SUMMARY
Most E3 ligases use a RING domain to activate a thioester-linked E2~ubiquitin-like protein (UBL) intermediate and promote UBL transfer to a remotely bound target protein. Nonetheless, RING E3 mechanisms matching a specific UBL and acceptor lysine remain elusive, including for RBX1, which mediates NEDD8 ligation to cullins and >10% of all ubiquitination. We report the structure of a trapped RING E3-E2~UBL-target intermediate representing RBX1-UBC12~NEDD8-CUL1-DCN1, which reveals the mechanism of NEDD8 ligation and how a particular UBL and acceptor lysine are matched by a multifunctional RING E3. Numerous mechanisms specify cullin neddylation while preventing noncognate ubiquitin ligation. Notably, E2-E3-target and RING-E2~UBL modules are not optimized to function independently, but instead require integration by the UBL and target for maximal reactivity. The UBL and target regulate the catalytic machinery by positioning the RINGE2~UBL catalytic center, licensing the acceptor lysine, and influencing E2 reactivity, thereby driving their specific coupling by a multifunctional RING E3.
INTRODUCTION
Ubiquitin-like protein (UBL) modification is a key eukaryotic mechanism for regulating protein function. For example, ubiquitin (UB) and SUMO are ligated either individually or as polyUB or polySUMO chains to a massive segment of the proteome, transforming target properties such as half-life, subcellular localization, or intermolecular interactions. By contrast, the UBL NEDD8 is exceptionally selective and chiefly modifies closely related cullin proteins (CULs) on a single conserved lysine. CULs constitutively associate with an RBX RING E3 and nucleate the Cullin-RING UB Ligase (CRL) superfamily. By stim ulating CRL activity and assembly, NEDD8 ligation to CULs controls ≈ 10%–20% of all cellular ubiquitination (Soucy et al., 2009). Notably, an inhibitor of NEDD8 conjugation is in anti-cancer clinical trials (Soucy et al., 2009) and also counteracts Vif-dependent HIV infectivity (Stanley et al., 2012).
Given the distinct functions of different UBL modifications, and the therapeutic potential for modulating their conjugation, a central challenge is to determine how a particular UBL is matched with a specific target. This involves cascades of E1, E2, and E3 enzymes. An E1-activated UBL is loaded onto an E2 catalytic cysteine, producing a transient thioester-bonded E2~UBL intermediate (here, covalent interactions are denoted with “~,” noncovalent complexes with “p=n-”). Most E3s, including ≈ 600 predicted RING E3s in humans, interact with dedicated subsets amongz30 E2~UBL intermediates to promote transfer of a UBL's C terminus from an E2 active site to a target's acceptor lysine or N terminus (here, this aminolysis reaction producing an isopeptide-bonded UBL~target complex is termed “ligation”; the UBL to be transferred is “donor” and site of ligation is “acceptor”) (Deshaies and Joazeiro, 2009; Metzger et al., 2014).
Current models posit that RING E3s are modular molecular machines (Deshaies and Joazeiro, 2009; Metzger et al., 2014): a protein interaction domain engages a motif distal from the acceptor lysine in the target protein, and a RING domain recruits and activates an E2~UBL intermediate. E3 RING and non-RING elements, the E2, and the donor UB interact with each other through surfaces remote from the active site to stabilize a closed E2~UB conformation that immobilizes and primes the thioester bond for nucleophilic attack (Dou et al., 2012b, 2013; Plechanovová et al., 2012; Pruneda et al., 2012). Within a RING E3-substrate complex, the RING domain, substrate-binding domain, and different domains within a substrate can rotate relative to each other. Thus, RING E3s are thought to loosely connect the remotely bound substrate to the activated RINGE-E2~UBL intermediate (Deshaies and Joazeiro, 2009; Metzger et al., 2014).
Paradigms for E2 selection of target lysines have been established by a few studies of SUMOylating and polyubiquitinating E2s that generally choose acceptor lysines by recognizing surrounding side chains. Structures of the SUMO E2, UBC9, bound to the target RanGAP also revealed E2 side chains directly binding the acceptor lysine and accelerating catalysis (Bernier-Villamor et al., 2002; Reverter and Lima, 2005; Yunus and Lima, 2006). One of these, an aspartate, is missing from the polyubiquitinating E2 UBE2S and the corresponding function is instead mediated by a glutamate proximal to the acceptor Lys11 in its target Ub (Wickliffe et al., 2011). A different mechanism is used by the E2 UBC13: a rigid adaptor protein places UB's acceptor Lys63 at the active site (Eddins et al., 2006). This raises the question of whether the numerous uncharacterized RING E3s and E2s use similar or divergent mechanisms for acceptor lysine targeting. Also, many RING E3s are multifunctional, interacting with different E2s to modify distinct targets, to transfer different UBLs, and/or to separately initiate and elongate UB chains (Deshaies and Joazeiro, 2009; Metzger et al., 2014). How a multifunctional RING E3 could steer a particular E2~UBL toward its specific substrate acceptor lysine(s) remains elusive.
RBX1 is a multifunctional RING E3 that acts sequentially with three E2s (UBC12, UBCH5, CDC34) to modify distinct targets with either NEDD8 or UB (Duda et al., 2011; Zimmerman et al., 2010). For simplification, we describe activities of human RBX1 associated with CUL1 to represent RBX-CUL complexes. First, RBX1 promotes NEDD8 ligation to Lys720 in the “WHB subdomain” of CUL1's C-terminal domain (CTD). Here, RBX1's N-terminal domain, a β strand constitutively anchored in CUL1's CTD, is the substrate-binding domain. RBX1's C-terminal RING, essential for cullin neddylation, binds and activates the UBC12~NEDD8 intermediate. A co-E3, DCN1, enhances this reaction by its PONY domain (DCN1P) binding CUL1 and UBC12's acetylated N terminus (Kim et al., 2008; Kurz et al., 2008; Monda et al., 2013; Scott et al., 2010, 2011). When assembled with NEDD8-modified CUL1 and one of 69 different human F-box proteins into a neddylated “SCF” type of CRL E3, RBX1 promotes UB transfer from the Cys of either UBCH5 or CDC34 to a substrate recruited to the F-box protein (Jin et al., 2004; Wu et al., 2010). Finally, in the NEDD8-modified SCF, RBX1 collaborates with CDC34ȈUB to mediate processive substrate polyubiquitination (Kleiger et al., 2009b; Pierce et al., 2009).
CRL regulation, and NEDD8 and UB ligase activities, depends on a flexible linker connecting RBX1's N-terminal and RING domains, allowing different relative domain orientations for different functions. Six prior structures showed the RING packing against the CUL's WHB subdomain (Angers et al., 2006; Duda et al., 2008; Fischer et al., 2011; Goldenberg et al., 2004; Zheng et al., 2002). While this RING conformation enables binding to the neddylation inhibitor/F-box protein exchange factor CAND1, it is catalytically inactive: docking an E2 on RBX1's RING places the E2 active site >30Å from CUL1's acceptor Lys720 and >50Å from an F-box protein bound substrate (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008). For neddylation, modeling UBC12 on a CUL1CTD-RBX1 structure (3RTR) with a reoriented RING revealed UBC12's active site close to but displaced from CUL1's acceptor Lys720 and did not provide a mechanism juxtaposing CUL1's Lys720 and UBC12's active site (Calabrese et al., 2011). Structures with neddylated CUL5CTD (3DQV) showed relative relocation of the neddylated cullin WHB and rotation of RBX1's RING, although it is not known how the RBX1-E2~UB module would be directed to a ubiquitination substrate (Duda et al., 2008). Thus, how any active RBX1-E2~UBL-substrate conformation is achieved remains incompletely understood. Indeed, there is no structure of any RING or RING-like E3-E2~UBL-substrate complex—cullin or otherwise—showing structural mechanisms by which a RING E3 promotes adjoining a specific donor UBL and substrate acceptor lysine.
Here, we address this problem and define the mechanism of NEDD8 ligation to CUL1 with a crystal structure representing an RBX1-UBC12~NEDD8-CUL1-DCN1 intermediate. This structure of a RING E3 trapped as if mediating UBL ligation also provides a framework for understanding RING E3 ligation specificity.
RESULTS
UBL and Acceptor Lys Specificity in RBX1-Mediated CUL1 Modification
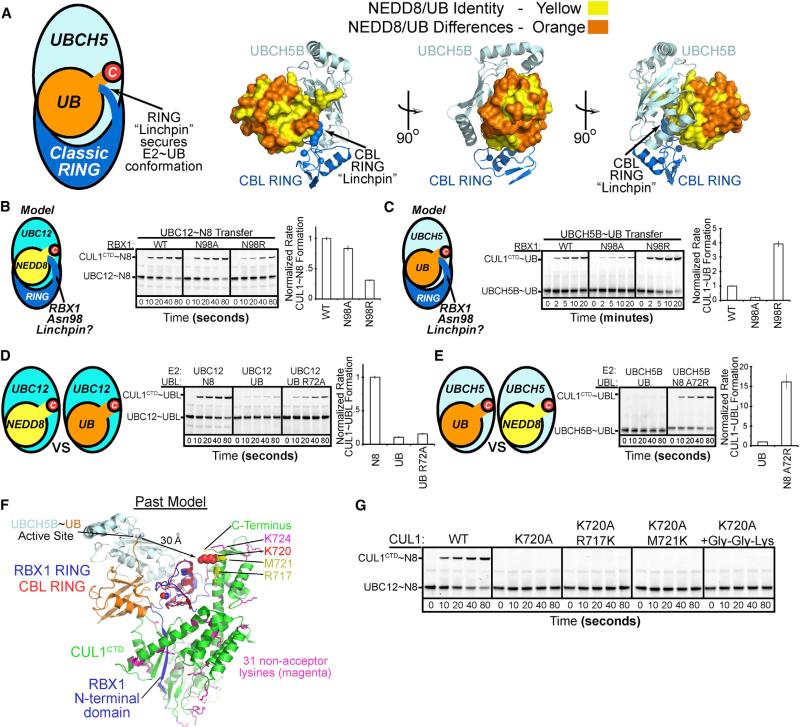
NEDD8 ligation to the acceptor Lys720 is recapitulated for RBX1 bound to CUL1CTD, even in the absence of DCN1 (Huang et al., 2009). We used this to test paradigms for RING E3 ligation. We first asked if RBX1 stimulates UBC12~NEDD8 ligation via a canonical catalytic “linchpin” identified in E3-UBCH5 UB structures, where a RING arginine cements the E2 UBCH5 and UB in the closed, active conformation (Figure 1A) (Dou et al., 2012b, 2013; Plechanovová et al., 2012; Pruneda et al., 2012). Ala mutation of RBX1's corresponding Asn98 had no effect in pulse-chase assays of NEDD8 transfer from UBC12 to CUL1CTD, while N98R impaired neddylation (Figure 1B). To test if this is specific for neddylation and if RBX1 activates UBCH5 UB through canonical mechanisms, we performed parallel experiments based on RBX1 promoting UB ligation from UBCH5 to CUL1 in vitro, albeit over minutes under conditions where CUL1 is neddylated within seconds (Duda et al., 2008; Wu et al., 2003a). Interestingly, reactions with UBCH5~UB displayed opposite mutational effects: N98A reduced and N98R stimulated UB ligation efficiency, consistent with RING-UBCH5~UB structures (Figure 1C). Comparing RBX1-mediated CUL1 modification with UBC12 suggested: NEDD8 and UBCH5 side-by-side (1) RBX1 uses a variant UB mechanism to stabilize an active UBC12ȈNEDD8 intermediate, (2) the RBX1 RING is not optimized for UBCH5~UB ligation to a cullin, and (3) RBX1 is more efficient at promoting CUL1 modification by UBC12~NEDD8 than UBCH5~UB.
Figure 1. Specificity of RBX1-Mediated NEDD8 Ligation to CUL1.
(A) Canonical RING-E2~UB architecture, highlighting the linchpin and conservation of E2 and RING binding residues in UB and NEDD8, shown for CBL-UBCH5~UB (Dou et al., 2013).
(B) Mutational analysis of RBX1's Asn98, corresponding to canonical RING linchpin, in RBX1-mediated pulse-chase fluorescent NEDD8 transfer from UBC12 to CUL1CTD. Graph, rate compared to wild-type (WT) RBX1; error, 1 SD.
(C) Mutational analysis of RBX1's Asn98, aka canonical RING linchpin, in RBX1-mediated pulse-chase fluorescent UB transfer from UBCH5B to CUL1CTD. Graph, rate compared to WT RBX1; error, 1 SD.
(D) Role of UBL in RBX1-mediated CUL1 modification assayed by comparing pulse-chase NEDD8, UB, or UB R72A transfer from NEDD8's E2 UBC12. The noncognate UBL UB was loaded on UBC12 in the pulse reaction either through its R72A mutation or use of E1 mutant UBA3 R190Q. Graph, rate compared to WT UBC12~NEDD8; error, 1 SD.
(E) Role of UBL in RBX1-mediated CUL1 modification assayed by comparing UB or NEDD8 A72R transfer from UBCH5B to CUL1CTD. The NEDD8 A72R mutation allows loading this noncognate UBL on UBCH5B in the pulse reaction. Graph, rate compared to WT UBCH5~UB; error, 1 SD.
(F) CUL1CTD Lys locations. Docking RBX1's RING from prior structures with that from CBL-UBCH5~UB revealed a >30Å gap between the active site and K720 (red) or other lysines (native in magenta, introduced in yellow) in CUL1 (Dou et al., 2013; Zheng et al., 2002).
(G) Acceptor Lys specificity for NEDD8 transfer from UBC12 to CUL1CTD or Lys mutants. Gly-Gly-Lys, appended to C terminus of CUL1CTD K720A.
Are the different transfer efficiencies of UBC12~NEDD8 and UBCH5~UB toward CUL1 explained by a “central dogma” of the UB/UBL field—that cognate E1-E2-E3 enzymes are responsible for matching a UBL with its specific target? RBX1 and UBC12 sequences are distinctive among RINGs and E2s, respectively, whereas NEDD8 is 100% identical to UB in key residues defined by RING-UBCH5~UB structures (Figure 1A). However, we obtained surprising results upon assaying RBX1-mediated CUL1CTD modification by a series of noncognate E2~UBL intermediates—UBC12~UB, UBC12~UB (R72A), and UBCH5~NEDD8 (A72R)—generated with altered-specificity mutants. (NEDD8's E1 or the R190Q mutant load UBC12 with UB R72A or UB, respectively, and UB's E1 loads UBCH5 with NEDD8 A72R; see Supplemental Information) Ligation efficiencies of UBC12~UB and UBC12~UB (R72A) were substantially reduced (Figure 1D). In striking contrast, NEDD8 (A72R) enabled efficient ligation from UBCH5 (Figure 1E). Thus, counter to current dogma, the identity of the UBL dictates specificity of RBX1-mediated ligation from UBC12 or UBCH5 to a cullin.
Finally, we considered the basis for lysine targeting by NEDD8. Although mutation of CUL1's Lys720 suppresses neddylation, one trivial explanation may be that other CUL1 lysines are spatially inaccessible (Figure 1F). However, introduction of lysines surrounding the K720A substitution, or appending Gly-Gly-Lys to the C terminus nearby, also fails to rescue CUL1CTD neddylation (Figure 1G). Thus, mechanisms precisely matching a particular UBL and target remain elusive.
RBX1-UBC12~NEDD8-CUL1-DCN1 Structure Trapped in Action
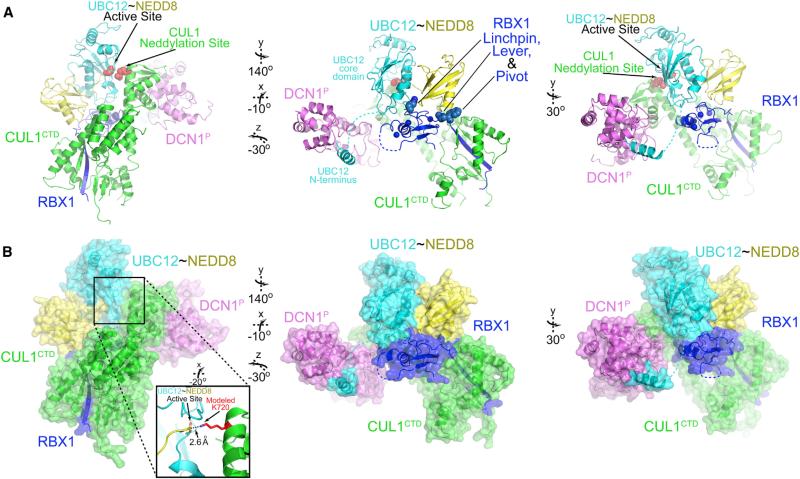
To decipher a mechanism for RING E3-mediated UBL-substrate pairing and the basis for cullin neddylation, we determined the crystal structure of a stabilized version of a NEDD8 ligation intermediate at 3.1Å resolution (Figure 2, Figure S1 available online; Movie S1). NEDD8's C terminus is covalently linked to a Ser replacing the catalytic Cys111 in a modified UBC12 (Scott et al., 2011; Wu et al., 2003b). The oxyester-bonded UBC12ȈNEDD8 intermediate was stable in complex with RBX1, DCN1P, and CUL1CTD harboring a K720R mutation to prevent NEDD8 transfer. Notably, relative to prior RBX1-CUL structures, the RING domain is reoriented in a unique architecture for NEDD8 ligation.
Figure 2. Structure of RBX1-UBC12~NEDD8-CUL1CTD-DCN1P —A RING E3 in Action.
(A) Structure of neddylation complex as cartoon, with active site and CUL1 acceptor residue 720 in red, and RBX1 “linchpin, lever, and pivot” spheres in center panel.
(B) Surface representation. Inset: model of catalytic center.
The heart of the complex is the active site, in which the UBC12~NEDD8 covalent linkage and CUL1 target site are juxtaposed and sequestered within UBC12's catalytic center (Figure 2A). A modeled CUL1 acceptor lysine's ε-amino group is poised for ligation at 2.6Å from NEDD8's C-terminal carbon (Figure 2B).
The means by which UBC12~NEDD8 is steered to the CUL1 acceptor differs from previously described mechanisms where an E2 directly recruits a substrate (Bernier-Villamor et al., 2002; Wickliffe et al., 2011). Here, side chain contacts between UBC12 and CUL1 are sparse and there was little effect of mutating side chains at the UBC12-CUL1 interface, with the exception of UBC12's Arg116 hydrogen bonding to the backbone of CUL1's penultimate Leu775 (Figure S1D).
Below, we compare catalytic modules for neddylation with those defined for SUMOylation and ubiquitination and discuss how the ligation architecture is achieved by NEDD8 steering the E3-E2~UBL active site to CUL1, CUL1 structure licensing the acceptor Lys720, and the properly positioned acceptor lysine toggling E2 reactivity to promote NEDD8 transfer.
Distinctive Features of Catalytic Modules Contribute to Ligation and Specificity
The RBX1 RING assembly with the catalytic core domain from UBC12 and its covalently linked NEDD8 broadly resembles other E3-E2~UBL complexes primed for ligation (Dou et al., 2012b, 2013; Plechanovová et al., 2012; Pruneda et al., 2012; Reverter and Lima, 2005), with distinct features contributing specificity. UBC12~NEDD8 adopts its own variation of the closed conformation (Figure 3A). UBC12's Asn113 fixes NEDD8's C-terminal tail, and UBC12's Tyr130 in the E2 core domain contacts NEDD8's Ile44 (Figures 3B and 3C). Although RING-UBCH5~UB intermediates display analogous alignment of UB's C-terminal tail by UBCH5's Asp87, and similar covalent linkage to UBCH5, UB's globular domain is relatively displaced: UB's Ile44 contacts UBCH5's Ser108, which corresponds to UBC12's Tyr134 a helical turn from Tyr130. Each E2~UBL intermediate's own closed conformation is important for ligation, as revealed by the relative locations of compensatory mutant pairs: E2 Leu mutants projecting methyl groups into the interface can compensate for those removed by corresponding UBL I44A mutations (Saha et al., 2011). UBC12 Y130L and UBCH5B S108L mutations rescue NEDD8 or UB I44A mutations, respectively (Figures 3D and 3E).
Figure 3. RING-E2 UBL-Acceptor Modules for Neddylation.
(A) Different RING-E2~UBL closed conformations shown with the E2 catalytic core domain aligned for RBX1-UBC12~NEDD8-CUL1CTD-DCN1P and CBL-UBCH5~UB (Dou et al., 2013).
(B) Close-up of E2~UBL interfaces from (A).
(C) Variation in E2~UBL interfaces shown with UBLs aligned from RBX1-UBC12~NEDD8-CUL1CTD-DCN1P and CBL-UBCH5~UB.
(D) Pulse-chase assays for compensation between UBC12 Leu mutants and NEDD8 I44A in RBX1-mediated CUL1 modification.
(E) Pulse-chase assays testing compensation between UBCH5 Leu mutant and UB I44A in RBX1-mediated CUL1 modification.
(F) Different RING-E2~UBL active site presentations highlighted by relative placement of UBC12 D143 and UBCH5B D117 side chains, viewed with UBLs aligned from RBX1-UBC12~NEDD8-CUL1CTD-DCN1P (CUL1 acceptor K720 modeled in place of Arg) and CBL-UBCH5~UB. UBC12 D143 carbonyl is also shown in sticks.
(G) Different E2~UBL active site presentations in UBC12~NEDD8-CUL1 and UBC9-SUMO~RANGAP1 (Reverter and Lima, 2005), oriented by aligning UBC9 on UBCH5 from (F) and highlighting different relative positions of UBC12 D143 and UBC9 D127. UBC12 D143 carbonyl is also shown in sticks.
(H) RBX1-mediated pulse-chase fluorescent NEDD8 transfer from UBC12 to CUL1CTD testing roles of side chains from RBX1's N113, which aligns NEDD8's C-terminal tail, and D143, opposite the UBC12~NEDD8 bond.
(I) RBX1's distinct linchpin Arg46 in spheres, shifted across RING domain from location of canonical linchpin as typified in CBL. RING-E2~UBL portions of RBX1-UBC12~NEDD8-CUL1CTD-DCN1P and CBL-UBCH5~UB structures shown with UBLs aligned.
(J) Linchpins (blue) in human RING domain sequences. Red, zinc ligands; green, E2-binding residue; purple, RBX1 pivot; yellow, lever.
(K) RBX1-mediated pulse-chase fluorescent NEDD8 transfer from UBC12 to CUL1CTD testing role of RBX1 R46 linchpin.
(L) Pulse-chase assays testing role of NEDD8's Leu8 and Thr9 from the b1-b2 loop in RBX1-mediated transfer from UBC12 to CUL1CTD.
See also Figure S2.
Similar binding between NEDD8 and UB to RING domains, coupled with different relative orientations of the UBL globular domains and E2s, leads to subtle variation in active site presentations, which appears to influence acceptor Lys recognition. Comparing different E2~UBL active sites (typically cysteines but in structures mutated to Ser or Lys) shows UBL C-termini aligned but the opposing surface relatively displaced (Figures 3F and 3G) (Dou et al., 2012b; Plechanovová et al., 2012; Reverter and Lima, 2005; Yunus and Lima, 2006). A modeled CUL1 acceptor lysine's ε-amino group contacts the UBC12~NEDD8 oxyester bond and the backbone carbonyl but not the side chain from UBC12's Asp143. However, prior structures showed UBC9's Asp side chain contacting the acceptor Lys, a configuration paralleled in models with UBCH5. As with the important role for UBCH5's Asp87 in ubiquitination, mutation of UBC12's Asn113, which aligns NEDD8's C-terminal tail, impairs ligation. Neddylation is not substantially affected for UBC12 D143A (Figure 3H), whereas the corresponding Asp is essential for ligation of SUMO by UBC9 and of UB by UBCH5.
The RBX1 RING binds UBC12 NEDD8 via a largely canonical RING-E2~UBL interface, consistent with mutation and NMR data (Calabrese et al., 2011; Huang et al., 2009). However, RBX1's Asn98 corresponding to the canonical “linchpin” does not contact UBC12. Furthermore, a modeled canonical linchpin Arg would repel UBC12's Lys120, explaining why the RBX1 N98R mutation hinders neddylation (Figures 1B, 3I, and S2A).
How then, does RBX1's RING activate? A distinct linchpin is located on the opposite side of the RING domain: RBX1's Arg46, conserved among RBXs across evolution, “glues” NEDD8's β1-β2 loop (residues 7–10) to the back and base of UBC12 (Figures 3I and 3J). Indeed, mutating RBX1's Arg46, or NEDD8's Leu8 and Thr9 from the b1-b2 loop, hinders neddylation (Figures 3K and 3L). The alternative linchpin location may enable RBX1 to cater to individual features of multiple cognate E2s (Figure S2B). Although future studies will be required to overcome challenges of neddylating the RBX1 R46A mutant to test its effects on SCF ubiquitination, this RBX1 region was implicated by NMR as binding to CDC34~UB (Spratt et al., 2012). Furthermore, modeling an RBX1 complex with CDC34 in place of UBC12 shows potential for an acidic loop essential for CDC34-mediated UB ligation to clash with a canonical linchpin (Duda et al., 2012; Huang et al., 2014; Petroski and Deshaies, 2005; Spratt et al., 2012; Ziemba et al., 2013). Accordingly, despite its role in activating UBCH5~UB, the N98R substitution in RBX1 impairs UB ligation from CDC34 (Figures S2C–S2E).
NEDD8 Directs an RBX1 Lever to Steer the Active Site to the CUL1 Acceptor Lysine
The catalytic assembly is organized by three-way interactions between RBX1, UBC12's E2 core domain, and its covalently linked NEDD8. A key feature is that NEDD8 positions the active site via interactions more than 30Å away, by coordinating RBX1's N-terminal strand anchored to CUL1, the subsequent linker, and C-terminal RING domain (Figures 4A–4C). RBX1's linker contacts NEDD8 and aids the RING in stabilizing the closed UBC12~NEDD8 conformation. Perhaps more importantly, the linker is reciprocally regulated by the donor NEDD8. The carbonyl from NEDD8's Lys33 forms a hydrogen bond with the amide from RBX1's Val38, and the backbone spanning NEDD8's Glu34 to Gly35 makes van der Waals contacts with RBX1's Ile37 to rigidify RBX1's linker into a unique conformation that has not been previously observed.
Figure 4. NEDD8 Pushes an RBX1 Lever and Directs an RBX1 Pivot to Juxtapose the Active Site and Acceptor Lys.
(A) RBX1 RING domain and W35 “pivot” positions in neddylation complex compared with CUL1-RBX1-CAND1 (Goldenberg et al., 2004; Zheng et al., 2002), with CUL1 aligned.
(B) Close-up of contacts between UBC12-linked donor NEDD8 and RBX1 linker. Zinc atoms bound to RING are shown as spheres.
(C) As in (B) except with NEDD8 in surface colored by identity with UB. RING and UBC12-binding residues are identical between NEDD8 and UB (yellow), but exposed surfaces and contacts to RBX1 pivot differ (orange).
(D) Comparison of effects of Ala mutations in place of RBX1 “lever” (I37) or other linker residues in RBX1-mediated NEDD8 transfer from UBC12 to CUL1CTD. Graph, rate compared to WT RBX1; error, 1 SD.
(E) Same as (D) but with variants of RBX1 Trp35 pivot.
(F) Same as (E) but monitoring fluorescent UB transfer from UBCH5B.
(G) Immunoblot comparing RBX1-mediated transfer of WT NEDD8, or variants with corresponding E31Q and E32D from UB, from UBC12 to CUL1CTD. Graph, rate compared to WT UBC12~NEDD8; error, 1 SD.
(H) RBX1-mediated transfer of fluorescent UB, or “neddylized” variants with Q31E and D32E, from UBCH5B to CUL1CTD. Graph, rate compared to WT UBCH5~UB; error, 1 SD.
(I) Schematic of roles and orientations of RBX1 elements and interactions with UBC12Pulse-chase assays forNEDD8, oriented as in prior RBX1-CUL1 structures or RBX1-UBC12~NEDD8-CUL1CTD-DCN1P.
(J and K) Pulse-chase assays for NEDD8-modifed SCFFbw7ΔD testing roles of RBX1 W35 “pivot” (J) and I37 “lever” (K) in directing WT or “neddylized” Q31E/D32E UB from UBCH5B to either an F-box-bound substrate (Cyclin E phosphopeptide) or neddylated CUL1.
Here, NEDD8 essentially “pushes” on the RBX1 linker, which in turn acts as a lever that positions the RING-UBC12~NEDD8 portion of the complex (Movie S2). As a result, NEDD8 directs juxtaposition of the UBC12~NEDD8 active site and CUL1's acceptor site. In agreement with this important role as a lever, mutation of Ile37 to Ala or simultaneous mutation with Val38 to Gly impairs cullin neddylation, whereas there is no effect of individual side chain replacements for the adjacent Asp36 or Val38 (Figure 4D).
The distal border of the NEDD8 binding site is RBX1's Trp35, which is the pivot around which the linker and RING rotate in different RBX1-cullin complex structures (Figure 4A; Movie S2). In the neddylation complex, RBX1's Trp35 is enwrapped by side chains from NEDD8's Glu31 and Glu32 (Figures 4B and 4C). Consistent with the structure, mutation of Trp35 to Ala, Tyr, or Phe, which would be tolerated by the donor NEDD8, only slightly decreases NEDD8 ligation to a cullin, whereas mutation to Asp, which would repel NEDD8's Glu31 and 32, is deleterious (Figure 4E).
The RBX1 Pivot Disfavors CUL1 Targeting by UB
The structure provides a rationale for why the UBL, rather than E2, is critical for switching RBX1 RING E3 specificity toward CUL1 (Figures 1D and 1E). Notably, this is not dictated by direct contacts between the donor NEDD8 and its target, CUL1. Rather, specificity comes from distinctive features of NEDD8's interactions with RBX1's pivot and lever, which position the RBX1-UBC12~NEDD8 assembly. NEDD8 residues that differ from UB and contact RBX1 are the above mentioned Glu31 and Glu32, which correspond to UB's Gln31 and Asp32 (Figure 4C). Despite seemingly subtle side chain differences, the aliphatic portion of NEDD8's Glu32 makes hydrophobic contacts with RBX1's Trp35 pivot, whereas UB's shorter Asp32 would repel the Trp35 pivot. Also, the hydrogen bond from NEDD8's Glu31 to RBX1's Trp35 side chain would be less favored by UB's Gln31. Indeed, shortening the pivot with RBX1 W35A, W35Y, and W35F mutations relieve repulsion and increase UB ligation to CUL1CTD (Figure 4F). Swapping identities of residues 31 and 32 is sufficient to transform the efficiencies of RBX1-mediated NEDD8 or UB ligation to CUL1CTD from UBC12 and UBCH5 (Figures 4G and 4H). The data suggest that subtle side chain differences between NEDD8 and UB orient the Trp35 pivot to direct the E2~UBL catalytic center toward CUL1, thereby promoting neddylation (Figure 4I).
Following neddylation, UBC12 would be released so RBX1 could bind an E2~UB intermediate for UB ligation to an F-box-protein bound target (Duda et al., 2011; Zimmerman et al., 2010). This shift in substrate preference upon neddylation of CUL1-RBX1 is manifested in a pulse-chase assay for UB transfer from UBCH5 to a Cyclin E phosphopeptide bound to the F-box protein FBW7ΔD (Figure 4J) (Duda et al., 2012; Hao et al., 2007). Although there is no structure of a neddylated SCF mediating UB ligation to a substrate, our data indicate requirement for a conformational change: the immediate product of NEDD8 ligation would resemble the RBX1-UBC12~NEDD8-CUL1-DCN1 structure, except with NEDD8's covalentȈlinkage transferred to CUL1 and UBC12 released. Modeling a RING-UBCH5~UB structure on RBX1's RING shows clashing between the donor UB and NEDD8~CUL1 (Dou et al., 2012b, 2013; Plechanovová et al., 2012; Pruneda et al., 2012) (Figure S3A). This provides a rationale for conformational flexibility observed in prior NEDD8-CUL5CTD-RBX1 structures, in which the RBX1 RING and NEDD8-linked CUL1 WHB domain appear to swing around relative to each other, via several points of rotation (Figures S3B and S3C) (Duda et al., 2008).
Given this conformational freedom, we wondered why UB is not substantially ligated to neddylated CUL1 and asked if this involves UB repelling RBX1's pivot in a neddylated CRL. In ubiquitination assays with the RBX1 W35A mutation, or with UB bearing the “neddylizing” D31E and Q32E mutations, NEDD8~CUL1 undergoes ubiquitination, with a concomitant decrease in UB ligation to the SCF substrate Cyclin E (Figure 4J). However, use of RBX1 with an I37A mutation in the “lever” eliminated NEDD8~CUL1 targeting by the “neddylized” UB, without substantially impacting SCF-bound Cyclin E ubiquitination (Figure 4K).
The simplest explanation for the data is that a neddylated CRL is sufficiently flexible to allow self multi- or polyubiquitination, confirmed by mass spec (Figure S3D). However, an E2-linked donor UB is deflected from NEDD8~CUL1 by UB's residues 31 and 32 and RBX1's Trp35. Also, F-box protein-bound substrate ubiquitination does not require RBX1's Ile37. With UB mutated to resemble NEDD8, UBCH5~UB Q31E/D32E likely binds RBX1 as in the neddylation intermediate. This includes interactions between the donor “neddylized” UB and RBX1's “Ile37 lever,” which stabilize and position the activated UBCH5~UB in such a way that the active site accesses lysines from NEDD8~CUL1 and its linked UB. With RBX1's Ile37 mutated, a catalytic structure mimicking RBX1-UBC12~NEDD8 is unattainable, while elements required for CRL substrate ubiq-uitination remain. Future studies will be required to visualize a neddylated CRL in the act of ligating UB to a substrate.
CUL1 Structurally Licenses Acceptor Lys for Projection into UBC12~NEDD8 Active Site
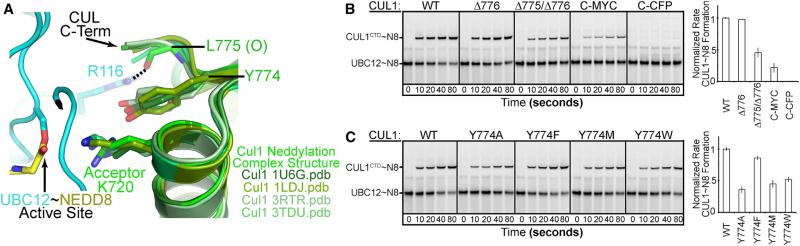
Two key CUL1 features influence selection of the Lys720 acceptor. First, surface complementarity between CUL1 and UBC12 enables their juxtapositioning and explains why nearby lysines failed to rescue CUL1 K720A neddylation (Figures 1G, 2, 5A, and S1D). Accordingly, deleting the two C-terminal residues that contribute to this interface, or imposing a barrier by fusing CUL1's Ala776 to cyan-fluorescent protein or a Myc-tag impairs neddylation (Figure 5B).
Figure 5. CUL1 Features Contributing to NEDD8 Acceptor Lys Selection.
(A) Close-up of complementary UBC12-CUL1 interface. Note UBC12 proximity to CUL1's C terminus and interactions with carbonyl from CUL1's penultimate Leu775. Tyr774 structurally aligns CUL1's Lys720, even in the absence of neddylation enzymes as in superposition of prior CUL1 WHB domain structures.
(B) Role CUL1 C terminus forming a complementary surface with UBC12, assayed by pulse-chase RBX1-mediated transfer of fluorescent NEDD8 from UBC12 to the indicated CUL1CTD C-terminal deletion or extension variants. Graph, rate compared to CUL1; error, 1 SD.
(C) Effects of substituting the CUL1 Lys720 stabilizing residue Tyr774 in experiments as in (B).
Second, ligation can be driven by projection of a structurally ordered acceptor Lys into the active site of an E2~UBL intermediate. Whereas a prior study of SUMOylation identified E2 residues guiding the acceptor Lys (Yunus and Lima, 2006), we find the CUL1 substrate contributing to this role in neddylation. CUL1's Tyr774 aligns and directs CUL1's acceptor (here an Arg, but normally Lys720) into the active site (Figure 5A). The importance of rigid support for CUL1's acceptor Lys720 is verified by wild-type neddylation with a Y774F mutant, which could maintain stacking even without a terminal hydroxyl (Figure 5C). By contrast, neddylation is impaired by the smaller Y774A substitution, the somewhat flexible Y774M mutant, or the too bulky Y774W mutant that would reposition Lys720. Thus, the substrate—CUL1—plays a role in licensing its acceptor Lys720 by structurally supporting projection into the UBC12~NEDD8 active site.
DCN1 Synergizes with RBX1-UBC12~NEDD8-CUL1 Catalytic Architecture
The structure reveals how DCN1 stimulates NEDD8 ligation in a manner that depends on RBX1 RING activity. Intriguingly, there are no apparent contacts between DCN1P and RBX1. Rather, DCN1P and CUL1-RBX1-UBC12~NEDD8 are optimized to interact in the catalytic architecture. DCN1P increases recruitment of UBC12 to CUL1-RBX1 by engaging both CUL1's WHB and UBC12's acetylated N terminus (Scott et al., 2011) (Figure 6A). Flexibility of some residues connecting UBC12's N terminus and catalytic domain, reflected by lack of electron density, might imply that DCN1 engages the RBX1-RING-bound UBC12~NEDD8 and CUL1 in several different relative orientations. However, docking DCN1 and the activated RBX1 RING-UBC12~NEDD8 on prior CUL1-RBX1 structures reveals substantial clashing (Figures 6B and 6C) (Angers et al., 2006; Calabrese et al., 2011; Duda et al., 2008; Fischer et al., 2011; Goldenberg et al., 2004; Zheng et al., 2002). Although rotation could allow RBX1's RING to bind and activate UBC12~NEDD8 in alternative RING orientations, docking models with RING positions in a neddylated CUL-RBX1 (Duda et al., 2008) would place UBC12's N terminus too far away from DCN1-CUL1 (Figure 6D). Thus, modeling suggests DCN1 tethers RBX1-bound UBC12ȈNEDD8 to CUL1 to synergize with the catalytic RBX1-UBC12Pulse-chase assays forNEDD8-CUL1 architecture. Accordingly, I37A and I37D mutations in the “lever” decrease DCN1-dependent NEDD8 ligation to a cullin's C-terminal domain, as does a donor UB that repels the RBX1 pivot (Figures 6E–6G).
Figure 6. DCN1P Synergizes with Catalytic RBX1-UBC12~NEDD8-CUL1 Architecture to Promote Neddylation.
(A) RBX1-UBC12~NEDD8-CUL1CTD-DCN1P structure, highlighting complementarity of NEDD8's surface with structure of RBX1 and UBC12 core domain, and unstructured UBC12 region linking UBC12's catalytic E2 core domain with the acetylated N terminus bound to DCN1P.
(B) Incompatibility of DCN1-UBC12~NEDD8 binding to RBX1-CUL1-CAND1 due to NEDD8 clashing with RBX1 and CUL1. Model generated by docking RING domains from the present and prior 1U6G Protein Data Bank (PDB) structures, with RBX1NTD-CUL1CTD oriented as in (A).
(C) Incompatibility of DCN1-UBC12~NEDD8 binding to RBX1-CUL1 in conformation from 3RTR PDB due to NEDD8 clashing with RBX1-CUL1, and DCN1-UBC12 clashing with RBX1. Model generated by docking RING domains from the present and prior 3RTR PDB structures, with RBX1NTD-CUL1CTD oriented as in (A).
(D) Incompatibility of DCN1-UBC12~NEDD8 binding to RBX1-CUL1~NEDD8 in conformation from 3DQV PDB due to too great a distance between UBC12's N terminus bound to DCN1 and UBC12's core domain bound to RBX1 RING. Oriented with RBX1NTD-CUL1CTD as in (A).
(E) Mutations testing role of RBX1 “lever” I37 in rapid quench-flow pulse-chase NEDD8 transfer from UBC12 to CUL1CTD in the presence or absence of DCN1P. NEDD8 detected by immunoblot.
(F) Mutations testing role of UBL identity (fluorescent “neddylized” [Q31E/D32E/R72A] or UB [R72A]) in transfer from UBC12 to CUL1CTD-RBX1 in the presence or absence of DCN1P, in rapid quench-flow pulse-chase assay. The UB R72A mutation allows loading on UBC12 in pulse reaction.
(G) Role of UBL identity (fluorescent NEDD8 or UB [R72A]) in transfer from UBC12 to CUL2CTD-RBX1 in the presence or absence of DCN1P.
Reactivity of E2~UBL Intermediate Is Triggered by Properly Positioned Acceptor Lys
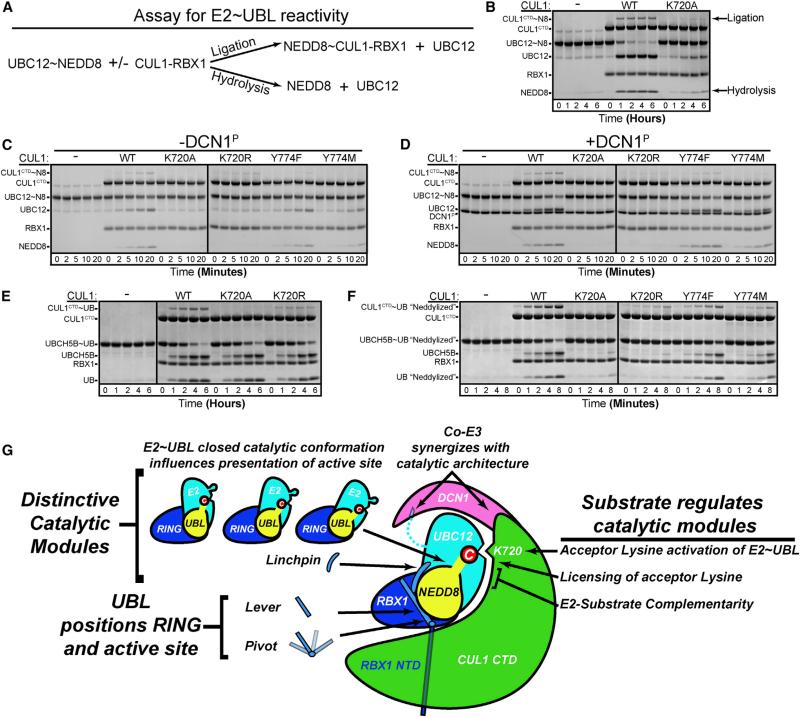
An additional factor potentially influencing reactivity of an E2~UBL intermediate emerged from our efforts to trap a stable complex for crystallography. Thioester linkages are optimally reactive toward properly positioned primary amines. Accordingly, adding RBX1-CUL1 to thioester-linked UBC12 NEDD8 results in rapid NEDD8 ligation to CUL1 (Figures S4A and S4B). Comparatively, ligation from oxyester-linked UBC12~NEDD8 is slowed, and only a fraction of NEDD8 is ligated to CUL1. The majority of the oxyester-linked UBC12~NEDD8 undergoes hydrolysis, discharging NEDD8 to solvent (Figures 7A and 7B). We do not know if the differences in substrate partitioning arise from subtle structural variations between the thioester and oxyester-linked complexes, or because an ester has expanded potential to react with 55 M water over an amine. Nonetheless, the relatively greater potential for the oxyester-linked complex to discharge via hydrolysis versus aminolysis offered the opportunity to assay for a role of the target lysine in stimulating reactivity of a UBC12~NEDD8 intermediate.
Figure 7. Potential for CUL1's Acceptor Lys720 to Toggle E2~UBL Reactivity and Summary of Elements Contributing to Neddylation.
(A) Mechanisms discharging an E2~UBL intermediate such as UBC12~NEDD8. Reaction with an acceptor Lys leads to ligation. Reaction with solvent leads to hydrolysis of the intermediate.
(B) Role of CUL1-RBX1 and CUL1's acceptor K720 on reactivity of oxyester-linked UBC12 (C111S)~NEDD8 complex, as assayed by hydrolysis and detected by Coomassie-stained SDS-PAGE.
(C and D) Role of factors influencing proper placement of CUL1 acceptor K720 (absence/presence of DCN1P; replacements for Y774) on reactivity of oxyester-linked UBC12 (C111S)~NEDD8.
(E and F) Role of factors influencing placement of RBX1-activated UBCH5~UB toward CUL1 acceptor K720 (WT UB or “Neddylized” Q31E/D32E that orients the RBX1 “pivot”) on reactivity of oxyester-linked UBCH5B (C85S)~UB, as assayed by hydrolysis. Note timescales (hours versus minutes).
(G) Mechanisms influencing reactivity and enzyme conformation that together drive neddylation and establish specificity for the multifunctional RING E3 RBX1. See also Figure S4.
Consistent with studies showing that RING E3-E2~UBL intermediates are activated for nucleophilic attack, RBX1-CUL1CTD harboring a K720A substitution stimulated hydrolysis of the oxyester-linked UBC12~NEDD8. Intriguingly, however, this discharge was massively accelerated in the presence of the CUL1 acceptor Lys720 (Figure 7B). Several observations suggest that the target Lys influences reactivity of the UBC12~NEDD8 bond. First, adding DCN1P to support juxtaposition of the active site and CUL1 target Lys720 increased both NEDD8 ligation to CUL1 and hydrolysis of the oxyester-linked UBC12~NEDD8 intermediate (Figures 7C and 7D). As with DCN1P-independent reactivity, DCN1P-dependent hydrolysis was stimulated by CUL1's Lys720. Second, the levels of hydrolysis correlate with projection of CUL1's Lys720 to the active site, as probed by mutations for CUL1 Tyr774 (Figures 7C and 7D). Third, stimulation by the acceptor Lys depends on a known catalytic element: hydrolysis is slowed with a Ser substitution in place of the E2 “catalytic Asn” (Figure S4C) (Wu et al., 2003b), which ultimately enabled crystallizing the neddylation complex.
Notably, the effect of the acceptor Lys is not restricted to UBC12: reactivity of an oxyester-linked UBCH5~UB intermediate is also stimulated through features that support projecting the CUL1 target Lys720 into the catalytic center of the activated, conformationally closed E2~UB intermediate. CUL1's Lys720 stimulates hydrolysis for the oxyester-linked UBCH5 complex with UB Q31E/D32E, where NEDD8-like interactions with the RBX1 lever and pivot can stabilize the catalytic architecture and juxtaposition the E2~UBL intermediate and CUL1 target. Furthermore, hydrolysis is decreased by the CUL1 Y774M mutant, with lesser effects for Y774F. In contrast, both ligation and hydrolysis are slow (hours versus minutes), and there is no obvious effect of CUL1's Lys720 for the oxyester-linked complex with wild-type UB, which repels RBX1's pivot (Figures 7E and 7F).
The data imply that the target Lys may toggle the E2~UBL active site, either indirectly or directly. The target Lys could potentially help hold the RING-UBC12~NEDD8 assembly in the activated position, which in turn stabilizes the closed UBC12~NEDD8 conformation that immobilizes the oxyester (and presumably thioester) bond to increase reactivity. It is also possible that insertion of the target Lys modulates the structure of the UBC12~NEDD8 active site. Indeed, superimposing various UBC12 structures reveals potential fluctuation in the loop clamping NEDD8's C terminus and CUL1's target site into the active site (Figure S4D). Irrespective of the mechanism, it seems likely that projection of CUL1's acceptor Lys720 into the catalytic center tweaks the UBC12~NEDD8 active site and stimulates reactivity.
DISCUSSION
Multimodal Mechanism for RING E3 Ligation and Specificity
A fundamental question in protein regulation is: how is a particular UBL matched with a specific target? We provide, to our knowledge, the first structure of a RING E3-E2~UBL-substrate complex representing a ligation intermediate. Similarity among RING-E2~UBL structures and E2 UBL-substrate complexes suggest overall related mechanisms of neddylation, ubiquitination, and SUMOylation, with distinct features establishing UBL and acceptor Lys specificity. We found that for neddylation, the catalytic modules are not optimized to function independently; instead, conformational activation of a RING-E2~UBL intermediate, stimulation of E2~UBL reactivity, and acceptor lysine recognition are intertwined.
Numerous mechanisms contribute to NEDD8 ligation and specificity (Figure 7G): (1) RBX1 uses a unique linchpin for RING-mediated allosteric activation of the UBC12~NEDD8 intermediate, (2) RBX1 and UBC12~NEDD8 regulate each other, with the RBX1 linker and RING synergizing to stabilize the closed, activated UBC12~NEDD8 conformation, and NEDD8 reciprocally pushing the RBX1 linker lever to steer the active site to CUL1's acceptor, (3) the unique UBC12~NEDD8 closed conformation presents a distinctive active siteȈconfiguration to the acceptor Lys, (4) shape complementarity at the UBC12-CUL1 interface targets the CUL1 acceptor, (5) CUL1 structure licenses the acceptor Lys in an extended conformation, (6) DCN1 tethers UBC12 to CUL1 in the catalytic architecture for neddylation, and (7) the CUL1 acceptor Lys toggles E2~UBL reactivity. Also, misdirected UB ligation to a cullin by the multi-functional RBX1 is prevented by (1) CDC34 structure (Huang et al., 2014) being incompatible with architecture for CUL1 modification, (2) RBX1 having a suboptimal linchpin for UBCH5~UB, and (3) UB repelling the RBX1 pivot, thereby deflecting RBX1's RING from targeting CUL1.
Specificity among Seemingly Similar Catalytic Modules
The RBX1-UBC12~NEDD8-CUL1CTD-DCN1P and prior structures provide insights into how specificity is established for similar RING E3-E2 catalytic modules. Diversity in RING-E2~UBL geometries influences interactions with acceptor lysines (Figures 3F, 3G, 7G, and S4D) (Dou et al., 2012b, 2013; Plechanovová et al., 2012; Pruneda et al., 2012; Reverter and Lima, 2005). Each E2~UBL combination is activated through its own closed conformation, with different angles between the E2 and UBL globular domain achieved by a variety of mechanisms. Multifunctional RING E3s may use different or even multiple linch-pins to cement their various E2~UBL partners. RBX1's Arg46 linchpin enables distinctive RING-E2 contacts and may support CDC34~UB as well as UBC12~NEDD8. However, the “canonical” linchpin Asn98 contributes to ubiquitination by UBCH5~UB, and suboptimal activation by Asn98 relative to Arg is one factor favoring NEDD8 over UB modification of CUL1. It seems likely that in multifunctional E3s, RING sequences reflect compromises to direct many E2~UBL partners toward their specific targets, rather than optimization of some functions.
Although a variety of ligation mechanisms have been proposed, common features include placement and deprotonation of the acceptor Lys within the active site and stabilization of the negatively charged transition state arising from Lys attack of the E2~UBL intermediate (Komander and Rape, 2012). Prior studies had implicated an E2 or substrate Asp in either guiding the acceptor Lys into the active site, or directly mediating deprotonation via ion-pairing (Plechanovová et al., 2012; Wickliffe et al., 2011; Yunus and Lima, 2006). For neddylation, the substrate and E2 both guide the acceptor Lys to the UBC12 active site without obvious acidic groups, and the corresponding Asp143 neither contacts the acceptor nor is required for ligation. Instead, the RBX1-UBC12~NEDD8-CUL1CTD-DCN1P structure is consistent with a mechanism for deprotonation akin to that proposed for SUMOylation, whereby CUL1 Lys720 contacts with elements dispersed throughout UBC12's active site— perhaps the backbone carbonyl of Asp143 or other carbonyls within the same loop, and/or the thioester bond itself—replace interactions between the ε-amino group and a proton from bulk solvent (Yunus and Lima, 2006). These same interactions may enable CUL1's Lys720 to reciprocally stabilize the catalytic architecture to activate UBC12. Although future studies will be required to visualize the ensuing intermediate, we note that UBC12's so-called “catalytic Asn” (Ser103 in the structure) would be ideally poised to stabilize the intermediate and surrounding active site loops in UBC12 (Figure S4E) (Wu et al., 2003b).
Integration of E3-E2~UBL-Target Modules by UBL-Guided Lever and Acceptor Lys Switch
A key finding is that the overall architecture and catalytic activity are influenced by substrates. The donor UBL NEDD8 steers its RING E3, RBX1, to position the active site at the acceptor (Figure 4). RBX1's linker stabilizes the activated UBC12~NEDD8 conformation in a manner analogous to non-RING elements priming the closed conformation of UBCH5~UB (Dou et al., 2012b, 2013; Plechanovová et al., 2012). The NEDD8-RBX1 linker interaction reciprocally positions the catalytic machinery to juxtapose the RING-UBC12 NEDD8 catalytic center and CUL1 acceptor (Figure 4). Thus, the donor UBL dictates targeting specificity allosterically. Subtle side chain differences between NEDD8 and UB—E31Q and E32D—repel an RBX1 pivot and divert the RBX1 RING E3 from CUL1.
Specificity is also imparted by the substrate, with CUL1 complementing UBC12's surface and projecting the acceptor Lys720 into the active site (Figures 5 and 7). E2-substrate interactions are reciprocal, with the properly positioned CUL1 acceptor Lys720 toggling reactivity of a covalent (albeit oxyester-linked) UBC12~NEDD8 or UBCH5~“neddylized” UB complex. DCN1P further synergizes with the active architecture (Figure 6). Thus, all components of the RBX1-UBC12~NEDD8-CUL1-DCN1 intermediate provide elements that interlock, with RBX1-UBC12~NEDD8 resembling a spring ready to discharge and CUL1's Lys720 a trigger for the reaction. In this way, optimal catalysis is integrated with UBL and acceptor targeting specificity.
Principles underlying neddylation also may apply to CRL-mediated ubiquitination. Although a “lever” directing RBX1 RING-E2~UB to ubiquitination targets remains to be identified, we speculate that NEDD8, CUL1, an F-box protein-bound substrate, or other E3 or E2 features could help steer the active site toward a ubiquitination target. For example, the E2 CDC34 has distinctive acidic loops and a C-terminal tail that may coordinate the donor or acceptor UB and anchor the RBX1-CDC34-UB orientation for target ubiquitination, much like DCN1 restrains the orientation of RBX1 RING-activated UBC12~NEDD8 toward CUL1 (Choi et al., 2010; Kleiger et al., 2009a; Petroski and Deshaies, 2005; Spratt and Shaw, 2011; Ziemba et al., 2013).
Other RING E3-E2 ligases probably also achieve specificity by the acceptor and UBL modulating enzyme conformation and reactivity. Many E2~UBL intermediates may be toggled by the acceptor lysine, particularly as a modeled CUL1 acceptor lysine contacts a UBC12 loop (Figure S4D) that is dynamic in other E2s (Berndsen et al., 2013; Dou et al., 2012b; Yunus and Lima, 2006). Several RING E3s couple substrate binding with E2~UB activation (Du et al., 2002; Mattiroli et al., 2014; Rojas-Fernandez et al., 2014). Also, other E3s undergo RING domain rotation to achieve activity, and for IAP and CBL E3s, the interdomain linkers that move during RING rotation stabilize the activated UBCH5~UB closed conformation by contacting UB (Dou et al., 2012a, 2012b, 2013; Dueber et al., 2011; Kobashigawa et al., 2011). For these and other RING E3s, we anticipate future studies will reveal that the E2-bound donor UB reciprocally stabilizes active architectures, steering the associated E2 active site toward ubiquitination substrates much like NEDD8 steers its linked E2 via interaction with RBX1's linker “lever.” The stage is now set for broadly understanding how targets and UBLs serve as switches by toggling E3/E2 conformations to establish ligation specificity.
EXPERIMENTAL PROCEDURES
Protein Expression
Proteins are human and were expressed either untagged or as GST- or Hisfusions in Escherichia coli or insect cells and purified as described (Calabrese et al., 2011; Duda et al., 2012; Huang et al., 2009; Scott et al., 2011). Assays with DCN1 used insect cell expressed N-terminally acetylated UBC12 (Scott et al., 2011). See the Extended Experimental Procedures for details.
Crystallography
The crystal structure used UBC12 (N-acetylated, N103S, C111S)~NEDD8 linked by an oxyester bond and stabilized by an E2 catalytic Asn mutation. UBC12 variant (60 μM), APPBP1-UBA3 (4 μM), and NEDD8 (80 μM) were incubated in 25 mM HEPES, 200 mM NaCl, 2.5 mM MgCl2, 1 mM ATP, pH 7.5, 30°C for 18 hr. UBC12~NEDD8 was purified by ion exchange and gel filtration chromatography. UBC12~NEDD8, RBX1-CUL1CTD (K720R), and DCN1P were mixed at 40 μM:35 μM:40 μM and crystals grown in 4°C hanging drops in ≈20% PEG3350, 0.2 M ammonium citrate, pH 7.0 with seeding, and cryoprotected by soaking in mother liquor with 12% and then 24% ethylene glycol. Data were collected at APS ID-24E. The structure was determined by molecular replacement. See the Extended Experimental Procedures for details.
Enzyme Assays
Pulse-chase assays exclusively monitoring the ligation reaction were adapted from (Scott et al., 2011) for UBL detection by fluorescence or immunoblotting. Indicated E2~UBL complexes were formed in the pulse reaction. After quenching the pulse, UBL was “chased” from E2 by adding variants of RBX1-CULCTD (Figures 1, 3, 4, 5, 6, 7, S1, and S4), neddylated SCF components and CyclinE peptide (Figures 4J, 4K, and S2), or RBX1-CULCTD and DCN1P (Figures 6E–6G, 7D, and S4), as indicated. Aliquots were taken at different times and products separated by SDS-PAGE and visualized by fluorescence or immunoblot. See the Extended Experimental Procedures for details.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Bozeman, D.W. Miller, J. Bollinger, D.J. Miller, and I. Kurinov for support. Funding was provided by the Howard Hughes Medical Institute (HHMI), the American Lebanese Syrian Associated Charities (ALSAC), NIH (5P30CA021765, R01GM069530 to B.A.S. and R01AG011085 to J.W.H.), and Damon Runyon (DRG 2061-10 to J.R.L.). B.A.S. is an Investigator of Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) accession code for the structure reported in this paper is 4P5O.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, four figures, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.04.037.
REFERENCES
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 2013;9:154–156. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Calabrese MF, Scott DC, Duda DM, Grace CR, Kurinov I, Kriwacki RW, Schulman BA. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat. Struct. Mol. Biol. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Wu K, Jeong K, Lee D, Jeon YH, Choi BS, Pan ZQ, Ryu KS, Cheong C. The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 2010;285:17754–17762. doi: 10.1074/jbc.M109.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 2012a;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012b;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Tron AE, Hammel M, Lambert LJ, Waddell MB, Mittag T, DeCaprio JA, Schulman BA. Structure of a glomulin-RBX1-CUL1 complex: inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell. 2012;47:371–382. doi: 10.1016/j.molcel.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, Zobel K, Maurer B, Varfolomeev E, Wu P, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Ceccarelli DF, Orlicky S, St-Cyr DJ, Ziemba A, Garg P, Plamondon S, Auer M, Sidhu S, Marinier A, et al. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat. Chem. Biol. 2014;10:156–163. doi: 10.1038/nchembio.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AY, Bommeljé CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J. Biol. Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 2009a;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009b;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. USA. 2011;108:20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat. Commun. 2014;5:3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW, Bennett EJ, Schulman BA. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure. 2013;21:42–53. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2ȈUb complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Fernandez A, Plechanovová A, Hattersley N, Jaffray E, Tatham MH, Hay RT. SUMO chain-induced dimerization activates RNF4. Mol. Cell. 2014;53:880–892. doi: 10.1016/j.molcel.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Spratt DE, Shaw GS. Association of the disordered C-terminus of CDC34 with a catalytically bound ubiquitin. J. Mol. Biol. 2011;407:425–438. doi: 10.1016/j.jmb.2011.01.047. [DOI] [PubMed] [Google Scholar]
- Spratt DE, Wu K, Kovacev J, Pan ZQ, Shaw GS. Selective recruitment of an E2Ȉubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 2012;287:17374–17385. doi: 10.1074/jbc.M112.353748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jäger S, Mason-Herr J, et al. Inhibition of a NEDD8 Cascade Restores Restriction of HIV by APOBEC3G. PLoS Pathog. 2012;8:e1003085. doi: 10.1371/journal.ppat.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell. 2003a;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003b;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/ Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Ziemba A, Hill S, Sandoval D, Webb K, Bennett EJ, Kleiger G. Multimodal mechanism of action for the Cdc34 acidic loop: a case study for why ubiquitin-conjugating enzymes have loops and tails. J. Biol. Chem. 2013;288:34882–34896. doi: 10.1074/jbc.M113.509190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.