Abstract
Growth is an important trait in animal breeding. However, the genetic effects underpinning fish growth variability are still poorly understood. QTL mapping and analysis of candidate genes are effective methods to address this issue. We conducted a genome-wide QTL analysis for growth in tilapia. A total of 10, 7 and 8 significant QTLs were identified for body weight, total length and standard length at 140 dph, respectively. The majority of these QTLs were sex-specific. One major QTL for growth traits was identified in the sex-determining locus in LG1, explaining 71.7%, 67.2% and 64.9% of the phenotypic variation (PV) of body weight, total length and standard length, respectively. In addition, a candidate gene GHR2 in a QTL was significantly associated with body weight, explaining 13.1% of PV. Real-time qPCR revealed that different genotypes at the GHR2 locus influenced the IGF-1 expression level. The markers located in the major QTL for growth traits could be used in marker-assisted selection of tilapia. The associations between GHR2 variants and growth traits suggest that the GHR2 gene should be an important gene that explains the difference in growth among tilapia species.
Tilapia is the common name of a group of cichlid fishes native to Africa and Middle East. Several tilapia species such as Nile tilapia, blue tilapia and Mozambique tilapia have been introduced into more than 90 countries for aquaculture and fisheries1. The annual global yield of farmed tilapia reached 3.1 million tons in 20092, ranking the second after carps in all farmed fishes. Some important traits, including growth and salt tolerance, are significantly different among tilapia species. For example, Nile tilapia is the most popular farmed tilapia species in fresh water due to its outstanding growth but hardly survives in sea water, while Mozambique tilapia could grow well in full strength sea water3. Different tilapia species could cross between each other and produce fertile offspring4, making them perfect resources for breeding new commercial strains. Tilapias and their hybrids are ideal objects for mapping quantitative trait loci (QTL) for important traits and for studying the associations between genotypes and phenotypes.
Quantitative trait loci (QTL) are genomic regions containing the genes or DNA fragments that determine quantitative traits. These loci could be identified and mapped onto the genome by using whole genome scans with suitable DNA markers and reference populations5. Microsatellites and SNPs are more popular for performing whole genome scans due to their high polymorphism and ease to genotype and analyze6. The identification of QTL is an important step to discover the genes determining major economic traits, such as growth5. Markers closely linked to QTL could be used in selective breeding6.
The purpose of this study was to conduct a genome-wide scan for QTL for growth traits, to identify DNA markers linked to growth, to facilitate selective breeding of marine tilapia, and to understand the mechanisms underlying phenotypic variations.
Results
Values of growth traits in the family for QTL mapping
Body weight, total length and standard length were measured at 140 days post hatch (dph) for the family OmR1 for QTL mapping, and were measured at 90 dph for three other families OmR2, ROm1 and ROm2, respectively. The average body weight, total length and standard length were 116.5 ± 48.9 g, 185.7 ± 19.1 mm and 152.6 ± 24.1 mm at 140 dph in OmR1, respectively. The average body weights were 54.9 ± 19.4, 59.7 ± 16.3 and 60.8 ± 13.4 g at 90 dph in OmR2, ROm1 and ROm2, respectively. The correlation coefficients were 0.98 between body weight and total length, 0.97 between body weight and standard length, and 0.99 between total length and standard length at 140 dph in OmR1, respectively. The correlations between paired traits were all significant (P < 0.01).
Genotyping DNA markers and constructing a linkage map
We genotyped 282 individuals with 169 DNA markers almost evenly covered 22 linkage groups of the tilapia genome. In addition, sex was used as one marker in the linkage mapping. A linkage map was constructed by using 154 DNA markers and one phenotypic marker (i.e. sex) first. Additional 15 markers were added into this new map by the second and third round of mapping, respectively. At last, the re-constructed linkage map contained 170 markers, spanning 1079.5 cM (Figure S1). It consisted of 22 linkage groups, corresponding to 22 groups of the former linkage map7. The average inter-marker distance was 6.58 cM. The marker orders are basically identical between the reconstructed and original linkage maps.
QTL analysis in the mixed sex population
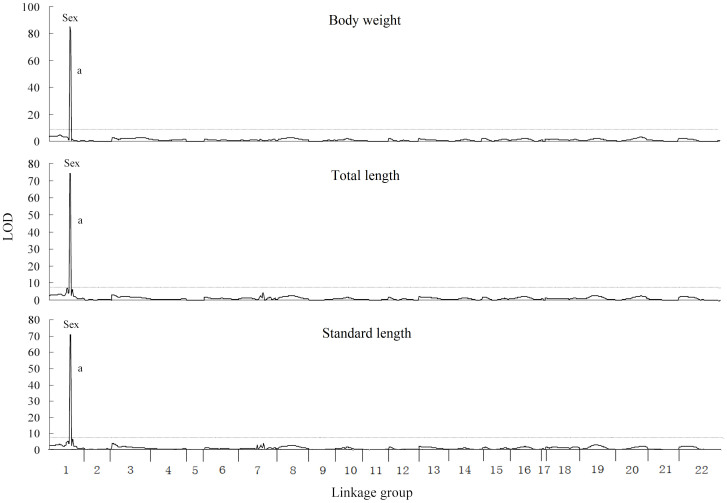
When all females and males were analyzed together, only one significant QTL was identified in the same region of linkage group (LG) 1 for body weight, total length and standard length at 140 dph, respectively (Figure 1). The significant QTL for body weight, total length and standard length were named as bw-a, tl-a and sl-a. These QTL were supported by LOD values of 85.39, 74.18 and 70.57, and explained 71.7%, 67.2% and 64.9% of variation of the traits, respectively. The marker “sex” was located at the peak of all these QTL.
Figure 1. QTL for growth traits mapped on the tilapia genome in a sex-mixed population.
The QTL surpassed the significant LOD threshold (P < 0.05) are genome-wide significant. The length of each linkage group is proportional to the genetic length (cM). The lower case letters indicate the identified QTL, which are described in Table 1. The names of traits are presented above their respective figures.
Fine mapping of the major QTL for growth
Ten new markers were used to refine the LG1 in which the major QTL for growth located. The marker S1543 was found to completely link with sex (Figure S1). The new round of QTL analysis was performed for body weight, total length and standard length based on the refined linkage map. The QTL bw-a, tl-a and sl-a were all refined to the same region of 1 cM (Table 1). The markers “sex” and S1543 were co-located at the peaks of bw-a, tl-a and sl-a.
Table 1. Significant QTL for growth traits at 140 dph in tilapia.
| Traits | LOD threshold | QTL ID | LOD (Significant level) | Linkage group | QTL interval | Peak position (cM) | PVE (%) |
|---|---|---|---|---|---|---|---|
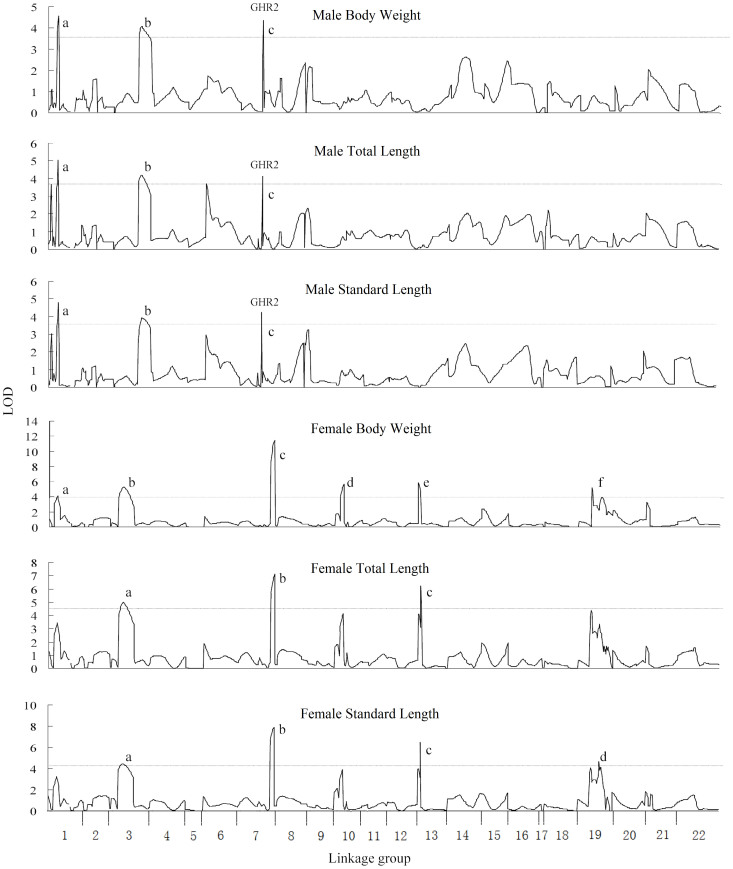
| M-BW | 3.6 | mbw_a | 4.56 (*) | LG1 | 9.9–11.3 | 11.3 | 23.8 |
| mbw_b | 4.06 (*) | LG3 | 40–53.1 | 44 | 9.4 | ||
| mbw_c | 4.35 (*) | LG7 | 32.6 | 32.6 | 13.1 | ||
| M-TL | 3.8 | mtl_a | 5.05 (*) | LG1 | 10.3–11.3 | 11.3 | 17.3 |
| mtl_b | 4.21 (*) | LG3 | 41–53.1 | 45 | 9.2 | ||
| mtl_c | 4.13 (*) | LG7 | 32.6 | 32.6 | 6.6 | ||
| M-SL | 3.7 | msl_a | 4.81 (*) | LG1 | 11.3 | 11.3 | 15.7 |
| msl_b | 3.93 (*) | LG3 | 45–51.1 | 46.1 | 6.9 | ||
| msl_c | 4.24 (*) | LG7 | 32.6 | 32.6 | 10.4 | ||
| F-BW | 3.9 | fbw_a | 4.12 (*) | LG1 | 13.3 | 13.3 | 6.3 |
| fbw_b | 5.29 (**) | LG3 | 12.4–30.8 | 19.8 | 8.2 | ||
| fbw_c | 11.44 (***) | LG7 | 52–58.5 | 58.5 | 19.6 | ||
| fbw_d | 5.67 (**) | LG10 | 9.0–13.9 | 13.9 | 9.3 | ||
| fbw_e | 5.86 (**) | LG13 | 0–4 | 0 | 9.6 | ||
| fbw_f | 5.16 (**) | LG19 | 18.9–19.5 | 18.9 | 6.6 | ||
| F-TL | 4.5 | ftl_a | 4.97 (**) | LG3 | 15.4–26.8 | 19.8 | 8.8 |
| ftl_b | 7.09 (***) | LG7 | 52–58.5 | 58.5 | 13.2 | ||
| ftl_c | 6.21 (**) | LG13 | 4.2–5.2 | 4.2 | 8.6 | ||
| F-SL | 4.3 | fsl_a | 4.42 (*) | LG3 | 17.4–23.8 | 19.8 | 7.6 |
| fsl_b | 7.86 (***) | LG7 | 52–58.5 | 58.5 | 14.8 | ||
| fsl_c | 6.5 (**) | LG13 | 4.2 | 4.2 | 8.6 | ||
| fsl_d | 4.64 (*) | LG19 | 30.4 | 30.4 | 5.8 | ||
| BW | 8.1 | bw_a | 85.39 (***) | LG1 | 32.1–33.1 | 32.1 | 71.7 |
| TL | 8 | tl_a | 74.18 (***) | LG1 | 32.1–33.1 | 32.1 | 67.2 |
| SL | 8.2 | sl_a | 70.57 (***) | LG1 | 32.1–33.1 | 32.1 | 64.9 |
QTL with genome-wide significance are presented. “*”, P < 0.05; “**”, P < 0.01; “***”, P < 0.001. PVE: the proportion of phenotypic variance explained. M-BW, M-TL and M-SL represent the body weight, total length and standard length of male; F-BW, F-TL and F-SL represent the body weight, total length and standard length of female; BW, TL and SL represent the body weight, total length and standard length of all fish, respectively.
Comparison of sex-determining locus between two tilapia species
The marker S1543 was found to completely link with “sex” marker in the refined linkage map, and its genotypes completely corresponded with the sex in the reference family OmR1, which has been used for QTL analysis and linkage mapping. Therefore, the S1543 marker was used to represent the sex-determining locus from Mozambique tilapia in BLAST analysis. BLAST was also performed for the microsatellite marker BJ702072 and SNP marker BJ675743, which are from the sex-determining locus of Nile tilapia. Markers BJ702072 and BJ675743 hit the regions of 16.49 Mb and 15.34 Mb in LG1 of Nile tilapia genome with E values of 0, respectively. The interval region between these two markers spanned about 1.15 Mb, presenting the reported sex-determining locus in Nile tilapia. The marker S1543 hit the region of 16.20 Mb in LG1 with an E value of 0, locating between markers BJ702072 and BJ675743.
Separate analysis of QTL in all female and male subpopulations
A total of 10, 7 and 8 significant QTL were identified for body weight, total length and standard length at 140 dph, respectively (Table 1). Among them, 6, 3 and 4 significant QTL were identified for body weight, total length and standard length in the all-female subpopulation, whereas 3, 3 and 3 significant QTL were identified for these three traits in the all-male subpopulation, respectively. QTL profiles were similar between body weight, total length and standard length in both females and males (Figure 2). For example, the QTL for female body weight fbw-b and fbw-c overlapped with the QTL for female total length ftl-a and ftl-b, respectively. The QTL for male body weight mbw-a, mbw-b and mbw-c overlapped with the QTL for male standard length msl-a, msl-b and msl-c, respectively.
Figure 2. QTL for growth traits mapped on the tilapia genome in male and female subpopulations.
The QTL surpassed the significant LOD threshold (P < 0.05) are genome-wide significant. The length of each linkage group is proportional to the genetic length (cM). The lower case letters indicate the identified QTL, which are described in Table 1. The names of traits are presented above their respective figures.
Significant differences were detected between the QTL profiles of females and males for each trait. For example, the QTL fbw-b, fbw-c, fbw-d from female weren't detected at the same regions in male. The most significant QTL for body weight in female was fbw-c which was located in LG7. Its LOD value of peak position was 11.44, explaining 19.9% of the phenotypic variation. In male, the QTL mbw-a in LG1 and mbw-c in LG7 explained 23.8% and 13.1% variation of body weight, respectively.
Fine mapping of QTL mbw-c for body weight
The LG7 was refined using new markers developed, and the second round of QTL analysis was performed based on the updated linkage map. QTL mbw-c was refined to a region of 1 cM between Omo649 and Omo652 by fine mapping. The genomic sequence of this region was downloaded and analyzed. Thirty-three genes were predicted (Table S1). Among them, one growth-related gene, GHR2, was identified.
The GHR2 gene was amplified and sequenced for the parents of the reference family. Four missense mutations were identified in the exons of one red tilapia parent, and were further genotyped for all individuals of the reference family. The linkage map was re-constructed based on the genotypes of all markers, including the SNPs in GHR2. The QTL analysis was re-performed using the updated linkage map and genotype data. The GHR2 gene was found to be located at the peak of QTL mbw140d-c.
Variations of GHR2
A total of 14 SNPs were found in the exons of GHR2 from the red tilapia parent of OmR1 reference family (Figure S2), but no SNP was found in the Mozambique tilapia parent. Two GHR2 alleles of red tilapia were identified based on the sequencing results in the parents and offspring, and were verified by cloning and sequencing of GHR2 cDNA. These two red tilapia alleles were named as GHR2red1 and GHR2red2 respectively.
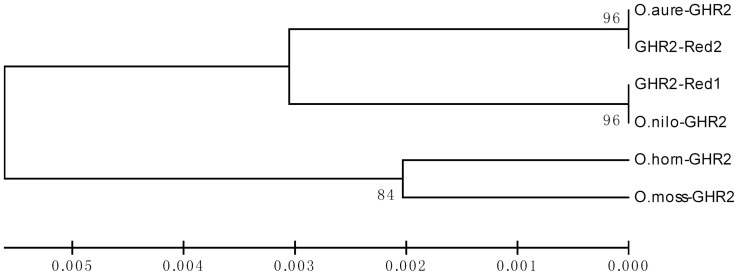
To trace the origin of alleles GHR2red1 and GHR2red2, a comparison of GHR2 between different tilapia species and alleles was conducted. Surprisingly, the deduced amino acid sequence of allele GHR2red1 was identical to that of GHR2 in Nile tilapia, and GHR2red2 was identical to GHR2 in blue tilapia (Figure S3). Similar results were found in the alignment between nucleotide sequences. UPGMA and NJ phylogenetic trees were constructed based on amino acid sequences using the MEGA 5.0 software, respectively. The topology of two trees was identical. The allele GHR2red1 was clustered with the Nile tilapia, and GHR2red2 was clustered with blue tilapia. Mozambique tilapia and Hornorum tilapia formed an independent branch (Figure 3).
Figure 3. The UPGMA phylogenetic tree of the amino acid sequences of GHR2s from different tilapias.
The GHR2 sequences are from O. mossambicus (EF452496), O. aureus (KJ845729), O. niloticus (NM_001279455), O. hornorum (EF371467) and two alleles of red tilapia identified in this study (KJ845730, KJ845731). The scale indicates the ratio of Amino acid substituted. The values beside branches represent the bootstrap scores and indicate the credibility of each branch.
The SNPs between two the red tilapia alleles were located in the exon 1, 4, 5 and 8 (Figure S2), and four of them led to the change of the 18th, 165th, 184th and 493rd amino acids, respectively. Further protein analysis indicated that the 18th amino acid located in the signal peptide, the 165th and 184th amino acids located in extracellular region, and the 493rd one located in the intracellular region. The extracellular region of GHR2 contained a fibronectin type-III domain in which the 165th and 184th amino acids were located. The 184th amino acid was an N-glycosylated site in Nile tilapia, and the 165th, 184th and 493rd ones closely neighbored to three phosphorylation sites in all tilapia species analyzed, respectively. Comparing to the allele GHR2red2 and blue tilapia GHR2, the allele GHR2red1 and Nile tilapia GHR2 obtained one N-glycosylated site in fibronectin type-III domain due to the change of the 184th amino acid (Figure S3).
Associations between GHR variations and growth traits
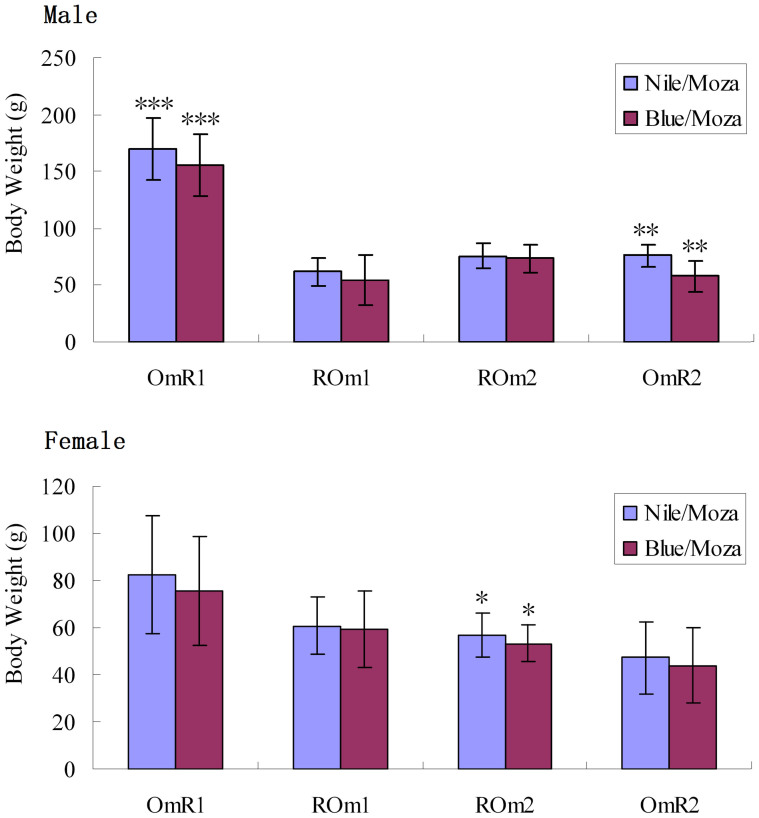
All SNPs in GHR2 gene have been genotyped for 580 individuals from families OmR1, OmR2, ROm1 and ROm2 by sequencing. These SNPs were completely linked, and no recombination was found between them. The genotypes were determined for all individuals. The four hybrid families only had two GHR2 genotypes, Nile/Moza type, which contained one Nile tilapia allele and one Mozambique tilapia allele, and Blue/Moza type which contained one blue tilapia allele and one Mozambique tilapia allele. Among the individuals of all four families, the average body weight of Nile/Moza type was higher than that of Blue/Moza type for each sex. The differences of body weight were statistically significant (P < 0.05) in OmR1 male, OmR2 male and ROm2 female (Figure 4). In the reference family OmR1 used for QTL analysis, the average body weight of males with Nile/Moza genotype was 170.2 g, 9.24% higher than that of males with Blue/Moza genotype (155.8 g) at 140 dph (Table S2).
Figure 4. The body weights of tilapia with different GHR2 genotypes in four families.
“Blue/Moza” represents the tilapia with one blue tilapia GHR2 allele and one Mozambique tilapia GHR2 allele; “Nile/Moza” represents the tilapia with one Nile tilapia GHR2 allele and one Mozambique tilapia GHR2 allele. “***” indicates P < 0.001, “**” indicates P < 0.01, and “*” indicates P < 0.05.
No significant interaction between GHR2 and other markers located in QTL was identified for growth traits when two-way and multiway ANOVAs were performed. Potential interaction between GHR2 and sex was found for body weight in family OmR1, but it was not significant (F = 2.516, P = 0.114).
Expression of GHR2, IGF1 and IGF2 in tilapia with different GHR2 genotypes
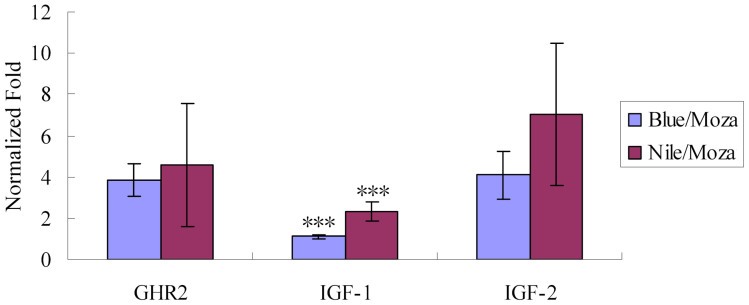
The real-time qPCR analysis was performed for GHR2, IGF-1 and IGF-2 in the individuals with Nile/Mozam or Blue/Moza genotype of GHR2, respectively. The expression level of IGF1 in Nile/Moza type doubled of that in Blue/Moza type, and the difference was statistically significant (P < 0.001). For GHR2 and IGF-2, though their average expression levels in Nile/Mozam type were higher than those in Blue/Moza type, respectively, the differences were not significant (Figure 5).
Figure 5. Temporal expressions of GHR2, IGF-1 and IGF-2 in tilapia with two GHR2 genotypes.
“Blue/Moza” represents the tilapia with one blue tilapia GHR2 allele and one Mozambique tilapia GHR2 allele; “Nile/Moza” represents the tilapia with one Nile tilapia GHR2 allele and one Mozambique tilapia GHR2 allele. The expression levels of genes are indicated using fold normalized by internal control. “***” indicates P < 0.001.
Discussion
QTL mapping, GHR2 variations and their associations with growth traits
As an important approach to identify the underlying genes of the quantitative traits and develop the DNA markers associated with traits, QTL mapping has been conducted in some fish species, including Atlantic salmon8, rainbow trout9, Japanese flounder10 and Asian seabass11. Some of these studies were performed for growth, the most important traits in aquaculture. QTL analysis about growth traits has been conducted with 42 markers and 114 individuals in tilapia before12. In the present study, more individuals were analyzed using 170 markers with an average inter-marker distance of 6.58 cM, providing more precise QTL profiles in tilapias.
In most of the tilapia species, males grow faster than females. Many factors such as nutrient loss during spawning and fasting during hatching have been claimed to result in the sexual growth dimorphism of tilapias13. In the present study, QTL analysis was performed at 140 dph, and the growth traits were also measured at 90 dph for the reference family OmR1 (data not shown). The body weight of males was doubled that of females at two stages. Spawning was not observed at 90 dph, and only occasional spawning was found at 140 dph, indicating that spawning and mouth-brooding may not explain the major growth differences between female and male in tilapia at early stages.
In the present study, QTL analysis was performed based on a mixed sex population, female and male population, respectively. More QTL were identified in females and males than in mixed sex population. The QTL analysis based on different sexes has been reported before14,15. Our results indicate that this strategy could increase the power to detect potential QTL for the traits with significant sexual dimorphism, may due to the elimination of interference of the sex-determining loci. The majority of the QTL for growth traits identified in this study were sex specific. Similar to the previous study in which majority of the markers associated with growth were located near the sex-determining loci in tilapia12, our results also suggest that sex determining loci play an important role in tilapia growth. A few studies about sex specific QTL have been reported for other traits such as body composition16 and pigmentation17 recently. In animals, many traits, including growth and pigmentation have undergone sex selection in millions of years of evolution18,19, and the selection pressure on these traits is different for females and males. The growth traits are determined by complex mechanism involving multiple pathways20. The sex-specific QTL detected may reflect the different selection pressure on different pathways and genes between females and males in tilapia.
Growth hormone and its related pathways play crucial roles in growth of vertebrates20,21. By binding with specific cell membrane receptors (GHR1 and GHR2), the growth hormone triggers a phosphorylation cascade20,21,22 and regulates the expression of insulin-like growth factors (IGF-1 and IGF-2) which stimulate the cell growth of target tissues23,24. The variations of GH, GHR and other genes in the growth hormone pathway could lead to serious abnormal growth such as dwarfism25,26,27 and adiposity28, and the over-expression of growth hormone is the major cause for gigantism in human29. The associations between the variations of these genes and growth traits have also been identified in livestock30,31,32,33, poultry34 and a few fishes35. It is generally accepted that the mutations of genes related to the growth hormone could cause significant differences of growth. In the present study, we identified two GHR2 alleles in red tilapia, and found that the hybrid individuals with the Nile tilapia GHR2 alleles had a higher expression level of IGF-1 and grew more quickly than those with the blue tilapia GHR2 alleles. Though no significant expression difference of the GHR2 gene was detected between two types of individuals, the amino acid differences in the extracellular region may alter the binding capacity of GHR2 with growth hormone, and further affect the expression level of downstream genes and growth rate. Significant differences of growth traits exist in different tilapia species. Among them, Nile tilapia grows faster than other tilapia species, including blue tilapia and Mozambique tilapia in freshwater3,36,37. Our results indicate that the GHR2 gene from the growth hormone pathway may be involved in the differentiation of growth traits in different tilapia species.
Sex determining mechanism is complex in fish38. QTL mapping is an approach to mapping sex-determining loci. In this study, we identified one major sex-determining locus in LG1 of Mozambique tilapia. In Nile tilapia, previous studies detected two sex-determining loci in LG1 and LG2339,40. These results suggest that the two tilapia species may share a major sex determining locus in LG1 whereas other loci with small effects on sex also exist in different tilapia species. Further detailed analysis of the interactions of the major and other sex determining loci will shed new insights into sex determining mechanisms in fish.
Potential applications of growth-associated markers in tilapia breeding
Marker-assisted selection (MAS) has been widely used in the breeding of plants41,42. However, a few MAS were reported in food fish due to the lack of the markers associated with important traits6. In the present study, 10, 7 and 8 significant QTL were identified for body weight, total length and standard length at 140 dph, respectively, and these QTL explained 5.8%–71.7% of the phenotypic variation. The growth-associated markers located in QTL could be used in the marker-assisted selection for breeding. The hybrids with the GHR2 alleles from Nile tilapia grew faster than those with the GHR2 alleles from blue tilapia, indicating that the individuals carrying the Nile tilapia GHR2 allele instead of the blue tilapia GHR2 allele should be kept in crossbreeding between tilapia species.
Since males grow much quicker than females in tilapia, all male populations are preferred in tilapia production. However, male populations are usually generated by sexual reverse using hormones, which may cause serious issues in health and environmental pollution43. The cross of YY males with XX females to produce all XY males is an ideal way to all males. However, producing YY males using traditional methods takes 4–5 years and needs a lot of spaces43. The markers located in the refined sex-determining locus from Mozambique tilapia identified in this study would be useful in the marker-assisted selection for rapid breeding of YY super male tilapia to produce all male populations. Using markers linked to the sex-determining loci could substantially shorten the time to produce YY males, as the time-consuming progeny tests are not required.
Conclusion
A whole genome scan identified a total of 10, 7 and 8 significant QTL for body weight, total length and standard length at 140 dph in tilapia, respectively. These data provide a useful resource for marker-assisted selection in tilapia. The association between GHR2 variants and growth traits implies that GHR2 gene is involved in the difference of growth among tilapia species.
Methods
Ethics statement
All experiments in this study were approved by the IACUC Committee of Temasek Life Sciences Laboratory (Approval number: TLL(F)-11-001), and the experiments were performed according to the regulations and guidelines established by this committee.
Fish and reference families
One Mozambique tilapia male and one red tilapia female were crossed to produce the reference family OmR1 for QTL analysis. For verifying the associations between markers and traits, three additional families were generated. The family OmR2 was produced by crossing one Mozambique tilapia male and one red tilapia female. The families ROm1 and ROm2 were produced by different single pair of Mozambique tilapia female and red tilapia male, respectively. The Mozambique tilapias were F1 offspring of one wild population from South Africa, and the red tilapias were from one local strain in Malaysia and Singapore. The families OmR1, OmR2, ROm1 and ROm2 consisted of 282, 134, 107 and 57 individuals, respectively. Each family was raised in one independent tank with circulating water in the marine fish facility of Temasek Life Sciences Laboratory. Fish were fed with tilapia pellets (BioMar) twice a day. The water temperature was 25–28°C during the experiments.
The body weight, total length and standard length were measured for all fish from four reference families at 90 dph. The fish from family OmR1 were injected with PIT tags for individual identification, and the traits were re-measured for them at 140 dph. The sex was distinguished according to the appearance of gonopore and gonad for each fish during the last measuring. Fin clips were collected for each parent and offspring, and stored in 75% ethanol for DNA extraction using the method described by Yue and Orban44.
Genotyping of markers and sequencing of the GHR2 gene
One hundred and fifty-four microsatellite markers uniformly distributed were selected from one tilapia linkage map published recently7 for the first round of linkage mapping and QTL mapping in the family OmR1. For fine mapping of the QTL, 10 and 4 new microsatellite markers were developed in LG1 and LG7 of the genome sequences of Nile tilapia following the method described previously7, respectively. The 5′ end of forward primers of these microsatellites was labeled with Fam or Hex fluorescent dye. PCR amplifications were performed in a 25 µL volume containing 10 ng genomic DNA, 1 x PCR buffer, 50 µM of each dNTPs, 0.2 µM forward and reverse primers and 1 unit Taq DNA polymerase (Fermentas). The reactions were conducted in BioRad cycler (Bio-Rad) using the following program: one cycle of 3 min at 94°C, 38 cycles of 30 s at 94°C, 30 s at 50°C, 55°C or 60°C and 45 s at 72°C, followed by a final extension of 5 min at 72°C. PCR products were resolved on ABI3730xl Genetic Analyzer (ABI) and genotyped using software GeneMapper 4.1 (ABI). All 168 microsatellite markers were genotyped for 282 individuals from reference family OmR1.
The GHR2 gene was genotyped by sequencing all individuals of four reference families OmR1, OmR2, ROm1 and ROm2. At first, PCRs were performed for all exons of GHR2 in the parents of reference families by using 6 pairs of primer (Table S4), and sequencing was conducted for the PCR products using BigDye (ABI) on ABI3730xl Genetic Analyzer (ABI). The SNPs in GHR2 gene were identified, and were further genotyped in offspring using PCR and sequencing. The haplotypes of GHR2 were determined based on the linkage relationship between all SNPs in offspring, and verified by cloning and sequencing in parents. The genotype of GHR2 gene was represented by two haplotypes for each fish.
The reference family OmR1 has been verified that it possessed a XY sex determining system, and its sex was determined by a single locus in LG1 in our previous study7. To map the sex-determining locus more precisely, sex also was used as one marker in the linkage mapping analysis. The genotype of “sex” marker was determining based on male heterogametic for each individual. All primers used in this study were listed in Table S3 and S4.
Re-constructing a linkage map in the family OmR1
The linkage map was re-constructed based on the reference family OmR1 according to the recommendation in the manual of MapQTL 5.0 software for excluding the interference of differences of recombination rate between families. Three rounds of linkage mapping were performed to provide precise reference maps for three rounds of QTL analysis. The first round of re-constructing was conducted using 155 markers. 14 markers from LG1 and LG7 were added to the genotype dataset in the second round of linkage mapping. The GHR2 gene was mapped in the third round of linkage mapping. In each round of mapping, the genotypes of markers in 282 individuals from OmR1 family were imported into the JoinMap 4.0 software, and the Kosambi mapping function was used to perform the linkage analysis. The linkage maps were constructed with a threshold of LOD 4.0.
QTL analysis, fine mapping and identification of candidate gene
MapQTL 5.0 software was used to perform the QTL analysis. The re-constructed linkage map and the genotyping data were exported from JoinMap software, and imported into MapQTL software along with the phenotype data for following analysis. The Kruskal-Wallis analysis (a nonparametric mapping method based on the rank sum test of Kruskal-Wallis) and IM analysis (an interval mapping method based on maximum likelihood approach to the segregation of a mixture of probability distributions) was used to identify the potential QTL. Permutation Test with 10000 times was conducted, and the genome-wide significant LOD thresholds were determined according to the interval at the relative cumulative value of 0.95. Cofactors were selected by using the automatic cofactor selection analysis, and MQM analysis (a mapping method based on multiple-QTL models) was performed using the selected cofactors. The significant QTL were identified, and the confidence interval of each QTL was defined according to the threshold levels of genome wide. To exclude the interference of sex on the identification of QTL, QTL analysis was performed based on the mixed sex population, female subpopulation and male subpopulation, respectively. Two-way and multiway ANOVAs were performed for identifying potential interactions between QTL using SPSS software, and the markers nearest to the peak of QTL were used to represent the QTL in the analysis.
Fine mapping was conducted for two major QTL, one was the major sex-linked QTL in LG1, which was identified in mixed sex population, and another one was the male-specific QTL mbw-c in LG7, which was identified in male subpopulation. The genome sequences of LG1 and LG7 of Nile tilapia were downloaded from NCBI, and microsatellites were screened in the regions which were beside or in the QTL using SiRoKo software45. Primers were designed for amplifying the microsatellites, and genotyping was performed using labeled primers for the parents of reference family. The informative primers were further used in genotyping 282 individuals from the family OmR1. New linkage map was constructed using the genotypes of all markers, and QTL analysis were re-performed based on the new linkage map and renewed genotypes.
Genome sequences in the refined QTL were analyzed using GENSCAN Web Server (http://genes.mit.edu/GENSCAN.html) and BLAST, and the potential candidate genes were identified according to their functions and locations. SNP markers were developed in the candidate genes, and were genotyped in the reference family. Then a new round of analysis, including re-constructing of the linkage map and QTL analysis was performed for mapping the genes in the linkage map and QTL.
Three rounds of QTL analysis were performed. First round of analysis was conducted using 154 microsatellite markers and “sex” marker. The second round of analysis was carried out using 168 microsatellite markers and “sex” marker. The genotypes of GHR2 gene were added to the genotype dataset in the third round of analysis. A total of 170 markers were used in the final round of QTL analysis.
Comparison of sex-determining locus among tilapias
The sequences of marker BJ702072 and BJ675743 were downloaded from NCBI. These two markers have been proved that they located at the upstream and downstream ends of the sex-determining locus in Nile tilapia, respectively46. BLAST analysis was performed against the whole genome sequences of Nile tilapia for the markers BJ702072, BJ675743 and S1543. The last one (i.e. S1543) was found to completely link with the sex-determining locus from Mozambique tilapia. The locations of the sex-determining loci from Nile tilapia and Mozambique tilapia were compared.
Sequence analysis of GHR2
The cDNA and amino acid sequences of GHR2 were downloaded from NCBI for Nile tilapia, Mozambique tilapia and Hornorum tilapia. All eight exons of GHR2 gene were amplified and sequenced for the parents of reference families and four blue tilapia individuals using six pairs of primers, and the SNPs and haplotypes were verified by cloning and sequencing. The cDNA and amino acid sequences of GHR2 were predicted using DNAstar (GeneCodes) software for the parents of reference families and blue tilapia, respectively. Sequence alignments of cDNA and protein were performed using Clustal X2.1 software47. The UPGMA and NJ phylogenetic trees were constructed using MEGA 5.0 software based on amino acid sequences48. The functional domain was predicted using ScanProsite (http://prosite.expasy.org/scanprosite/). The signal peptide, transmembrane region, N-glycosylated site and Phosphorylation site were predicted by using SignalP 4.1 Server, TMHMM Server 2.0, NetNGlyc 1.0 Server and NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/), respectively49,50.
Real-time analysis of GHR2 expression
Ten tilapias with different GHR2 genotypes were randomly selected from family OmR2 at 90 dph. 5 of them had a Nile/Moza genotype which contained one Nile tilapia allele and one Mozambique tilapia allele, and another 5 fish had a Blue/Moza genotype which contained one blue tilapia allele and one Mozambique tilapia allele. Livers were collected from each fish, quickly frozen in liquid nitrogen and kept in a -80°C freezer for the RNA isolation.
Total RNA was isolated for each sample using TRIZOL (Invitrogen), and treated with RNase-free DNase enzyme (Fermentas) following the instruction manuals. Approximately 500 ng of RNA for each sample was reverse-transcribed using RevertAid First Strand cDNA Synthesis Kit (Fermentas) with random primers. Primers were designed and selected for performing the real-time analysis of GHR2, IGF-1, IGF-2 and β-actin according to the recommendations in the manual of iQ SYBR Green Supermix (Bio-Rad), respectively (Table S4). Real-time PCR reactions were conducted using iQ SYBR Green Supermix in an iQ5 Real Time PCR System (Bio-Rad). Each reaction contained 20 μl components: 2 μl cDNA sample (twenty times dilution), 10 μl SYBR Master Mix, 0.4 μl forward primer, 0.4 μl reverse primer and 7.2 μl ddH2O. The program for real-time PCR was: 95°C for 15 s, 40 cycles of 95°C for 5 s and 60°C for 30 s, followed by melting curve analysis. Three replicates were performed for each reaction. The expression levels of GHR2, IGF-1 and IGF-2 were calculated by normalizing to the internal control β-actin using ΔΔCT method51. For assessing the significant level of expression differences between two groups, T-tests were conducted using SPSS software.
Statistical analysis of growth traits
Individuals from reference families were grouped according to sex and genotype. The average values and standard deviations (sd) were calculated for each group using Excel software. The significant levels of differences between different groups were analyzed, and the correlation coefficients between different traits were calculated using SPSS software.
Author Contributions
G.H.Y. supervised the project. F.L. designed the experiments. F.L., F.S., J.L., J.H.X., G.H.F., G.L., R.J.T., Z.Y.W. and D.Q. conducted the experiments. F.L. conducted the data analyses and wrote the manuscript. Y.G.H. finalized the manuscript.
Supplementary Material
Supplementary imformation
Acknowledgments
This research is supported by the National Research Foundation, Prime Minister's Office, Singapore under its Competitive Research Program (CRP Award No. NRF-CRP002-001). We thank Mr. Huiming Liu and members of our animal house for taking care of our fish. The linkage and QTL analysis were conducted using software JoinMap and MapQTL in Dr. Jiang's laboratory in the Chinese Academy of Agricultural Sciences.
References
- Pullin R. S. V., Palomares M. L., Casal C. V., Dey M. M. & Pauly D. Environmental impacts of tilapias. In Tilapia aquaculture, proceedings of the 4th International Symposium on Tilapia in Aquaculture. Fitzsimmons, K. (ed.) 554–570 (N.E. Regional Agricultural Engineering Service, Ithaca, New York, USA, 1997). [Google Scholar]
- FAO. Fishery and aquaculture statistics. (Rome., 2009). [Google Scholar]
- Kamal A. H. M. M. & Mair G. C. Salinity tolerance in superior genotypes of tilapia, Oreochromis niloticus, Oreochromis mossambicus and their hybrids. Aquaculture 247, 189–201 (2005). [Google Scholar]
- Lovshin L. L. Tilapia hybridization. In The biology and culture of tilapias. Pullin, R. S. V. and Lowe, R. H. (eds.) 279–308 (International Center for Living Aquatic Resource Management Manila, Philippines, 1982). [Google Scholar]
- Borevitz J. O. & Chory J. Genomics tools for QTL analysis and gene discovery. Curr Opin Plant Biol 7, 132–136 (2004). [DOI] [PubMed] [Google Scholar]
- Yue G. H. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish and Fisheries 15, 376–396 (2014). [Google Scholar]
- Liu F. et al. A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus x Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 14, 58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranski M., Moen T. & Vage D. I. Mapping of quantitative trait loci for flesh colour and growth traits in Atlantic salmon (Salmo salar). Genet Sel Evol 42, 17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wringe B. F. et al. Growth-related quantitative trait loci in domestic and wild rainbow trout (Oncorhynchus mykiss). BMC Genet 11, 63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki A. et al. Linkage analysis of resistance to Streptococcus iniae infection in Japanese flounder (Paralichthys olivaceus). Aquaculture 308, S62–S67 (2010). [Google Scholar]
- Xia J. H. et al. Whole genome scanning and association mapping identified a significant association between growth and a SNP in the IFABP-a gene of the Asian seabass. BMC Genomics 14, 295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaani A., Zilberman N., Tinman S., Hulata G. & Ron M. Genome-scan analysis for quantitative trait loci in an F2 tilapia hybrid. Mol Genet Genomics 272, 162–172 (2004). [DOI] [PubMed] [Google Scholar]
- Schreiber S., Focken U. & Becker K. Individually reared female Nile tilapia (Oreochrornis niloticus) can grow faster than males. J Appl Ichthyol 14, 43–47 (1998). [Google Scholar]
- Abasht B. et al. Fatness QTL on chicken chromosome 5 and interaction with sex. Genet Sel Evol 38, 297–311 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrenberg D. L., Wang S., Hannon R. M., Garland T. Jr & Pomp D. QTL underlying voluntary exercise in mice: interactions with the "mini muscle" locus and sex. J Hered 101, 42–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankra-Badu G. A. et al. Mapping main, epistatic and sex-specific QTL for body composition in a chicken population divergently selected for low or high growth rate. BMC Genomics 11, 107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. S., Hamilton A. M., Frank A., Barmina O. & Kopp A. Genetic basis of sex-specific color pattern variation in Drosophila malerkotliana. Genetics 180, 421–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. W. & Hixon M. A. Sexual and lifetime selection on body size in a marine fish: the importance of life-history trade-offs. J Evol Biol 24, 1653–1663 (2011). [DOI] [PubMed] [Google Scholar]
- Svensson E. I. & Waller J. T. Ecology and sexual selection: evolution of wing pigmentation in calopterygid damselflies in relation to latitude, sexual dimorphism, and speciation. Am Nat 182, E174–195 (2013). [DOI] [PubMed] [Google Scholar]
- Pilecka I., Whatmore A., Hooft van Huijsduijnen R., Destenaves B. & Clayton P. Growth hormone signaling: sprouting links between pathways, human genetics and therapeutic options. Trends Endocrinol Metab 18, 12–18 (2007). [DOI] [PubMed] [Google Scholar]
- Herrington J. & Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab 12, 252–257 (2001). [DOI] [PubMed] [Google Scholar]
- Fukamachi S., Yada T. & Mitani H. Medaka receptors for somatolactin and growth hormone: phylogenetic paradox among fish growth hormone receptors. Genetics 171, 1875–1883 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews L. S., Norstedt G. & Palmiter R. D. Regulation of insulin-like growth factor I gene expression by growth hormone. Proc Natl Acad Sci USA 83, 9343–9347 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali O. et al. Hormonal regulation of hepatic IGF-I and IGF-II gene expression in the marine teleost Sparus aurata. Mol Reprod Dev 71, 12–18 (2005). [DOI] [PubMed] [Google Scholar]
- Meyer C. W. et al. A novel missense mutation in the mouse growth hormone gene causes semidominant dwarfism, hyperghrelinemia, and obesity. Endocrinology 145, 2531–2541 (2004). [DOI] [PubMed] [Google Scholar]
- Arman A. et al. Novel splice site mutation in the growth hormone receptor gene in Turkish patients with Laron-type dwarfism. J Pediatr Endocrinol Metab 21, 47–58 (2008). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Identification of a novel splicing mutation in the growth hormone (GH)-releasing hormone receptor gene in a Chinese family with pituitary dwarfism. Mol Cell Endocrinol 313, 50–56 (2009). [DOI] [PubMed] [Google Scholar]
- Mong J. L. et al. Associations of the growth hormone receptor (GHR) gene polymorphisms with adiposity and IGF-I activity in adolescents. Clin Endocrinol 73, 313–322 (2010). [DOI] [PubMed] [Google Scholar]
- Goldenberg N. et al. Treatment of pituitary gigantism with the growth hormone receptor antagonist pegvisomant. J Clin Endocrinol Metab 93, 2953–2956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M. M., Antunes R. C., Silva H. D. & Goulart L. R. Association of PIT1, GH and GHRH polymorphisms with performance and carcass traits in Landrace pigs. J Appl Genet 46, 195–200 (2005). [PubMed] [Google Scholar]
- Hua G. H. et al. Polymorphism of the growth hormone gene and its association with growth traits in Boer goat bucks. Meat Sci 81, 391–395 (2009). [DOI] [PubMed] [Google Scholar]
- Kim N. K. et al. A previously unreported DraI polymorphism within the regulatory region of the bovine growth hormone gene and its association with growth traits in Korean Hanwoo cattle. Anim Genet 35, 152–154 (2004). [DOI] [PubMed] [Google Scholar]
- Estany J. et al. Association of CA repeat polymorphism at intron 1 of insulin-like growth factor (IGF-I) gene with circulating IGF-I concentration, growth, and fatness in swine. Physiol Genomics 31, 236–243 (2007). [DOI] [PubMed] [Google Scholar]
- Huang N., Cogburn L. A., Agarwal S. K., Marks H. L. & Burnside J. Overexpression of a truncated growth hormone receptor in the sex-linked dwarf chicken: evidence for a splice mutation. Mol Endocrinol 7, 1391–1398 (1993). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Genotypes, haplotypes and diplotypes of IGF-II SNPs and their association with growth traits in largemouth bass (Micropterus salmoides). Mol Biol Rep 39, 4359–4365 (2011). [DOI] [PubMed] [Google Scholar]
- Pullin R. S. V. Tilapia genetic resources for aquaculture. (International Center for Living Aquatic Resources Management, Bangkok, Thailand, 1988). [Google Scholar]
- EI-Hawarry W. N. Growth performance, proximate muscle composition and dress-out percentage of Nile tilapia (Oreochromis niloticus), blue tilapia (Oreochromis aureus) and their interspecific hybrid (♂ O. aureus X ♀ O. niloticus) cultured in semi-intensive culture system. World's Vet J 2, 17–22 (2012). [Google Scholar]
- Gui J. F. & Zhu Z. Y. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin Sci Bull 57, 1751–1760 (2012). [Google Scholar]
- Cnaani A. et al. Genetics of sex determination in tilapiine species. Sex Dev 2, 43–54 (2008). [DOI] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Weller J. I., Hulata G. & Ron M. Linkage and physical mapping of sex region on LG23 of Nile tilapia (Oreochromis niloticus). G3 2, 35–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M., Akram N. A., Mehboob Ur R. & Foolad M. R. Marker-assisted selection in plant breeding for salinity tolerance. Methods Mol Biol 913, 305–333 (2012). [DOI] [PubMed] [Google Scholar]
- Sureshkumar S. et al. Marker assisted selection of low phytic acid trait in maize (Zea mays L.). Hereditas 151, 20–27 (2014). [DOI] [PubMed] [Google Scholar]
- Mair G. C., Abucay J. S., Skibinski D. O. F., Abella T. A. & Beardmore J. A. Genetic manipulation of sex ratio for the large scale production of all-male tilapia Oreochromis niloticus L. Can J Fish Aquat Sci. 54, 396–404 (1997).
- Yue G. H. & Orban L. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis 26, 3081–3083 (2005). [DOI] [PubMed] [Google Scholar]
- Kofler R., Schlotterer C. & Lelley T. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23, 1683–1685 (2007). [DOI] [PubMed] [Google Scholar]
- Lee B. Y. & Kocher T. D. Comparative genomics and positional cloning. In Aquaculture genome technologies. Liu, Z. J. (ed.) 323–335 (Blackwell Publishing Professional Ames, USA, 2007). [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J. & Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9, 299–306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S. & Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294, 1351–1362 (1999). [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & T D. S. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary imformation