Abstract
Background: MicroRNA (miRNA) is small endogenous, single strand RNA molecules that regulate gene expression at post-transcriptional level through several mechanisms to affect key cellular event including male germ cells differentiation, proliferation, development and apoptosis. Mutation and/or aberrant expression of miRNAs have been associated with progression of various disorders, including infertility.
Objective: The purpose of this research was to study the estrogen receptor beta (ERβ(, hsa-mir-21 and, hsa-mir-22 expression level in oligospermic infertile and control fertile men and correlation between them.
Materials and Methods: In this study, the change in mir-21, mir-22 expression and their common target gene (ERβ) expression levels were evaluated in oligospermic infertile men (n= 43) compared with 43 age matched healthy control by Real-Time PCR methods.
Results: Expression analysis by qRT-PCR test on miRNA have identified that mir-21, mir-22 levels were significantly higher than those in normal controls (p<0.0001) and ERβ expression level significantly decreased in comparison with the normal group (p<0.0001).
Conclusion: Our study showed that mir-21 and mir-22 are indirectly involved in spermatogenesis by regulating of the estrogen receptor and might have a diagnostic and prognostic value in men infertility.
Key Words: mir-21, mir-22, microRNA, Fertility, Oligospermia
Introduction
Spermatogenesis consists of a complex procedure of proliferation and differentiation of germ cells that leads undifferentiated diploid cells to differentiate into haploid male gamete cells. It is regulated at the transcriptional and the post-transcriptional level (1). microRNAs (miRNAs) are an essential regulator of spermatogenesis at post-transcriptional level. miRNAs are single-stranded, small (18-25 nucleotides) non-coding RNAs, which act as a regulator of genes in spermatogenesis (2).
miRNAs were first detected in human spermatozoa by Ostermeier et al they are abundant in spermatozoa however, the molecular features of miRNA in spermatogenesis and male fertility are not well defined (3). Understanding of these mechanisms is needed for providing high reproductive efficiency and prognostic value in fertility. Infertility affects 10-15% of couples about 50% of these cases are because of male factors (4). Oligospermia (sperm count <15×106 ml) is a common cause of infertility in males, but the mechanisms that cause disease are not well known (5).
Sperm-born mRNAs have an essential role in normal spermatogenesis. Aberrant in gene expression have a negative impact on spermatozoa differentiation and development (6, 7). Estrogen has a positive effect on sperm capacitating, fertilizing ability, as well as has a special role in male reproductive regulation (8). Cell signaling of estrogen is mediated through the estrogen receptors (ERs) that are present in the male reproductive tract, sperm and germ cells (9). It has been demonstrated that estrogen receptor gene knockout (ERKO) causes decrease in mice spermatozoa count and viability (10). Previous studies showed that mir-21 and mir-22 were predicted to target ERβ gene (11, 12).
In this study we investigated mir-21, mir-22 and ERβ gene expression in oligospermic infertile (n= 43) compared with that of control fertile men (n= 43) to detect any difference that could provide new insights in the molecular mechanism happening during spermatogenesis.
Materials and methods
Study design
The design of this study is a cross-sectional survey which was conducted in period of one year (2013). From infertile men (n=723) conferred to the Tabriz Alzahra Infertility Center (mean age 27.5±4.8 years), which despite of continuous intercourse they had a background of infertility for more than two years, 43 oligospermic infertile men were selected. Control samples (n=43), selected from normal fertile volunteers (mean age 26±3.3) had a child in the last two years and their semen analysis was normal. Neither control subjects nor patients groups did not treated with the drug for two months before sampling. The written agreement of the subjects was given according to the rules of medical ethics. This research was approved by the ethics Committees of Tabriz medical University.
Exclusion criteria
The volunteers with infertile partner, genital tract infection, alcohol and drug consumption, autoimmune disease, smoking and abnormality in reproductive tract were excluded from the study.
Hormone Analysis
The blood was placed at 37oC for 10 minutes to clot formation. The clot from the test tube was slightly removed then supernatant was centrifuged at 1000 × g for 10 minutes at 4oC. samples were analyzed for 17β-estradiol (estrogen), testosterone, follicle stimulating hormone (FSH) and luteinizing hormone (LH) using an enzyme-linked immunosorbent assay (ELISA) by commercial ELISA kit (AccuBind ELISA, Monobind, USA) according to the manufacturer's instructions.
Isolation of spermatozoa from seminal fluid
Semen samples were collected in a sterile flask and incubated at 37oC for 30 min to get the fluid then according to WHO guidelines, semen analysis was accomplished (13). Spermatozoa were purified by Goodrich methods (14). In brief, the samples were washed two times in 1× PBS buffer solution, then somatic cells were absent in SCLB solution (0.1% SDS, 0.5% TX-100 in Diethylpyrocarbonate (DEPC) water). The cells were counted, if somatic cells were, present the process was repeated. Finally the solution was frozen at -80oC. In this study papanicolaou staining and strict criteria were used to assess sperm morphology (15, 16).
RNA isolation
Total RNA was isolated using exiqon miRCURY RNA isolation kit (Exiqon, Denmark) according to the manufacturer’s instructions. Quantity and quality of the isolated RNA was measured by Nanodrop 1000 (NanoDrop ND-1000 spectrophotometer; Thermo Fisher Scientific, Waltham, MA).Total RNAs were reversed to cDNA using LNA universal RT miRNA PCR kit (Exiqon, Denmark). Briefly, 20 ng of total RNA was reverse transcribed. cDNA Synthesis was performed by thermal cycler (Eppendorf, Germany) with the following condition, value, 60 min at 42oC, 5 min at 95oC and immediately cool to 4oC until use.
Real-time PCR analysis
Quantitative real-time reverse transcriptase-PCR was carried out by using the Corbett Rotor-Gene 6000 Real-Time PCR system (Qiagen, Germany). miRNA’s quantification was performed using MiRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Denmark). Mir-16 was used as the endogenous control miRNA. ERβ relative expression was measured by qPCR with primers (ERβ: 5′-AGCCTGTTCGATCAAGTG -3′ and 5′-CCTCATCACTGTCCAGAA-3′) using SYBR Green PCR Kit (Qiagen, Germany). The expression levels were normalized to β-actin as housekeeping gene with the following primers (5′-TGGACTTCGA GCAAGAGATG-3′ and 5′-GAAGGAAGGCTG GAAGAGTG-3′). The reactions were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS software (version 18). The results were expressed as mean±SD. Genes relative expression level were calculated by using the 2DDCq model (17). Unpaired Student's t-test was used to analyze the differences in gene expression between oligospermic and control group. Correlation analysis was performed using the spearman rank correlation test. In all analysis p<0.05 was considered as significant.
Results
mir-21, mir-22 and ERβ expression levels in oligospermic and control group
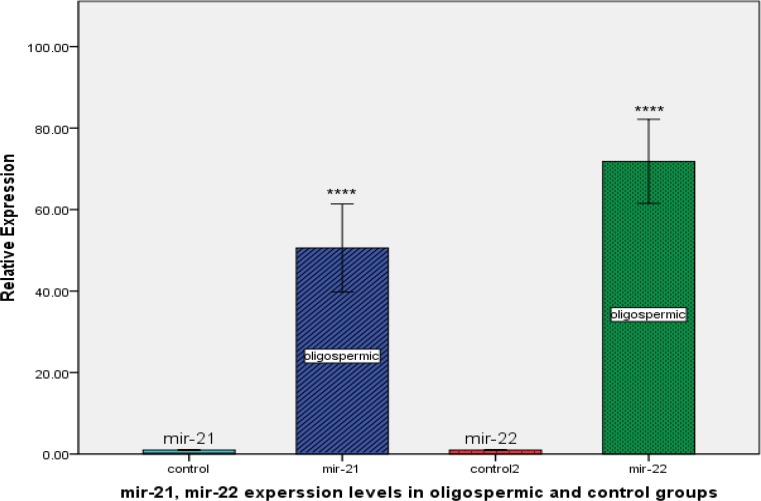
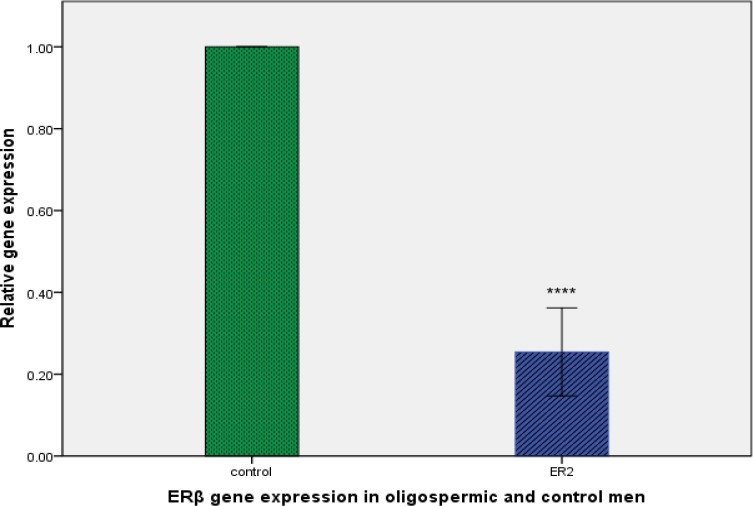
We determined the expression levels of mir-21, mir-22 and ERβ in oligospermic and control group (Figure 1). By real-time quantitative RT-PCR analysis, we found that expression levels of mir-21 and mir-22 were much higher in oligospermic than control group (p<0.0001 and p<0.0001, respectively). Inversely, ERβ expression was significantly lower in oligospermic than control group (p<0.0001; Figure 2).
Figure 1.
Relative expression levels of mir-21 and mir-22 in oligospermic and control group. ***P<0.0001compared with control group.
Figure 2.
Relative expression levels of the estrogen receptor beta (ERβ) gene in control and oligospermic group. ***P<0.0001 compared with control group
Correlation between expression levels of ERβ and seminal plasma parameters
Correlation between ERβ expression and semen were analyzed using spearman׳s rank correlation test (Table I). Expression levels of ERβ were strongly and positively correlated with those of sperm count, quick progressive, slow progressive and normal morphology (spearman׳s correlation coefficient; +0.849, +0.758, +0.753 and +0.805, respectively) and negatively correlated with immotile (spearman’s correlation coefficient; -0.735).
Table I.
Correlation analysis between ERβ, mir-21, mir-22 and seminal plasma parameters
| Variable | mir-21 | mir-22 | ERβ |
|---|---|---|---|
| Volume | 0.1778a | 0.04610 | -0.1449 |
| 0.6231b | 0.8994 | 0.6896 | |
| Sperm count | -0.9014* | -0.7717* | 0.8495* |
| 0.0004 | 0.0089 | 0.0019 | |
| Quick progressive | -0.7581* | -0.7781* | 0.7581* |
| 0.0111 | 0.0121 | 0.0111 | |
| Slow progressive | -0.8655* | -0.7796* | 0.7532* |
| 0.0012 | 0.0078 | 0.0119 | |
| Non-progressive | -0.9131* | -0.8661* | 0.7587* |
| 0.0002 | 0.0012 | 0.0110 | |
| Immotile | 0.8781* | 0.8001* | -0.7350* |
| 0.0008 | 0.0054 | 0.0154 | |
| Normal morphology | -0.9064* | -0.8064* | 0.8057* |
| 0.0003 | 0.0004 | 0.0049 | |
| pH | -0.6530* | -0.7047* | 0.4590 |
| 0.0407 | 0.0229 | 0.1821 |
Spearman correlation coefficient
P, spearman’s rank correlation test
P<0.05 is considered significant
Correlation between expression levels of mir-21 and seminal plasma parameters
Relation between miRNAs expression and semen parameters, such as volume, sperm count, quick progressive, slow progressive, non-progressive, immotile, normal morphology and pH, was performed using Spearman׳s rank correlation test (Table I). Expression levels of mir-21 were negatively correlated with those of sperm count, quick progressive, slow progressive, normal morphology and pH (spearman’s correlation coefficient; -0.901, -0.758, -0.865, -0.906, and -0.653, respectively) and positively correlated with immotile (spearman׳s correlation coefficient; +0.878).
Correlation between expression levels of mir-22 and seminal plasma parameters
Relation between expression levels of miRNAs and semen parameters, such as volume, sperm count, quick progressive, slow progressive, non-progressive, immotile, normal morphology and pH, was performed using Spearman׳s rank correlation test (Table I).
Expression levels of mir-22 were negatively correlated with those of sperm count, quick progressive, slow progressive, normal morphology and pH (spearman׳s correlation coefficient; -0.771, -0.778, -0.779, -0.806, and -0.704, respectively) and positively correlated with immotile (spearman׳s correlation coefficient; +0.8)
Correlation between hormones and ERβ, mir-21, mir-22
Expression levels of mir-21 were not significantly correlated with hormones but mir-22 expression levels were strongly and positively correlated with estrogen (spearman׳s correlation coefficient; +0.665). Expression levels of ERβ were negatively correlated with estrogen (spearman׳s correlation coefficient; -0.730) (Table II).
Table II.
Correlation analysis between ERβ, mir-21, mir-22 and hormones
| Hormones | mir-21 | mir-22 | ERβ |
|---|---|---|---|
| FSH | 0.4332a | 0.3039 | -0.3039 |
| 0.2111b | 0.3934 | 0.3934 | |
| LH | 0.4720 | 0.2651 | -0.4590 |
| 0.1685 | 0.4592 | 0.1821 | |
| E2 | 0.4720 | 0.6659* | -0.7306* |
| 0.1685 | 0.0356 | 0.0164 | |
| Test | -0.1616 | -0.2263 | -0.05819 |
| 0.6555 | 0.5296 | 0.8731 |
Spearman correlation coefficient
P, spearman’s rank correlation test
P<0.05 is considered significant
Correlation between expression levels of mir-22, mir-21 and ERβ
We found that expression levels of mir-21, mir-22 were strongly and negatively correlated with ERβ (spearman׳s correlation coefficient; -0.834, -0.0820 respectively (Table III).
Table III.
Correlation analysis between mir-21, mir-22 and ERβ
Spearman correlation coefficient
P, spearman’s rank correlation test
P<0.05 is considered significant
Discussion
Spermatogenesis is a complex procedure of male gem cells differentiation and proliferation that many genes are involved in. Disruption of the gene expression is impairs spermatogenesis and fertility. Almost mature sperm’s mRNAs are similar to those found in testis that it can show normal spermatogenesis, despite recognizing a complex population of mRNA in human mature sperm, their role and aspects affecting their expression is remain unknown. miRNAs regulate genes expression at the post- transcriptional level. They have an essential role during spermatogenesis, increase or decrease expression of miRNAs in mature sperm impair development and/or fertility (16).
Lian et al showed that significant changes in miRNA expression were observed in non-abstractive azoospermic infertile patients compared with control fertile men (17). In this study we evaluated mir-21, mir-22 and their target gene (ERβ) expression levels in oligospermic infertile compared with control fertile men. We showed that mir-21 and mir-22 were significantly over expressed in oligospermic compared with the control group. We also showed that high mir-21 and mir-22 expression were associated with significant decreases in ERβ gene expression level in oligospermic group. It has been proved that mir-21 and mir-22 were predicted to target ERβ, which have been implicated in ERβ regulation (11, 12).
Bhat-Nakshatri et al reported significant inverse association between the expression levels of mir-21, mir-22 and ERβ in breast cancer cells (11). Similarly, in the present study, we have showed that mir-21, mir-22 expression levels were increased unlike ERβ expression, which was decreased in sperm cells. ERβ localization in germ cell, spermatozoa, epithelial cells, Sertoli and Leydig cells which suggested the important role of ERβ in spermatogenesis (18). Lucas et al showed that activation of ERβ by estrogen (E2) increased proliferation of immature Sertoli cells (19). Sertoli cells are the somatic cells of the testis that are important for spermatogenesis. Sertoli cells accelerate differentiation of germ cells to spermatozoa, Sertoli cells dysfunction impairs spermatogenesis and fertility (20, 21).
miRNA might be involved in spermatogenesis through other genes. Possible targets of mir-21 are MEST, AKT2, AKT1, phosphatase and tensin homolog deleted on chromosome ten (PTEN) and PLAG1 (-). PTEN and AKT1 are the targets of mir-22 (25, 26). Defect in MEST processing or action is associated with low sperm counts. MEST dysfunction was seen in idiopathic infertile men with sperm morphology below normal spermatozoa and progressive sperm motility below 40%. It can be used as a biomarker for sperm quality (27, 28). AKT1 is a serin/theronin kinas enzyme. It is the moderator of cellular growth, survival, metabolism and proliferation, in different cell types (29).
It is proved that in Akt2-/- male mice apoptotic spermatozoa in null mice were more than wild-type mice, and sperm concentration and motility, in the null mice were significantly lower than those wild-type (30). PTEN has an important role in distinct cellular procedures, including cell survival, transformation, migration, proliferation, and moderate the germ cell differentiation (31). The pleomorphic adenoma gene 1 (Plag1) disruption in mouse affects pre- and postnatal growth and development of organs, with reproductive effects (32, 33).
Specific miRNAs are differentially expressed in oligospermic sperm samples in comparison with normal control ejaculates. miRNAs are developing as key players in germ cell act Detailed information about miRNA expression and function will be essential to understanding the miRNA’s roles in reproductive procedures. The diverse facets of post-transcriptional regulation in male germ cells differentiation opens up new ways for contraceptive development and the survey of male fertility. Further study is required to determine the specific roles to all miRNA species involved in spermatogenesis. miRNAs could become novel targets for male contraception and for gene therapy in male infertility.
Acknowledgments
We would like to thank authorities of Tabriz University of Medical Science for the scientific and ethical approval and financial support of this research. This study has been done as a Ph.D. thesis of Alireza Abhari in Women’s Reproductive Health Research Center, Tabriz University of Medical Sciences.
Conflict of interest
There is no conflict of interest in this article
Footnote
This article extracted from Ph.D. thesis. (Alireza Abhari)
References
- 1.Idler RK, Yan W. Control of messenger RNA fate by RNA-binding proteins: an emphasis on mammalian spermatogenesis. J Androl. 2012;33:309–337. doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajender S, Meador C, Agarwal A. Small RNA in spermatogenesis and male infertility. Frontiers Bioscie. 2012;4:1266–1274. doi: 10.2741/s330. [DOI] [PubMed] [Google Scholar]
- 3.Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl. 2005;26:70–74. [PubMed] [Google Scholar]
- 4.Swan SH, Elkin EP, Fenster L. The question of declining sperm density revisited: an analysis of 101 studies published 1934-1996. Environ Health Perspect. 2000;108:961–966. doi: 10.1289/ehp.00108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoysman R. Oligospermia associated with normal testicular function and epididymal lesions or malpositions. Acta Eur Fertil. 1992;23:117–121. [PubMed] [Google Scholar]
- 6.Lalancette C, Platts AE, Johnson GD, Emery BR, Carrell DT, Krawetz SA. Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med. 2009;87:735–748. doi: 10.1007/s00109-009-0485-9. [DOI] [PubMed] [Google Scholar]
- 7.Hamatani T. Spermatozoal RNA profiling towards a clinical evaluation of sperm quality. Reprod Biomed Online. 2011;22:103–105. doi: 10.1016/j.rbmo.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Carreau S, Silandre D, Bois C, Bouraima H, Galeraud-Denis I, Delalande C. Estrogens: a new player in spermatogenesis. Folia Histochem Cytobiol. 2007;45:5–10. [PubMed] [Google Scholar]
- 9.Ellmann S, Sticht H, Thiel F, Beckmann MW, Strick R, Strissel PL. Estrogen and progesterone receptors: from molecular structures to clinical targets. Cell Mol Life Sci. 2009;66:2405–2426. doi: 10.1007/s00018-009-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy EM, Washburn TF, Bunch DO, Goulding EH, Gladen BC, Lubahn DB, et al. Targeted disruption of the estrogen receptor gene in male mice causes alteration of spermatogenesis and infertility. Endocrinology. 1996;137:4796–4805. doi: 10.1210/endo.137.11.8895349. [DOI] [PubMed] [Google Scholar]
- 11.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delic D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75:998–1004. doi: 10.1016/j.steroids.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Menkveld R. Clinical significance of the low normal sperm morphology value as proposed in the fifth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen. Asian J Androl. 2010;12:47–58. doi: 10.1038/aja.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53:161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- 15.Check JH, Adelson HG, Schubert BR, Bollendorf A. Evaluation of sperm morphology using Kruger's strict criteria. Arch Androl. 1992;28:15–17. doi: 10.3109/01485019208987674. [DOI] [PubMed] [Google Scholar]
- 16.Singh S, Sharma S, Jain M, Chauhan R. Importance of papanicolaou staining for sperm morphologic analysis: comparison with an automated sperm quality analyzer. Am J Clin Pathol. 2011;136:247–251. doi: 10.1309/AJCPCLCSPP24QPHR. [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- 19.Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunawan A, Cinar MU, Uddin MJ, Kaewmala K, et al. Investigation on association and expression of ESR2 as a candidate gene for boar sperm quality and fertility. Reprod Domest Anim. 2012;47:782–790. doi: 10.1111/j.1439-0531.2011.01968.x. [DOI] [PubMed] [Google Scholar]
- 21.Lucas TF, Siu ER, Esteves CA, Monteiro HP, Oliveira CA, Porto CS, et al. 17beta-estradiol induces the translocation of the estrogen receptors ESR1 and ESR2 to the cell membrane, MAPK3/1 phosphorylation and proliferation of cultured immature rat Sertoli cells. Biol Reprod. 2008;78:101–114. doi: 10.1095/biolreprod.107.063909. [DOI] [PubMed] [Google Scholar]
- 22.Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls PK, Harrison CA, Rainczuk KE, et al. Retinoic acid promotes Sertoli cell differentiation and antagonises activin-induced proliferation. Mol Cell Endocrinol. 2013;377:33–43. doi: 10.1016/j.mce.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 24.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nature Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 25.Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Nat Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Jiang Y, Zhang H, Greenlee AR, Yu R, Yang Q. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo [a] pyrene-7,8-diol-9,10-epoxide. Toxicol InVitro. 2010;24:1168–1175. doi: 10.1016/j.tiv.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Zhang Y, Zhao J, Kong F, Chen Y. Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol Cell Biochem. 2011;357:31–38. doi: 10.1007/s11010-011-0872-8. [DOI] [PubMed] [Google Scholar]
- 29.Poplinski A, Tuttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 30.Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 31.Stabile V, Russo M, Chieffi P. 17beta-estradiol induces Akt-1 through estrogen receptor-beta in the frog (Rana esculenta) male germ cells. Reproduction. 2006;132:477–484. doi: 10.1530/rep.1.01107. [DOI] [PubMed] [Google Scholar]
- 32.Kim ST, Omurtag K, Moley KH. Decreased spermatogenesis, fertility, and altered Slc2A expression in Akt1-/- and Akt2-/- testes and sperm. Reprod Sci. 2012;19:31–42. doi: 10.1177/1933719111424449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIver SC, Stanger SJ, Santarelli DM, Roman SD, Nixon B, McLaughlin EA. A unique combination of male germ cell miRNAs coordinates gonocyte differentiation. PloS One. 2012;7:e35553. doi: 10.1371/journal.pone.0035553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hensen K, Braem C, Declercq J, Van Dyck F, Dewerchin M, Fiette L, et al. Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev Growth Differ. 2004;46:459–470. doi: 10.1111/j.1440-169x.2004.00762.x. [DOI] [PubMed] [Google Scholar]