Abstract
The ENU-induced repro57 mutation was identified in an unbiased screen for the discovery of novel genes for fertility. Male repro57 homozygous mice are infertile, and exhibit significantly reduced testis weight compared to wild type. Histological examination of mutant testes revealed that spermatocytes degenerated during late prophase, and no mature spermatozoa were found in the seminiferous epithelium, suggesting that infertility is caused by arrest of spermatogenesis at late meiotic prophase. Consistent with this hypothesis, the number of foci with MLH1, a protein essential for crossing over, is greatly reduced in repro57 mutant spermatocytes, which also lack chiasmata between homologs and exhibit premature dissociation of XY chromosomes. In repro57 mutant mice, we identified a mutation in the Rnf212 gene, encoding Ring Finger Protein 212. The overall phenotype of repro57 mouse is consistent with the recently reported phenotype of the Rnf212 knockout mouse; slight differences may be due to genetic background effects. Thus, the repro57 nonsense mutation provides a new allele of the mouse Rnf212 gene.
Keywords: Meiosis, repro57, Crossing over, Spermatogenesis, Mouse
INTRODUCTION
During meiosis, homologous chromosomes undergo highly programmed and evolutionarily conserved processes, including pairing of homologous chromosomes, synapsis, recombination to form chiasmata between homologs, and chromosome segregation mediated by chiasmata resolution. At the onset of meiosis, recombination is initiated by DNA double-strand breaks (DSBs) formation by SPO11 in most organisms (Phadnis et al. 2011). DNA ends at the DSB sites are loaded with the recombinases RAD51 and DMC1, promoting homolog interaction leading to synapsis, which is mediated by formation of a unique structural scaffold, the synaptonemal complex (SC) (Cromie & Smith 2007). During recombination, DSBs are repaired by either crossing over (CO) or non-crossing over processes. CO sites are created by exchange of non-sister chromatids between homologs, and formation of at least one CO on each chromosome is essential for the accurate segregation of homologous chromosomes (Baudat et al. 2013). However, the precise mechanisms and full repertoire of genes responsible for mammalian CO recombination are still unclear (Handel & Schimenti 2010, Bolcun-Filas & Schimenti 2012). Rnf212 has been shown to be essential for meiotic recombination in C. elegans (Jantsch et al. 2004, Bhalla et al. 2008), and in humans, RNF212 has been associated with variation in the genome-side recombination rate (Kong et al. 2008, Chowdhury et al. 2009). Moreover, the infertility and CO failure of mice bearing a targeted knockout of the mouse gene, Rnf212tm1Nhtr (herein referred to as Rnf212 KO) provide evidence for an essential role of RNF212 in mammalian CO recombination (Reynolds et al. 2013).
To discover genes involved in meiosis and reproduction, an unbiased phenotype-driven approach using N-ethyl-N-nitrosourea (ENU)-induced mutagenesis and fertility screening was conducted by the Reproductive Genomics Program at The Jackson Laboratory (JAX) (Lessard et al. 2004, Handel et al. 2006). From this program, we identified homozygous repro57 mice as infertile, and report here that the male repro57 mutant mice exhibit meiotic arrest with defective CO. The repro57 mice have a nonsense mutation in exon 4 of the Rnf212 gene, encoding a Ring Finger Protein 212, RNF212 (also known as Zip3 in budding yeast and ZHP3 in worms), thereby identifying a new allele of Rnf212.
MATERIALS AND METHODS
Mice
The repro57 mice used in this study were produced by the NIH-supported Reproductive Genomics program at JAX (http://reproductivegenomics.jax.org/), where each mutant line was designated by a “repro” number in order of appearance. Mice of the JF1/Ms (JF1) strain were obtained from the National Institute of Genetics (Mishima, Japan), and C3HeB/FeJ (C3H) mice were obtained from CLEA Japan (Tokyo, Japan). The repro57 mutation was induced in a C57BL/6J (B6) background and mutagenized mice were subsequently outcrossed to C3HeB/FeJ (C3H) (Handel et al. 2006), with creation of a repro57-C3H congenic line. For experimental analyses, repro57 homozygous mice were obtained by mating of repro57 heterozygous mice. Mice were euthanized using CO2 at 10 to 12 weeks (or at 0 to 5 weeks of age to follow the first wave of spermatogenesis). All experiments using animals were approved by the animal care and use regulatory committees of both Okayama University and JAX.
Mapping the repro57 mutation
Adult B6 mice were mutagenized with ENU and the repro57 infertility phenotype was identified in the third generation by mating with normal mice for three weeks. More precise phenotypes were subsequently determined from in vitro fertilization (IVF) and histological examination of reproductive tissues (Handel et al. 2006). To establish the chromosomal linkage of the repro57 mutation, genome scans using polymorphic microsatellite markers were performed on DNA obtained from affected and unaffected mice. For fine mapping analysis, 740 F2 mice were produced by mating the F1 mice derived from an intercross between the repro57 mouse and JF1 mouse. The F2 offspring were phenotyped by histology and genotyped using polymorphic microsatellite markers to narrow the candidate region.
DNA sequencing was performed to identify the site of the repro57 mutation. Genomic DNA was isolated from liver of repro57 mutants and founder strain B6 mice. DNA samples were reextracted once with phenol-chloroform; the 260/280 ratio was about 2.0. Whole-genome libraries were prepared and subjected to sequencing on the illumina HiSeq 2000 (illumine, CA, USA); sequence analysis was focused on the candidate region identified from mapping.
Bioinformatics Analysis
The dataset was subjected to quality control using NGSQCtoolkit v2.3 and reads with base qualities greater ≥ 30 over 70% of total read length were used in the downstream analysis (Patel & Jain 2012). High quality reads were mapped to the mouse genome (build-mm10) using BWA v0.5.10-tpx (Li & Durbin 2009). The resulting alignment was sorted by coordinates and further converted to a binary alignment map (BAM) format by Picard v 1.8.4- SortSam utility (http://picard.sourceforge.net). Picard-MarkDuplicates module was used to remove duplicates from the data. Subsequently, Genome Analysis tool kit (GATK) v 2.2–16 (McKenna et al. 2010, DePristo et al. 2011) module IndelRealigner and BaseRecalibrator were used to pre-process the alignments. The realigned and recalibrated BAM alignment file was used as an input to GATK-UnifiedGenotyper (variant caller) at parameters, -stand_call_conf 50.0, -stand_emit_conf 30.0 and -dcov 200, and variant calls were restricted to the target region. Finally, variant calls were annotated by snpEff v2.0.5 (Cingolani et al. 2012).
Cytological preparations and immunostaining
Histology
Testes from mice were fixed in Bouin's solution, and paraffin-embedded. Sections (5 m) were stained with hematoxylin and eosin (HE) for evaluation of spermatogenesis. Immunohistochemistry was performed as previously described (Fujiwara et al. 2013). Mouse monoclonal anti-human PCNA primary antibody and anti-rat IgG HRP labeled secondary antibody (Table 1) were used to label the histological sections. For fluorescence immunostaining, goat polyclonal anti-human DMC1 (Table 1) was used in phosphate buffer saline (PBS) containing 5% skim milk for 3 h at 37C. Donkey anti-goat IgG-DyLight 488 secondary antibody (Table 1) was applied with the same buffer as the primary antibody and incubated for 1 h at RT. Nuclei were counterstained with DAPI, mounted using VECTASHIELD®Mounting Medium (H-1200, Vector Laboratories Inc., CA, USA), and observed using a ZEISS Axio Imager.A1 (ZEISS, Jena, Germany). A TUNEL assay was performed according to the manufacturer’s instructions (11684817910, Roche Applied Science, Upper Bavaria, Germany).
Table 1.
Antibodies for fluorescence immunostaining
| Antibody | Host | Cat. Number or reference | Dilution |
|---|---|---|---|
| Anti-βactin | rabbit | Abcam, ab8227 | 1:1000 |
| Anti-DMC1 | goat | Santa Cruz, sc-8973 | 1:50 |
| Anti-γH2AX | mouse | Upstate, JBW301 | 1:500 |
| Anti-H1T | guinea pig | Handel Lab (Cobb et al. 1999) | 1:1000 |
| Anti-MLH1 | mouse | BD Pharmingen 551091 | 1:50 |
| Anti-PCNA | mouse | Santa Cruz, sc-25280 | 1:500 |
| Anti-SYCP1 | rabbit | Novous Biologicals, NB300-229 | 1:100 |
| Anti-SYCP3 | guinea pig | de Massy Lab | 1:1500 |
| Anti-SYCP3 | rabbit | Chuma Lab | 1:1000 |
| Anti-RNF212 | rabbit | Hunter Lab (Reynolds et al. 2013) | 1:1000 |
| Anti-goat IgG-DyLight 488 | donkey | Jackson Immuno Research, 705-485-003 | 1:500 |
| Anti-guinea pig IgG Alexa Fluor 594 | goat | Invitrogen, A11076 | 1:200 |
| Anti-guinea pig IgG AMCA | goat | Jackson Immuno Research, 106-156-003 | 1:500 |
| Anti-mouse IgG Alexa Fluor 488 | goat | Invitrogen, A21121 | 1:200 |
| Anti-mouse IgG HRP | goat | Santa Cruz, sc-2005 | 1:400 |
| Anti-rabbit IgG Cy3 | donkey | Jackson Immuno Research, 711-165-152 | 1:500 |
| Anti-rabbit IgG HRP | goat | Santa Cruz, sc-2004 | 1:20000 |
| anti-rabbit IgG Alexa Fluor 488 | goat | Invitrogen, A11008 | 1:200 |
Surface-spread chromatin Preparation
Meiotic cell spreads were prepared as previously described (Peters et al. 1997) with some modifications. Briefly, testes were immersed in PBS, and the retrieved seminiferous tubules were washed and immersed in ice-cold hypotonic extraction buffer (30 mmol/L Tris, 50 mmol/L sucrose, 17 mmol/L trisodium citrate dihydrate, 5 mmol/L ethylenediamineteraacetic acid (EDTA), 0.5 mmol/L dithiothreitol (DTT), and 0.5 mmol/L phenylmethyl-sulphonyl fluoride (PMSF), pH 8.2) for 1 h. Segments of the tubules were minced in 100 mmol/L sucrose solution (pH 8.2), and the cell suspension collected was spread onto glass slides, previously immersed in 1% paraformaldehyde containing 0.15% Triton X-100; the slides were placed in a moisture box for 2 h. The slides were washed twice for 2 min with PBS with 0.4% DRIWEL (Fuji film co., LTD, Tokyo, Japan). After blocking with 5% skim milk in PBS, the preparation slides were incubated with primary antibodies diluted in the blocking buffer for 3 h at RT. Secondary antibodies were then incubated for 1 h at RT. The antibodies used were listed in Table 1. The slides were then mounted with VECTASHIELD Mounting Medium with DAPI (H-1200, Vector Laboratories Inc.) and observed using a ZEISS Axio Imager.A1 (ZEISS).
Air-dried chromosome Preparation
The procedure for chromosomes preparation was as previously described (Fujiwara et al. 2013).
Western Blot Analysis
Testes removed from repro57 mutant and control (wild type, WT) mice were homogenized in radioimmunoprecipitation assay buffer (RIPA: 150 mM NaCl, 10 mM Tris-HCl [pH 7.2], 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate, and 5 mM EDTA). The protein extract was applied on 10% SDS-polyacrylamide gels for electrophoresis and transferred onto Immobilon-P Transfer Membranes (Millipore, MA, USA). The membrane was incubated with anti-RNF212 antibody and HRP-conjugated secondary antibody. Subsequently, the same membrane was incubated with anti-ACTIN antibody after the previous antibodies were removed using Stripping Solution (193-16375; Wako, Osaka, Japan). The antibodies used are listed in Table 1. The membrane was developed using an ECL Advance Western Blotting Detection Kit (RPN2135; Amersham Biosciences, Amersham, UK) and observed.
Gene expression analysis by RT-PCR
mRNAs were isolated from the testes of WT and repro57 mutant mice at 10 weeks of age using TRIzol (Invitrogen, CA, USA) following the manufacturer’s instructions. Genomic DNA in the RNA sample was digested using DNaseI (Takara Bio Inc., Shiga, Japan), and the RNA solutions were purified by phenol/chloroform treatment. cDNAs were synthesized by reverse transcription reaction using Superscripts III reverse transcriptase (Invitrogen) and Oligo dT primer (Invitrogen). RNAs in the cDNA solution were then digested with RNase H (Toyobo, Osaka, Japan). Given evidence for splice variants of Rnf212 (Reynolds et al. 2013) (http://www.ncbi.nlm.nih.gov/nuccore/XM_006535331.1, http://www.ncbi.nlm.nih.gov/nuccore/XM_006535332.1, http://www.ncbi.nlm.nih.gov/nuccore/XM_001476621.5), the sequences of the variant-specific primers, how they align to the exonic structure, and the PCR cycles are shown in Supplementary Figure 1.
Statistical analyses
Data were expressed as a mean ± standard errors. The statistical significance of differences in mean values was assessed by the Student’s t-test.
RESULTS
Infertility and meiotic defects in the testes of repro57 mutant mice
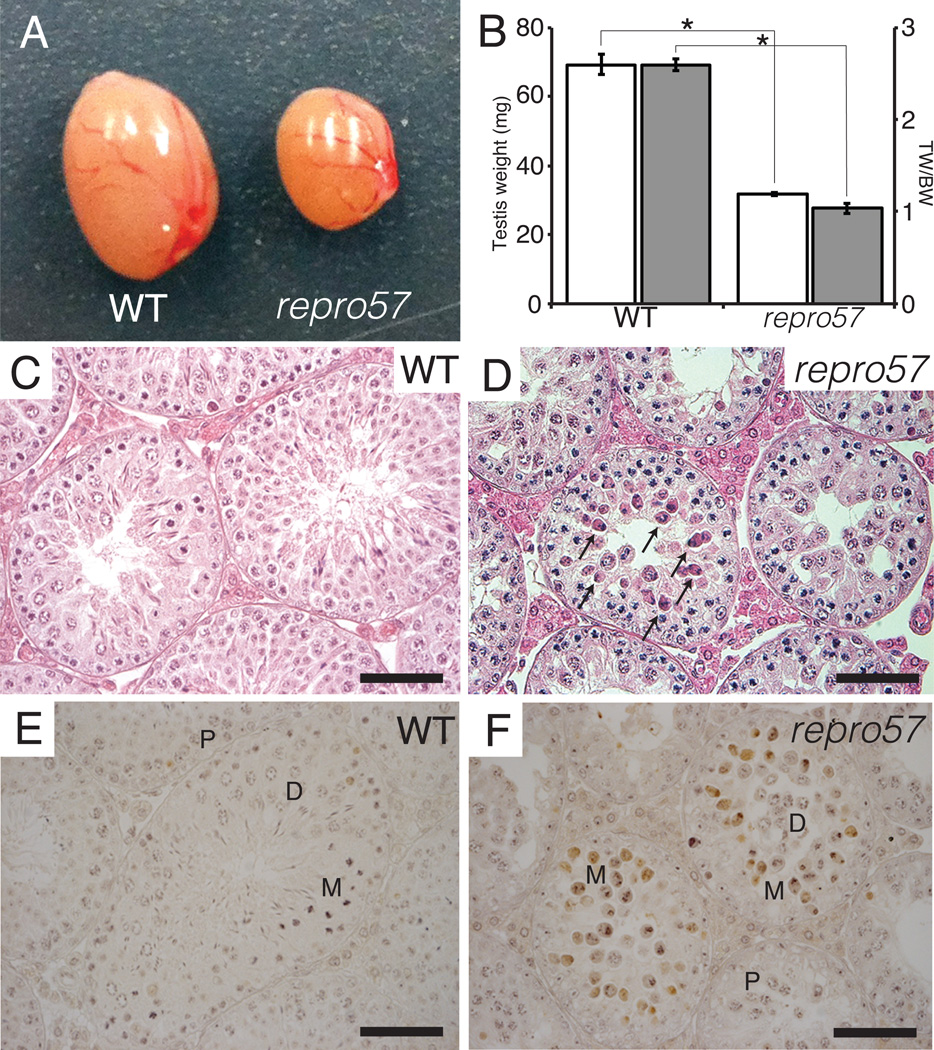
Both female and male repro57 mutant mice on a congenic C3H background failed to produce offspring after natural mating with normal heterozygous or WT individuals for more than 2 months. Heterozygotes were fully fertile with an average litter size of 6.9±1.9 (Table 2). Adult homozygous repro57 mice of both sexes showed no apparent morphological defects in non-reproductive organs, and there was no difference in body weight between the mutant and WT males. However, the mutant testis weight was approximately 50% of WT (Fig. 1A,B), suggesting germ-cell depletion. Histological examination of testes from mutants at 12 weeks of age revealed many degenerating spermatocytes at late prophase stages, with no mature spermatozoa observed in the seminiferous tubules of repro57 mice (Fig. 1D), in contrast to WT testes (Fig. 1C). A TUNEL assay confirmed increased numbers of apoptotic germ cells in testes of repro57 mouse compared to WT (Fig. 1E), with cell death specific to metaphase spermatocytes (Fig. 1F).
Table 2.
Mating results
| Female | Male | N | # of pups |
|---|---|---|---|
| repro57/repro57 | +/repro57 | 4 | 0 |
| +/repro57 | repro57/repro57 | 3 | 0 |
| +/repro57 | +/repro57 | 10 | 6.9 ± 1.9 |
N = number of mating pairs
# of pups per litter is presented as mean ± S.D.
Figure 1. Histology of the testis of WT and repro57 mice.
(A) Testes of WT (left) and repro57 (right) mice. (B) The paired testis weights (TW in mg, white bars) and testis-body weight ratios (TW/BW, gray bars) were significantly reduced in repro57 mice (N=14) compared to those of WT (N=8). Bars indicate standard error; asterisk indicates p < 0.005. (C,D) HE-stained testis sections show degenerating spermatocytes in the seminiferous tubules of repro57 mice (arrows in D). (E,F) Apoptotic cell death was observed only in metaphase spermatocytes in the testis of repro57 mice. P: pachynema, D: diplonema, M: metaphase spermatocyte. Scale bars, 50 µm in C, D, G and H, and 200 µm in E and F.
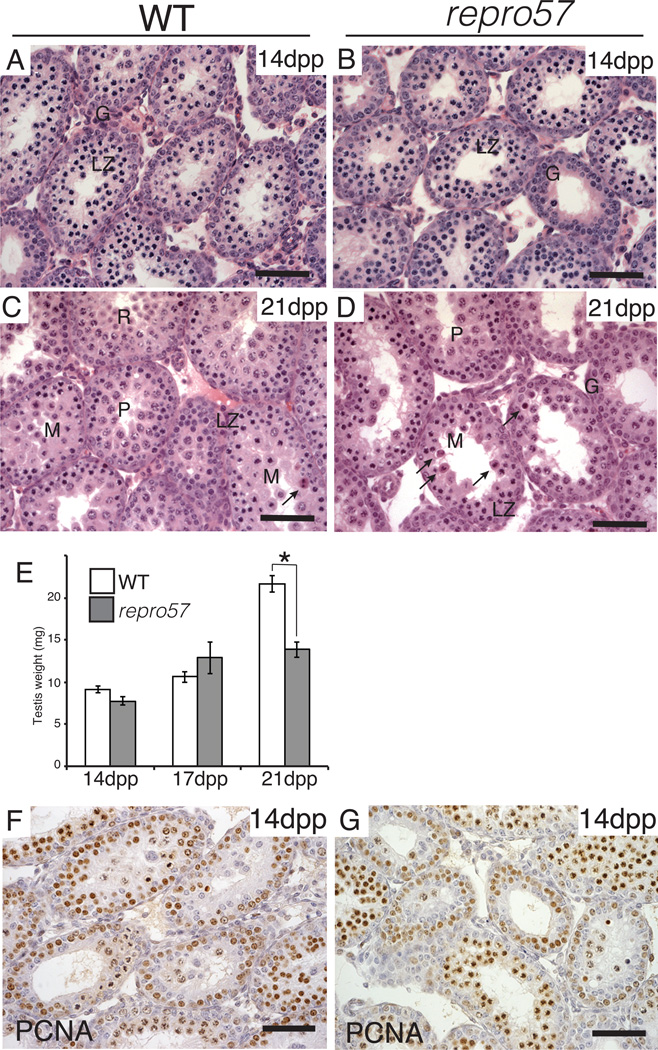
To determine developmental onset of the phenotype, we examined juvenile WT and repro57 testis histology. At 14 day postpartum (dpp), there were no abnormalities in either WT or repro57 mice (Fig. 2A,B). However, by 21 dpp, when the seminiferous tubules of WT mouse were filled with round spermatids (Fig. 2C), those of repro57 mouse exhibited degenerating germ cells and no spermatids (Fig. 2D). Reflecting this, testis weights of repro57 mice were significantly lower than WT at 21 dpp (Fig. 2E). We examined proliferating cell nuclear antigen (PCNA), which plays an important role in both the DNA synthesis machinery of proliferating mouse spermatogonia (Yazawa et al. 2000) and meiotic progression (Roa et al. 2008). The distribution of PCNA was similar in the testes of WT and repro57 mice (Fig. 2F,G).
Figure 2. Developmental analysis of testes in WT and repro57 mice.
(A–D) HE-stained sections of the testis from WT (A and C, N=5 and 6 mice at 14 and 21 dpp, respectively) and repro57 (B and D, N=4 and 3 mice at 14 and 21 dpp, respectively) mice during the first-wave of spermatogenesis. G: spermatogonia, L/Z: leptonema/zygonema, P: pachynema, R: round spermatid. Arrows indicate degenerating germ cells. (E) Testis weights for WT and repro57 mice during the peri-pubertal period of development. Bars indicate standard error; asterisk indicates p < 0.05. (F,G) Immunolabeling for PCNA on testis sections from prepuberal WT (A) and repro57 (B) mice at 14 dpp. Scale bar, 50 µm.
Cytological crossing over in repro57 spermatocytes
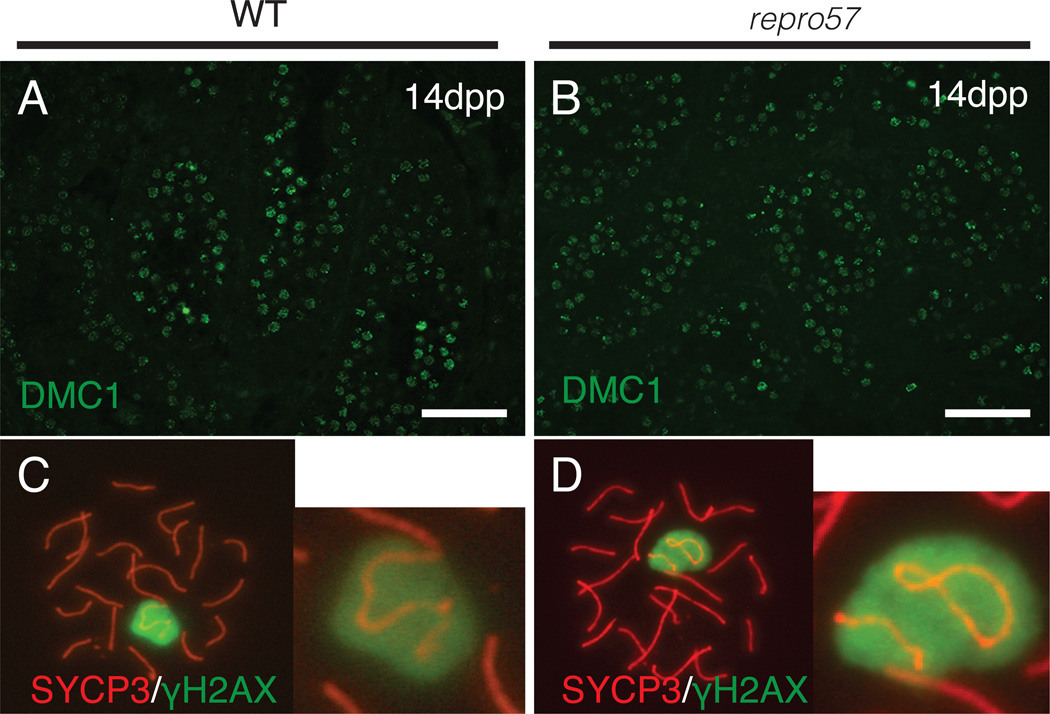
We assessed marker proteins of meiotic prophase progression to determine abnormalities of meiotic chromosomal dynamics in repro57 mutant testes. The distribution of DMC1, an early recombination repair protein, is similar in WT and repro57 germ cells (Fig. 3A–B). We determined pattern of staining for γH2AX (phosphorylated histone H2AFX), which accumulates at the sites of meiotic DNA DSBs in early prophase, and later is restricted to the XY body (Fig. 3C). The repro57 mutant spermatocytes exhibited normal distribution of γH2AX during pachytene stage (Fig. 3D). However, 30% of repro57 mutant pachytene spermatocytes showed XY dissociation (Fig. 3D, N=260 cells), while only 3.7% of WT pachytene spermatocytes exhibited XY dissociation (Fig. 3C, N=135 cells).
Figure 3. Analysis of DNA damage repair and XY body dynamics in WT and repro57 spermatocytes.
(A,B) Sections from 14 dpp testes were immunolabeled for DMC1. Scale bars, 50 µm. (C,D) Spread spermatocyte nuclei were immunolabeled for SYCP3 (red) and γH2AX (green), with magnified views of the XY bodies (insets), revealing XY chromosome dissociation in repro57 mutant spermatocytes. (Original magnification of E, F = X1000)
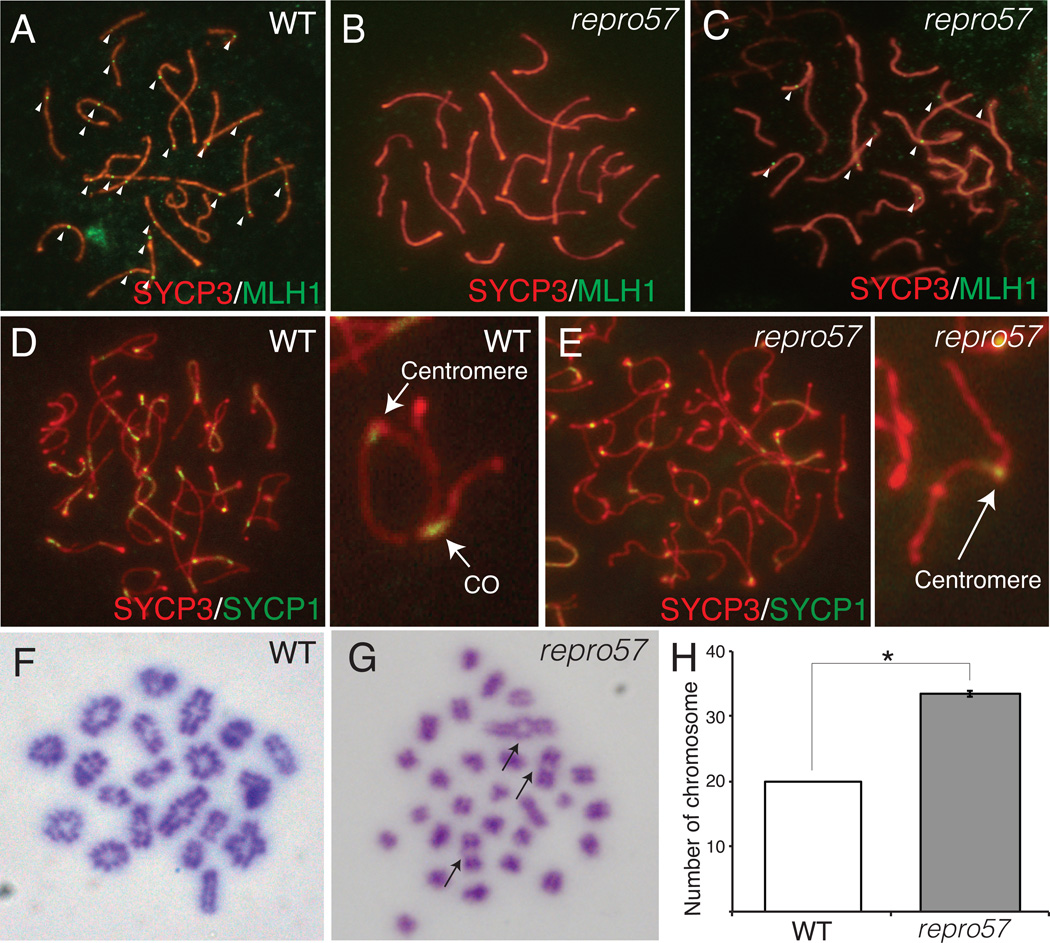
To identify COs, we immunolabeled for MLH1 foci, which mark COs during mid- to late-pachytene substages and for the testis-specific histone H1 variant HIST1H1T (herein referred to by its common designation of H1t), which is expressed in spermatocytes at post-mid-pachytene stages (Cobb et al. 1999). H1t-positive spermatocytes in the WT mice contained normal numbers of MLH1 foci (1–2 foci on each chromosome) (Fig. 4A). However, almost all H1t-positive homozygous mutant spermatocytes showed complete absence of MLH1 foci (Fig. 4B), although a few had 1–8 MLH1 foci per cell (Fig. 4C). We also scored COs by labeling diplotene spermatocytes for co-localization of SYCP3 and SYCP1 in partially desynapsed diplotene spermatocytes. In contrast to WT spermatocytes, the repro57 mutant spermatocytes lacked SYCP1 – SYCP3 co-localization (a mark of CO between homologs). However, homologs in mutant germ cells were partially associated at centromere (Fig. 4E), as were homologs in WT diplotene spermatocytes (Qiao et al. 2012) (Fig. 4D). At diakinesis/metaphase I, all 20 chromosomes exhibited crossing over in WT spermatocytes (Fig. 4F), but only a few chromosomes in repro57 mutant spermatocytes exhibited chiasmata (Fig. 4G). Consistent with lack of chaismata, there was a significantly increased number of chromosomes (univalents) in mutant spermatocytes compared to WT (Fig. 4H).
Figure 4. Analysis of crossing-over in WT and repro57 spermatocytes.
(A–C) Pachytene spermatocytes from WT (A) and repro57 homozygous mice (B,C) immunolabeled with SYCP3 (red), MLH1 (green). MLH1 foci are absent in most H1T-positive repro57 spermatocytes (B), however some mutant spermatocytes exhibited 1–8 weak MLH1 foci (C). Arrowheads indicate MLH1 focus. (D,E) Diplotene spermatocytes immunolabeled with SYCP3 (red) and SYCP1 (green) from D. WT, with a magnified view of a chromosome with chiasma and E. repro57, with a magnified view of a chromosome lacking CO. (F–H) Giemsa-stained chromosome of diakinesis/metaphase I spermatocyte from WT (F) and repro57 mice (G). Arrows in G indicate bivalent chromosomes with chiasma, but many lack chiasmata. Original magnification X1000. (H) The number of chromosomes, including both bivalent and univalent, was compared (N=21 cells for WT and N=60 cells for repro57). Bars indicate standard error; asterisk indicates p < 0.005. Data are reported as mean ± standard error.
A mutation in the ring finger protein 212 gene (Rnf212) in repro57 mice
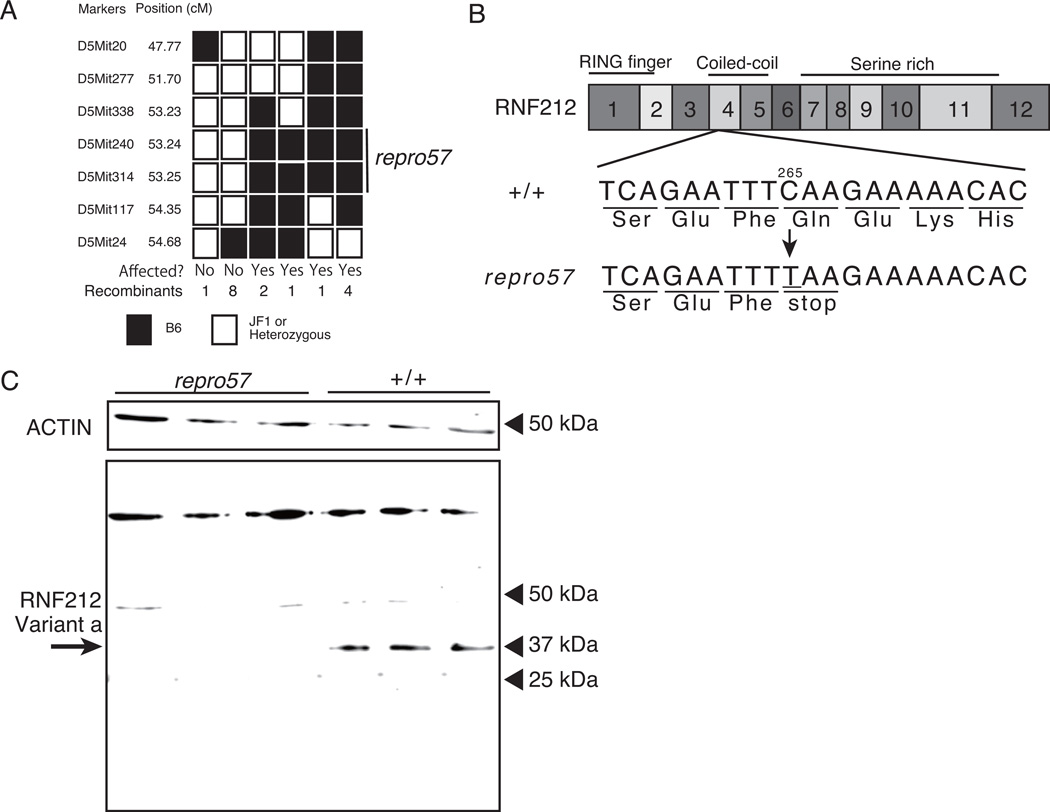
The repro57 mutation was mapped to ~3 Mb region of Chr. 5 between D5Mit338 and D5Mit117 by linkage analysis using 82 affected F2 male mice (Fig. 5A). This region includes 70 genes; among these, Rnf212 (ring finger protein 212) was a strong candidate gene for harboring the repro57 mutation. Similar to repro57 mutant mice, mice with a targeted gene knockout of Rnf212 exhibit male and female infertility, with male germ cells arrested at late meiotic prophase (Reynolds et al. 2013). High-through-put sequencing of DNA from homozygous repro57 and WT mice revealed a C to T transversion at nucleotide 265 of the Rnf212 mRNA (at 108757107 bp in Chr5, GRCm38.p2), creating a premature stop codon within the coiled-coil domain of RNF212 (Fig. 5B). This mutation was common to all mice with the repro57 phenotype, and, moreover, no other exonic mutation was detected within the repro57 region. We assessed expression of Rnf212 transcript variants in WT and repro57 testes. Three variants contain the RING finger domain (Reynolds et al. 2013); bioinformatics analysis revealed that 5’ end of two other transcripts contain variant-specific sequences, lacking the RING finger domain (GRCm38.p2, C57BL/6J). RT-PCR analysis of WT and repro57 testis RNA revealed expression of variants a, b, c and e in both WT and repro57, while variant d was not detected in either WT or repro57 (Supplementary Figure 1).
Figure 5. Fine mapping of repro57 mutation and loss of RNF212 expression.
(A) Segregation of alleles of 7 microsatellite markers in 82 affected mice of the F2 progeny obtained from crosses between +/repro57 and JF1 mice. The positions of the microsatellite markers in the critical region are indicated in cM according to the mouse genome sequence (NCBI Build 38) and the region that contains the repro57 locus is indicated by a bar. (B) Schematic diagram of the Rnf212 transcript indicating the position of the nonsense mutation in the exon 4 of Rnf212. (C) Western blot analysis revealed no expression of RNF212 in the testis of adult repro57 mice, and a single band was detected only in the testes of WT. Non-specific bands were detected in the testes of both WT and repro57. ACTIN was used as an internal control.
To determine if RNF212 protein is absent in repro57 mice, we performed western blot analysis using whole testis lysates, with a polyclonal antibody raised against the full-length mouse RNF212. In WT testes, a single RNF212 band (correlating to splice variant a) was detected at about 34 kDa, while no RNF212, or or even a truncated protein, were detected in repro57 mutant testes (Fig. 5C). Non-specific bands at higher molecular sizes were observed in both WT and repro57 lysates.
DISCUSSION
A moderate-scale unbiased mutagenesis effort coupled with phenotype screening is a powerful tool to create animal models for human disease. We identified the repro57 fertility model by a fertility screen in such an ENU mutagenesis program. We find that there is a nonsense mutation in the Rnf212 gene in repro57 mutant mice. Similar to the Rnf212 knockout phenotype, the repro57 mutant mice exhibit male infertility, arrest of spermatogenesis in meiosis and defects in cytological markers of recombination and chiasma formation. These observations identify a new allele of the mouse Rnf212 gene, Rnf212repro57.
A nonsense mutation in Rnf212, encoding a meiotic recombination factor, in repro57 mutants
Deep sequencing revealed a nonsense mutation in exon 4 of the Rnf212 gene in repro57 mutant. This mutation could truncate the RNF212 protein, ablating the coiled-coil and serine-rich domains. Interestingly, both this study and the previous study (Reynolds et al. 2013) find that only a single RNF212 isoform is expressed in the testis, in spite of evidence for several splicing variants. This could suggest that the variants are not translationally competent.
There is considerable evidence for essential function of RNF212 in meiosis in a variety of species. Rnf212 was shown to be essential for meiotic recombination in C. elegans (Jantsch et al. 2004, Bhalla et al. 2008). In human, variants of RNF212 have been associated with different genome-wide recombination rates (Kong et al. 2008, Chowdhury et al. 2009, Kong et al. 2014). Creation of a targeted mutation, Rnf212tm1Nhtr, revealed that mouse RNF212 was essential for CO formation; the Rnf212 KO spermatocytes lacked chiasmata, and exhibit depletion of spermatids and mature spermatozoa (Reynolds et al. 2013). These phenotypes of Rnf212 KO mutant mouse resemble those of the repro57 mutant mouse, providing evidence that the new Rnf212 allele, Rnf212repro57, is responsible for infertility of the repro57 mice. Final confirmation that the new Rnf212repro57 allele is solely causative of the repro57 infertility phenotype awaits a formal genetic complementation test.
Spermatogenic and meiotic defects in Rnf212repro57 mice
Many aspects of the spermatogenesis and meiotic phenotype of Rnf212repro57 mutant males mirror those found in the Rnf212 KO males. Even during the juvenile onset of spermatogenesis, degenerating and apoptotic spermatocytes were observed (at 21 dpp). However, normal expression pattern of PCNA in the mutant testes suggests that mitotic proliferation and meiotic initiation are normal in the absent of RNF212. Similar to mice with the Rnf212 KO mutation (Reynolds et al. 2013), Rnf212repro57 spermatocytes form the SC and exhibit synapsis. Interestingly, another RNF212 homologue ZHP3 is also not required for SC formation (Jantsch et al. 2004); however yeast Zip3 mutants exhibited defective SC formation (Agarwal & Roeder 2000). Evidence that DNA DSBs are repaired in Rnf212repro57 spermatocytes includes apparently normal localization of DMC1, which is essential for DNA strand invasion (Moens et al. 2002), and redistribution of γH2AX by mid-prophase in mutant spermatocytes. However, mutant Rnf212repro57 spermatocytes, like Rnf212 KO spermatocytes, fail to form COs as marked by MLH1 foci, exhibiting lack of chiasmata in most homologous chromosome pairs. Together, observations on mutant spermatocytes for either of two different Rnf212 alleles suggest that mouse RNF212 plays a role in maturation of CO events in meiotic recombination, but the mechanism is still not clear. Recent evidence suggests that small ubiquitin-like protein (SUMO) plays an important role in assembly and disassembly of SC by regulating protein-protein interaction during meiosis (Watts & Hoffmann 2011). In this context, it is interesting that the mouse RNF212 mediates SUMOylation, with SUMO modification stabilizing the association of MutSγ with nascent CO intermediates (Reynolds et al. 2013). Human RNF212 and yeast homologue Zip3 contain a RING-finger domain, ubiquitin ligating activity for post-translational protein modification (Deshaies & Joazeiro 2009), and Zip3 was also reported to be involved in SUMO modification (Perry et al. 2005, Cheng et al. 2006). In this context, it is relevant that the repro57 mutation in the Rnf212 gene may still lead to a protein with an intact RING domain. Loss of RING domain in the targeted Rnf212 KO allele resulted in absence of MLH1 foci in germ cells (Reynolds et al. 2013), and although the majority of Rnf212repro57 spermatocytes also lacked MLH1 foci, a few had a markedly reduced number of MLH1 foci and chiasmata, suggesting that the Rnf212repro57 allele could be hypomorphic in this respect. Because much of the value of different gene alleles lies in the ability to tease apart protein domain function, further experimental comparisons of the two mutated alleles are warranted.
Rnf212 infertility models
These results establish the Rnf212repro57 mice as a single-base change model for male infertility, suggesting the possibility that some human male infertility could be attributed to RNF212 mutation. Although the initial screening of the repro57 mice suggested only male infertility (http://reproductivegenomics.jax.org/mutants/G1-586-2.html), female repro57 homozygous mice on the congenic C3H background also exhibited infertility. Interestingly, the C3H congenic Rnf212repro57 males exhibited a remarkably higher rate of XY dissociation than that of the Rnf212 KO mutants on a B6 background (Reynolds et al. 2013). Given also recent studies showing that genomic variations between subspecies cause alteration of phenotypes (Keane et al. 2011, Yang et al. 2011), our findings suggest that phenotypic differences between Rnf212 mutants could be due to not only the allelic difference but also to genetic background effects.
In summary, this study identifies a new allele of the mouse RNf212 gene, Rnf212repro57, and shows its association with male fertility. These results bolster genetic evidence that RNF212 is essential for CO and chiasma formation during meiosis. Because COs promote the accurate segregation of homologs, thereby avoiding aneuploidy, the mouse Rnf212 mutations provide important models for investigating the requirements for accurate gametic chromosome segregation. In humans, chromosome aneuploidy leads to infertility, especially in males (Egozcue et al. 2000), congenital defects such as Down syndrome, or high rates of miscarriage due to defective embryo development (Ambartsumyan & Clark 2008). Thus, genetic models such as these continue to be valuable for human genetic disease.
Supplementary Material
(A) Expression of Rnf212 transcript variants in the testis by RT-PCR. Variant d was not detected in both WT and repro57. Expected sizes of amplified PCR fragment of Rnf212 variants a, b, c, d, e and Gapdh were 185, 189, 129, 359, 402 and 416 bp, respectively. Gapdh was used as an internal control. Primer sequences used for RT-PCR were shown. (B) Schematic view of transcript variants of mouse Rnf212. Exons are numbered, and positions of domains are indicated. Circled STOP marks indicate a premature stop codon. Asterisks indicate variant d or e specific exons. The position of repro57 mutation (RNF212_Q87*) is indicated by arrowhead. Arrows show the positions of the primers for RT-PCR, and expected product sizes were also indicated. Variants a-c are cited from Reynolds et al. (2013).
Acknowledgements
We thank Lucy Rowe and Mary Barter (The Jackson Laboratory, USA) for preliminary mapping analysis, Drs. Bernard de Massy (National Center for Scientific Research, France), Neil Hunter (University of California, Davis, USA), and Shinichiro Chuma (Kyoto University, Japan) for gifts of SYCP3 (guinea pig), RNF212 and SYCP3 (rabbit) antibodies, and Dr. Junko Noguchi (National Institute of Agrobiological Sciences, Japan) for comments on histology. We acknowledge the Scientific Services of The Jackson Laboratory for outstanding support.
Funding: Supported by Japan Society for the Promotion of Science (JSPS), Strategic Young Researcher Oversea Visits Program for Acceleration Brain Circulation to Y.F., Sasakawa Scientific Research Grant 22-426 (Japan Science Society) to Y.F., and the NIH, HD42137 to the Reproductive Genomics Program at The Jackson Laboratory. Research reported in this publication was also partially supported by the National Cancer Institute under award number P30 CA034196 to JAX; the content is solely the responsibility of the authors and does not necessarily the official views of the NIH.
Footnotes
Declaration of interest: The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Ambartsumyan G, Clark AT. Aneuploidy and early human embryo development. Human Molecular Genetics. 2008;17:R10–R15. doi: 10.1093/hmg/ddn170. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nature Reviews. Genetics. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genetics. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Schimenti JC. Genetics of meiosis and recombination in mice. International Review of Cell and Molecular Biology. 2012;298:179–227. doi: 10.1016/B978-0-12-394309-5.00005-5. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes and Development. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genetics. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J, Reddy RK, Park C, Handel MA. Analysis of expression and function of topoisomerase I and II during meiosis in male mice. Molecular Reproduction and Development. 1999;46:489–498. doi: 10.1002/(SICI)1098-2795(199704)46:4<489::AID-MRD7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Smith GR. Branching out: meiotic recombination and its regulation. Trends in Cell Biology. 2007;17:448–455. doi: 10.1016/j.tcb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual Review of Biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Egozcue S, Blanco J, Vendrell JM, Garcia F, Veiga A, Aran B, Barri PN, Vidal F, Egozcue J. Human male infertility: chromosome anomalies, meiotic disorders, abnormal spermatozoa and recurrent abortion. Human Reproduction Update. 2000;6:93–105. doi: 10.1093/humupd/6.1.93. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Ogonuki N, Inoue K, Ogura A, Handel MA, Noguchi J, Kunieda T. t-SNARE Syntaxin2 (STX2) is implicated in intracellular transport of sulfoglycolipids during meiotic prophase in mouse spermatogenesis. Biology of Reproduction. 2013;88:141. doi: 10.1095/biolreprod.112.107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Lessard C, Reinholdt L, Schimenti J, Eppig JJ. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Molecular and Cellular Endocrinology. 2006;250:201–205. doi: 10.1016/j.mce.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nature Reviews. Genetics. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Molecular and Cellular Biology. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Masson G, Gudbjartsson DF, Villemoes R, Magnusdottir E, Olafsdottir SB, Thorsteinsdottir U, Stefansson K. Common and low-frequency variants associated with genome-wide recombination rate. Nature Genetics. 2014;46:11–16. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM, Gudjonsson SA, Sverrisson S, Thorlacius T, Jonasdottir A, Hardarson GA, Palsson ST, Frigge ML, Gulcher JR, Thorsteinsdottir U, Stefansson K. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- Lessard C, Pendola JK, Hartford SA, Schimenti JC, Handel MA, Eppig JJ. New mouse genetic models for human contraceptive development. Cytogenetic and Genome Research. 2004;105:222–227. doi: 10.1159/000078192. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. Journal of Cell Science. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N, Borner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Research. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Hyppa RW, Smith GR. New and old ways to control meiotic recombination. Trends in Genetics. 2011;27:411–421. doi: 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Hoog C, Paddy M, Hunter N. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genetics. 2012;8:e1002790. doi: 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, Hoog C, Cohen PE, Hunter N. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nature Genetics. 2013;45:269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, Edelmann W, Scharff MD. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts FZ, Hoffmann E. SUMO meets meiosis: an encounter at the synaptonemal complex: SUMO chains and sumoylated proteins suggest that heterogeneous and complex interactions lie at the centre of the synaptonemal complex. Bioessays. 2011;33:529–537. doi: 10.1002/bies.201100002. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP. Subspecific origin and haplotype diversity in the laboratory mouse. Nature Genetics. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa T, Yamamoto T, Nakayama Y, Hamada S, Abe S. Conversion from mitosis to meiosis: morphology and expression of proliferating cell nuclear antigen (PCNA) and Dmc1 during newt spermatogenesis. Development Growth and Differentiation. 2000;42:603–611. doi: 10.1046/j.1440-169x.2000.00544.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Expression of Rnf212 transcript variants in the testis by RT-PCR. Variant d was not detected in both WT and repro57. Expected sizes of amplified PCR fragment of Rnf212 variants a, b, c, d, e and Gapdh were 185, 189, 129, 359, 402 and 416 bp, respectively. Gapdh was used as an internal control. Primer sequences used for RT-PCR were shown. (B) Schematic view of transcript variants of mouse Rnf212. Exons are numbered, and positions of domains are indicated. Circled STOP marks indicate a premature stop codon. Asterisks indicate variant d or e specific exons. The position of repro57 mutation (RNF212_Q87*) is indicated by arrowhead. Arrows show the positions of the primers for RT-PCR, and expected product sizes were also indicated. Variants a-c are cited from Reynolds et al. (2013).