Abstract
As a key element in the T-helper 17 (Th17) cell-mediated inflammatory process, interleukin-23 receptor (IL-23R) may play a crucial role in the pathogenesis of atherosclerosis. Single nucleotide polymorphisms (SNPs) in IL-23R have been studied in several diseases. However, its association with coronary artery disease (CAD) remains unclear. This study examined whether genetic polymorphisms in IL-23R were associated with susceptibility to CAD. Two IL-23R SNPs, rs1884444 and rs6682925, were genotyped in 462 CAD patients and 486 healthy controls. Data showed that percentages of rs6682925TC and CC genotypes were significantly higher in patients than in controls (odds ratio [OR]=1.23, 95% confidence interval [CI]: 1.04–1.79, p=0.022; OR=2.35, 95% CI: 1.52–3.43, p<0.001, respectively). Frequency of the rs1884444 polymorphism did not reveal any significant differences between patients and healthy donors. Further analyses demonstrated a significantly increased number of rs6682925CC genotype in patients with hypertension. Moreover, we investigated the effect of IL-23R polymorphisms on gene expression by assessing mRNA level of IL-23R in peripheral blood mononucleated cells (PBMCs). Results showed that subjects carrying rs6682925TC and CC genotypes had significantly increased mRNA level of IL-23R in PBMCs than those with TT genotype. These data suggest that IL-23R rs6682925T/C polymorphism may act as a risk factor of CAD.
Introduction
More and more evidence have confirmed the essential role of inflammatory processes in the development of atherosclerotic lesions. In coronary artery disease (CAD), a clinical form of coronary atherosclerosis, the activation of inflammatory factors may influence various stages of the development and progression of lesions within coronary vessels (Giacoppo et al., 2014). Macrophages and T-lymphocytes may be of particular significance for the formation of atherosclerotic plaques, in which the cells are engaged in the immune response, which also comprises enhanced expression of adhesion molecules and increased release of cytokines (Roy, 2014). Cytokines constitute a network with positive and negative feedback mechanisms and thus they exert miscellaneous and pleiotropic effects. The final result of particular cytokine activation depends on receptor representation, circulating soluble cytokine receptors, and cytokine antagonists (Ussher and Drucker, 2014).
Interleukin (IL)-23 is a heterodimeric proinflammatory cytokine composed of a p19 subunit and a p40 subunit, which is also part of IL-12 (Oppmann et al., 2000; Wiekowski et al., 2001). IL-23 is predominantly produced by antigen-presenting cells in response to microbial or host immune stimuli and is involved in the regulation of immune responses against infections and tumor development through the engagement of the IL-23 receptor (IL-23R) (Parham et al., 2002). The IL-23R is composed of the IL-23R subunit and the IL-12Rβ1 subunit, and is essential for the Th17 cell-mediated immune response (Kuka et al., 2010; Miller et al., 2010; Peral de Castro et al., 2012). Th17 cells are a recently discovered proinflammatory CD4+ effector T cell population that contributes to pathogen clearance and tissue inflammation by expressing high levels of the proinflammatory cytokine IL-17 in response to stimulation (Gao et al., 2010; Kocieda et al., 2012). Moreover, the novel inflammation pathway axis - IL23/IL17 axis has been shown to play a pivotal role in inflammatory and autoimmune diseases (Belladonna et al., 2002; D'Elios et al., 2010). Thus, IL-23R plays an important role in the initiating, maintaining, and accelerating the IL-23/IL-17 inflammatory signal transduction pathway (Tang et al., 2012). Studies have indicated that IL-23 can be involved in the development of atherosclerosis. Researchers have shown that Chlamydia pneumoniae (Cp)-matured dendritic cells may contribute to T-cell mediated immunopathology of atherosclerosis through IL-12 and IL-23 (Ausiello et al., 2006). Another report has presented that IL-23 gene expression is significantly decreased in unstimulated peripheral blood mononuclear cells (PBMCs) of CAD patients compared with those healthy controls (Khojasteh-Fard et al., 2012). As an important part of the IL-23 pathway, IL-23R may also play critical roles in atherosclerosis and atherosclerosis-related diseases.
Several clinically relevant polymorphic sites have been reported in the IL-23R gene. However, little is known about the relationship between genetic polymorphisms in the IL-23R gene and the susceptibility to CAD. In the current study, we examined two IL-23R SNPs, rs1884444T/G and rs6682925T/C, in 462 CAD patients and 486 healthy controls, and investigated the effects of these polymorphisms on gene expression.
Materials and Methods
Patients and controls
The study group included 462 CAD patients and 486 controls recruited from Changhai Hospital and Shanghai East Hospital. The diagnosis of CAD was confirmed by coronary angiography performed with the Judkins technique using a quantitative coronary angiographic system, and it was defined by angiography with at least one main coronary vessel >50% luminal narrowing or with a history of acute myocardial infarction. In the same period, 486 outpatients who underwent regular physical examinations at the same hospital were recruited as controls. They were diagnosed free of CAD by their medical history of CAD or angiography, free of clear ischemic changes by electrocardiography and without chest pain symptoms. Individuals with congestive heart failure, peripheral vascular disease, rheumatic heart disease, pulmonary heart disease, tumor, chronic kidney or hepatic disease were excluded from the study. All individuals enrolled were from the Han population in China. Social demographic information, family history of CAD, past history, and lifestyle factors were obtained through questionnaire interview. Subjects with more than 15 pack-year history were defined as smoker. Written informed consent was obtained from each participant. The study was approved by the Review Boards of Changhai Hospital and Shanghai East Hospital. Each study participant provided a peripheral blood sample.
DNA extraction and genotyping
Genomic DNA was extracted from 5 mL frozen whole blood using the DNA Extraction Kit (Fastagen, China) according to the manufacturer's protocol. The rs6682925 T/C and rs1884444T/G polymorphisms were genotyped by a primer-introduced restriction analysis (PIRA)-PCR assay. For rs6682925, a mismatched G was introduced to the sense primer to replace T at −3 bp from the polymorphic site to create a MboI restriction site (sense 5′-ATGGAAATAACATTTTAAGTGCCGAT-3′ and antisense 5′-CATCTATGTTACCGCAATGGCA-3′). Similarly, for rs1884444, a mismatched G was introduced to the antisense primer to replace T at +2 bp from the polymorphic site to create a HaeIII restriction site (sense 5′-TCTTAGGGAAAAATGTTATGCTTTTT-3′ and antisense 5′-GCATCCCATTGAATAGTGGC-3′). The restriction enzymes of the two loci (rs6682925 and rs1884444) were MboI and HaeIII (Fermentas International, Inc., Burlington, Canada), respectively. Genotyping was performed without knowing the subject's case and control status. Different genotype patterns by the PIRA-PCR assay were randomly selected for direct sequencing to determine the genotypes by using an automated sequencer (Perkin-Elmer ABI model 377 genetic analysis; Applied Biosystems, Foster City, CA), and the results were 100% concordant.
Isolation of RNA and real-time qPCR for IL-23R
Total cellular RNA was extracted from PBMCs of healthy controls using the Tripure reagents (Roche, Sandy, UT). RNA pellets were dissolved in 30 μl nuclease-free water and were stored at −70°C. The purity and concentration of RNA were determined by measuring OD260/280 ratio and OD260, respectively on a Nano-Drop spectrophometer. Preparations with a ratio of OD260/280 that was lower than 1.6 were discarded. Synthesis of cDNA and real-time qPCR for IL-23R were conducted, RT2 qPCR Primer Assay for Human IL23R (QIAGEN, Valencia, CA).
Statistical analysis
Genotype and allele frequencies of IL-23R polymorphisms were compared between CAD cases and controls using the Chi-square test and odds ratios (OR), and 95% confidence intervals (CIs) were calculated to assess the relative risk conferred by a particular allele and genotype. Demographic and clinical data between groups were compared by Chi-square test and Student's t-test. The mRNA level of IL-23R was compared by Student's t-test. The linkage disequilibrium between these polymorphisms and the haplotypes were conducted using the SHEsis software, from the website http://analysis.bio-x.cn/ (Bio-X, Inc., Shanghai, China). Statistical significance was assumed at the p<0.05. The SPSS statistical software package version 13.0 was used for the statistical analyses.
Results
Clinical characteristics of the study subjects
The clinical characteristics of all the subjects are shown in Table 1. There was no significant difference in age (p>0.05) and gender (p>0.05) between the CAD patients and the healthy adults. Compared to the controls, CAD patients exhibited higher level of body mass index (p<0.05), increased number of smokers, increased frequency of hypertension (p<0.05), and increased frequency of diabetes (p<0.05), all of which are established CAD risk factors (Table 1).
Table 1.
General Characteristics of the CAD Patients and Control Group
| Characteristics | CAD (n=462) | Control (n=486) | p-Value |
|---|---|---|---|
| Age (years, mean±SD) | 55.2±9.8 | 56.4±10.1 | >0.05 |
| Age range | 39–79 | 32–78 | >0.05 |
| Gender (M/F) | 270/192 | 288/198 | >0.05 |
| BMI (kg/m2) | 25.6±5.2 | 23.2±4.6 | <0.05 |
| Smoking, n (%) | 173 (37.4) | 92 (18.9) | <0.05 |
| Hypertension, n (%) | 245 (53.0) | 79 (16.3) | <0.05 |
| Diabetes, n (%) | 146 (31.6) | 82 (16.9) | <0.05 |
Data are mean±SD
CAD, coronary artery disease; BMI, body mass index.
IL-23R polymorphisms and susceptibility to CAD
Distributions of the IL-23R rs1884444 and rs6682925 polymorphisms in the CAD cases and controls are summarized in Table 2. Genotypes of these two polymorphisms among the controls were in agreement with the Hardy–Weinberg equilibrium (p>0.05). As for the rs1884444T/G polymorphism, percentages of TT, TG, and GG genotypes were 46.1%, 43.2%, and 10.7% in the controls, and were 47.4%, 43.1%, and 9.5% in the patients. No significant differences were identified between CAD patients and controls (Table 2). As for the rs6682925T/C SNP, prevalence of TT, TC, and CC genotypes were 41.2%, 46.5%, and 12.3% in the controls, whereas were 30.1%, 47.8%, and 22.1% in the patients. Frequencies of TC and CC genotypes were significantly higher in CAD patients than in controls (OR=1.23, 95% CI: 1.04–1.79, p=0.022 and OR=2.35, 95% CI: 1.52–3.43, p<0.001, respectively). Also, C allele of the rs6682925T/C SNP showed clearly increased percentage in patients compared with controls (46.0% vs. 35.6%, p<0.001). In addition, we analyzed the linkage disequilibrium of the two SNPs, and no linkage between these two polymorphisms was observed (D′ <0.1). Investigation of haplotypes showed that T-C and G-C (rs1884444-rs6682925) haplotypes had significantly higher proportions in the patients than in controls (OR=1.48, 95% CI: 1.17–1.89, p<0.001 and OR=1.33, 95% CI: 1.05–1.92, p=0.016, respectively, Table 2). These results suggest that the rs6682925T/C SNP is associated with increased susceptibility to CAD.
Table 2.
IL-23R Polymorphisms in CAD Patients and Controls
| Polymorphism | Cases n=462 (%) | Controls n=486 (%) | OR (95% CI)a | p-Valuea |
|---|---|---|---|---|
| rs1884444T/G | ||||
| Genotype | ||||
| TT | 219 (47.4) | 224 (46.1) | 1.00 | |
| TG | 199 (43.1) | 210 (43.2) | 0.96 (0.76–1.23) | 0.764 |
| GG | 44 (9.5) | 52 (10.7) | 0.78 (0.43–1.35) | 0.431 |
| Allele | ||||
| T | 637 (68.9) | 658 (67.7) | 1.00 | |
| G | 287 (31.1) | 314 (32.3) | 0.82 (0.71–1.12) | 0.452 |
| rs6682925T/C | ||||
| Genotype | ||||
| TT | 139 (30.1) | 200 (41.2) | 1.00 | |
| TC | 221 (47.8) | 226 (46.5) | 1.23 (1.04–1.79) | 0.022b |
| CC | 102 (22.1) | 60 (12.3) | 2.35 (1.52–3.43) | <0.001b |
| Allele | ||||
| T | 499 (54.0) | 626 (64.4) | 1.00 | |
| C | 425 (46.0) | 346 (35.6) | 1.51 (1.21–1.64) | <0.001b |
| Haplotype | ||||
| rs1884444-rs6682925 | ||||
| T-T | 346 (37.4) | 427 (43.9) | 1.00 | |
| T-C | 291 (31.5) | 231 (23.8) | 1.48 (1.17–1.89) | <0.001b |
| G-T | 153 (16.6) | 199 (20.5) | 0.92 (0.69–1.17) | 0.672 |
| G-C | 134 (14.5) | 115 (11.8) | 1.33 (1.05–1.92) | 0.016b |
Data were adjusted for age and sex.
p-Value is statistically significant (<0.05).
Association between rs6682925T/C SNP and hypertension or diabetes in CAD
Hypertension is a critical risk factor of CAD. We, therefore, investigated whether rs6682925T/C was associated with hypertension in CAD. In the patient group, 245 subjects were diagnosed with hypertension. Prevalence of rs6682925CC genotype and C allele were significantly elevated in patients with hypertension than those without hypertension (OR=1.73, 95% CI: 1.04–3.03, p=0.039 and OR=1.33, 95% CI: 1.03–1.76, p=0.031, respectively. Table 3). To understand whether rs6682925T/C SNP is independently correlated with hypertension, we examined this polymorphism with hypertension in the control group. Interestingly, we did not observe any significant variations in the genotypes and alleles of the SNP between controls with hypertension and those without hypertension (Table 4). Diabetes is another important risk factor of CAD. We evaluated the correlation between rs6682925T/C SNP and diabetes in CAD. However, no significant differences were identified between patients with and without diabetes (Table 4). These data indicate rs6682925T/C SNP is involved in the development of hypertension-associated CAD.
Table 3.
rs6682925T/C and Hypertension Status in CAD Patients and Controls
| rs6682925T/C | Hypertension (+) | Hypertension (−) | OR (95% CI)a | p-Valuea |
|---|---|---|---|---|
| CAD patients | N=245 | N=217 | ||
| Genotype | ||||
| TT | 65 (26.5) | 74 (34.1) | 1.00 | |
| TC | 118 (48.2) | 103 (47.5) | 1.29 (0.82–2.02) | 0.248 |
| CC | 62 (25.3) | 40 (18.4) | 1.73 (1.04–3.03) | 0.039b |
| Allele | ||||
| T | 248 (50.6) | 251 (57.8) | 1.00 | |
| C | 242 (49.4) | 183 (42.2) | 1.33 (1.03–1.76) | 0.031b |
| Healthy controls | N=79 | N=407 | ||
| Genotype | ||||
| TT | 30 (38.0) | 170 (41.8) | 1.00 | |
| TC | 35 (44.3) | 191 (46.9) | 1.03 (0.60–1.73) | 0.992 |
| CC | 14 (17.7) | 46 (11.3) | 1.69 (0.83–3.47) | 0.162 |
| Allele | ||||
| T | 95 (60.1) | 531 (65.2) | 1.00 | |
| C | 63 (39.9) | 283 (34.8) | 1.21 (0.77–1.72) | 0.236 |
Data were adjusted for age and sex.
p-Value is statistically significant (<0.05).
Table 4.
rs6682925T/C and Diabetes Status in CAD Patients and Controls
| rs6682925T/C | Diabetes (+) | Diabetes (−) | OR (95% CI)a | p-Valuea |
|---|---|---|---|---|
| CAD patients | N=146 | N=316 | ||
| Genotype | ||||
| TT | 46 (31.5) | 93 (29.4) | 1.00 | |
| TC | 69 (47.3) | 152 (48.1) | 0.82 (0.53–1.41) | 0.629 |
| CC | 31 (21.2) | 71 (22.5) | 0.74 (0.42–1.63) | 0.433 |
| Allele | ||||
| T | 161 (45.1) | 338 (53.5) | 1.00 | |
| C | 131 (44.9) | 294 (46.5) | 0.82 (0.65–1.19) | 0.578 |
| Healthy controls | N=82 | N=404 | ||
| Genotype | ||||
| TT | 35 (42.7) | 165 (40.8) | 1.00 | |
| TC | 36 (43.9) | 190 (47.0) | 0.73 (0.47–1.39) | 0.512 |
| CC | 11 (13.4) | 49 (12.2) | 1.02 (0.52–2.45) | 0.779 |
| Allele | ||||
| T | 106 (64.6) | 520 (64.4) | 1.00 | |
| C | 58 (35.4) | 288 (35.6) | 0.96 (0.63–1.41) | 0.979 |
Data were adjusted for age and sex.
Effect of IL-23R polymorphisms on gene expression
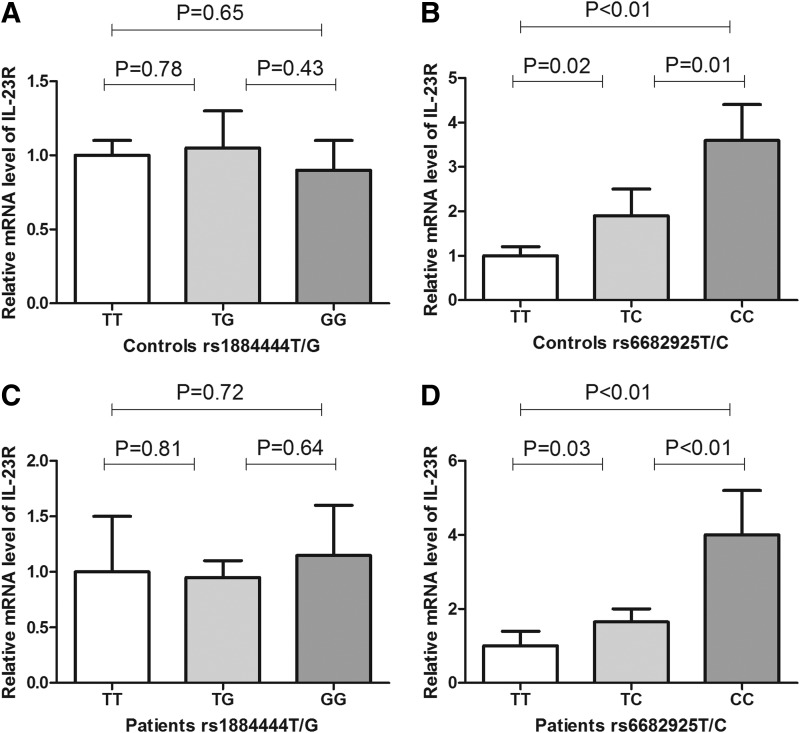
Researches on the functions of IL-23R rs1884444 and rs6682925 polymorphisms remain limited. In this study, we analyzed the effect of IL-23R polymorphisms on gene expression by assessing mRNA of IL-23R in PBMCs. As for the rs1884444T/G polymorphism, mRNA of IL-23R were examined from 62 healthy controls, in which 21 subjects were with TT genotype, 21 subjects were with TG genotype, and 20 subjects were with GG genotype. Data showed that the mRNA levels of IL-23R were not significantly changed among subjects carrying different rs1884444 genotypes (Fig. 1A). As for the rs6682925T/C polymorphism, mRNA of IL-23R were examined from 85 healthy controls, in which 29 subjects were with TT genotype, 30 subjects were with TC genotype, and 26 subjects were with CC genotype. Results demonstrated that the mRNA levels of IL-23R were significantly elevated in subjects carrying TC and CC genotypes than in those with TT genotype (Fig. 1B). We also evaluated the effect of IL-23R polymorphisms on gene expression in CAD patients. Similar to the controls, mRNA levels of IL-23R were not significantly changed among patients carrying different rs1884444 genotypes (Fig. 1C), whereas the mRNA levels of IL-23R were significantly elevated in CAD patients carrying rs6682925TC and CC genotypes than in those with TT genotype (Fig. 1D). These data suggest that the rs6682925 polymorphism may increase the expression of IL-23R.
FIG. 1.
(A) Relative mRNA level of IL-23R in peripheral blood mononuclear cells (PBMCs) from controls with different rs1884444 genotypes. A total of 62 healthy controls were included, in which 21 subjects were with TT genotype, 21 subjects were with TG genotype, and 20 subjects were with GG genotype. (B) Relative mRNA level of IL-23R in PBMCs from controls with different rs6682925 genotypes. A total of 85 healthy controls were included, in which 29 subjects were with TT genotype, 30 subjects were with TC genotype, and 26 subjects were with CC genotype. (C) Relative mRNA level of IL-23R in PBMCs from coronary artery disease (CAD) patients with different rs1884444 genotypes. A total of 43 patients were included, in which 15 subjects were with TT genotype, 15 subjects were with TG genotype, and 13 subjects were with GG genotype. (D) Relative mRNA level of IL-23R in PBMCs from CAD patients with different rs6682925 genotypes. A total of 52 patients were included, in which 17 subjects were with TT genotype, 19 subjects were with TC genotype, and 16 subjects were with CC genotype. The levels of β-actin were used as an internal control, and the IL-23R mRNA expression was calculated by 2[−Delta Delta C(T)] method.
Discussion
Studies have implied that IL-23 can be involved in the development of CAD (Ausiello et al., 2006; Khojasteh-Fard et al., 2012). For example, IL-23 may promote atherosclerosis through affecting dendritic cells. IL-23 gene expression is downregulated in PBMCs of CAD patients. However, effects of IL-23R on CAD have not been reported. In this molecular epidemiological study, we sought to identify genetic factors that confer individual susceptibility to CAD. Our results were obtained by analyzing 462 CAD patients and 486 healthy controls, and showed that rs6682925T/C SNP in the IL-23R was associated with increased risk for CAD. Further data identified that the rs6682925T/C polymorphism was correlated with hypertension-associated CAD. Function analyses suggested that the rs6682925T/C polymorphism could upregulate mRNA level of IL-23R in PBMCs.
The IL-23R rs1884444T/G polymorphism is associated with various diseases. It has been reported that rs1884444 is correlated with reduced risk to schistosomiasis-associated immune reconstitution inflammatory syndrome in a Kenyan population (Ogola et al., 2014). This SNP can be found in the Chinese population as well. A recent study has shown that rs1884444T/G polymorphism is associated with Alzheimer's disease in Han Chinese (Liu et al., 2014). Peng et al. (2013) has reported a strong correlation between the SNP and increased risk of hepatocellular carcinoma. However, other studies have shown that rs1884444 is not associated with esophageal squamous cell carcinoma or systemic lupus erythematosus in the Chinese population (Chen et al., 2013; Ni et al., 2014). In the current study, we did not observe any significant association between rs1884444 and CAD (Table 2). The rs1884444T/G SNP is located at exon 2 of IL-23R and results in a change of amino acid from His to Gln, which is responsible for the signal peptide of IL-23R. According to the web-based SNP analysis tool, Pupa-Suite2, the T to G base change of rs1884444 may disrupt an exonic splicing enhancer, resulting in exon skipping, malformation, or transcript alternative splicing. In this study, we evaluated the polymorphism and gene expression, and found that the SNP did not affect the mRNA level of IL-23R (Fig. 1A).
The rs6682925 polymorphism is also correlated with different diseases. Rs6682925 TC/CC variant genotypes have shown a strong association with increased risk of acute myeloid leukemia (Qian et al., 2013). Chu et al. (2012) have reported that rs6682925 is associated with esophageal cancer. We found that rs6682925 TC and CC genotypes were correlated with increased susceptibility to CAD. The SNP rs6682925 is located at 907-bp upstream from the transcriptional start position of IL-23R. The web-based tool of TFSEARCH 1.3 shows that the T to G base change of rs6682925 might affect a predicted GATA-X transcription factor binding. Xu et al. (2013) found that the C allele (G allele in antisense strand) of rs6682925 may increase the promoter activity of IL-23R by luciferase assay. We identified upregulated mRNA level of IL-23R in subjects carrying TC and CC genotypes (Fig. 1B), which was consistent with the web-based prediction. In addition, our data revealed that prevalence of rs6682925CC genotype was not altered in hypertension, but significantly increased in hypertension-associated CAD (Table 3), indicating a special mechanism of the polymorphism and hypertension-associated CAD.
Interestingly, our data revealed that patients lacking hypertension presented significantly higher number of rs6682925CC genotype than healthy individuals without hypertension (18.4% vs. 11.3%, Table 3; p=0.011, data not shown). It might be because the effect of rs6682925T/C polymorphism is not limited to CAD-related hypertension. The SNP may have major effects on CAD and have independent effects on CAD-related hypertension. Therefore, patients lacking hypertension may still present significantly higher proportion of rs6682925CC genotype than healthy individuals without hypertension. In addition, the rs6682925T/C polymorphism seemed not to be correlated with diabetes (Table 4). However, since The SNP may have major effects on CAD, it is reasonable that percentage of C allele was still higher in patients without diabetes than controls without diabetes (46.5% vs. 35.6%, Table 4; p<0.001, data now shown).
In summary, the current study identified that rs6682925T/C polymorphism was associated with increased risk of CAD, especially hypertension-associated CAD. Also, the SNP may increase mRNA level of IL-23R. The results provide important knowledge for understanding the effect of IL-23 on the development of CAD.
Acknowledgment
This work was funded by the Academic Leaders Training Program of Pudong Health Bureau of Shanghai (Grant No.: PWRd2013-06).
Disclosure Statement
No competing financial interests exist.
References
- Ausiello C.M., Fedele G., Palazzo R., Spensieri F., Ciervo A., and Cassone A. (2006). 60-kDa heat shock protein of Chlamydia pneumoniae promotes a T helper type 1 immune response through IL-12/IL-23 production in monocyte-derived dendritic cells. Microbes Infect 8,714–720 [DOI] [PubMed] [Google Scholar]
- Belladonna M.L., Renauld J.C., Bianchi R., Vacca C., Fallarino F., Orabona C., Fioretti M.C., Grohmann U., and Puccetti P. (2002). IL-23 and IL-12 have overlapping, but distinct, effects on murine dendritic cells. J Immunol 168,5448–5454 [DOI] [PubMed] [Google Scholar]
- Chen G.M., Feng C.C., Ye Q.L., Wang J., Cen H., Li R., Peng H., Zhou M., Leng R.X., Fan Y.G., Tao J.H., Pan H.F., and Ye D.Q. (2013). Lack of association between IL-23R gene polymorphisms and systemic lupus erythematosus in a Chinese population. Inflamm Res 62,791–795 [DOI] [PubMed] [Google Scholar]
- Chu H., Cao W., Chen W., Pan S., Xiao Y., Liu Y., Gu H., Guo W., Xu L., Hu Z., and Shen H. (2012). Potentially functional polymorphisms in IL-23 receptor and risk of esophageal cancer in a Chinese population. Int J Cancer 130,1093–1097 [DOI] [PubMed] [Google Scholar]
- D'Elios M.M., Del Prete G., and Amedei A. (2010). Targeting IL-23 in human diseases. Expert Opin Ther Targets 14,759–774 [DOI] [PubMed] [Google Scholar]
- Gao X., Ding G., Wang Z., Fu H., Ni Z., Ma J., Song S., Liu F., and Fu Z. (2010). Adjuvant treatment suppresses IL-17 production by T cell-independent myeloid sources in nonobese diabetic mice. Mol Immunol 47,2397–2404 [DOI] [PubMed] [Google Scholar]
- Giacoppo D., Capodanno D., Dangas G., and Tamburino C. (2014). Spontaneous coronary artery dissection. Int J Cardio 175,8–20 [DOI] [PubMed] [Google Scholar]
- Khojasteh-Fard M., Abolhalaj M., Amiri P., Zaki M., Taheri Z., Qorbani M., Bazzaz J.T., and Amoli M.M. (2012). IL-23 gene expression in PBMCs of patients with coronary artery disease. Dis Markers 33,289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocieda V.P., Adhikary S., Emig F., Yen J.H., Toscano M.G., and Ganea D. (2012). Prostaglandin E2-induced IL-23p19 subunit is regulated by cAMP-responsive element-binding protein and C/AATT enhancer-binding protein beta in bone marrow-derived dendritic cells. J Biol Chem 287,36922–36935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuka M., Baronio R., Valentini S., Monaci E., Muzzi A., Aprea S., De Gregorio E., and D'Oro U. (2010). Src kinases are required for a balanced production of IL-12/IL-23 in human dendritic cells activated by Toll-like receptor agonists. PloS One 5,e11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu J.T., Zhang W., Zong Y., Lu R.C., Zhou J., and Tan L. (2014). Interleukin-23 receptor polymorphisms are associated with Alzheimer's disease in Han Chinese. J Neuroimmunol 271,43–48 [DOI] [PubMed] [Google Scholar]
- Miller J.M., Bidula S.M., Jensen T.M., and Reiss C.S. (2010). Vesicular stomatitis virus modified with single chain IL-23 exhibits oncolytic activity against tumor cells in vitro and in vivo. Int J Infereron Cytokine Mediator Res 2010,63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B., Chen S., Xie H., and Ma H. (2014). Functional polymorphisms in interleukin-23 receptor and susceptibility to esophageal squamous cell carcinoma in Chinese population. PloS One 9,e89111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogola G.O., Ouma C., Jura W.G., Muok E.O., Colebunders R., and Mwinzi P.N. (2014). A non-synonymous polymorphism in IL-23R Gene (rs1884444) is associated with reduced risk to schistosomiasis-associated Immune Reconstitution Inflammatory Syndrome in a Kenyan population. BMC Infect Dis 14,316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppmann B., Lesley R., Blom B., Timans J.C., Xu Y., Hunte B., Vega F., Yu N., Wang J., Singh K., Zonin F., Vaisberg E., Churakova T., Liu M., Gorman D., Wagner J., Zurawski S., Liu Y., Abrams J.S., Moore K.W., Rennick D., de Waal-Malefyt R., Hannum C., Bazan J.F., and Kastelein R.A. (2000). Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13,715–725 [DOI] [PubMed] [Google Scholar]
- Parham C., Chirica M., Timans J., Vaisberg E., Travis M., Cheung J., Pflanz S., Zhang R., Singh K.P., Vega F., To W., Wagner J., O'Farrell A.M., McClanahan T., Zurawski S., Hannum C., Gorman D., Rennick D.M., Kastelein R.A., de Waal Malefyt R., and Moore K.W. (2002). A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 168,5699–5708 [DOI] [PubMed] [Google Scholar]
- Peng Q., Qin Y., Chen Z., Deng Y., Xu J., Li S., and Qin X. (2013). Correlation between interleukin23 receptor gene polymorphisms and risk of hepatitis B virus infection in patients. Mol Med Rep 8,613–620 [DOI] [PubMed] [Google Scholar]
- Peral de Castro C., Jones S.A., Ni Cheallaigh C., Hearnden C.A., Williams L., Winter J., Lavelle E.C., Mills K.H., and Harris J. (2012). Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol 189,4144–4153 [DOI] [PubMed] [Google Scholar]
- Qian X., Cao S., Yang G., Pan Y., Yin C., Chen X., Zhu Y., Zhuang Y., Shen Y., and Hu Z. (2013). Potentially functional polymorphism in IL-23 receptor and risk of acute myeloid leukemia in a Chinese population. PloS One 8,e55473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. (2014). Atherosclerotic Cardiovascular Disease Risk and Evidence-based Management of Cholesterol. N Am J Med Sci 6,191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Chen S., Qian H., and Huang W. (2012). Interleukin-23: as a drug target for autoimmune inflammatory diseases. Immunology 135,112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussher J.R., and Drucker D.J. (2014). Cardiovascular actions of incretin-based therapies. Circ Res 114,1788–1803 [DOI] [PubMed] [Google Scholar]
- Wiekowski M.T., Leach M.W., Evans E.W., Sullivan L., Chen S.C., Vassileva G., Bazan J.F., Gorman D.M., Kastelein R.A., Narula S., and Lira S.A. (2001). Ubiquitous transgenic expression of the IL-23 subunit p19 induces multiorgan inflammation, runting, infertility, and premature death. J Immunol 166,7563–7570 [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu Y., Pan S., Liu L., Liu J., Zhai X., Shen H., and Hu Z. (2013). IL-23R polymorphisms, HBV infection, and risk of hepatocellular carcinoma in a high-risk Chinese population. J gastroenterol 48,125–131 [DOI] [PubMed] [Google Scholar]