Abstract
Loss of DNA mismatch repair (MMR) function, due to somatic or germline epi/genetic alterations of MMR genes leads to the accumulation of numerous mutations across the genome, creating a molecular phenotype known as microsatellite instability (MSI). In gastric cancer (GC), MSI occurs in about 15% to 30% of the cases. This review summarizes the current knowledge on the molecular mechanisms underlying the acquisition of MSI in GC as well as on the clinic, pathologic and molecular consequences of the MSI phenotype. Additionally, current therapeutic strategies for GC and their applicability in the MSI subset are also discussed.
Keywords: Gastric cancer, Microsatellite instability, Mismatch repair genes, Oncogenes, Helicobacter pylori
Core tip: This review summarizes the current knowledge on the molecular mechanisms underlying the acquisition of microsatellite instability (MSI) in gastric cancer (GC) as well as on the clinic, pathologic and molecular consequences of the MSI phenotype. Additionally, current therapeutic strategies for GC and their applicability in the MSI subset are also discussed.
MICROSATELLITE INSTABILITY AND THE MISMATCH REPAIR SYSTEM
Microsatellite instability (MSI) phenotype is characterized by the accumulation of numerous mutations across the genome mainly in repetitive sequences (microsatellites) due to a defective DNA mismatch repair (MMR) system[1].
The MMR system is composed of at least seven proteins, h-MLH1, h-MLH3, h-MSH2, h-MSH3, h-MSH6, h-PMS1 and h-PMS2, which associate with specific partners to form functional heterodimers that recognize base-pair mismatches and small nucleotide insertion/deletions (1-4 base pairs) that occur during DNA replication[2,3]. h-MLH1 and h-MSH2 are essential components of the MMR machinery and form five functional heterodimeric complexes: the MutS complex formed by h-MSH2/h-MSH3 (hMutSβ) or h-MSH2/h-MSH6 (hMutSα) heterodimers, and the MutL complex composed by h-MLH1/h-PMS2 (hMutLα), h-MLH1/h-PMS1 (hMutLβ), or h-MLH1/h-MLH3 (hMutLγ) heterodimers[2]. DNA MMR initiates with the assembling of hMutS complex to DNA. The type of MutS heterodimer formed depends on the type of DNA alteration to be corrected. h-MSH2/h-MSH6 heterodimer is required to correct both base-base mispairs and small insertion/deletion loops whereas h-MSH2/h-MSH3 heterodimer works to repair insertion-deletion loops only[4]. Following the initiation of DNA MMR by the MutS complex, recruitment of MutL heterodimer occurs[5,6]. MutL proteins function to connect the mismatch recognition complex to other downstream effectors of the repair machinery such as proliferating cell nuclear antigen, DNA polymerases δ and epsilon, single-stranded DNA-binding protein and possibly helicase(s), which are needed to complete the repair process[4,7,8]. h-MLH1/PMS2 heterodimer is the only hMutL complex shown to be linked to human MMR system and cancer. The role of the other two hMutL complexes is less well understood. In vitro studies showed that h-MLH1/h-MLH3 heterodimer participates in the repair of base-base mispairs and one-nucleotide insertion/deletion loops but the studies have failed to show the in vivo functionality of the complex[5]. In addition, biochemical studies support the existence of h-MLH1/h-PMS1 heterodimers in human cells, unlike in vitro and in vivo studies that do not support their role in neither MMR and MSI induction nor in cancer predisposition[5,9,10].
TYPE OF MMR SYSTEM ALTERATIONS UNDERLYING MSI IN GASTRIC CANCER
Genetic and epigenetic alterations occurring at the MMR system effectors, namely in h-MLH1 and h-MSH2, and less frequently in h-MSH6 and h-PMS2, are the main mechanism by which MMR system failure occurs in MSI gastrointestinal cancers[4].
In stomach cancer, MSI occurs in about 15%-30% of the cases. MSI gastric cancer (GC) can occur in the context of hereditary syndromes, such as in the Lynch syndrome, but most of them arise in a sporadic form and only a small fraction show familial clustering (10%)[11]. Lynch families are characterized by having an excess of synchronous and metachronous colorectal cancer (CRC) but frequently show extra-colonic tumours, including GC[12,13]. Most of Lynch syndrome-associated cancers have h-MLH1, h-MSH2 germline mutations as the causal genetic event underlying MMR deficiency, and only a small fraction of them harbor alterations in h-MSH6 and h-PMS2 genes[14,15]. In addition, loss of MMR system function may also be caused by mechanisms other than germline mutations in MMR genes. This is the case of deletions of the terminal end of the EPCAM gene that have been identified in a small number of families with Lynch syndrome whose tumours demonstrate loss of h-MSH2[16]. In these cases, a failure in transcriptional termination of EPCAM results in the generation of fusion transcripts with the adjacent h-MSH2 gene, giving rise to methylation of the h-MSH2 promoter, particularly in epithelial tissues where EPCAM is expressed at high levels[16]. Constitutional epimutations of the h-MLH1 gene have also been identified in mutation-negative individuals with a clinical diagnosis of Lynch syndrome[17-22]. This defect is characterized by soma-wide promoter methylation and transcriptional silencing of a single allele of the h-MLH1 gene[19,20,22]. The frequencies of germline epimutations of h-MLH1 and h-MSH2 seem to be quite high in the genetically proven Lynch-syndrome cases (about 16% of all mutations) although rather infrequent in a cohort of Lynch-syndrome suspected patients (0.6% and 0.9%, respectively)[21]. Additionally, the 944C>T germline mutation of TGFBRII has also been associated to Lynch-syndrome[23].
Somatic mutations in MMR genes have also been described in sporadic MSI GC. However, in contrast to Lynch syndrome-associated cancers, these mutations were shown to constitute a molecular effect rather than a cause of the mutator phenotype[24]. Epigenetic silencing of h-MLH1 by promoter hypermethylation is the main mechanism leading to MMR deficiency in both sporadic and familial MSI GC cases[25-28]. In addition, Helicobacter pylori (H. pylori) infection may have a role in the impairment of nuclear MMR activity, a subject that will be further discussed in this review[29,30].
MSI AND H. PYLORI INFECTION
H. pylori is the most common chronic infection worldwide and the major etiologic factor for GC[31]. The fact that only about 1% of all infected individuals develop GC is explained by the interplay between environmental factors, host-inflammatory genetic susceptibility and variations in the pathogenicity of the bacterial strains[32-35].
The molecular mechanisms by which H. pylori induces GC are not fully elucidated, but the chronic inflammation that accompanies the infection is an important trigger, since it induces cellular and DNA damage, and creates an environment rich in cytokines and growth factors that contribute to carcinogenesis[36,37]. The persistence and combination of bacterial virulence factors and inflammatory factors acting on host gastric epithelial cells during the long-lasting H. pylori infection leads to epi/genetic mutations, microRNA (miRNA) gene expression changes, and alterations in cell signaling pathways[29,37,38]. H. pylori infection generates an oxidative microenvironment due to an increased production of reactive oxygen species and reactive nitrogen species, which leads to the oxidative DNA damage of the host cells and thus to mutagenesis[39-45]. Moreover, H. pylori stimulates the production of pro-inflammatory mediators, either by epithelial or immune cells, such as IL-1, IL-6, IL-8, TNF-α, IFN-γ, RANTES, COX-2, 5-LOX, and growth factors such as granulocyte-macrophage colony stimulating factors (GM-CSF) which are well-known factors involved in the different steps of tumorigenesis, such as cellular transformation, promotion, survival, proliferation, invasion, angiogenesis, and metastasis[38,46,47].
Another mechanism through which H. pylori may contribute to neoplastic transformation of the gastric cells is by inducing genomic instability[29]. It has been demonstrated that H. pylori induces an increased level of mutations in both the nuclear DNA (nDNA) and mitochondrial DNA (mtDNA)[30,43,48-50]. Genomic instability may be mediated by an impairment of the MMR pathway. In fact, it has been shown that H. pylori decreases the expression of MLH1, PMS1, PMS2, MSH2 and MSH6 in GC cell lines and in a mouse model of infection[30,48,51,52], and also decreases the MMR activity[30]. Concordantly, clinical studies have shown that MLH1 levels are lower in H. pylori-infected individuals in comparison with those that do not harbor the bacteria[53]. Furthermore, MLH1 and MSH2 expression increases in the gastric mucosa after H. pylori eradication treatment[51]. The H. pylori-induced defective nDNA repair might have repercussions in mtDNA repair, due to sharing of some components of the nDNA repair that act in the mitochondria, partly explaining the increased level of mtDNA mutations in gastric cells infected by H. pylori[30,49,50,54]. These data suggest that H. pylori impairs central DNA repair mechanisms, inducing a transient mutator phenotype, which renders gastric epithelial cells vulnerable to the accumulation of genetic instability, thus contributing to gastric carcinogenesis in infected individuals[29].
MSI AND TARGET GENE MUTATIONS IN GC
As previously mentioned, cells with a deficient MMR system accumulate mutations throughout the genome. These mutations, typically insertions or deletions, occur mainly in microsatellite-bearing genes, and affect both coding and non-coding regions. When affecting microsatellites of coding genes, MSI-associated insertion/deletion mutations result in frameshift mutations leading to truncated proteins with impaired or no function. If these mutations affect genes that confer any tumorigenic advantage, they will likely appear at high frequency due to selection during tumour development. In contrast, when affecting non-coding intronic or intragenic regions, they are likely silent and present at low frequencies, unless they occur in gene regulatory regions (promoter regions and 3’ UTR region, for example) that may control gene expression[55-57]. Since MSI GCs show widespread somatic mutations, it is difficult to disclose which are the real target genes whose mutations drive MSI gastric carcinogenesis and which are the bystander genes whose mutations have little or no contribution to malignancy. In this regard, the frequency of mutations and their in vitro or in vivo functionality were proposed as relevant criteria to distinguish between drivers from bystander mutant genes. Additionally, inactivation of the other repeat tract by other molecular mechanism, and the involvement of the candidate MSI target gene in a bona fide growth suppressor pathway should also be taken into consideration[55,58,59]. A database that gathers all mononucleotide microsatellite mutations in human MSI tumours of different organs, SelTarbase (http://www.seltarbase.org/), was created, allowing the identification of relevant genes for tumorigenesis based on their mutation frequency[60]. Nevertheless, to date, several genes have been identified to be critical targets of the defective MMR and to be specifically altered in GC displaying MSI as listed in Table 1. These comprise genes involved in DNA repair, chromatin structure regulation, apoptosis, cell cycle progression, transcription regulation and signal transduction. A new class of target genes that show frameshifts mutations in MSI GC has recently been identified and include genes involved in the processing machinery of miRNA, which harbor mononucleotide repeats in their coding sequences[61]. More recently, whole genome and exome sequencing of GC samples revealed novel genes, ARID1A and RNF43, to be mutated in 83% and 55%, of MSI cases, respectively[62,63].
Table 1.
Target genes in gastric tumours with microsatellite instability
| Gene pathway | Target gene |
| DNA repair/chromatin structure regulation | ATR |
| BLM | |
| CHK1 | |
| MED1 | |
| MRE11 | |
| MSH2 | |
| MSH3 | |
| MSH6 | |
| RAD50 | |
| Signal transduction | DP2 |
| IGFIIR | |
| RIZ | |
| TGF-βRII | |
| Transcriptional regulation | TCF4 |
| E2F4 | |
| microRNA regulation | AGO2 |
| TNRC6A | |
| Cell death | APAF1 |
| BAX | |
| BCL10 | |
| CASPASES | |
| FAS | |
| UVRAG | |
| Other | BHD |
| PAI-1 |
ONCOGENIC MUTATIONS IN MSI GC
In recent years, a number of studies contributed to better understand gastric tumour development demonstrating that MSI tumours are more prone to exhibit mutations in specific genes, in contrast to tumours with distinct types of genomic instability[64-66]. Of particular relevance are members of the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways that have been found to be mutated and activated in the progression of gastric carcinogenesis. Specifically, mutations in the epithelial growth factor receptor (EGFR), KRAS, PIK3CA and mixed lineage kinase 3 (MLK3) have been described in a number of studies[64,65].
EGFR is a transmembrane tyrosine kinase receptor that in response to extracellular stimuli leads to the activation of two major signalling cascades, the MAPK and PI3K pathways, which are critical in controlling cellular proliferation, differentiation and survival[67]. Therefore, deregulation of this complex network of signalling pathways is known to contribute to the development of GC[64]. EGFR overexpression has been reported in GC in several studies but the underlying mechanisms of aberrant expression remain poorly understood[64,68]. EGFR structural alterations as amplifications and mutations have been described by many as contributing to EGFR overexpression. For instance, Deng et al[69] reported EGFR amplification in about 8% in a series of primary GC samples analysed. EGFR increased copy number was also observed in approximately 13% of 77 primary GC, which was mainly attributed to polysomy of chromosome 7[70]. Somatic mutations of EGFR have also been described in about 5% of a set of gastric adenocarcinomas[71]. However, other studies have shown EGFR mutations to rarely occur in GC[70,72]. In the MSI subset of GC, however, data is very limited. Our group has recently investigated somatic hotspot mutations of the EGFR gene as well as structural alterations on the A13 repeat within the 3’-untranslated region of EGFR (3’-UTR polyA repeat) in a cohort of 63 MSI GC. Results revealed that although no pathogenic mutations were found in the hotspot regions of EGFR, deletions at the 3’-UTR polyA repeat were found in a high proportion (48%) of MSI GC[65]. Mutations in the 3’-UTR polyA repeat of EGFR have been found to be associated with EGFR overexpression in colon carcinomas through enhancement of EGFR mRNA stability[73] suggesting a putative role for these mutations also in GC development. Furthermore, these EGFR alterations were found isolated or in concomitance with mutations in KRAS and/or PIK3CA genes suggesting a cumulative effect of both oncogenic events in MSI GC[65].
Downstream of EGFR, KRAS, BRAF and PIK3CA have also been investigated for mutations in GC. KRAS mutations in codons 12 and 13 have been detected in GC in several studies and frequencies were shown to be around 4%[74,75]. In most cases, however, KRAS mutations are observed in the MSI subset of GC[65,74-76]. Indeed, our group has analysed a panel of GC samples and KRAS mutations were detected in about 18% of the MSI cases[65]. Furthermore, Brennetot et al[76] described KRAS mutations in GC samples only in the MSI subset in about 30% of the cases. A recent large international multicentre study also corroborates the idea that KRAS mutations are related to DNA MMR in GC[75]. In contrast to KRAS, BRAF mutations are rarely observed in GC, as demonstrated by others and our group[74,77-80]. PIK3CA, a gene that encodes for the catalytic subunit p110-alpha of PI3K, is frequently mutated in many human cancers including GC leading to constitutive activation of the PI3K-Akt signalling pathway[81]. More specifically, Samuels et al[81] initially described a high frequency of PIK3CA mutations (25%) in GC, although that could be the result of a small sample size. Further studies, including those from our group, subsequently identified PIK3CA mutations in GC specimens that ranged from 4% to 16%[82-87]. As for KRAS, PIK3CA mutations were also demonstrated to occur preferentially in the MSI subset of GC[82-84]. Furthermore, PIK3CA and KRAS mutations were described as alternative oncogenic events in this subset of MSI GC[83]. Our group also evaluated PIK3CA mutations in a series of MSI GC samples and identified PIK3CA mutations in about 14% of the samples[65]. More recently, a meta-analysis evaluating PI3K aberrations identified PIK3CA mutations in 7%-15% and PIK3CA amplification in 46% of the GC[88]. PIK3CA was also evaluated by Shi et al[86] reporting that 67% of GC had amplification of the gene. In accordance with the role of PI3K pathway in MSI GC alterations in other genes besides PIK3CA have also been significantly associated with the MSI subset of GC[66].
In addition to KRAS and BRAF genetic alterations, mutations in MLK3, a gene also involved in the MAPK pathway, were described to mainly occur in the MSI subset[89]. Indeed, our group investigated MLK3 mutations in gastrointestinal tumours and described these mutations to be functionally relevant[90]. In particular, in MSI GC samples MLK3 mutations were found in a range 3%-17%[65,90].
Overall, the incidence of mutations in members of the EGFR-MAPK-PI3K signalling pathway could be proved useful for prognostic and therapeutic strategies, a subject that is discussed thereafter.
MSI IN GC - PROGNOSIS AND THERAPEUTIC APPROACHES
GC patients are often diagnosed at advanced stages of the disease mostly due to the late onset of symptoms and poor diagnostic tools. Therefore, patients diagnosed with GC are usually associated with a poor prognosis[91]. In recent years, however, efforts have been made to identify better molecular prognostic markers as well as provide novel and more specific targeted therapies to improve overall survival of GC patients.
The different patterns of genomic instability are associated with specific subsets of GC patients having distinct clinico-pathological and molecular characteristics and subsequently have implications at the prognostic and therapeutic levels as summarized in Figure 1[90,92]. Indeed, the overall survival of patients with GC displaying MSI phenotype is better than that of patients with MSS phenotype[11,93]. In particular, in respect to the clinic-pathological features of the MSI GC, most are of the intestinal histotype, located in the distal part of the stomach and occur more frequently in older women[11,94-96]. More interestingly, MSI tumours usually have an overall long-term prognosis that is favourable even in patients with advanced disease due to the fact that these tumours have a lower ability to invade serosal layers that preferentially spread to the periphery of the stomach via the lymphatic stream to the nodes[11,94-96]. In addition, analysis of long term survival data of patients revealed higher survival rates of patients with advanced MSI GC in comparison to patients with other types of GC even if at the same disease stage[97]. Further, evaluation of MSI and MSS GC patients revealed a correlation of MSI at multiple loci with long term survival in advanced GC suggesting that this particular subset of MSI tumours are less aggressive and subsequently associated to a favourable prognosis[11]. Interestingly, our group also found patients with MSI GC with familial history and patients with sporadic MSI GC to display similar clinico-pathologic characteristics[11,26].
Figure 1.
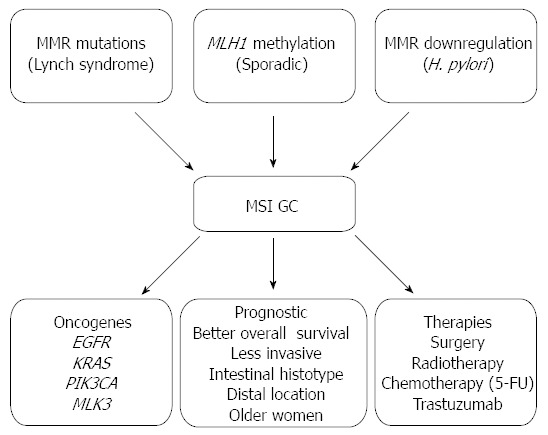
Summary of microsatellite instability gastric cancer associated clinico-pathologic and molecular aspects. This figure summarizes the current knowledge on the molecular mechanisms underlying the acquisition of MSI in GC as well as the clinic, pathologic and molecular consequences of the MSI phenotype. MSI: Microsatellite instability; MMR: Mismatch repair; GC: Gastric cancer; H. pylori: Helicobacter pylori; 5-FU: 5-fluorouracil.
Molecular biomarkers have also been put forward as putative candidates with prognostic value, including EGFR, HER2 and VEGFA as recently reviewed in Durães et al[98]. Indeed, EGFR has been throughout investigated, although its role as prognostic factor remains controversial. In several studies the expression of EGFR was shown to be related with the survival of GC patients and associated with an adverse prognostic value[99-102]. However, recent studies found that positive EGFR expression is not prognostic of patient outcome in GC patients[103-105]. Similarly, the prognostic value of HER2, a tyrosine kinase receptor, is also uncertain as demonstrated through the evaluation of HER2 expression by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH)[106,107]. In contrast, VEGF-A over-expression was suggested to be associated with a poor prognosis for overall survival and disease-free survival in patients with GC[102,108,109]. Nonetheless, information is scarce as to the prognostic value of EGFR, HER2 or VEGFA expression in the MSI subset of GC.
In addition to the clinico-pathologic characteristics and molecular biomarkers, other inflammation-related factors have been associated with GC prognosis[110].
Despite the many advances in the development of new lines of therapy for cancer in general, GC patients have had little benefit. The conventional therapies for GC patients include surgery, radio- and chemo-therapy regimens but the overall outcome of GC patients remains poor, in part due to the diagnosis at an advanced stage[91]. In addition, 5-fluorouracil (5-FU) and cisplatin-based chemotherapy regimens are frequently used in patients at an advanced stage of the disease[111]. Noteworthy, there is still controversy as to the benefits of 5-FU based adjuvant therapy in the MSI subset of GC. Early studies using CRC cells have determined that, in contrast to MSS, MSI cells were insensitive to 5-FU[112], suggesting the same could be valid for GC cells. In fact, a recent large-scale study in GC patients with stage II and III, revealed that 5-FU-based adjuvant chemotherapy showed better disease-free survival in the MSS/MSI-low group but showed no benefits in the MSI-high group[113]. However, conflicting data exist as other reports have shown that the survival of GC patients after the administration of 5-FU did not correlate with MSI status[114].
In the past few years, novel targeted therapies have been tested and approved for GC patients. Regrettably, the successful rates in GC patients are not as encouraging as expected. At present, the only targeting agent approved for GC patients is trastuzumab, a recombinant humanized monoclonal antibody that targets HER2, which efficacy has been demonstrated in HER2 positive GC patients in a phase III large multicentric trial (ToGA study)[115]. Several other targeted agents are currently being investigated or already in clinical trials, most of them focusing on the EGFR pathway or angiogenesis[116]. More specifically, antibodies against EGFR are been evaluated in GC patients in clinical trials including cetuximab and panitumumab, though with disappointing results. Data from the phase III trial EXPAND revealed that the addition of cetuximab to capecitabine-cisplatin provided no additional benefit to chemotherapy alone in the first line treatment of advanced GC[117]. Similarly, the addition of panitumumab to epirubicin, oxaliplatin, and capecitabine chemotherapy did not increase the overall survival of oesophagogastric adenocarcinoma in the REAL3 phase III trial[118]. Anti-VEGF and VEGFR agents as bevacizumab, ramucirumab, apatinib, sorafenib, sunitinib and cediranib have also been evaluated in GC patients in clinical trials with variable outcomes[116]. Furthermore, examples of other targeting agents being tested in GC include everolimus, an mTOR targeting agent; onartuzumab, an antibody against HGFR; vorinostat, an HDAC inhibitor; AZD4547, an FGFR inhibitor; and BYL719, a PIK3A inhibitor[98,116]. Yet again, data on the effects of targeted therapies in the MSI subset of GC is scarce and warrant further studies.
CONCLUSION
The subset of GC with MSI display specific clinic, pathologic and molecular features and therefore are associated to distinct molecular signalling pathways of tumour development[90,92]. The available data indicates that MSI status evaluation is critical for appropriate prognosis assessment in GC patients. Despite all the recent advances, GC remains a challenging cancer. Thus, a better understanding of the molecular aspects of MSI GC is required to further develop new diagnostic and prognostic tools as well as novel therapeutic targets and strategies.
Footnotes
Supported by FEDER through Programa Operacional Factores de Competitividade - COMPETE, and National Funds, No. FCOMP-01-0124-FEDER-000022; and Grants from Fundação para a Ciência e a Tecnologia (FCT), No. IF/00136/2013 (to Velho S), No. SFRH/BPD/63716/2009 (to Fernandes MS) and No. SFRH/BPD/33420/2008 (to Leite M)
P- Reviewer: Fukuhara S, Park SH S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
References
- 1.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 4.Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 5.Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischer F, Cejka P, Jiricny J. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 6.Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, Fishel R, Lee JB. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plys AJ, Rogacheva MV, Greene EC, Alani E. The unstructured linker arms of Mlh1-Pms1 are important for interactions with DNA during mismatch repair. J Mol Biol. 2012;422:192–203. doi: 10.1016/j.jmb.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivatsan A, Bowen N, Kolodner RD. Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. J Biol Chem. 2014;289:9352–9364. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 10.Räschle M, Marra G, Nyström-Lahti M, Schär P, Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 11.Pedrazzani C, Corso G, Velho S, Leite M, Pascale V, Bettarini F, Marrelli D, Seruca R, Roviello F. Evidence of tumor microsatellite instability in gastric cancer with familial aggregation. Fam Cancer. 2009;8:215–220. doi: 10.1007/s10689-008-9231-7. [DOI] [PubMed] [Google Scholar]
- 12.Castells A, Castellví-Bel S, Balaguer F. Concepts in familial colorectal cancer: where do we stand and what is the future? Gastroenterology. 2009;137:404–409. doi: 10.1053/j.gastro.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 14.Phelan CM, Iqbal J, Lynch HT, Lubinski J, Gronwald J, Moller P, Ghadirian P, Foulkes WD, Armel S, Eisen A, et al. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: results from a follow-up study. Br J Cancer. 2014;110:530–534. doi: 10.1038/bjc.2013.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 17.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 18.Goel A, Nguyen TP, Leung HC, Nagasaka T, Rhees J, Hotchkiss E, Arnold M, Banerji P, Koi M, Kwok CT, et al. De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int J Cancer. 2011;128:869–878. doi: 10.1002/ijc.25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Miyakura Y, Sugano K, Akasu T, Yoshida T, Maekawa M, Saitoh S, Sasaki H, Nomizu T, Konishi F, Fujita S, et al. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol. 2004;2:147–156. doi: 10.1016/s1542-3565(03)00314-8. [DOI] [PubMed] [Google Scholar]
- 21.Niessen RC, Hofstra RM, Westers H, Ligtenberg MJ, Kooi K, Jager PO, de Groote ML, Dijkhuizen T, Olderode-Berends MJ, Hollema H, et al. Germline hypermethylation of MLH1 and EPCAM deletions are a frequent cause of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:737–744. doi: 10.1002/gcc.20678. [DOI] [PubMed] [Google Scholar]
- 22.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 23.Lu SL, Kawabata M, Imamura T, Akiyama Y, Nomizu T, Miyazono K, Yuasa Y. HNPCC associated with germline mutation in the TGF-beta type II receptor gene. Nat Genet. 1998;19:17–18. doi: 10.1038/ng0598-17. [DOI] [PubMed] [Google Scholar]
- 24.Pinto M, Wu Y, Mensink RG, Cirnes L, Seruca R, Hofstra RM. Somatic mutations in mismatch repair genes in sporadic gastric carcinomas are not a cause but a consequence of the mutator phenotype. Cancer Genet Cytogenet. 2008;180:110–114. doi: 10.1016/j.cancergencyto.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 25.Gu M, Kim D, Bae Y, Choi J, Kim S, Song S. Analysis of microsatellite instability, protein expression and methylation status of hMLH1 and hMSH2 genes in gastric carcinomas. Hepatogastroenterology. 2009;56:899–904. [PubMed] [Google Scholar]
- 26.Leite M, Corso G, Sousa S, Milanezi F, Afonso LP, Henrique R, Soares JM, Castedo S, Carneiro F, Roviello F, et al. MSI phenotype and MMR alterations in familial and sporadic gastric cancer. Int J Cancer. 2011;128:1606–1613. doi: 10.1002/ijc.25495. [DOI] [PubMed] [Google Scholar]
- 27.Pinto M, Oliveira C, Machado JC, Cirnes L, Tavares J, Carneiro F, Hamelin R, Hofstra R, Seruca R, Sobrinho-Simões M. MSI-L gastric carcinomas share the hMLH1 methylation status of MSI-H carcinomas but not their clinicopathological profile. Lab Invest. 2000;80:1915–1923. doi: 10.1038/labinvest.3780201. [DOI] [PubMed] [Google Scholar]
- 28.Bevilacqua RA, Simpson AJ. Methylation of the hMLH1 promoter but no hMLH1 mutations in sporadic gastric carcinomas with high-level microsatellite instability. Int J Cancer. 2000;87:200–203. doi: 10.1002/1097-0215(20000715)87:2<200::aid-ijc7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 29.Machado AM, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection generates genetic instability in gastric cells. Biochim Biophys Acta. 2010;1806:58–65. doi: 10.1016/j.bbcan.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Machado AM, Figueiredo C, Touati E, Máximo V, Sousa S, Michel V, Carneiro F, Nielsen FC, Seruca R, Rasmussen LJ. Helicobacter pylori infection induces genetic instability of nuclear and mitochondrial DNA in gastric cells. Clin Cancer Res. 2009;15:2995–3002. doi: 10.1158/1078-0432.CCR-08-2686. [DOI] [PubMed] [Google Scholar]
- 31.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 32.Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 33.Machado JC, Pharoah P, Sousa S, Carvalho R, Oliveira C, Figueiredo C, Amorim A, Seruca R, Caldas C, Carneiro F, et al. Interleukin 1B and interleukin 1RN polymorphisms are associated with increased risk of gastric carcinoma. Gastroenterology. 2001;121:823–829. doi: 10.1053/gast.2001.28000. [DOI] [PubMed] [Google Scholar]
- 34.Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology. 2007;133:659–672. doi: 10.1053/j.gastro.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, Capelinha AF, Quint W, Caldas C, van Doorn LJ, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst. 2002;94:1680–1687. doi: 10.1093/jnci/94.22.1680. [DOI] [PubMed] [Google Scholar]
- 36.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 37.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Sepulveda AR. Helicobacter, Inflammation, and Gastric Cancer. Curr Pathobiol Rep. 2013;1:9–18. doi: 10.1007/s40139-013-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies GR, Simmonds NJ, Stevens TR, Sheaff MT, Banatvala N, Laurenson IF, Blake DR, Rampton DS. Helicobacter pylori stimulates antral mucosal reactive oxygen metabolite production in vivo. Gut. 1994;35:179–185. doi: 10.1136/gut.35.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smoot DT, Elliott TB, Verspaget HW, Jones D, Allen CR, Vernon KG, Bremner T, Kidd LC, Kim KS, Groupman JD, et al. Influence of Helicobacter pylori on reactive oxygen-induced gastric epithelial cell injury. Carcinogenesis. 2000;21:2091–2095. doi: 10.1093/carcin/21.11.2091. [DOI] [PubMed] [Google Scholar]
- 41.Obst B, Wagner S, Sewing KF, Beil W. Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis. 2000;21:1111–1115. [PubMed] [Google Scholar]
- 42.Khanzode SS, Khanzode SD, Dakhale GN. Serum and plasma concentration of oxidant and antioxidants in patients of Helicobacter pylori gastritis and its correlation with gastric cancer. Cancer Lett. 2003;195:27–31. doi: 10.1016/s0304-3835(03)00147-2. [DOI] [PubMed] [Google Scholar]
- 43.Touati E, Michel V, Thiberge JM, Wuscher N, Huerre M, Labigne A. Chronic Helicobacter pylori infections induce gastric mutations in mice. Gastroenterology. 2003;124:1408–1419. doi: 10.1016/s0016-5085(03)00266-x. [DOI] [PubMed] [Google Scholar]
- 44.Izzotti A, De Flora S, Cartiglia C, Are BM, Longobardi M, Camoirano A, Mura I, Dore MP, Scanu AM, Rocca PC, et al. Interplay between Helicobacter pylori and host gene polymorphisms in inducing oxidative DNA damage in the gastric mucosa. Carcinogenesis. 2007;28:892–898. doi: 10.1093/carcin/bgl208. [DOI] [PubMed] [Google Scholar]
- 45.Handa O, Naito Y, Yoshikawa T. Redox biology and gastric carcinogenesis: the role of Helicobacter pylori. Redox Rep. 2011;16:1–7. doi: 10.1179/174329211X12968219310756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S, Han SU, Lee KM, Park KH, Cho SW, Hahm KB. 5-LOX inhibitor modulates the inflammatory responses provoked by Helicobacter pylori infection. Helicobacter. 2007;12:49–58. doi: 10.1111/j.1523-5378.2007.00469.x. [DOI] [PubMed] [Google Scholar]
- 47.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and cancer: how hot is the link? Biochem Pharmacol. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 48.Yao Y, Tao H, Park DI, Sepulveda JL, Sepulveda AR. Demonstration and characterization of mutations induced by Helicobacter pylori organisms in gastric epithelial cells. Helicobacter. 2006;11:272–286. doi: 10.1111/j.1523-5378.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- 49.Machado AM, Desler C, Bøggild S, Strickertsson JA, Friis-Hansen L, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection affects mitochondrial function and DNA repair, thus, mediating genetic instability in gastric cells. Mech Ageing Dev. 2013;134:460–466. doi: 10.1016/j.mad.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Hiyama T, Tanaka S, Shima H, Kose K, Tuncel H, Ito M, Kitadai Y, Sumii M, Yoshihara M, Shimamoto F, et al. Somatic mutation in mitochondrial DNA and nuclear microsatellite instability in gastric cancer. Oncol Rep. 2003;10:1837–1841. [PubMed] [Google Scholar]
- 51.Park DI, Park SH, Kim SH, Kim JW, Cho YK, Kim HJ, Sohn CI, Jeon WK, Kim BI, Cho EY, et al. Effect of Helicobacter pylori infection on the expression of DNA mismatch repair protein. Helicobacter. 2005;10:179–184. doi: 10.1111/j.1523-5378.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim JJ, Tao H, Carloni E, Leung WK, Graham DY, Sepulveda AR. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology. 2002;123:542–553. doi: 10.1053/gast.2002.34751. [DOI] [PubMed] [Google Scholar]
- 53.Mirzaee V, Molaei M, Shalmani HM, Zali MR. Helicobacter pylori infection and expression of DNA mismatch repair proteins. World J Gastroenterol. 2008;14:6717–6721. doi: 10.3748/wjg.14.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Shin MG, Jo WH, Kim MJ, Kim HR, Lee WS, Park DH, Won JH, Shin JH, Suh SP, et al. Association between Helicobacter pylori-related peptic ulcer tissue and somatic mitochondrial DNA mutations. Clin Chem. 2007;53:1390–1392. doi: 10.1373/clinchem.2007.088047. [DOI] [PubMed] [Google Scholar]
- 55.Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, Keller G, Bork P, Doeberitz Mv, Gebert JF. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative Real Common Target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 56.Perucho M. Tumors with microsatellite instability: many mutations, targets and paradoxes. Oncogene. 2003;22:2223–2225. doi: 10.1038/sj.onc.1206580. [DOI] [PubMed] [Google Scholar]
- 57.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 58.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 59.Duval A, Rolland S, Compoint A, Tubacher E, Iacopetta B, Thomas G, Hamelin R. Evolution of instability at coding and non-coding repeat sequences in human MSI-H colorectal cancers. Hum Mol Genet. 2001;10:513–518. doi: 10.1093/hmg/10.5.513. [DOI] [PubMed] [Google Scholar]
- 60.Woerner SM, Yuan YP, Benner A, Korff S, von Knebel Doeberitz M, Bork P. SelTarbase, a database of human mononucleotide-microsatellite mutations and their potential impact to tumorigenesis and immunology. Nucleic Acids Res. 2010;38:D682–D689. doi: 10.1093/nar/gkp839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim MS, Oh JE, Kim YR, Park SW, Kang MR, Kim SS, Ahn CH, Yoo NJ, Lee SH. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol. 2010;221:139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 62.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 63.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 64.Velho S, Corso G, Oliveíra C, Seruca R. KRAS signaling pathway alterations in microsatellite unstable gastrointestinal cancers. Adv Cancer Res. 2010;109:123–143. doi: 10.1016/B978-0-12-380890-5.00004-1. [DOI] [PubMed] [Google Scholar]
- 65.Corso G, Velho S, Paredes J, Pedrazzani C, Martins D, Milanezi F, Pascale V, Vindigni C, Pinheiro H, Leite M, et al. Oncogenic mutations in gastric cancer with microsatellite instability. Eur J Cancer. 2011;47:443–451. doi: 10.1016/j.ejca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 66.Liu J, McCleland M, Stawiski EW, Gnad F, Mayba O, Haverty PM, Durinck S, Chen YJ, Klijn C, Jhunjhunwala S, et al. Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer. Nat Commun. 2014;5:3830. doi: 10.1038/ncomms4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 68.Becker JC, Müller-Tidow C, Stolte M, Fujimori T, Tidow N, Ilea AM, Brandts C, Tickenbrock L, Serve H, Berdel WE, et al. Acetylsalicylic acid enhances antiproliferative effects of the EGFR inhibitor gefitinib in the absence of activating mutations in gastric cancer. Int J Oncol. 2006;29:615–623. doi: 10.3892/ijo.29.3.615. [DOI] [PubMed] [Google Scholar]
- 69.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moutinho C, Mateus AR, Milanezi F, Carneiro F, Seruca R, Suriano G. Epidermal growth factor receptor structural alterations in gastric cancer. BMC Cancer. 2008;8:10. doi: 10.1186/1471-2407-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z, Liu L, Li M, Wang Z, Feng L, Zhang Q, Cheng S, Lu S. Epidermal growth factor receptor mutation in gastric cancer. Pathology. 2011;43:234–238. doi: 10.1097/PAT.0b013e328344e61b. [DOI] [PubMed] [Google Scholar]
- 72.Mammano E, Belluco C, Sciro M, Mencarelli R, Agostini M, Michelotto M, Marchet A, Nitti D. Epidermal growth factor receptor (EGFR): mutational and protein expression analysis in gastric cancer. Anticancer Res. 2006;26:3547–3550. [PubMed] [Google Scholar]
- 73.Yuan Z, Shin J, Wilson A, Goel S, Ling YH, Ahmed N, Dopeso H, Jhawer M, Nasser S, Montagna C, et al. An A13 repeat within the 3’-untranslated region of epidermal growth factor receptor (EGFR) is frequently mutated in microsatellite instability colon cancers and is associated with increased EGFR expression. Cancer Res. 2009;69:7811–7818. doi: 10.1158/0008-5472.CAN-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliveira C, Pinto M, Duval A, Brennetot C, Domingo E, Espín E, Armengol M, Yamamoto H, Hamelin R, Seruca R, et al. BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene. 2003;22:9192–9196. doi: 10.1038/sj.onc.1207061. [DOI] [PubMed] [Google Scholar]
- 75.van Grieken NC, Aoyama T, Chambers PA, Bottomley D, Ward LC, Inam I, Buffart TE, Das K, Lim T, Pang B, et al. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer. 2013;108:1495–1501. doi: 10.1038/bjc.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brennetot C, Duval A, Hamelin R, Pinto M, Oliveira C, Seruca R, Schwartz S. Frequent Ki-ras mutations in gastric tumors of the MSI phenotype. Gastroenterology. 2003;125:1282. doi: 10.1016/j.gastro.2003.02.002. [DOI] [PubMed] [Google Scholar]
- 77.Gylling A, Abdel-Rahman WM, Juhola M, Nuorva K, Hautala E, Järvinen HJ, Mecklin JP, Aarnio M, Peltomäki P. Is gastric cancer part of the tumour spectrum of hereditary non-polyposis colorectal cancer? A molecular genetic study. Gut. 2007;56:926–933. doi: 10.1136/gut.2006.114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SH, Lee JW, Soung YH, Kim HS, Park WS, Kim SY, Lee JH, Park JY, Cho YG, Kim CJ, et al. BRAF and KRAS mutations in stomach cancer. Oncogene. 2003;22:6942–6945. doi: 10.1038/sj.onc.1206749. [DOI] [PubMed] [Google Scholar]
- 79.Wu M, Semba S, Oue N, Ikehara N, Yasui W, Yokozaki H. BRAF/K-ras mutation, microsatellite instability, and promoter hypermethylation of hMLH1/MGMT in human gastric carcinomas. Gastric Cancer. 2004;7:246–253. doi: 10.1007/s10120-004-0300-9. [DOI] [PubMed] [Google Scholar]
- 80.Zhao W, Chan TL, Chu KM, Chan AS, Stratton MR, Yuen ST, Leung SY. Mutations of BRAF and KRAS in gastric cancer and their association with microsatellite instability. Int J Cancer. 2004;108:167–169. doi: 10.1002/ijc.11553. [DOI] [PubMed] [Google Scholar]
- 81.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 82.Li VS, Wong CW, Chan TL, Chan AS, Zhao W, Chu KM, So S, Chen X, Yuen ST, Leung SY. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer. 2005;5:29. doi: 10.1186/1471-2407-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Duval A, Carneiro F, Machado JC, Hamelin R, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 84.Sukawa Y, Yamamoto H, Nosho K, Kunimoto H, Suzuki H, Adachi Y, Nakazawa M, Nobuoka T, Kawayama M, Mikami M, et al. Alterations in the human epidermal growth factor receptor 2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer. World J Gastroenterol. 2012;18:6577–6586. doi: 10.3748/wjg.v18.i45.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J, van Hummelen P, Go C, Palescandolo E, Jang J, Park HY, Kang SY, Park JO, Kang WK, MacConaill L, et al. High-throughput mutation profiling identifies frequent somatic mutations in advanced gastric adenocarcinoma. PLoS One. 2012;7:e38892. doi: 10.1371/journal.pone.0038892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, Ji M, Xu L, He N, Shi B, et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barbi S, Cataldo I, De Manzoni G, Bersani S, Lamba S, Mattuzzi S, Bardelli A, Scarpa A. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res. 2010;29:32. doi: 10.1186/1756-9966-29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chong ML, Loh M, Thakkar B, Pang B, Iacopetta B, Soong R. Phosphatidylinositol-3-kinase pathway aberrations in gastric and colorectal cancer: meta-analysis, co-occurrence and ethnic variation. Int J Cancer. 2014;134:1232–1238. doi: 10.1002/ijc.28444. [DOI] [PubMed] [Google Scholar]
- 89.Chadee DN, Kyriakis JM. A novel role for mixed lineage kinase 3 (MLK3) in B-Raf activation and cell proliferation. Cell Cycle. 2004;3:1227–1229. doi: 10.4161/cc.3.10.1187. [DOI] [PubMed] [Google Scholar]
- 90.Velho S, Oliveira C, Paredes J, Sousa S, Leite M, Matos P, Milanezi F, Ribeiro AS, Mendes N, Licastro D, et al. Mixed lineage kinase 3 gene mutations in mismatch repair deficient gastrointestinal tumours. Hum Mol Genet. 2010;19:697–706. doi: 10.1093/hmg/ddp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ottini L, Falchetti M, Lupi R, Rizzolo P, Agnese V, Colucci G, Bazan V, Russo A. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol. 2006;17 Suppl 7:vii97–vi102. doi: 10.1093/annonc/mdl960. [DOI] [PubMed] [Google Scholar]
- 93.Kim H, An JY, Noh SH, Shin SK, Lee YC, Kim H. High microsatellite instability predicts good prognosis in intestinal-type gastric cancers. J Gastroenterol Hepatol. 2011;26:585–592. doi: 10.1111/j.1440-1746.2010.06487.x. [DOI] [PubMed] [Google Scholar]
- 94.dos Santos NR, Seruca R, Constância M, Seixas M, Sobrinho-Simões M. Microsatellite instability at multiple loci in gastric carcinoma: clinicopathologic implications and prognosis. Gastroenterology. 1996;110:38–44. doi: 10.1053/gast.1996.v110.pm8536886. [DOI] [PubMed] [Google Scholar]
- 95.Oliveira C, Seruca R, Seixas M, Sobrinho-Simões M. The clinicopathological features of gastric carcinomas with microsatellite instability may be mediated by mutations of different “target genes”: a study of the TGFbeta RII, IGFII R, and BAX genes. Am J Pathol. 1998;153:1211–1219. doi: 10.1016/s0002-9440(10)65665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seruca R, Santos NR, David L, Constância M, Barroca H, Carneiro F, Seixas M, Peltomäki P, Lothe R, Sobrinho-Simões M. Sporadic gastric carcinomas with microsatellite instability display a particular clinicopathologic profile. Int J Cancer. 1995;64:32–36. doi: 10.1002/ijc.2910640108. [DOI] [PubMed] [Google Scholar]
- 97.Beghelli S, de Manzoni G, Barbi S, Tomezzoli A, Roviello F, Di Gregorio C, Vindigni C, Bortesi L, Parisi A, Saragoni L, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery. 2006;139:347–356. doi: 10.1016/j.surg.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 98.Durães C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464:367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 99.Inokuchi M1, Murayama T, Hayashi M, Takagi Y, Kato K, Enjoji M, Kojima K, Kumagai J, Sugihara K. Prognostic value of co-expression of STAT3, mTOR and EGFR in gastric cancer. Exp Ther Med. 2011;2:251–256. doi: 10.3892/etm.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matsubara J, Yamada Y, Nakajima TE, Kato K, Hamaguchi T, Shirao K, Shimada Y, Shimoda T. Clinical significance of insulin-like growth factor type 1 receptor and epidermal growth factor receptor in patients with advanced gastric cancer. Oncology. 2008;74:76–83. doi: 10.1159/000139127. [DOI] [PubMed] [Google Scholar]
- 101.Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H, Sasako M. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18:5992–6000. doi: 10.1158/1078-0432.CCR-12-1318. [DOI] [PubMed] [Google Scholar]
- 102.Lieto E, Ferraraccio F, Orditura M, Castellano P, Mura AL, Pinto M, Zamboli A, De Vita F, Galizia G. Expression of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) is an independent prognostic indicator of worse outcome in gastric cancer patients. Ann Surg Oncol. 2008;15:69–79. doi: 10.1245/s10434-007-9596-0. [DOI] [PubMed] [Google Scholar]
- 103.Atmaca A, Werner D, Pauligk C, Steinmetz K, Wirtz R, Altmannsberger HM, Jäger E, Al-Batran SE. The prognostic impact of epidermal growth factor receptor in patients with metastatic gastric cancer. BMC Cancer. 2012;12:524. doi: 10.1186/1471-2407-12-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jácome AA, Wohnrath DR, Scapulatempo Neto C, Carneseca EC, Serrano SV, Viana LS, Nunes JS, Martinez EZ, Santos JS. Prognostic value of epidermal growth factor receptors in gastric cancer: a survival analysis by Weibull model incorporating long-term survivors. Gastric Cancer. 2014;17:76–86. doi: 10.1007/s10120-013-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong L, Han Y, Yang J, Zhang H, Jin Y, Brain L, Li M, Zhao Q. Prognostic value of epidermal growth factor receptor in patients with gastric cancer: a meta-analysis. Gene. 2013;529:69–72. doi: 10.1016/j.gene.2013.07.106. [DOI] [PubMed] [Google Scholar]
- 106.Barros-Silva JD, Leitão D, Afonso L, Vieira J, Dinis-Ribeiro M, Fragoso M, Bento MJ, Santos L, Ferreira P, Rêgo S, et al. Association of ERBB2 gene status with histopathological parameters and disease-specific survival in gastric carcinoma patients. Br J Cancer. 2009;100:487–493. doi: 10.1038/sj.bjc.6604885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grabsch H, Sivakumar S, Gray S, Gabbert HE, Müller W. HER2 expression in gastric cancer: Rare, heterogeneous and of no prognostic value - conclusions from 924 cases of two independent series. Cell Oncol. 2010;32:57–65. doi: 10.3233/CLO-2009-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ji YN, Wang Q, Li Y, Wang Z. Prognostic value of vascular endothelial growth factor A expression in gastric cancer: a meta-analysis. Tumour Biol. 2014;35:2787–2793. doi: 10.1007/s13277-013-1371-1. [DOI] [PubMed] [Google Scholar]
- 109.Maeda K, Chung YS, Ogawa Y, Takatsuka S, Kang SM, Ogawa M, Sawada T, Sowa M. Prognostic value of vascular endothelial growth factor expression in gastric carcinoma. Cancer. 1996;77:858–863. doi: 10.1002/(sici)1097-0142(19960301)77:5<858::aid-cncr8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 110.Chang WJ, Du Y, Zhao X, Ma LY, Cao GW. Inflammation-related factors predicting prognosis of gastric cancer. World J Gastroenterol. 2014;20:4586–4596. doi: 10.3748/wjg.v20.i16.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010;(3):CD004064. doi: 10.1002/14651858.CD004064.pub3. [DOI] [PubMed] [Google Scholar]
- 112.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.An JY, Kim H, Cheong JH, Hyung WJ, Kim H, Noh SH. Microsatellite instability in sporadic gastric cancer: its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int J Cancer. 2012;131:505–511. doi: 10.1002/ijc.26399. [DOI] [PubMed] [Google Scholar]
- 114.Oki E, Kakeji Y, Zhao Y, Yoshida R, Ando K, Masuda T, Ohgaki K, Morita M, Maehara Y. Chemosensitivity and survival in gastric cancer patients with microsatellite instability. Ann Surg Oncol. 2009;16:2510–2515. doi: 10.1245/s10434-009-0580-8. [DOI] [PubMed] [Google Scholar]
- 115.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 116.Liu L, Wu N, Li J. Novel targeted agents for gastric cancer. J Hematol Oncol. 2012;5:31. doi: 10.1186/1756-8722-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490–499. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- 118.Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, Wotherspoon A, Saffery C, Middleton G, Wadsley J, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:481–489. doi: 10.1016/S1470-2045(13)70096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


