Abstract
The recent re-annotation of the transcriptome of human and other model organisms, using next-generation sequencing approaches, has unravelled a hitherto unknown repertoire of transcripts that do not have a potential to code for proteins. These transcripts have been largely classified into an amorphous class popularly known as long noncoding RNAs (lncRNA). This discovery of lncRNAs in human and other model systems have added a new layer to the understanding of gene regulation at the transcriptional and post-transcriptional levels. In recent years, three independent studies have discovered a number of lncRNAs expressed in different stages of zebrafish development and adult tissues using a high-throughput RNA sequencing approach, significantly adding to the repertoire of genes known in zebrafish. A subset of these transcripts also shows distinct and specific spatiotemporal patterns of gene expression, pointing to a tight regulatory control and potential functional roles in development, organogenesis, and/ or homeostasis. This review provides an overview of the lncRNAs in zebrafish and discusses how their discovery could provide new insights into understanding biology, explaining mutant phenotypes, and helping in potentially modeling disease processes.
Introduction
Understanding the complexity of the mammalian transcriptome has hugely improved over the past decade owing to the technologies that revolutionized the throughputs of nucleotide sequencing and, thus, enabling genome-scale annotation of transcriptome at single base pair resolution. The scrupulous investigation of the mammalian RNA repertoire has revealed a large number of transcripts that do not get translated into functional proteins and, thus, do not seem to have any “recognizable purpose.”1,2 These transcripts were initially thought to constitute transcriptional noise and were largely annotated as junk transcripts.3,4 Over the past decade, evidence from sequencing of full-length cDNA libraries, as well as dedicated high-throughput sequencing activities such as the FANTOM project for mouse, ENCODE project for human, and projects modeled on similar lines such as the modENCODE for model systems such as the fly and worm have systematically annotated an increasingly dynamic and hitherto uncharacterized set of transcripts with no obvious potential to encode for functional proteins.2,5,6 These transcripts have been shown to play important roles in regulation and maintenance of homeostasis. Hence, these nonprotein-coding transcripts have largely been categorized in a diverse and mostly poorly characterized class of RNAs called the noncoding RNAs (ncRNAs). This has also significantly strengthened the concept of a larger role of ncRNAs as crucial functional elements in eukaryotic genomes.
ncRNAs can be largely classified into the housekeeping ncRNAs and regulatory ncRNAs depending on their functions in the cell.7–9 Housekeeping ncRNAs are expressed constitutively in cells and are necessary for vital cellular functions. The best-known examples for this class of ncRNAs are the tRNAs.7 The regulatory ncRNAs form a larger class and are specifically expressed during certain developmental stages and in specific tissues or disease conditions.7,8 Based on their transcript size, regulatory ncRNAs can be further grouped into two subclasses: small ncRNAs (20–200 nt) and long noncoding RNAs (lncRNAs, >200 nt).10,11 The small regulatory ncRNAs are exemplified by short interfering RNAs (siRNA), small nucleolar RNA (snoRNA), piwi-interacting RNAs (piRNAs), and microRNAs (miRNAs). siRNAs form complexes with argonaute proteins and are involved in post-transcriptional gene regulation; snoRNAs direct the methylation and pseudouridylation of ribosomal RNA; and piRNAs are mostly restricted to germline, associate with PIWI-clade Argonaute proteins, and regulate the silencing of transposable elements in the germline.10,12 Among the small ncRNAs, miRNA are the best-studied class, having vital functions in normal physiological processes of an organism.13–17 On the other hand, lncRNA forms the biggest class of ncRNAs.18 The discovery of lncRNA H-19 and Xist in the early 1990s with their roles in genomic imprinting revealed the involvement of lncRNA in regulation of genome as well as in development and functioning of an organism.19,20 In later years, several independent reports have identified lncRNA expression in mammalian and non mammalian organisms, including humans.21–24 The dedicated high-throughput lncRNA annotation projects such as GENCODE have identified 13,870 lncRNA loci and 23,898 lncRNA transcripts in humans.25 Initiatives such as the NONCODE have already catalogued 210,831 lncRNA in mammals.26 Similarly, the FANTOM consortium has identified more than 11,000 lncRNA in the mouse genome.27 With greater availability of transcriptome datasets in the public domain, the repertoire of vertebrate lncRNAs has been constantly increasing over the last few years.
In recent years, three independent studies have discovered a number of lncRNAs expressed in zebrafish using high-throughput RNA deep sequencing approaches.28–30 The initial two studies revealed lncRNAs in early developmental stages with potential functions in vertebrate embryogenesis.28,29 The latter study catalogued the lncRNA expression in specific tissues or cell types during adult stages and suggested a potential role for these lncRNA transcripts in tissue maintenance and repair.30 In this article, we provide a concise overview of the field on lncRNAs with specific emphasis on zebrafish lncRNAs and its implications in understanding and modeling phenotypic variations. This article also discusses the potential towards understanding the functional roles of lncRNAs in development and disease.
lncRNAs: Definition and Genomic Context
By definition, lncRNAs are transcripts with more than 200 nucleotides in length and do not have an obvious potential to code for a functional protein.31 The former criterion based on length differentiates it from the smaller housekeeping or regulatory RNAs. Some recent reports have also considered transcript length as a parameter to classify lncRNA as small lncRNA (200–950 nt), medium lncRNA (950–4800 nt), and large lncRNA (>4800 nt).32 Based on such classification, it appears that the majority of human lncRNAs consists of small-length lncRNAs unlike the mouse that has medium-length lncRNAs, but these findings need to be further validated.32 By definition, lncRNA class also encompasses ncRNAs previously annotated as antisense transcripts, intronic transcripts, processed pseudogenes, long intergenic noncoding RNAs (lincRNAs), and isoforms of protein-coding transcripts that do not encode for a functional protein.33–43 The long or large intergenic noncoding RNAs, popularly called the lincRNAs, form the largest class of lncRNAs discovered in higher organisms.38
lncRNAs are generally thought to be transcribed such as messenger RNAs by RNA Polymerase II, but with exceptions where they have been shown to be potentially encoded by RNA Polymerase III.31,32,44 Similar to mRNA, the lncRNA transcripts have features such as 5′-capping, splicing, and poly-adenylation.31,32,44 On the basis of their origin and orientation with regard to protein-coding genes, lncRNA are classified into (a) Intronic lncRNAs that are transcribed from the introns of protein-coding genes, for example, lncRNA COLDAIR, which is induced by cold temperature for epigenetic silencing of floral repressors and subsequent flowering in spring and lncRNA “51A,” whose levels have been found to be upregulated in Alzheimer's patients34; (b) Sense overlapping lncRNAs, which are noncoding transcript variants of messenger RNA with absent or non functional open reading frames (ORFs), for example, lncRNA “FMR5” that has a potential application in the diagnosis of fragile X syndrome and Fragile X-associated tremor/ataxia syndrome9,25,35; (c) natural antisense transcripts that are transcribed from the opposite strand of either protein-coding genes or the noncoding genes, for example, lncRNA ALC-1 that is induced in hypertropic ventricles and β-MHC which is induced during pathophysiological condition and reduces the contraction force of the heart37,38; (d) lincRNA that are polyadenylated ncRNA transcribed by RNA pol II with a transcript length of approximately 20 kb. They have characteristic histone methylation signatures in their promoter and transcribed regions, exhibit greater sequence conservation, and display tissue-specific expression; for example, Cyrano that is known to function during neurogenesis in zebrafish and megamind, which is involved in the brain morphogenesis and eye development in zebrafish21,28,38; and (e) Ultraconserved region encoding lncRNAs (T-UCRs) originating from exonic and inter-genic ultraconserved regions that are 100% conserved between the orthologus regions of human, rat, and mouse. They act as co-activators within transcriptionally active sites and as enhancers in splicing, for example, T-UCR “uc.73” and “uc.388” whose expression levels have been correlated with the patho-physiological features of colorectal cancer39,40; (f) LncRNAs encoded by 3′ untranslated regions (uaRNAs) that originate from full-length transcripts of 3′ untranslated regions but are incapable of independent transcription due to lack of the pol II occupancy sites in their promoter, for example, uaRNA from 3′ UTR region of the myocyte enhancer factor 2C41; (g) LncRNAs with enhancer-like functions (eRNAs) that are produced by the RNA pol II activity on the enhancer sequences bi-directionally, are mostly non poly adenylated with a short half life, and are actively involved in promoting mRNA synthesis, for example, eRNA “Nctc1” that originates from insulin-like growth factor 2/H19 enhancers and is known to derive promoter transcription as well as its association with tissue-specific transcription factors.42,43
Biological Function and Disease Associations
LncRNA are envisaged to regulate gene expression in numerous ways and by several mechanisms. Nevertheless, the present understanding of lncRNA-mediated regulation is derived from a few well-studied candidate examples, while the entire spectrum of biological mechanisms through which lncRNAs mediate their functional roles remains largely uncharacterized.31,32,44 Briefly, the well-studied examples of lncRNA functions could be summarized as (a) chromatin regulation, in which an lncRNA such as Xist and Air acts in either cis or trans to orchestrate epigenetic changes by recruiting chromatin modifying complexes and/or DNA methylases to bring about gene activation or repression;31,45 (b) LncRNAs such as PANDA and DHFR, which can affect transcriptional regulation directly by regulating the activity of RNA pol II, or by associating with transcriptional co-activators or co-repressor complexes;44,45 (c) LncRNAs such as MALAT1 beget post-transcriptional regulation of mRNAs by modulating their splicing, transportation, translation, and degradation.44,46,47
lncRNA functions could also be summarized as an outcome of its molecular interactions with other biomolecules in the cell such as the DNA, other RNA species, and proteins. A number of distinct examples of each of these interactions and their biological regulatory outcomes have been reviewed in the literature.48,49 A number of recent papers also suggest that a subset of lncRNAs could interact with smaller RNAs, including miRNAs, and modulate their regulatory effect.50,51 Our group has validated one such regulatory interaction between a conserved pair of miRNA and lncRNA in zebrafish.50
Conservation and Variation in lncRNAs
Deciphering lncRNA function based on comparative sequence analysis has been largely limited by the fact that a very few lncRNAs display sequence conservation across species.28,31 Recent reports have suggested that lncRNAs follow different criteria of conservation than their protein-coding counterparts.21,22,31 Protein-coding genes have stringent functional restrictions in terms of their length and number as well as sequence of amino acids, in addition to the preservation of the ORF.31 On the contrary, lncRNAs exhibit high conservation over short stretches of their length to preserve their functionality as well as secondary structures.9,31,52 Researchers have also estimated the rates of sequence variation, in terms of change in the nucleotide numbers in lncRNA sequences over evolutionary timescales and have suggested that lncRNA sequences evolve rapidly in comparison to the sequences of protein-coding genes.31,53 This provided the basis for the general notion that nucleotide sequence conservation is not a fundamental necessity for preservation of lncRNA functionality. Recently, our group has analyzed functional elements in lncRNAs, characterized by experimental datasets of RNA-protein, RNA–RNA interaction sites, and small RNA processing sites.49,54 Our analysis hints that the functional elements in lncRNA are characterized by a lower variation frequency, almost comparable to that of protein-coding genes, suggesting that a comparison of sequence divergence based on whole lncRNA transcripts may be misleading. Nevertheless, other studies have also identified a small number of lncRNAs with recognizable degrees of sequence conservation between vertebrates.28 In addition, a number of lncRNAs also show close sequence conservation in the promoter regions, suggesting a conserved mechanism of regulation of lncRNAs.9,31 Overall, the evidence seems to suggest conservation rather than variation being a dominant force shaping the functional and regulatory domains of lncRNAs, and a further examination of this new proposition could be gained immensely from analysis of structural, regulatory, and interacting elements of lncRNAs.
In a recent study, Kapusta et al. highlighted the importance of transposable elements in evolutionary origin and diversification of lncRNA genes, which, in addition, could also explain to a fair extent the reason behind the low nucleotide sequence conservation of lncRNA. It revealed that transposable elements, because of their inherent characteristic to introduce regulatory sequences on chromosomal insertion and to move and spread in genomes in a lineage-specific fashion, have played an important role in the lineage-specific diversification of lncRNA pool across human, mouse, and zebrafish.55
Zebrafish lncRNAs
The existing mammalian lncRNAs have been largely annotated by genomic studies on cultured cell lines and adult tissues.21,22,56 Although these studies effectively discovered a large number of cell type-specific lncRNAs, they overlooked the ones being expressed during specific developmental stages. Comparative functional studies and sequence investigations of protein-coding and non protein-coding genes in zebrafish have greatly augmented our knowledge about their mammalian counterparts. Therefore, zebrafish has emerged to be a competent model organism for decoding the mysteries of the mammalian lncRNA functions and mechanisms.
A handful of lncRNA transcripts have been previously studied in zebrafish. This includes antisense transcript tyrosine kinase containing immuno-globulin and epidermal growth factor homology domain-1 antisense (tie-1AS) involved in vascular development.57 The antisense transcript binds to tie-1 mRNA, thereby regulating the gene expression leading to defects in endothelial cell contact junctions and resulting in vascular anomalies.57 Recently, another antisense lncRNA PU.1 AS has been shown to regulate the expression of genes involved in development of the immune system.58 Tcl1 upstream neuron-associated lincRNA (TUNA) has also been reported to be important for its neurological and locomotor role in the organism, as knockdown of TUNA resulted in impaired locomotor functions.59
Apart from the antisense transcripts, a number of lncRNAs have been documented to be involved in zebrafish embryogenesis-like sox2-ot lncRNA and cyrano lncRNA, both of which are involved in the neurogenesis in zebrafish.28,60 LncRNA megamind has been studied in relation to brain morphogenesis and eye development.28 Similarly, lncRNA MALAT1 was found to be essential for vertebrate development and also shows association with cancers, corroborating with a number of evidences from human studies on its association with cancer metastasis.28,47 Another conserved class of lncRNAs studied in zebrafish has been the Y RNAs, a component of the Ro ribo-nucleoprotein that are conserved across the vertebrate species.61 Zebrafish zY1 RNA shows conservation at structure as well as sequence level with that of human Y1 RNA. The zY1 RNA could substitute the function of initiating chromosomal DNA replication similar to that of hY1 RNA.61
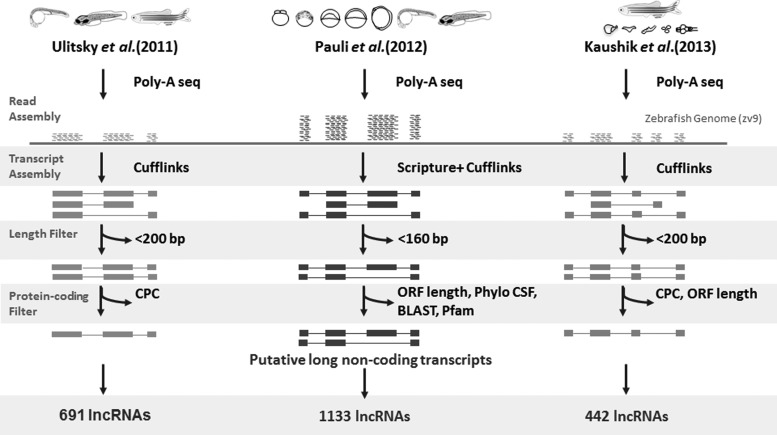
The availability of the reference genome of zebrafish along with sequences of wild-type strains has enabled a much required template and impetus for annotation of transcriptome at a genome scale.62–65 The corpus of annotated lncRNAs in zebrafish has been majorly derived from three recent publications, with each characterizing a distinct subset of the lncRNome.28–30 While Pauli et al. characterized the lncRNome of eight developmental time points,29 Ulitsky et al. annotated the lncRNome from three developmental time points of zebrafish.28 In a very recent analysis, Kaushik et al. annotated the lncRNome derived from five major tissues from adult zebrafish.30 However, the studies have largely relied on RNA-sequencing approach, with major differences in the time points or tissues considered and limited differences in the experimental protocols and analysis pipelines. Nevertheless, the three studies have a few overlapping lncRNA candidates, suggesting the existence of a potentially larger uncharacterized lncRNA repertoire in zebrafish.
LncRNA Expression in Early Zebrafish Development
The process of organism development is intriguing and is driven by a complex and dynamic network of gene expression and regulation in a spatiotemporal fashion. Recently, with the discovery of pervasive noncoding transcription, the role of ncRNA in early vertebrate development has also been contemplated. Since the hallmarks of early and late embryonic development are significantly different, one expects a discrete change in the coding and noncoding transcriptome expression profiles. With this background, Pauli et al. performed RNA deep sequencing to explore the lncRNA expression profiles in eight early embryonic developmental stages of zebrafish, that is, 2–4 cell, 1000 cell, dome, shield, bud, 28, 48, and 120 hour post fertilization (hpf).29 It is important to note that each selected stage corresponded to a developmental hallmark. A total of 56,535 high-confidence transcripts across 28,912 loci were assembled, out of which 1133 unique multi-exonic embryonic lncRNAs, including 397 lincRNAs, 184 intronic overlapping lncRNAs, and 566 antisense exonic overlapping lncRNAs, were identified. Another subset of 41 lncRNA was identified as potential precursors for generation of miRNAs, snoRNAs, and other small RNA of an unknown category.29
The time-dependent expression profiles of lncRNA and protein-coding transcript loci were subdivided into three categories (a) transcripts present specifically in two to four cell embryos that were deposited by the parents and undergo decay within the first few hours of embryogenesis; (b) transcripts that were transcribed zygotically and were present at low levels during early cleavage and at high levels in the dome, shield, and bud stages; and (c) transcripts which were induced during organogenesis and were transcribed during 1 day post fertilization. Expression dynamics of lncRNA and protein-coding genes showed stark differences.29 First, lncRNA transcripts were more likely to be parentally supplied than the protein-coding genes; that is, 42% of lncRNAs were found to be provided by parents as compared with 34% of the protein-coding transcripts.29 Second, the lncRNAs were found to have stringent temporal expression dynamics in comparison to the protein-coding genes and relative alterations in transcript levels of lncRNA between two consecutive stages were greater than the protein-coding genes.29 The study also evaluated the association between the expression dynamics of each protein-coding gene with each lncRNA locus in order to allocate putative functional roles to them. The study identified 33% of the lncRNA linked to the loci which are rich in developmental functions and also linked to other important loci that are involved in signaling as well as cell cycle, indicating the developmental roles of lncRNA.29
In order to determine the tissue-restricted spatial expression of the lncRNA during early developmental stages, in situ hybridization was performed for a selected set of 32 lncRNAs.29 Specific expression patterns were observed for hoxAa-lncRNA in a fertilized two-cell stage embryo and for myo18a-lncRNA in developing somites at 28 hpf. LncRNAs such as miR-9-7 lncRNA and st18 lncRNA had restricted expression in cells of the developing nervous system at 48 hpf. Similarly, the myo18a-lncRNA was distinctly expressed at the myoseptum, overlapping the expression domain of dystrophin, and was anticipated to have a function in cell–cell contact formation and a structural role in cell adhesion. The hoxAa lncRNA was observed in nuclear regions in early cleavage stage embryos and was later found to be associated with chromatin in mitotically dividing cells at 4 cell stage and 16 cell stage, respectively. The lncRNA mprip was observed at the bud stage embryo and was later found to accumulate specifically around the large nuclei of the yolk syncytical layer and the least around the small nuclei of neighboring cells.29
The RNA-seq experiments were complemented by ChIP-sequencing in the shield stage to annotate chromatin-wide presence of tri-methylated lysine 4 on histone 3 (H3K4me3) marks on promoters and tri-methylated lysine 27 on histone H3 (H3K27me3) marks on transcribed regions and compared them with protein-coding transcripts as well as mammalian lncRNA.29 Only 29% of lncRNA were marked with both H3K4me3 and H3K27me3 domains as compared with 63% of zebrafish protein-coding genes. Analysis also revealed that the fraction of H3K4me3-marked zebrafish lncRNA genes was close to that observed in human lncRNA, suggesting a conserved mechanism of epigenetic regulation in vertebrate lncRNAs.29
LincRNA Expression Profile During Zebrafish Late Developmental Stages
Ulitsky et al. discovered a large number of lncRNA transcripts across three developmental stages in zebrafish and also deciphered the functional roles for a few of these transcripts.28 Precisely, they performed RNA deep sequencing with poly(A)-site mapping and genome-wide chromatin histone H3 modifications mapping on 24 and 72 hpf zebrafish embryos as well as the adult zebrafish to identify 550 distinct lincRNA loci with 691 transcripts and 66,895 poly(A)-sites. Out of these 550 distinct lincRNA loci, only a small fraction had a detectable sequence similarity with mammalian lncRNAs.28 This study also provided the comparative analysis of expression correlation between zebrafish and mammalian lincRNAs with their neighboring protein-coding genes. It was also observed that zebrafish lincRNAs such as mammalian lncRNAs were present within <10 kb of protein-coding genes and the distances being approximately equivalent to two adjacent protein-coding genes.28 The lincRNAs also showed relatively high expression and conservation as per the RNA seq and phastCons analysis. LincRNAs such as linc-mipep, linc-tbx2b, linc-gtf2f2b, and linc-arid4a were found to be specifically expressed in different parts of the central nervous system, whereas linc-trpc7 and linc-cldn7a expression was found to be restricted to non-neuronal tissues such as notochord and pronephros, respectively.28
Since tissue-restricted expression is often linked to fundamentally important function, the study further explored the functional roles of two lincRNAs “cyrano” and “megamind” during zebrafish development.28 These two lincRNAs were chosen for experiments based on their tissue-specific expression profiles and conservation with mammalian lincRNAs. The lincRNA olp5 “cyrano” was explicitly expressed in the nervous system and notochord of zebrafish, whereas lincRNA birc6 “megamind” was expressed in eyes and the brain. For functional analysis, morpholino antisense oligos (MO) were targeted against the cyrano and megamind splice sites as well as its conserved sites. Zebrafish embryos injected with these MOs displayed developmental defects because of diminished levels of cyrano and megamind transcripts. The developmental defects produced by splice site MOs were rescued by spliced cyrano/megamind RNA and also by mature human or mouse cyrano/megamind RNA.28 These results implicated that mammalian orthologues could also function in zebrafish, suggesting conserved functions of lincRNAs in vertebrate embryonic development.
LncRNA Expression in Adult Zebrafish
Embryogenesis and early development is more about how regulatory networks act in time and space to contribute to the development of various organs and tissues. In contrast, adulthood demands a different set of gene expression, more to do with the maintenance of form and function of what is already established by developmental programs. It is, thus, expected that adult tissue transcriptome would have distinct patterns significantly different from the developmental profiles. Previous microarray based studies comparing developmental and adult tissue profiles of messenger RNAs corroborate this difference in gene expression profiles.66,67 In order to discover lncRNA players in adult tissues, our group performed an RNA-seq-based study of adult zebrafish tissues.30 We used Poly-A RNA sequencing followed by computational analysis to identify tissue-restricted lncRNA transcript signatures from five different tissues of adult zebrafish, namely, brain, heart, blood, liver, and muscle. The five tissues were chosen based on their cell type constituency.30 The brain was selected, as it is a heterogeneous organ with varied cell types, and also an organ in which most transcripts are expressed. The heart and blood being a part of the cardiovascular system have lesser cell types than the brain, while the liver and muscle are relatively more homogeneous organs, dominated mostly by one or two cell types and specialized for a single physiological activity. Our analysis revealed 442 lncRNA transcripts from adult zebrafish tissues, out of which 419 were novel lncRNA transcripts. Of these, 77 lncRNAs showed predominant tissue-restricted expression across the five major tissues investigated. As expected, the brain as a tissue with diverse cell types consisted of the largest number of tissue-restricted lncRNAs (n=47), while the heart and blood constituted 12 lncRNA transcripts each. Very few lncRNAs were found to be expressed in homogeneous tissues such as the liver (n=2) and muscle (n=4). A subset of these tissue-specific lncRNAs was further validated by real-time polymerase chain reaction confirming the predominant expression of lncRNAs in corresponding tissues. Two of the lncRNAs expressed in the brain were qualitatively evaluated by whole mount in situ hybridization in early developmental stage and adult brain. The transcript, lncBrHM_035, displayed distinct localization in the eye, mid and hind brain of 24 hpf zebrafish embryos; whereas in adult zebrafish brain, the expression was restricted to the cerebellum.30 Another lncRNA transcript, lncBrM_002 could be detected in the mid and hind brain of 24 hpf zebrafish embryos and in adult zebrafish brain, the expression was restricted to the cerebellum and eminentia granularis.30 Both the lncRNAs showed overlapping but varied expression in the developmental stage (24 hpf), while in the adult brain they were present in very specific domains, suggesting that a lncRNA can have diverse and specific functions in embryonic as well as adult tissues. This body of work constitutes a useful genomic resource towards understanding the expression of lncRNAs in various tissues in adult zebrafish in the context of maintenance of organ form and function.30
Similarities and Differences
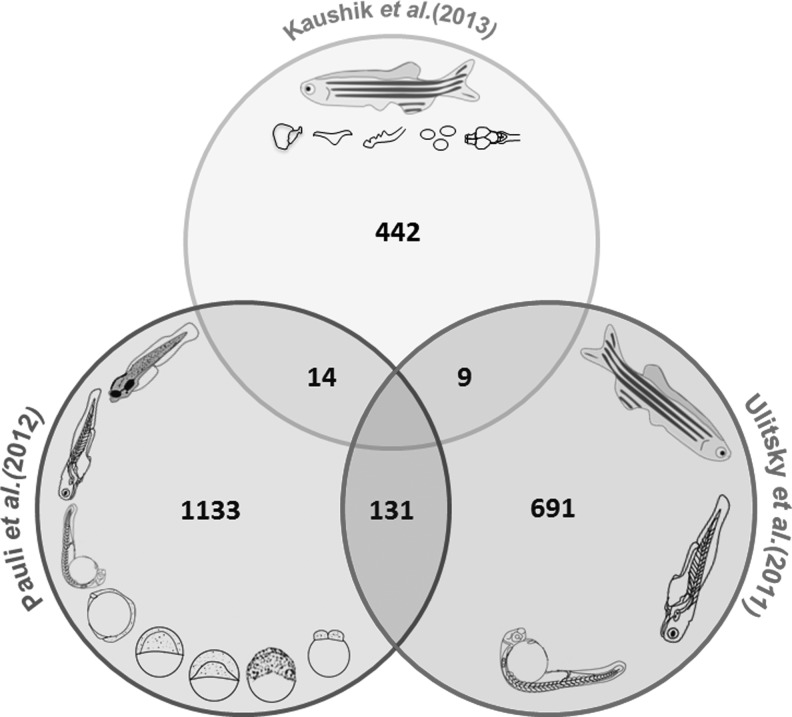
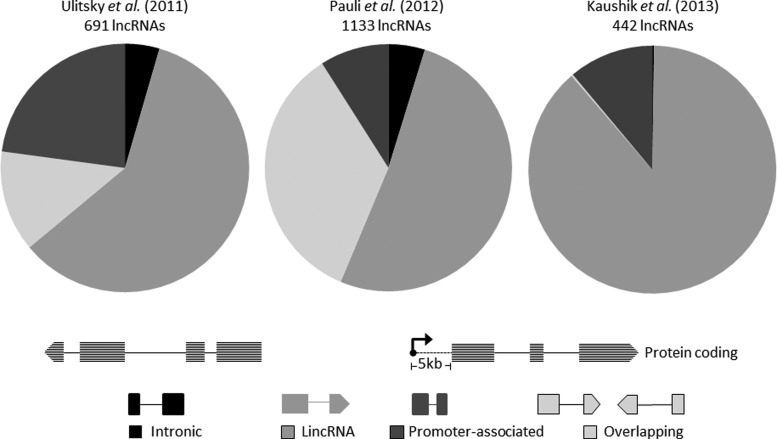
While the three studies uncovered a hitherto unknown repertoire of zebrafish lncRNAs, there have been major similarities and differences in the analysis outcomes. A number of lncRNAs overlap between the three studies. A total of 131 lncRNAs overlapped between Pauli and Ulitsky datasets, while 14 overlapped between Kaushik and Pauli and 9 between Kaushik and Ulitsky datasets, respectively (Fig. 1). None of the lncRNAs overlapped in all the three datasets. All the datasets put together, a total of 2,266 lncRNAs were discovered, many of which have distinct spatiotemporal profiles. The major differences stem from the tissues/time points considered, the stringency of the analysis protocols, and therefore the outcomes. These studies have covered themes ranging from documenting the conserved function of lncRNAs,28 their roles in early vertebrate development29 and their occurrence in adult zebrafish tissues as well.30 While Ulitsky et al. identified the conserved lincRNAs across mammalian genomes by synteny block analysis, Pauli et al. have catalogued lncRNAs in eight developmental stages of zebrafish using a slightly altered approach. They chose all transcripts of a length more than 160 bp and used four different filters for distinguishing noncoding transcripts from coding transcripts. First, they aligned all transcripts phylogenetically and calculated their phylogenetic codon substitution frequency; then, they retained the transcripts of less than 20 PhyloCSF only. The retained transcripts were further checked for their similarity to known protein-coding domains using blastx, blastp, and pfam; consequentially, the transcripts with similarity to the protein-coding domain were eliminated from the study. The remaining transcripts were checked for their ORF length. Only those transcripts that have ORF <100 were selected at this stage. In addition, out of all the transcripts that did not show sequence alignments in PhyloCSF and had ORF <30, amino acids were also considered. Finally, both the datasets were combined and the transcripts that overlapped with sense exons of protein-coding transcripts were eliminated.29 In a recent study by our group, we have assembled and merged five tissue-specific datasets obtained after poly-A seq. All the known protein-coding transcripts were removed and transcripts with a length of >200 bp only were retained, which were further checked for their protein-coding capacity by coding potential calculator (CPC) and ORF length. At this stage, the transcripts that had a CPC score of less than −1 were retained and checked further for their ORF length. The majority of the protein-coding transcripts have an ORF >100 amino acids; however, since there are 405 known human functional proteins that have an ORF of 30–100 amino acids,68 we retained transcripts with an ORF length of <30 amino acids only. These transcripts were further checked for any overlap with protein-coding transcripts and known lncRNAs in zebrafish. Finally, the retained transcript pool was checked for unique expression in only one of the tissues giving 77 tissue-specific lncRNAs.30 Albeit the major differences, ample similarities exist between the analysis pipelines considered, which includes transcript assembly and utilization of almost similar cut-offs for length of transcripts and ORFs. The highlights of the analysis protocols followed by each of the individual studies are summarized in Fig. 2. A comparative analysis of the tissue lncRNome using the ORF criteria used by other groups revealed a significantly larger repertoire of lncRNAs.30 Analysis of the three datasets also revealed that the majority of lncRNAs were intergenic to the protein-coding genes, whereas a small proportion of these transcripts were mapped to introns on annotated protein-coding genes. We also found that ∼35% lncRNA transcripts discovered by Pauli et al. overlapped with known protein-coding genes, while 13% of the lncRNA transcripts identified by Ulitsky et al. overlapped with protein-coding genes (Fig. 3).
FIG. 1.
Illustration of long noncoding RNA (lncRNA) overlaps in three recent reports of zebrafish.
FIG. 2.
Summary of the RNAseq analysis pipeline for lncRNA discovery utilized by the three recent reports.
FIG. 3.
Genomic mappings and annotations of zebrafish lncRNA transcripts discovered by the three studies.
The lncRNAs discovered by these studies in zebrafish have a number of similarities to lncRNAs discovered in other mammalian species. First, similar to mammalian lncRNAs, Zebrafish lncRNAs are about a third in length compared with their protein-coding counterparts and have a comparatively lesser number of exons per transcript. Second, they have been shown to be expressed at lower levels than the protein-coding genes.21,22,28,29 Third, the preferential locations of zebrafish antisense exonic lncRNAs were observed to be in close proximity to genes with important developmental functions such as transcription factor activity, cell fate specification, as well as embryonic development and morphogenesis.29 On the contrary, certain classes of lncRNA such as lincRNAs and intronic overlapping lncRNAs did not show this preference.28 However, the physical propinquity of the lncRNAs with genes of important developmental functions, in general, neither has any functional link nor shows a relationship in their expression. The only exception are the sense intronic overlapping lncRNAs that positively associate with neighboring protein-coding genes in terms of their expression.28,29
The identification of lncRNAs as well as the distinction of lncRNAs from protein-coding mRNAs largely depends on the computational and bioinformatics algorithms employed. These algorithms characterize transcripts by their ORF lengths, coding potential and synteny conservation but still a certain degree of uncertainty remains with regard to the true noncoding potential of these predicted lncRNA transcripts. The computational algorithms employed can also potentially misidentify the lncRNAs containing short conserved regions as protein-coding transcripts or protein-coding transcripts containing short or weakly conserved ORFs as noncoding transcripts. These uncertainties have been further intensified by some recent ribosome profiling studies that have identified the protein-coding characteristic in a number of putative lncRNA transcripts. Ingolia et al. have performed ribosome profiling of mouse embryonic stem cells and have identified a wide range of unannotated ORFs as well as highly translated short ORFs among previously annotated lincRNAs that had been earlier described as not having canonical ORFs.69 On similar lines, Chew et al. performed ribosome profiling on eight early developmental stages of zebrafish and identified that a number of earlier proposed developmental lncRNAs have protein-coding contaminants. They further validated the findings on embryonic stem cells and obtained similar results for numerous putative mammalian lncRNAs.70 They also developed translated ORF classifier (TOC) as a filter for distinguishing the ORFs of coding sequences from ORFs in 5′ leaders and from ORFs in 3′ trailers. TOC classifier, when applied to published developmental zebrafish lncRNome, predicted that ∼50% of lncRNAs could potentially be classified as protein-coding mRNA.70
These results also hint that RNA could potentially function at multiple levels both as ncRNA performing regulatory roles and as mRNA for protein synthesis. The steroid receptor RNA activator (SRA)/steroid receptor RNA activator protein (SRAP) has been documented to have overlapping function, as both RNA and protein. Further, these findings hint toward the dual functional roles of a transcript in different developmental stages.71,72 Nevertheless, these atypical protein-coding transcripts could be considered an important resource for identification of novel proteins. Recently, Bazzini et al. have used ribosomal foot printing to identify the micropeptide-encoding genes that code for transcripts with small ORFs (≤100 amino acids) in vertebrates. They identified numerous translated small ORFs across five developmental stages of zebrafish transcriptome and also detected several hundred small ORFs in the annotated lncRNAs with previously undefined coding sequences.73 These observations are further justified by the recent example of Toddler, which was annotated as ncRNA in zebrafish but was later found to encode a short, conserved, secreted peptide that activates APJ/Apelin receptor signaling, acts globally as a motogen, and promotes gastrulation movements.74 In summary, RNA-sequencing and ribosome profiling could be employed to derive high-confidence annotations for genuine noncoding lncRNAs. However, studies in recent years have also provided evidence for ribosomal associations with ncRNAs (see review by Ulitsky and Bartel38). Therefore, caution should be exercised when employing ribosomal profiling as the sole criterion for defining protein-coding function.
How the Understanding of lncRNAs Will Influence Zebrafish Biology
Historically, zebrafish has been employed for understanding functions of protein-coding genes. A number of reports in recent years have utilized the power of next-generation sequencing toward understanding the landscape of zebrafish transcriptome.28–30,64,75,76 The recent discovery of a large number of ncRNAs in zebrafish, including small ncRNAs and lncRNAs, has opened up a completely new repertoire of biological regulation, which is yet to be comprehensively understood. The discovery of lncRNAs in zebrafish is suggested to impact the understanding of zebrafish biology in a variety of ways. The first obvious way is that it extends the possibility of explaining phenotypes of mutants, which occurred in previously un-annotated loci in the zebrafish genome. Second, it could potentially influence zebrafish biology through understanding novel regulatory pathways involved in embryonic development, organogenesis, and tissue maintenance. Many of the lncRNAs discovered from the genome-wide screens show extremely restricted expression levels, suggesting a well co-ordinated regulatory mechanism operating at the transcriptional or the post-transcriptional levels. A better understanding of the landscape of lncRNA transcription in the zebrafish genome would also enable one to draw parallels from the Human lncRNome, and provide a useful opportunity to model ncRNA mutations in zebrafish. This gains more importance in the light of the recent discovery of a number of variants with disease/human traits from genome-wide association studies mapping to potential lncRNA loci, suggesting a hitherto unexplored significance of lncRNAs in disease biology.77,78 Zebrafish is also poised to be a fantastic complement to other vertebrate model systems for understanding the biology of ncRNAs, given the ease and economy of generation of mutant animals using either chemical, insertional, or genome editing tools such as the transcription activator-like effector nuclease (TALEN) and clustered, regularly interspaced, short palindromic repeat (CRISPR) technology.79–82 TALEN and CRISPR technologies have emerged as robust genome editing tools for generating precise and heritable genomic deletions as well as knockout of both protein-coding and noncoding genes, including lncRNAs with high accuracy, opening up new avenues for study of noncoding genome. It has not escaped our notice that a number of zebrafish retroviral insertions map83,84 to currently annotated lncRNAs (Kapoor et al., unpublished results), offering a readily available template to study lncRNA biology in zebrafish. This provides an unprecedented opportunity to link zebrafish phenomics to ncRNA biology, as currently most of the functionality assays for lncRNAs are largely restricted to cell lines. Phenotypic effects have not been largely explored for lncRNAs, with a few exceptions in mouse and zebrafish. Recent initiatives toward assembly of the genomic variations, epigenome, and transcriptomes in line with the human ENCODE project would definitely add to the understanding of zebrafish transcriptome and its regulatory correlates.64,65,85 Thus, understanding zebrafish lncRNome would open up new possibilities toward understanding its effects in modulating the developmental processes and homeostasis.
Acknowledgments
This work was funded by the Council of Scientific and Industrial Research (CSIR), India through grant BSC0123 (GENCODE-C). K.K. and S.K. acknowledge Senior Research Fellowships from CSIR, India and UGC, India, respectively. A.J. acknowledges fellowship from MLP1202 grant.
Disclosure Statement
The authors report no conflicts of interest and no competing financial interests for writing this article. The authors alone are responsible for the content and writing of this aricle.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. . The transcriptional landscape of the mammalian genome. Science 2005;309:1559–1563 [DOI] [PubMed] [Google Scholar]
- 2.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature 2003;422:633–637 [DOI] [PubMed] [Google Scholar]
- 4.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 2007;17:556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. . Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002;420:563–573 [DOI] [PubMed] [Google Scholar]
- 6.Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, et al. . modMine: flexible access to modENCODE data. Nucleic Acids Res 2012;40:D1082–D1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2001;2:919–929 [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006;15:R17–R29 [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–641 [DOI] [PubMed] [Google Scholar]
- 10.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol 2010;220:126–139 [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–874 [DOI] [PubMed] [Google Scholar]
- 12.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 2011;90:430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett 2005;579:5911–5922 [DOI] [PubMed] [Google Scholar]
- 14.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. . Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006;312:75–79 [DOI] [PubMed] [Google Scholar]
- 15.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. . A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010;328:1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012;336:233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalwani MK, Sharma M, Singh AR, Chauhan RK, Patowary A, Singh N, et al. . Reverse genetics screen in zebrafish identifies a role of miR-142a-3p in vascular development and integrity. PLoS One 2012;7:e52588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009;23:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991;351:153–155 [DOI] [PubMed] [Google Scholar]
- 20.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, et al. . The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992;71:527–542 [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. . Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res 2012;22:2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, et al. . Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol 2012;4:427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, et al. . NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res 2014;42:D98–D103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaji H, Severin J, Lizio M, Forrest AR, van Nimwegen E, Rehli M, et al. . Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res 2011;39:D856–D860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147:1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, et al. . Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 2012;22:577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik K, Leonard VE, Shamsudheen KV, Lalwani MK, Jalali S, Patowary A, et al. . Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One 2013;8:e83616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer TR, Dinger ME, Mattick JS. Long noncoding RNAs: insights into function. Nat Rev Genet 2009;10:155–159 [DOI] [PubMed] [Google Scholar]
- 32.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, et al. . Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 2011;10:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011;331:76–79 [DOI] [PubMed] [Google Scholar]
- 35.Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet 2014;133:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. . Antisense transcription in the mammalian transcriptome. Science 2005;309:1564–1566 [DOI] [PubMed] [Google Scholar]
- 37.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 2012;111:1349–1362 [DOI] [PubMed] [Google Scholar]
- 38.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. . Ultraconserved elements in the human genome. Science 2004;304:1321–1325 [DOI] [PubMed] [Google Scholar]
- 40.Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology 2012;82:114–118 [DOI] [PubMed] [Google Scholar]
- 41.Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, et al. . Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res 2011;39:2393–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. . Widespread transcription at neuronal activity-regulated enhancers. Nature 2010;465:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eun B, Sampley ML, Van Winkle MT, Good AL, Kachman MM, Pfeifer K. The Igf2/H19 muscle enhancer is an active transcriptional complex. Nucleic Acids Res 2013;41:8126–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012;40:6391–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. . The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhartiya D, Kapoor S, Jalali S, Sati S, Kaushik K, Sachidanandan C, et al. . Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov 2012;7:503–513 [DOI] [PubMed] [Google Scholar]
- 49.Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B, et al. . lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford) 2013;2013:bat034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One 2013;8:e53823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, et al. . microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep 2013;3:2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics 2009;93:291–298 [DOI] [PubMed] [Google Scholar]
- 53.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 2006;22:1–5 [DOI] [PubMed] [Google Scholar]
- 54.Jalali S, Jayaraj GG, Scaria V. Integrative transcriptome analysis suggest processing of a subset of long non-coding RNAs to small RNAs. Biol Direct 2012;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, et al. . Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 2013;9:e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. . Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. . A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010;115:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei N, Pang W, Wang Y, Xiong Y, Xu R, Wu W, et al. . Knockdown of PU.1 mRNA and AS lncRNA regulates expression of immune-related genes in zebrafish Danio rerio. Dev Comp Immunol 2014;44:315–319 [DOI] [PubMed] [Google Scholar]
- 59.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. . An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014;53:1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, et al. . Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 2009;15:2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collart C, Christov CP, Smith JC, Krude T. The midblastula transition defines the onset of Y RNA-dependent DNA replication in Xenopus laevis. Mol Cell Biol 2011;31:3857–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meli R, Prasad A, Patowary A, Lalwani MK, Maini J, Sharma M, et al. . FishMap: a community resource for zebrafish genomics. Zebrafish 2008;5:125–130 [DOI] [PubMed] [Google Scholar]
- 63.Bhartiya D, Maini J, Sharma M, Joshi P, Laddha SV, Jalali S, et al. . FishMap Zv8 update—a genomic regulatory map of zebrafish. Zebrafish 2010;7:179–180 [DOI] [PubMed] [Google Scholar]
- 64.Patowary A, Purkanti R, Singh M, Chauhan R, Singh AR, Swarnkar M, et al. . A sequence-based variation map of zebrafish. Zebrafish 2013;10:15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. . The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013; 496:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, et al. . Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 2005;1:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexander MS, Kawahara G, Kho AT, Howell MH, Pusack TJ, Myers JA, et al. . Isolation and transcriptome analysis of adult zebrafish cells enriched for skeletal muscle progenitors. Muscle Nerve 2011;43:741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, et al. . The abundance of short proteins in the mammalian proteome. PLoS Genet 2006;2:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147:789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 2013;140:2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, et al. . The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 2004;566:43–47 [DOI] [PubMed] [Google Scholar]
- 72.Cooper C, Vincett D, Yan Y, Hamedani MK, Myal Y, Leygue E. Steroid receptor RNA activator bi-faceted genetic system: heads or tails? Biochimie 2011;93:1973–1980 [DOI] [PubMed] [Google Scholar]
- 73.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, et al. . Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014;33:981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, et al. . Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 2014;343:1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Craig TA, Zhang Y, McNulty MS, Middha S, Ketha H, Singh RJ, et al. . Whole transcriptome RNA sequencing detects multiple 1α,25-dihydroxyvitamin D3-sensitive metabolic pathways in developing zebrafish. Mol Endocrinol 2012;26:1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Craig TA, Zhang Y, Magis AT, Funk C, Price ND, Ekker SC, et al. . Detection of 1,25-dihydroxyvitamin D-regulated miRNAs in zebrafish by whole transcriptome sequencing. Zebrafish 2014;11:207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354–361 [DOI] [PubMed] [Google Scholar]
- 78.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, et al. . LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013;41:D983–D986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sivasubbu S, Balciunas D, Davidson AE, Pickart MA, Hermanson SB, Wangensteen KJ, et al. . Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev 2006;123:513–529 [DOI] [PubMed] [Google Scholar]
- 80.Sivasubbu S, Balciunas D, Amsterdam A, Ekker SC. Insertional mutagenesis strategies in zebrafish. Genome Biol 2007;8Suppl 1:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS One 2013;8:e76387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, et al. . Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci U S A 2007;104:12428–12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varshney GK, Huang H, Zhang S, Lu J, Gildea DE, Yang Z, et al. . The Zebrafish Insertion Collection (ZInC): a web based, searchable collection of zebrafish mutations generated by DNA insertion. Nucleic Acids Res 2013;41:D861–D864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sivasubbu S, Sachidanandan C, Scaria V. Time for the zebrafish ENCODE. J Genet 2013;92:695–701 [DOI] [PubMed] [Google Scholar]