs and the MIC distributions of EUCAST.
Methods
From September 2006 to June 2007 and from February 2008 to August 2008, all C. difficile strains cultured from stool of patients with diarrhea were prospectively collected in the laboratory of the Institute of Medical Microbiology (IMM) at the University of Zurich. The first specimens were collected to evaluate for the presence of the ribotype 027 in Switzerland. The second set was used to compare various assays. Patients were hospitalized or seen on an outpatient basis at the University Hospital of Zurich (USZ), a tertiary care hospital. There were no repeat stool specimens from patients with recurrent CDI. The study was approved by the cantonal ethic committee of Zurich, Department of Health (#EK-1561). The ethics committee waived the need for informed consent from the patients for this retrospective analysis.
Charts were reviewed retrospectively for 94 patients who had a stool specimen positive for culture or toxin. Clinical details were entered into a case report form generated in the database system Access (Microsoft Schweiz GmbH, Wallisellen, Switzerland). The following data were collected: demographics (e.g., sex, age and ethnicity), duration of hospitalization, ward, health state (e.g., previous history, drug history with focus on anti-infective, gastric, cytostatic and immunosuppressive medication, procedures, such as surgery, tube feeding and their complications), and microbiological results as related to CDI. “Underlying disease” was defined as the leading diagnosis, and “complication” was defined as a disease engrafted on the lead diagnosis (Additional file 1: Table S1). Because antibiotic treatment may precede development of CDI for up to 8 weeks [13], we recorded all antibiotics for this time frame before collection of the stool specimen.
Isolation of C. difficile
Stool specimens were inoculated onto cycloserin-cefoxitin-fructose agar (CCFA, Difco, Detroit, MI) plates that were incubated under anaerobic conditions for at least 2 days at 37°C. Suspicious colonies were yellowish spreading colonies on CCFA (i.e., acid production from fructose) and were confirmed by conventional methods (i.e., gas chromatography of metabolic fatty acids) and an agglutination assay for the C. difficile somatic antigen (Microscreen C. difficile latex confirmation assay, Lab M Limited, Heywood, UK).
Detection of C. difficiletoxin
C. difficile toxin testing was conducted in a cytotoxicity assay [16]. Briefly, monolayers of human fibroblasts (MRC-5, courtesy of W. Bossart, Institute of Medical Virology) were maintained in tubes with 2 ml of 10% fetal calf serum in Dulbecco’s minimal essential medium (DMEM, Gibco Life Technologies, Zug, Switzerland) and inoculated with 200 μl of the supernatant of a centrifuged stool suspension (diluted 1:10 in phosphate buffered saline, centrifuged for 10 minutes at 3000 g). After an overnight incubation at 37°C, the monolayers were observed for cytotoxic effects. In cases of positive cytotoxicity but a negative culture for C. difficile, the specificity of the cytotoxic effect by C. difficile cytotoxin was proved by a neutralization assay. Briefly, supernatants (100 μl) of the centrifuged stool suspensions were preincubated (30’, room temperature) with an equal volume of C. sordellii antitoxin (TechLab, Blacksburg, VA) that neutralizes C. difficile cytotoxin and, in parallel, with phosphate buffered saline, and both mixtures were retested in the cell-culture cytotoxicity assay. Neutralization of the cytotoxic effect by the antitoxin was considered confirmation of a positive toxin result [16]. From September 2006 to June 2007, all isolates of C. difficile were frozen in skim milk and stored at −80°C. From February 2008 to August 2008, isolates were kept in a chopped meat suspension and stored at room temperature. C. difficile strains from patients with a negative cytotoxin assay were further characterized by PCR. To detect the toxin genes tcdA and tcdB, a crude DNA extract was obtained. Toxin genes were detected in a multiplex PCR approach with the primers TA1/TA2 and TB1/TB2 as described [17].
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was done on subcultures of the isolates of C. difficile on brucella sheep blood agar (Difco) with the Etest (Biomérieux, Marcy-l’Etoile, France). Plates were incubated for 48 h under anaerobic conditions [18]. The Etest, based on predefined gradients of antibiotic concentrations on a plastic strip, allowed us to determine the exact MIC of anti-infective drugs. The following antibiotics were selected for susceptibility testing because of their association with CDI: amoxicillin/clavulanate, ceftriaxone, ciprofloxacin, clindamycin, meropenem, metronidazole, piperacillin/tazobactam and vancomycin. The MIC results were categorized in susceptible, intermediate and resistant, following the guidelines of CLSI (Performance Standards for Antimicrobial Susceptibility Testing; 23rd Informational Supplement; http://www.clsi.org/) or, if those were not available, with the values of EUCAST (European Committee on Antimicrobial Susceptibility Testing; http://www.eucast.org/).
Compilation of literature as related to antibiotic susceptibility of C. difficilestrains
We used “Clostridium difficile” and “susceptibility” as key words to identify articles reporting data on antibiotic susceptibility (MIC50, MIC90 or MIC distributions) of C. difficile. We focused our literature research on papers published between 1980 and 2013 since the focus of our work was to get a most comprehensive idea about resistance development over time and to compare our data from Zurich to published data.
Statistical analysis
Statistical analyses used Stata/SE 13.1 (Stata, College Station, TX). Odds ratios were calculated by comparing the usage of antibiotics in relation to the duration from admission to C. difficile–positive testing in the study population to the overall usage of antibiotics at the USZ in 2006-2008. Differences in the MICs between the data from IMM and EUCAST were calculated using the Wilcoxon rank-sum test and the non-parametric equality-of-medians test. The Wilcoxon-rank-sum test determines if the MICs from the two groups are from populations with the same distribution, and the equality-of-medians test tests the null hypothesis that the samples of the two groups were drawn from populations with the same median.
Results
Characteristics of the study population
The patient cohort (n = 94) was predominantly of Caucasian origin (i.e., 84% Swiss, 12.8% from the European Community, 1.1% from the U.S. and only 2.1% of Asian origin). Of the patients, 82/94 (87.2%) were inpatients, and 34/92 (37%) underwent surgery. The most common underlying diseases were malignancies and cardiovascular diseases (Additional file 1: Table S1).
Diarrhea was explicitly documented in the charts in 81.9% of the cases. Hospital directives do not indicate testing for C. difficile with no evidence of diarrhea. Thus, most likely all patients included in this study suffered from diarrhea even when documentation was missing. Average duration of diarrhea was 6.8 days (median: 3 days). In 46/94 (inpatients: 40, outpatients: 6) stool specimens, C. difficile grew in the culture, and its toxin was detected by the cell-culture cytotoxin assay directly from the stool samples (Table 1). In 38/94 (inpatients: 33, outpatients: 5) stool specimens, C. difficile grew, but the specimen was negative in the cytotoxin assay. In 17 of those 38 cultures, the toxin was detected at the DNA level by PCR. Also 10/94 specimens (inpatients: 9, outpatients: 1) were culture negative, but had a positive cytotoxin result confirmed by the neutralization test. In five of those specimens with negative culture, another specimen sample taken a couple of days apart from the one analyzed for the C. difficile toxin was available, which was then used for antibiotic susceptibility testing.
Table 1.
Specimen collection available
| Total number of patients | n = 94 | |||||||
|---|---|---|---|---|---|---|---|---|
| "Inpatients" n = 82 | "Outpatients" n = 12 | |||||||
| Clinical data sets | Data complete of n = 78 | Charts missing n = 4 | Data complete | |||||
| Stool analysis positive for toxin/culture; n = 94 | +/+ | −/+ | +/−* | +/+ | −/+ | +/+ | −/+ | +/−* |
| 38 | 31 | 9 | 2 | 2 | 6 | 5 | 1 | |
| Antibiotic susceptibility testing done in n = 86 | 36** | 31 | 4 | 2 | 2 | 6 | 4** | 1 |
*a stool specimen taken a couple of days apart with growth of C. difficile was used for antibiotic susceptibility testing; an alternative stool specimens was only available in 4/9 cases in the inpatient cohort.
**in two and one cases, respectively, from in- and outpatients, cultures were lost and no antimicrobial susceptibility testing could be done.
Risk factors and clinical presentation
Many risk factors contribute to CDI [19]. Risk factors were evaluated retrospectively before the first positive culture of C. difficile (Additional file 1: Table S1). Inpatients were hospitalized for 17.2 days in average (median 9 days). During their current hospitalization, 25 patients (26.6%) were in the ICU for at least 1 day. Thirty patients (32.6%) had one or more complications: 20 patients (21.7%) suffered from infectious complications, and further complications spanned the entire spectrum of medicine (Additional file 1: Table S1). Seven patients (7.4%) died shortly thereafter of causes unrelated to CDI.
In the 8 weeks preceding the first positive culture, 83.3% of the patients (i.e., in- and outpatients) were prescribed antimicrobials, 37.8% antifungal medication, 73.3% gastrointestinal medication, 37.8% immunosuppressive medication, and 20.0% cytostatic medication (Additional file 1: Table S1).
Penicillins (i.e., amoxicillin, amoxicillin/clavulanate and piperacillin/tazobactam) were the most commonly prescribed antibiotics (i.e., 38.5% of our cohort took penicillins with an average duration of 4.6 days (median 4 days), followed by cephalosporins (29.5%) and quinolones (26.9%) with an average duration of 6.8 and 4.9 days, respectively (median 6 and 2 days, respectively)) (Table 2). Further antimicrobials were carbapenems (15.4%), glycopeptides (10.3%), nitromidazoles (5.1%), lincosamides (2.6%), aminoglycosides (3.8%) and others (11.5%). Within the hospitalized patients, 25 were given one class of antibiotics while 34 patients were given more than one class before diagnosis of CDI (Additional file 2: Table S2).
Table 2.
Antimicrobial use during hospitalization and prior to detection of Clostridium difficile in stool
| Antimicrobials | Number of patients (%) N = 78† | Duration of treatment | Duration of treatment |
|---|---|---|---|
| Mean (+/− SD) | Median (range) | ||
| All Antimicrobials | 59 (75.6%) | 10.8 (11.0) | 7 (1-33) |
| Penicillins* | 30 (38.5%) | 4.6 (3.8) | 4 (1-12) |
| Cephalosporins** | 23 (29.5%) | 6.8 (31.5) | 6 (1-26) |
| Fluoroquinolones*** | 21 (26.9%) | 4.9 (6.6) | 2 (1-12) |
| Carbapenems# | 12 (15.4%) | 9.8 (6.5) | 4 (1-21) |
| Glycopeptides## | 8 (10.3%) | 5 (8.2) | 2 (1-25) |
| Nitroimidazoles### | 4 (5.1%) | 5.8 (4.6) | 5 (1-12) |
| Aminoglycosides†† | 3 (3.8%) | 2.7 (1.5) | 3 (1-4) |
| Clindamycin | 2 (2.7%) | 4 (1.4) | 4 (3-5) |
| Macrolides††† | 1 (1.3%) | 3 (0) | 3 (3) |
| Trimethoprim/Sulfamethoxazole | 9 (11.5%) | 4.7 (5.0) | 2 (1-14) |
†4 charts of inpatients were missing.
The following antibiotics were recorded:
*Penicillins: amoxicillin (n = 2), amoxicillin/clavulanate (n = 13) acid, piperacillin/tazobactam (n = 18).
**Cephalosporins: cefazolin (n = 2), cefepime (n = 12), ceftazidime (n = 1), ceftriaxone (n = 3), cefuroxime (n = 5).
***Fluoroquinolones: ciprofloxacin (n = 19), levofloxacin (n = 1), norfloxacin (n = 1).
#Carbapenems: ertapenem (n = 3), imipenem/cilastatin (n = 2), meropenem (n = 8).
##Glycopeptides: teicoplanin (n = 2), vancomycin (n = 6); all glycopeptides were given intravenously.
###Nitroimidazoles: metronidazole (n = 3), ornidazole (n = 1).
††Aminogylcosides: garamycin (n = 1), tobramycin (n = 2).
†††Macrolides: azithromycin (n = 1).
Antibiotic susceptibility pattern of C. difficile
We observed a uniform susceptibility to metronidazole, meropenem, and piperacillin/tazobactam, with one outlier for vancomycin. The one outlier was a true vancomycin-resistant strain or an erroneous MIC determination. We believe that the observed resistance was a laboratory artifact. Unfortunately, the strain was lost. Over 90% of all strains were susceptible to amoxicillin/clavulanate. In contrast, more than 90% of the strains were intermediate or resistant to clindamycin, ceftriaxone and ciprofloxacin (Table 3).
Table 3.
Antimicrobial Susceptibility of CDAD Isolates
| Antimicrobials (n = 86) | MIC interpretative criteria (μg/ml) (CLSI) | Susceptible | Intermediate | Resistant | ||
|---|---|---|---|---|---|---|
| S | I | R | ||||
| Amoxicillin/Clavulanate | ≤4/2 | 8/4 | ≥16/8 | 81 (94.2%) | 1 (1.2%) | 4 (4.7%) |
| Ceftriaxone | ≤16 | 32 | ≥64 | 3 (3.5%) | 60 (65.1%) | 33 (30.4%) |
| Ciprofloxacin@ | ≤2 | 4 | ≥8 | 0 | 1 (1.2%) | 85 (98.8%) |
| Clindamycin | ≤2 | 4 | ≥8 | 7 (8.1%) | 34 (39.5%) | 45 (52.3%) |
| Meropenem | ≤4 | 8 | ≥16 | 86 (100%) | 0 | 0 |
| Metronidazole¶ | ≤8 | 16 | ≥32 | 86 (100%) | 0 | 0 |
| Piperacillin/Tazobactam | ≤32 | 64 | ≥128 | 86 (100%) | 0 | 0 |
| Vancomycin* | ≤2 | >2 | 85 (98.8%) | 0 | 1 (1.2%) | |
@CLSI and EUCAST did not define breakpoints for ciprofloxacin. Here instead are the breakpoints given for moxifloxacin defined by CLSI which should closely approximate the breakpoints of ciprofloxacin.
¶Breakpoints for metronidazole by EUCAST: S ≤ 2; R2 mg/L.
*No breakpoints defined by CLSI; breakpoints presented by EUCAST.
We wondered whether certain anti-infective drugs were over-represented in our patient cohort and, thus, preferentially associated with CDI. We compared the antimicrobial usage of our patient cohort to the overall usage of antimicrobials in the USZ during the same period as defined by daily dose/100 bed days. Carbapenems were given considerably more often to the study population (Figure 1). The numbers of prescriptions for glycopeptides and sulfonamides were about the same in those two groups. On the other side, penicillins, fluoroquinolones, lincosamides, aminoglycosides, and macrolides were prescribed significantly less often than the cumulative usage of the hospital.
Figure 1.
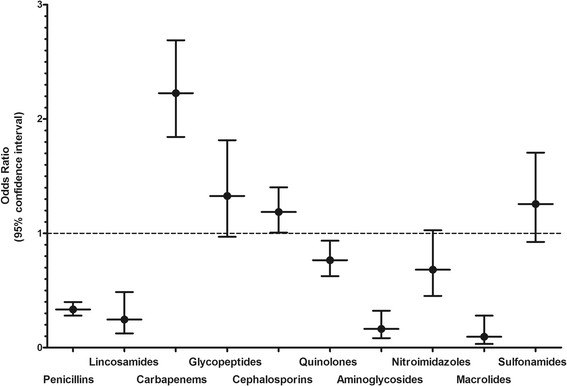
Patients treated with carbapenems or cephalosporins appear to have had a higher risk for developing CDI. Odds ratios were calculated by comparing the intake of the various antibiotics examined in our study with the defined daily intake of the antibiotics prescribed in the University Hospital of Zurich during the study period. The dashed line separates the antibiotics which are associated with a significant risk for CDI.
The MIC50 and 90 did not reveal a straightforward increase over the last three decades (Additional file 3: Table S3). A closer look, however, showed that the MIC is subtly increasing for amoxicillin/clavulanate, ceftriaxone, piperacillin/tazobactam and metronidazole. The MICs we measured, among all MICs reported, were always at the highest level. In the 1980s, ciprofloxacin and clindamycin already showed a sharp increase in the MIC50 and 90 [20]-[26].
MIC distribution of C. difficileto various antibiotics at the University Hospital of Zurich is similar to data from EUCAST
We compared the MIC distribution of the isolates described here with data from worldwide sources made available by EUCAST (http://mic.eucast.org/Eucast2/). The distributions of the epidemiological cut-off values (ECOFF) showed significant differences between the one obtained by the IMM and EUCAST (Figure 2) by the Wilcoxon rank-sum test for amoxicillin/clavulanate (p < 0.0001), clindamycin C p < 0.0001), ciprofloxacin (p < 0.0027), metronidazole (p = 0.0003), piperacillin/tazobactam (p < 0.00001) and vancomycin (p < 0.00001). Similar results were obtained for the difference of the medians of the two groups using the nonparametric equality-of-medians test (amoxicillin/clavulanate, clindamycin C, piperacillin/tazobactam, ciprofloxacin and vancomycin, p < 0.0001; meropenem, p = 0.012; metronidazole, p = .0.017).
Figure 2.
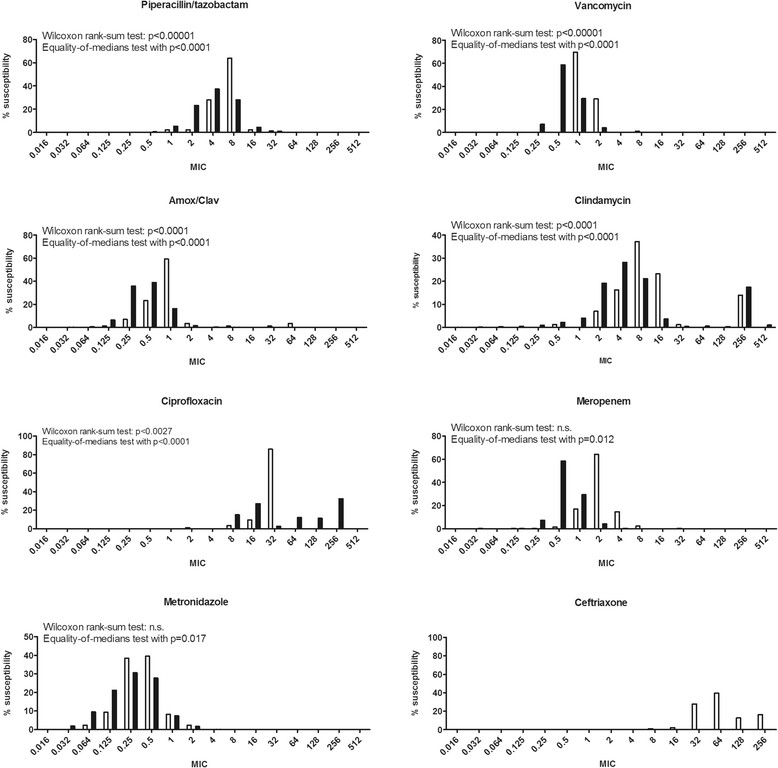
MIC from the collection of C. difficile from ZH and from EUCAST. The white bars were the data from ZH, the black bars from EUCAST. We compared the data from ZH with the data from EUCAST with the Wilcoxon rank-sum test and the nonparametric equality-of-medians test.
Discussion
Here, we determined the antibiotics that are associated with CDI in a tertiary hospital center in Switzerland and whether the antibiotic susceptibility of C. difficile strains influenced the occurrence of CDI. Our main findings are that i) the antibiotics mostly associated with CDI are carbapenems and cephalosporins, ii) the preserved microbial activity by some antibiotics to C. difficile does not prevent CDI, and iii) antibiotic susceptibility showed a trend to increases of the MIC50 and MIC90 of half of the tested antibiotics for the strains collected at the USZ. Very importantly, all C. difficile strains were susceptible to metronidazole and vancomycin, except one vancomycin-resistant strain that we believe was a laboratory artifact.
Patients with CDI had a wide variety of underlying diseases (Additional file 1: Table S1). Neoplastic, cardiovascular and gastrointestinal diseases were most common and probably reflect the frequency of hospitalization for these conditions. We observed no accumulation in any distinct division or ward, rendering highly unlikely that a local outbreak contributed to the number of CDI cases.
We made a major effort to assess complications that preceded the CDI. Infectious complications were by far the most frequent and self-explanatory complication as a trigger for CDI. All other complications occurred less frequently and were equally distributed with each representing 1-7% of the total (Additional file 1: Table S1).
Fifteen patients suffered from CDI although they had no antibiotic use in their history. These patients had a number of risk factors: 12/15 had proton pump inhibitors, and 7/15 suffered from malignancies treated with cytostatic and/or immunosuppressive drugs. Only one patient lacked identifiable risk factors. We know that proton pump inhibitors, for example, represents an independent risk factor for CDI [19]. Notably, 34/90 (37.8%) of patients received antifungal drugs. Some studies have reported that Candida in the GI-tract acts as protective factor against CDI, and that the use of antifungals drugs is an independent risk factor for CDI [27],[28]. We do not know the extent to which antifungal drugs contributed to the CDI in our cohort.
In our cohort, most cases of CDI occurred in patients treated with penicillins, followed by patients treated with cephalosporins, quinolones and carbapenems. However, we must take into account the rank order of prescribing the various antibiotics. Indeed, the defined daily dose/100 bed days was the highest for penicillins, followed by cephalosporins, quinolones and carbapenems (defined daily dose/100 bed days (DDD/100bd) in 2006, 2007, and 2008 for penicillins were 23.22, 25.77, and 26.10, for cephalosporins were 9.36, 10.45 and 9.67, for quinolones 8.78, 9.82, 9.9, for carbapenems 3.66, 4.45 and 4.08, respectively). Considering the rank order, we identified the administration of carbapenems and cephalosporins as a major risk factor for CDI in this cohort of patients. Glycopeptides and sulfonamides are most likely also a risk factor for CDI; however, the OR for these two antibiotic classes was not statistically significant. In a paradoxical finding, glycopeptides were identified as a potential trigger for CDI. The reason is most likely its co-administration with other broad-spectrum antibiotics and the fact that its intravenous application has no therapeutic role against C. difficile. Notably, in 9/11 cases where vancomycin was prescribed, the patients received a combination of antibiotics. It remains unclear whether penicillins (i.e., amoxicillin, amoxicillin/clavulanate and piperacillin/tazobactam) due to their inherent activity against C. difficile, and the aminoglycosides and macrolides due to their lack of activity against anaerobes are less likely associated with development of CDI. Our data agree with a recently published meta-analysis that reported the strongest association for cephalosporins and clindamycin with CDI and a lesser one for carbapenems, trimethoprim/sulphonamides, fluoroquinolones and penicillin combinations [29]. The differing results we observed for clindamycin, penicillins and for fluoroquinolones are surprising – it might be due to patient selection in the hospital or genetic differences in the C. difficile strains but in fact remains enigmatic.
A major goal was to assess the local susceptibility patterns of the strains isolated (Table 3) and to compare them to patterns of strains worldwide and over the last three decades (Additional file 3: Table S3). We observed uniform susceptibility of C. difficile vis-à-vis meropenem, tazobactam/piperacillin, and metronidazole. Only 1/86 isolates was resistant to vancomycin with an MIC measured of 6 μg/ml. This finding was striking since vancomycin resistant isolates are a rarity worldwide. We believe that this resistant strain is a laboratory artifact i.e., erroneous MIC determination. In any case, the reduced vancomycin activity would have no clinical relevance: vancomycin at the doses given perorally or rectally to treat CDI far exceeds the concentration of an MIC of 6 μg/ml. Most isolates were susceptible to amoxicillin/clavulanate. Note that EUCAST has defined MIC clinical breakpoints solely for metronidazole and vancomycin, and CLSI has them for most antibiotics but not for vancomycin and ciprofloxacin. The C. difficile strains showed mixed susceptibility to the other three antibiotics tested: the predominant majority of isolates were of intermediate susceptibility or resistant to ceftriaxone and clindamycin; less than 10% were still susceptible. All isolates but one was entirely resistant to ciprofloxacin.
Meropenem’ s antimicrobial activity against C. difficile does not appear to protect against CDI; patients on meropenem suffered more frequently from CDI. This is most likely due to the disruption of the GI flora and the overall morbidity of patients requiring carbapenems or co-medications, such as proton pump inhibitors. Very positively, the nitromidazoles and glycopeptides were still close to 100% active against C. difficile despite their frequent use.
We compared the local susceptibility pattern to the ECOFF (epidemiological cut-off value) data collection of EUCAST "European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org). The ECOFF aims to differentiate between wild-type and non-wild-type strains (acquired resistance mutations), based on MIC. Surprisingly, we found that the variance and the median of the MIC from ZH differ from the MICs from EUCAST. Overall, it appeared that the MIC for the antibiotics studied at ZH showed a shift to higher MIC apart from ciprofloxacin and metronidazole. MIC for ciprofloxacin was markedly higher for the data provided by EUCAST than for the data in ZH even though all strains in ZH were resistant. This difference in MIC might be related to ribotype 027, which is known for its particular high resistance to ciprofloxacin. We also observed two populations of strains when looking at the MIC for clindamycin and for amoxicillin/clavulanate. EUCAST does not report this separation in two populations for amoxicillin/clavulanate. The differences noted are most likely due to local prescription habits/directives; indeed, at the University Hospital of Zurich, the directives promote the use of amoxicillin/clavulanate and less of ciprofloxacin whenever appropriate, in particular because of the progressive increase of ciprofloxacin resistance of gram-negative bacteria.
The data collected by EUCAST, however, do not integrate the MIC evolution over time. A compilation of the literature since the first identification of C. difficile did not reveal a substantial change of antimicrobial susceptibility over time. In 1980, Dzink and Bartlett reported that over 95% of all strains tested were susceptible to vancomycin, penicillin G, ampicillin, and metronidazole, 60% of the strains were susceptible to clindamycin, and all strains were resistant to the cephalosporins tested (i.e., cephalotin and cefoxitin) [24]. Thus, the discord between the activities of penicillins and cephalosporins was observed already in the very first report. Looking at all data, there is tendency of increases in MIC for all antibiotics tested over the last three decades, but a clear picture is lacking (Additional file 3: Table S3).
Limitations of the study are the number of cases and the retrospective nature of this work. The number of cases we analyzed is in the range of cases presented in published work; nonetheless, we acknowledge that interpretations must be done cautiously due to the limited number of cases and the changes in microbiological assays over time. Our study was further limited by the lack of individual treatment information for the control patients. We therefore had to rely on hospital-wide prescribed DDDs of the different antimicrobials as a comparison group without information on which drugs were given simultaneously. This and the limited sample size precluded multivariable analyses including all antimicrobials which would have been preferable.
Conclusions
In summary, the antibiotic susceptibilities of the C. difficile samples from the University Hospital of Zurich are similar to those reported by others. Patients treated with carbapenems and cephalosporins had the highest risk of developing CDI, irrespective of the antimicrobial activity of carbapenems. Most likely, the underlying diseases or complications, in concert with the carbapenems’ activity on the GI-tract flora, rendered these patients more susceptible to CDI. Differences of MICs between the data from ZH vs EUCAST are minor and are most likely due to local prescriptions habits. Very importantly, all strains, but one, were fully susceptible to metronidazole and vancomycin the antibiotic cornerstone for the treatment of CDI.
Authors’ contributions
SKR, CR, RZ, SP and RFS designed the study; ACB did all the chart reviews and put the tables and figures together. SKR supervised the chart reviews. AS configured the initial data base for data collection. SS and BL did the statistics. Microbiology work was done by ACB, SP and RZ. ACB wrote the manuscript, SKR, RZ and RFS edited the manuscript. All authors read and approved the final manuscript.
Additional files
Electronic supplementary material
Additional file 1: Table S1: Patient characteristics prior to first C. difficile positive culture or toxin. (DOCX 36 KB)
Additional file 2: Table S2: Number of inpatients with multiple classes of antibiotics prior to CDI. (DOCX 18 KB)
Additional file 3: Table S3: Susceptibility of C. difficile to various antibiotics over time. (DOCX 125 KB)
Below are the links to the authors’ original submitted files for images.
Acknowledgements
We thank Ms. S. Blasowitsch for her excellent technical assistance in the laboratory.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Andrea C Büchler, Email: andrea.buechler@gmx.ch.
Silvana K Rampini, Email: Silvana.rampini@usz.ch.
Simon Stelling, Email: simon.stelling@ergon.ch.
Bruno Ledergerber, Email: infled@usz.uzh.ch.
Silke Peter, Email: silke.peter@med.uni-tuebingen.d.
Alexander Schweiger, Email: Alexander.Schweiger@spital-schwyz.ch.
Christian Ruef, Email: Christian.ruef@hirslanden.ch.
Reinhard Zbinden, Email: rzbinden@imm.uzh.ch.
Roberto F Speck, Email: Roberto.speck@usz.ch.
References
- 1.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298(10):531–534. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 2.Kuntz JL, Chrischilles EA, Pendergast JF, Herwaldt LA, Polgreen PM. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case–control study. BMC Infect Dis. 2011;11:194. doi: 10.1186/1471-2334-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor NS, Thorne GM, Bartlett JG. Comparison of two toxins produced by Clostridium difficile. Infect Immun. 1981;34(3):1036–1043. doi: 10.1128/iai.34.3.1036-1043.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan NM, Pellett S, Wilkins TD. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35(3):1032–1040. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noren T. Clostridium difficile and the disease it causes. Methods Mol Biol. 2010;646:9–35. doi: 10.1007/978-1-60327-365-7_2. [DOI] [PubMed] [Google Scholar]
- 6.Karas JA, Enoch DA, Aliyu SH. A review of mortality due to Clostridium difficile infection. J Infect. 2010;61(1):1–8. doi: 10.1016/j.jinf.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Tschudin-Sutter S, Widmer AF, Perl TM. Clostridium difficile: novel insights on an incessantly challenging disease. Curr Opin Infect Dis. 2012;25(4):405–411. doi: 10.1097/QCO.0b013e32835533a2. [DOI] [PubMed] [Google Scholar]
- 8.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 9.Shen A. Clostridium difficile toxins: mediators of inflammation. J Innate Immun. 2012;4(2):149–158. doi: 10.1159/000332946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 11.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353(23):2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 12.Gerding DN. Clostridium difficile 30 years on: what has, or has not, changed and why? Int J Antimicrob Agents. 2009;33(Suppl 1):S2–S8. doi: 10.1016/S0924-8579(09)70008-1. [DOI] [PubMed] [Google Scholar]
- 13.Baxter R, Ray GT, Fireman BH. Case–control study of antibiotic use and subsequent Clostridium difficile-associated diarrhea in hospitalized patients. Infect Control Hosp Epidemiol. 2008;29(1):44–50. doi: 10.1086/524320. [DOI] [PubMed] [Google Scholar]
- 14.Bignardi GE. Risk factors for Clostridium difficile infection. J Hosp Infect. 1998;40(1):1–15. doi: 10.1016/S0195-6701(98)90019-6. [DOI] [PubMed] [Google Scholar]
- 15.Phillips ST, Nagaro K, Sambol SP, Johnson S, Gerding DN. Susceptibility of hamsters to infection by historic and epidemic BI Clostridium difficile strains during daily administration of three fluoroquinolones. Anaerobe. 2011;17(4):166–169. doi: 10.1016/j.anaerobe.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Chang TW, Lauermann M, Bartlett JG. Cytotoxicity assay in antibiotic-associated colitis. J Infect Dis. 1979;140(5):765–770. doi: 10.1093/infdis/140.5.765. [DOI] [PubMed] [Google Scholar]
- 17.Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. 2002;40(9):3470–3475. doi: 10.1128/JCM.40.9.3470-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Citron DM, Ostovari MI, Karlsson A, Goldstein EJ. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J Clin Microbiol. 1991;29(10):2197–2203. doi: 10.1128/jcm.29.10.2197-2203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janarthanan S, Ditah I, Adler DG, Ehrinpreis MN. Clostridium difficile-associated diarrhea and proton pump inhibitor therapy: a meta-analysis. Am J Gastroenterol. 2012;107(7):1001–1010. doi: 10.1038/ajg.2012.179. [DOI] [PubMed] [Google Scholar]
- 20.Chow AW, Cheng N, Bartlett KH. In vitro susceptibility of Clostridium difficile to new beta-lactam and quinolone antibiotics. Antimicrob Agents Chemother. 1985;28(6):842–844. doi: 10.1128/AAC.28.6.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmee M, Avesani V. Comparative in vitro activity of seven quinolones against 100 clinical isolates of Clostridium difficile. Antimicrob Agents Chemother. 1986;29(2):374–375. doi: 10.1128/AAC.29.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clabots CR, Peterson LR, Gerding DN. Characterization of a nosocomial Clostridium difficile outbreak by using plasmid profile typing and clindamycin susceptibility testing. J Infect Dis. 1988;158(4):731–736. doi: 10.1093/infdis/158.4.731. [DOI] [PubMed] [Google Scholar]
- 23.Cheng SH, Chu FY, Lo SH, Lu JJ. Antimicrobial susceptibility of Clostridium difficile by E test. J Microbiol Immunol Infect. 1999;32(2):116–120. [PubMed] [Google Scholar]
- 24.Dzink J, Bartlett JG. In vitro susceptibility of Clostridium difficile isolates from patients with antibiotic-associated diarrhea or colitis. Antimicrob Agents Chemother. 1980;17(4):695–698. doi: 10.1128/AAC.17.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura S, Nakashio S, Mikawa M, Yamakawa K, Okumura S, Nishida S. Antimicrobial susceptibility of Clostridium difficile from different sources. Microbiol Immunol. 1982;26(1):25–30. doi: 10.1111/j.1348-0421.1982.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 26.Gianfrilli P, Luzzi I, Pantosti A, Occhionero M. In vitro susceptibility of Clostridium difficile isolates to 12 antimicrobial agents. Chemioterapia. 1984;3(1):41–44. [PubMed] [Google Scholar]
- 27.Hsu MS, Wang JT, Huang WK, Liu YC, Chang SC. Prevalence and clinical features of Clostridium difficile-associated diarrhea in a tertiary hospital in northern Taiwan. J Microbiol Immunol Infect. 2006;39(3):242–248. [PubMed] [Google Scholar]
- 28.Manian FA, Bryant A. Does Candida species overgrowth protect against Clostridium difficile infection? Clin Infect Dis. 2013;56(3):464–465. doi: 10.1093/cid/cis854. [DOI] [PubMed] [Google Scholar]
- 29.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother. 2014;69(4):881–891. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Patient characteristics prior to first C. difficile positive culture or toxin. (DOCX 36 KB)
Additional file 2: Table S2: Number of inpatients with multiple classes of antibiotics prior to CDI. (DOCX 18 KB)
Additional file 3: Table S3: Susceptibility of C. difficile to various antibiotics over time. (DOCX 125 KB)


