Figure 1. Specificity of RBX1-Mediated NEDD8 Ligation to CUL1.
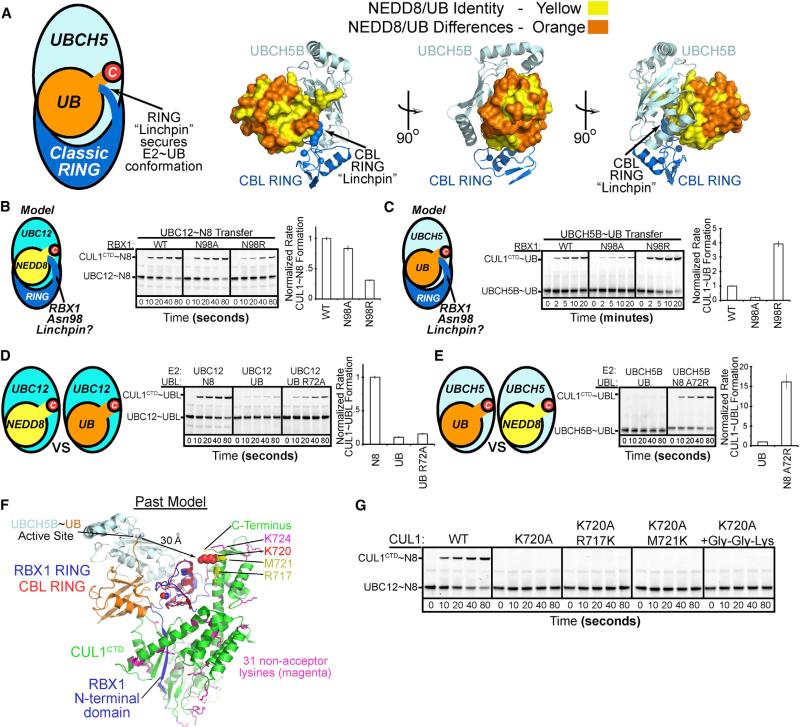
(A) Canonical RING-E2~UB architecture, highlighting the linchpin and conservation of E2 and RING binding residues in UB and NEDD8, shown for CBL-UBCH5~UB (Dou et al., 2013).
(B) Mutational analysis of RBX1's Asn98, corresponding to canonical RING linchpin, in RBX1-mediated pulse-chase fluorescent NEDD8 transfer from UBC12 to CUL1CTD. Graph, rate compared to wild-type (WT) RBX1; error, 1 SD.
(C) Mutational analysis of RBX1's Asn98, aka canonical RING linchpin, in RBX1-mediated pulse-chase fluorescent UB transfer from UBCH5B to CUL1CTD. Graph, rate compared to WT RBX1; error, 1 SD.
(D) Role of UBL in RBX1-mediated CUL1 modification assayed by comparing pulse-chase NEDD8, UB, or UB R72A transfer from NEDD8's E2 UBC12. The noncognate UBL UB was loaded on UBC12 in the pulse reaction either through its R72A mutation or use of E1 mutant UBA3 R190Q. Graph, rate compared to WT UBC12~NEDD8; error, 1 SD.
(E) Role of UBL in RBX1-mediated CUL1 modification assayed by comparing UB or NEDD8 A72R transfer from UBCH5B to CUL1CTD. The NEDD8 A72R mutation allows loading this noncognate UBL on UBCH5B in the pulse reaction. Graph, rate compared to WT UBCH5~UB; error, 1 SD.
(F) CUL1CTD Lys locations. Docking RBX1's RING from prior structures with that from CBL-UBCH5~UB revealed a >30Å gap between the active site and K720 (red) or other lysines (native in magenta, introduced in yellow) in CUL1 (Dou et al., 2013; Zheng et al., 2002).
(G) Acceptor Lys specificity for NEDD8 transfer from UBC12 to CUL1CTD or Lys mutants. Gly-Gly-Lys, appended to C terminus of CUL1CTD K720A.